Abstract
Cancer is the result of a complex multistep process that involves the accumulation of sequential alterations of several genes, including those encoding microRNAs (miRNAs). miRNAs are a class of 17- to 27-nucleotide single-stranded RNA molecules that regulate gene expression posttranscriptionally. A large body of evidence implicates aberrant miRNA expression patterns in most, if not all, human malignancies. This article reviews our current knowledge about miRNAs, focusing on their involvement in cancer and their potential as diagnostic, prognostic, and therapeutic tools.
Cancer, which develops because of a multistep process resulting in the accumulation of several genomic alterations, is characterized by unrestricted proliferation, invasion, and metastasis. In cancer, many molecular pathways are affected, involving canonical protein-coding genes as well as recently discovered noncoding genes. Noncoding RNAs include a class of small RNAs (17 to 27 nucleotides in length), microRNAs (miRNAs), that control gene expression by regulating mRNA translation.
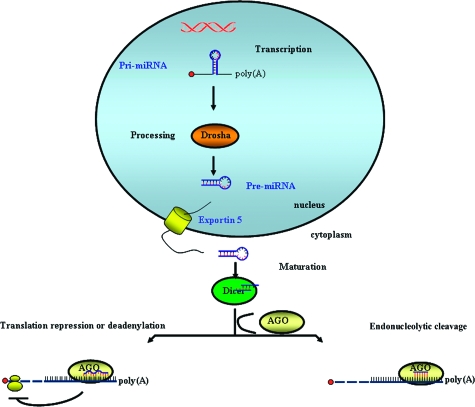
The biogenesis of miRNAs starts with the transcription of genomic regions located within or between protein-coding genes, resulting in the synthesis of miRNA precursor molecules (pri-miRNAs). Pri-miRNAs are currently thought to be transcribed primarily by RNA polymerase II and, less frequently by RNA polymerase III. Drosha, a specific ribonuclease of the RNase III endonuclease family, then enzymatically cuts the transcribed pri-miRNA in a smaller fragment (∼70 nucleotides). This hairpin pre-miRNA is then exported to the cytoplasm by Exportin-5 in a Ran-GTP-dependent manner and cleaved into an imperfect double-strand RNA (dsRNA), duplex-designated miRNA, which is termed miRNA/miRNA*. This process is performed by Dicer, an RNase III endonuclease composed of a helicase domain and a dsRNA-binding domain. One strand of the miRNA/miRNA* duplex is then selected to function as a mature miRNA and preferentially loaded into a miRNA ribonucleoprotein (miRNP) complex, whereas the other strand is likely degraded.1 As a part of the miRNP complex, single or multiple miRNA copies bind to mRNA 3′untranslated regions (3′UTR). Those that bind with perfect complimentarity lead to mRNA degradation; whereas imperfect binding leads to inhibition of translation (Figure 1).
Figure 1.
Biogenesis of miRNAs and assembly into protein complexes. miRNA precursor molecules (pri-miRNAs) fold into hairpin structures that contain imperfect base-paired stems in a two-step process catalyzed by two different RNase III-type endonucleases. Drosha first cleaves pri-miRNAs, forming ∼70 nucleotide hairpins (pre-miRNAs) in the nucleus. Subsequently, pre-miRNAs are transported to the cytoplasm by Exportin 5, where they are cleaved by Dicer to yield ∼20-bp miRNA duplexes. One miRNA strand is then selected to function as a mature miRNA, whereas the other strand is degraded. After processing, miRNAs are assembled into RNP (ribonucleic protein) complexes, called miRNPs, with proteins of the AGO family. miRNPs tether to the 3′UTR of a mRNA target to repress protein synthesis. In the case of perfect bp alignment, the miRNP complex cleaves the duplex miRNA-mRNA; however, multiple mechanisms2 are used on duplex miRNA:mRNA with imperfect complementary.
The numerous biochemical mechanisms that govern miRNA function, however, are likely dependent on the availability of local regulatory factors.2 An estimated one-third of protein-coding human mRNAs are susceptible to this complex miRNA regulatory network. Every cellular process is regulated by miRNAs, and an aberrant miRNA expression signature is a hallmark of several diseases, including cancer. These data suggest that miRNA genes could function as potential oncogenes and tumor repressors genes in the human body. Thus, an accurate evaluation of changes in miRNA expression could provide new insight into basic mechanisms of cancer.
Methods to Identify Deregulated Expression of miRNAs in Human Tumors
The first evidence of aberrant miRNA expression in human cancers was described in B-cell chronic lymphocytic leukemia, wherein hemizygous and/or homozygous chromosomal deletion at the 13q14 locus resulted in the loss or reduction of miR-15 and miR-16 expression.3 This discovery of new genes linked to cancer prompted investigation of miRNA expression in human tumors. Consequently, several techniques have been developed to support this research.
Two widely performed high-throughput techniques are used for miRNA profiling. The solid-phase array-based platform, developed first by Liu and colleagues,4 is semiquantitative, requires transcript amplification/labeling, and carries an inherent limitation of cross-hybridization among miRNAs of the same family. Conversely, flow-based, liquid-phase profiling has the advantage of increased specificity in discriminating the expression of closely related miRNAs as well as higher sensitivity in detecting modