Abstract
Mismatch repair mutations are the cause of generalized genomic instability and are particularly evident at microsatellite loci, which is known as microsatellite instability (MSI). MSI is present in 85% to 90% of colorectal cancers and occurs in hereditary non-polyposis colorectal cancer (HNPCC). The National Cancer Institute recommends the “Bethesda panel” for MSI screening. Recently, a novel T25 mononucleotide marker was described, termed CAT25. This microsatellite marker displays a quasi-monomorphic pattern in normal tissues. The aim of our study was to evaluate the performance of CAT25 in HNPCC patients and to compare its reliability with the results of the Bethesda panel. We tested 55 tumor tissues from HNPCC patients using both the Bethesda panel and the CAT25 mononucleotide marker. One hundred healthy blood donors were used as controls. The CAT25 microsatellite was found to be altered in all 13 colorectal cancers classified as MSI-H using the standard Bethesda panel. Colorectal tumors that showed a stable Bethesda pattern did not show altered CAT25 repeats. Additionally, CAT25 showed a monomorphic allele pattern in all tissue samples. In our series, the concordance between the Bethesda panel and CAT25 in identifying colorectal cancers with high MSI reached 100%. Our results suggest that the CAT25 microsatellite represents a sensitive and specific marker for MSI and could be, at least, included in the panel of markers for the identification of HNPCC patients.
Hereditary non-polyposis colorectal cancer (HNPCC) is an autosomal dominant, inherited disease caused by germline mutations in any of the mismatch repair genes: MLH1, MSH2, MSH6, MSH3, and PMS2.1,2
The inactivation of the mismatch repair system results in the accumulation of DNA replication errors, which participate in tumor development by affecting genes regulating critical cell functions.1
HNPCC patients are prone to develop colorectal cancers and several other malignancies, (endometrium, ovaries, stomach, small bowel, hepatobiliary, and urinary tract).3,4,5,6 HNPCC related colon cancers show a typical phenotype: tumors are frequently located in the right colon and show mucinous or medullar histology, lymphoid infiltrates, negative MLH1 or MSH2 protein expression, and microsatellite instability (MSI). Microsatellite instability is due to the accumulation of replication errors and is observed in more then 80% of HNPCC patients.6 MSI is characterized by the difference in length between normal and tumor tissues of short, repeated DNA sequences.7 In 1997, the “International Workshop on Microsatellite Instability and RER Phenotype in Cancer detection and Familial Predisposition,” sponsored by the National Cancer Institute (NCI), proposed a panel of five microsatellite markers (known as the Bethesda markers or Bethesda panel) to assess the presence of MSI-H (microsatellite instability-high): two of the markers are mononucleotides (BAT25 and BAT26) and three are dinucleotides (D17S250, D5S346, and D2S123).8
Tumors with instability at two or more of these five markers are defined as being high-frequency, MSI-H; tumors with instability in one marker or showing no instability are defined as low frequency MSI (MSI-L) or microsatellite stable (MSS), respectively.8 The NCI microsatellite markers are studied in DNA amplified from tumor and normal tissues and amplification may be hampered due to the limited amount of available tissues; furthermore, this analysis requires a considerable amount of time and money to perform.
MSI status represents a prognostic and predictive factor for colorectal cancers: patients with MSI-H have a better prognosis than microsatellite stable (MSS) ones, and recent studies suggest that MSI-H cancers could be less responsive to 5-fluorouracil based chemotherapy.9,10,11
The 2002 Bethesda Consensus meeting recognized that dinucleotide markers have lower sensitivity and specificity than mononucleotide ones to identify MSI-H tumors and suggested that mononucleotide microsatellites should replace dinucleotide ones in the panel.12,13
In 2005 Findeisen et al described a novel T25 mononucleotide marker located in the 3′ untranslated region of the CASP2 gene that displays a quasimonomorphic pattern in normal tissue: CASP2 gene (7q34-q35) (GenBank accession no. NM_032982; position nucleotides 2685 to 2709). The CAT25 microsatellite may be a useful and simple marker for MSI.14
The aim of the present study was to evaluate the sensitivity and specificity of the CAT25 microsatellite and to compare its reliability with the five markers of the NCI panel in 55 HNPCC colorectal tumors.
Materials and Methods
Study Population
Fifty-five unrelated patients were included in this analysis. They met, at least, one of the Revised Bethesda Guidelines.13 We also examined DNA from 100 healthy blood donors (control subjects) to study the CAT25 allelic profile.
Tumor DNA specimens were purified from formalin-fixed, paraffin-embedded tumor tissues with the QIAamp Mini Kit (Qiagen), according to the manufacturer. Tumor samples were microdissected under the supervision of a pathologist (I.B.) to ensure that only areas enriched in cancer cells (80% of all cells) were used for tumor DNA preparation.
Genomic DNA was purified peripheral-blood lymphocytes by the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer.
Microsatellite Analysis
To assess MSI status, we used the Bethesda panel, which includes three dinucleotide markers (D2S123, D5S346, D27S250) and two mononucleotide repeats (BAT-25 and BAT26) (Boland 1998). Microsatellite sequences were PCR amplified from tumor DNA (cancer cells) and normal DNA (lymphocytes) using the follow primers. (BAT25 FW-VIC-5′-TCGCCTCCAAGAATGTAAGT-3′ and REV-5′-CATTTTAACTATGGCTC-3′; BAT26 FW-NED-5′-TGACTACTTTTGACTTCA-3′ and REV-5′-AACCATTCAACATTTTTAACCC-3′; D2S123 FW-6-FAM-5′-AAACAGGATGCCTGCCTT-3′ and REV-5′-GGACTTTCCACCTATGGGAC-3′; D17S250 FW-6-FAM-5′-AATAGACAATAAAAATATGTGTGT-3′ and REV5′-TATATATTTAAACCATTTGAAAGTG-3′; and D5S346 FW-PET 5′-TACTCACTCTAGTGATAAATCGG-3′ and REV 5′-TTCAGCGAATTGAGAGTTACAG-3′). Oligonucleotide primers were fluorescently labeled with 5′-VIC, 5′-NED, 5′-6-FAM, 5′-6-FAM, 5′-PET, respectively. Multiplex PCR reactions were performed in 25-μl reaction volumes, containing 50 ng of DNA, 1× PCR buffer reaction [20 mmol/L Tris-HCl (pH 8.4), 50 mmol/L], 1.5 mmol/L MgCl2, 25 pmol of each labeled and unlabeled primer, and 0.25 U of Taq polymerase (Invitrogen, Germany). The PCR cycling conditions included the following cycles: 1 cycle at 95°C for 15 minutes; 40 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute; and 1 extension cycle at 72°C for 10 minutes. Pentaplex PCR was performed (in an Applied Biosystems 9700 thermocycler) starting with 10 minutes at 95°C followed by 35 cycles of 1 minute at 94°C, 30s at 55°C, and 30s at 72°C, with a final extension at 72°C for 7 minutes.
The CAT25 microsatellite was PCR amplified and single PCR performed, independently by panel Bethesda PCR, with the sense primer end-labeled with the fluorescent marker 5′-NED. PCR primers for the amplification of CAT25 marker (forward 5′-CCTAGAAACCTTTATCCCTGCTT-3′ and reverse 5′-GAGCTTGCAGTGAGCTGAGA-3′) were designed as previously described.14 The PCR mixes was performed in a total reaction volume of 25 μl containing 30 to 50 ng of DNA, 2 mmol/L concentration of each deoxynucleotide triphosphate, 0.3 pmol of of dye-labeled forward and unlabeled reverse primers, 1X PCR buffer [20 mmol/L Tris-HCl (pH 8.4), 50 mmol/L KCl], 1.5 mmol/L MgCl2, and 0.5 units of TaqDNA polymerase (Invitrogen, Germany).
Two μl of PCR products were mixed with 12 μl of formamide and 0.5 μl of 500 LIZ Size Standard (Applied Biosystems, Foster City, CA), denatured, and run on a ABI (Applied Biosystems) PRISM 310 automated capillary electrophoresis; allelic sizes were estimated using GeneScan 3.7 (Applied Biosystems, Foster City, CA). MSI was identified by the presence of additional peaks corresponding to small deletions or insertions in the microsatellite sequences in the tumor DNA compared with the matched normal DNA. Tumors were classified as highly unstable (MSI-H) if at least 40% of the markers showed instability. Tumors with instability in one marker (20%) were defined as low instability microsatellite (MSI-L), and tumors showing no alterations were defined as microsatellite stable (MSS).
To exclude the possibility of technical artifacts or contamination, all observed differences were confirmed by new PCR and by new GeneScan analysis.
Results
We evaluated MSI in 55 formalin-fixed, paraffin-embedded, colorectal, cancers from patients fulfilling the Revised Bethesda Guidelines. With the Bethesda panel 13 tumors (23.6%) showed instability at two or more markers and so they were classified as MSI-H.
Tumors showed instability at five (two cases), four (four cases), three (four cases), and two (three cases) markers (Table 1). BAT25, BAT26, D2S123, D17S250, and D5S346 showed instability, respectively, in 11 (84.6%), 12 (92.3%), six (46.1%), nine (69.2%), and five (38.5%) of MSI-H tumors (13 cases). Five patients were MSI-L because they showed instability only in one microsatellite: BAT25 (two cases), BAT26 (one case), D2S123 (one case), and D5S346 (one case). The remaining 37 tumors were classified as MSS, according to the NCI panel (Table 1).
Table 1.
Instability Profile of ICG-HNPCC Microsatellite Markers and CAT25 in 55 Colorectal Cancers Fulfilling the Bethesda Guidelines
| No. | BAT25 | BAT26 | D2S123 | D17S250 | D5S346 | CAT25 | CAT25 ALLELIC TYPING |
|---|---|---|---|---|---|---|---|
| 12 | − | − | − | − | − | − | 146/146 |
| 37 | − | − | − | − | − | − | 146/146 |
| 120 | − | − | − | − | − | − | 146/146 |
| 134 | − | − | − | − | − | − | 146/146 |
| 135 | − | − | − | − | − | − | 146/146 |
| 140 | + | + | − | + | − | + | 137/146 |
| 148 | + | + | − | + | + | + | 138/146 |
| 150 | − | − | − | − | − | − | 146/146 |
| 157 | − | − | − | − | − | − | 146/146 |
| 163 | − | − | − | − | − | − | 146/146 |
| 192 | − | − | + | − | − | − | 146/146 |
| 200 | − | − | − | − | − | − | 146/146 |
| 215 | + | + | − | − | − | + | 138/146 |
| 219 | − | − | − | − | − | − | 146/146 |
| 230 | + | + | − | + | − | + | 137/146 |
| 238 | + | + | − | + | + | + | 135/146 |
| 246 | − | − | − | − | − | − | 146/146 |
| 268 | − | − | − | − | − | − | 146/146 |
| 279 | − | + | + | + | − | + | 136/146 |
| 289 | + | + | + | − | − | + | 137/146 |
| 324 | − | − | − | − | − | − | 146/146 |
| 344 | + | − | − | − | − | − | 146/146 |
| 351 | − | − | − | − | − | − | 146/146 |
| 354 | + | − | − | − | − | − | 146/146 |
| 384 | + | + | + | + | − | + | 134/146 |
| 417 | − | − | − | + | + | + | 136/146 |
| 436 | + | + | + | + | − | + | 138/146 |
| 456 | − | − | − | − | − | − | 146/146 |
| 487 | − | − | − | − | − | − | 146/146 |
| 474 | − | − | − | − | − | − | 146/146 |
| 491 | − | − | − | − | − | − | 146/146 |
| 524 | − | − | − | − | + | − | 146/146 |
| 525 | − | + | − | − | − | − | 146/146 |
| 532 | − | − | − | − | − | − | 146/146 |
| 537 | − | − | − | − | − | − | 146/146 |
| 538 | − | − | − | − | − | − | 146/146 |
| 544 | − | − | − | − | − | − | 146/146 |
| 545 | − | − | − | − | − | − | 146/146 |
| 546 | − | − | − | − | − | − | 146/146 |
| 547 | − | − | − | − | − | − | 146/146 |
| 548 | − | − | − | − | − | − | 146/146 |
| 563 | − | − | − | − | − | − | 146/146 |
| 582 | + | + | + | + | + | + | 137/146 |
| 586 | − | − | − | − | − | − | 146/146 |
| 593 | − | − | − | − | − | − | 146/146 |
| 600 | − | − | − | − | − | − | 146/146 |
| 602 | − | − | − | − | − | − | 146/146 |
| 603 | − | − | − | − | − | − | 146/146 |
| 613 | − | − | − | − | − | − | 146/146 |
| 627 | − | − | − | − | − | − | 146/146 |
| 641 | − | − | − | − | − | − | 146/146 |
| 642 | − | − | − | − | − | − | 146/146 |
| 649 | + | + | + | − | + | + | 136/146 |
| 674 | + | + | − | − | − | + | 137/146 |
| 729 | − | − | − | − | − | − | 146/146 |
No. refers to patient registration in our data base.
We examined the CAT25 performance both in DNA from normal subjects and in colorectal cancer tissues. In normal subjects we also sequenced the CAT25 PCR products, to exclude single nucleotide polymorphisms. We did not detect any polymorphism.
In normal subjects, the PCR product most frequently observed was a fragment of 146 bp, which was found in 94 samples (94%). In five individuals (5%), we found a fragment of 147 bp and a product of 145 bp was detected in one case (1%). No additional alleles shorter than 145 bp or longer than 147 bp were observed (Table 2).
Table 2.
Allelic Distribution of CAT25 in 100 Normal Subjects
| Size (bp) | % |
|---|---|
| 145 | 1% |
| 146 | 94% |
| 147 | 5% |
Based on the CAT25 allelic profile, we marked, as wild-type allele, the PCR product of 146 bp; the 145 and 147 alleles were considered as wild-type too, and we considered indicative of MSI any allele of CAT25 <145 or >147 bp.
Colorectal cancer specimens, previously typed for NCI marker panel, were analyzed for instability at the CAT25 locus. All 13 MSI-H colorectal tumors (100%) showed CAT25 instability. Moreover, colorectal tumors, showing MSI-L by the Bethesda panel, did not show altered CAT25 repeats and also MSS tumors (typed for NCI markers) were CAT25 stable.
In tumor tissues, we also evaluated the CAT25 allelic profile. In MSI-H cancers, the most frequent allele was a 137 bp fragment, detected in 5/13 (38%). In these patients, the wild-type allele of normal DNA (lymphocyte DNA) was of 146 bp (Table 3) (Figure 1).
Table 3.
Allelic Distribution of CAT25 in 13 MSI-H Tumors
| CAT25 | |
|---|---|
| Size (bp) | % Tumors |
| 134 bp | 7.7 |
| 135 bp | 7.7 |
| 136 bp | 23.1 |
| 137 bp | 38.4 |
| 138 bp | 23.1 |
Figure 1.
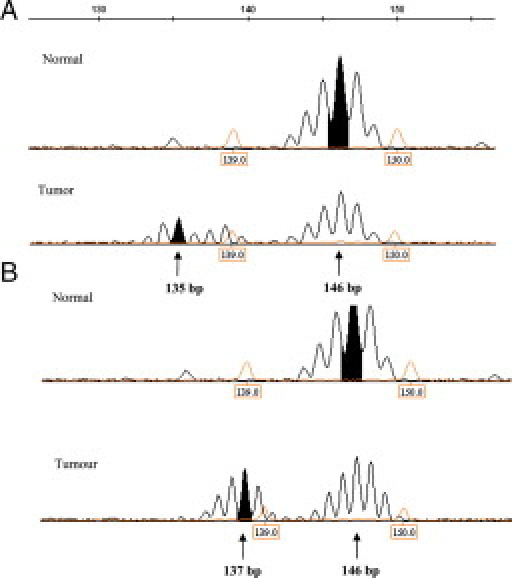
Examples of MSI-H profiles, (A–B), generated with the CAT25 quasi-monomorphic mononucletide marker.
The 37 MSS tumors (Bethesda panel) as well as the five MSI-L cases, showed a monomorphic pattern of 146 bp (Table 1).
In our experience CAT25 correctly classified 13 of 13 (100%) MSI-H colorectal cancers; with this marker, the matched analysis tumor/lymphocyte DNA was not necessary due to the monomorphic allele pattern.
Discussion
The NCI panel was proposed to assess the status of microsatellite instability in HNPCC colorectal cancers.13 The current standard method is time consuming and expensive: microsatellite markers are studied in DNA amplified from tumor and normal tissues, and amplification may be difficult due to the limited amount of available tissues.
Several groups have argued in favor of the use of mononucleotide repeats only for MSI analysis, and some authors suggest replacing the Bethesda panel with pentaplex quasimonomorphic mononucleotide repeats.15,16,17,18
The use of mononucleotide repeat microsatellites for MSI characterization has been shown to be advantageous over many dinucleotide microsatellites due to the quasi monomorphic nature of both loci and their high sensitivity to MSI. However, several studies have shown that BAT25 and BAT26 microsatellites exhibit a variability in allelic profiles in different populations, and this problem may lead to false positive results if tumor tissue only is studied for MSI analysis.8,14,19,20
Recently, it has been proposed a novel (quasi) monomorphic marker, the CAT25 microsatellite, which seems to show the same sensitivity and specificity of the NCI panel and may be studied in tumor DNA only. Some authors have suggested using CAT25 in combination with BAT25 and BAT26 mononucleotide microsatellites.14,21
The aim of this study was the characterization of the CAT25 mononucleotide marker and evaluation of its sensitivity and specificity in comparison with NCI Bethesda panel in colon cancer patients. The CAT25 allelic profile study, conducted in 100 healthy donor subjects, showed a (quasi) monomorphic allele pattern.
The analysis was conducted in 55 formalin-fixed, paraffin-embedded, colorectal cancers from HNPCC patients, previously analyzed for mismatch repair germline mutations; MSI status was evaluated by using both the Bethesda marker panel and the CAT25 microsatellite. Thirteen patients showed MSI-H with the Bethesda panel and all showed instability in CAT25 marker. No CAT25 repeat alterations were observed in tumors classified as MSS or MSI-L by the Bethesda panel.
In our small series, the concordance between the Bethesda panel and the CAT25 microsatellite in identifying MSI-H colorectal cancers reached 100%; CAT25 showed sensitivity and specificity equally to the NCI standard panel. One patient (#279), classified as MSI-H, showed BAT25 wild-type and CAT25 mutated and one more patient (#417) showed BAT25 and BAT26 wild-type with CAT25 mutated (Table 1). All 13 MSI-H tumors (Bethesda panel) have been correctly characterized by CAT25, suggesting that CAT25 locus may also be an excellent marker for early detection of MSI-H colorectal cancer in HNPCC patients.
Our results of the CAT25 marker analysis confirm previous reports by other authors: we detected a quasi-monomorphic allele pattern in normal subjects, no mutations in MSS or MSI-L specimens, and 100% mutation frequency in MSI-H tumors. We believe that to determine the MSI status of HNPCC patients, the CAT25 marker could be considered similar to the NCI panel in terms of accuracy, but with less expenditure of money and time, for determination of the MSI status of HNPCC patients. Based on these results, the CAT25 marker could be considered (together with Bethesda panel) a major tool to study HNPCC patients, especially if there are problems in finding the normal tissue for some patients. More studies are necessary to delineate the possible role of the CAT25 microsatellite analysis as in adjunct or as in alternative to the Bethesda panel.
Acknowledgements
We acknowledge Prof. Italo Bearzi, M.D., for supervision in tumor sample microdissection and Raffaella Bracci, M.D., for suggestions and review of the manuscript.
Footnotes
F.B and E.G. contributed equally to this work.
References
- 1.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 2.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 3.Watson P, Lynch HT. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993;71:677–685. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Merg A, Lynch HT, Lynch JF, Howe JR. Hereditary colon cancer–part I. Curr Probl Surg. 2005;42:195–256. doi: 10.1067/j.cpsurg.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Merg A, Lynch HT, Lynch JF, Howe JR. Hereditary colon cancer–part II. Curr Probl Surg. 2005;42:267–333. doi: 10.1067/j.cpsurg.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 6.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 7.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 8.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 9.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, Moslein G, Baker SM, Liskay RM, Burgart LJ, Honchel R, Halling KC. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- 12.Pedroni M, Roncari B, Maffei S, Losi L, Scarselli A, Di Gregorio C, Marino M, Roncucci L, Benatti P, Ponti G, Rossi G, Menigatti M, Viel A, Genuardi M, de Leon MP. A mononucleotide markers panel to identify hMLH1/hMSH2 germline mutations. Dis Markers. 2007;23:179–187. doi: 10.1155/2007/703129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, Benner A, Dondog B, Pawlita M, Dippold W, Wagner R, Gebert J, von Knebel Doeberitz M. T25 repeat in the 3′ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res. 2005;65:8072–8078. doi: 10.1158/0008-5472.CAN-04-4146. [DOI] [PubMed] [Google Scholar]
- 15.Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R. Quasimonomorphic mononucleotide repeats for high-level microsatellite instability analysis. Dis Markers. 2004;20:251–257. doi: 10.1155/2004/159347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong YF, Cheung TH, Lo KW, Yim SF, Chan LK, Buhard O, Duval A, Chung TK, Hamelin R. Detection of microsatellite instability in endometrial cancer: advantages of a panel of five mononucleotide repeats over the National Cancer Institute panel of markers. Carcinogenesis. 2006;27:951–955. doi: 10.1093/carcin/bgi333. [DOI] [PubMed] [Google Scholar]
- 17.Xicola RM, Llor X, Pons E, Castells A, Alenda C, Piñol V, Andreu M, Castellví-Bel S, Payá A, Jover R, Bessa X, Girós A, Duque JM, Nicolás-Pérez D, Garcia AM, Rigau J, Gassull MA. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99:244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 18.Bacher JW, Flanagan LA, Smalley RL, Nassif NA, Burgart LJ, Halberg RB, Megid WM, Thibodeau SN. Development of a fluorescent multiplex assay for detection of MSI-high tumors. Dis Markers. 2004;20:237–250. doi: 10.1155/2004/136734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyatt R, Chadwick RB, Johnson CK, Adebamowo C, de la Chapelle A, Prior TW. Polymorphic variation at the BAT-25 and BAT-26 loci in individuals of African origin. Implications for microsatellite instability testing. Am J Pathol. 1999;155:349–353. doi: 10.1016/S0002-9440(10)65131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 21.Deschoolmeester V, Baay M, Wuyts W, Van Marck E, Van Damme N, Vermeulen P, Lukaszuk K, Lardon F, Vermorken JBM, Wuyts W. Detection of microsatellite instability in colorectal cancer using an alternative multiplex assay of quasi-monomorphic mononucleotide markers. J Mol Diagn. 2008;10:154–159. doi: 10.2353/jmoldx.2008.070087. [DOI] [PMC free article] [PubMed] [Google Scholar]


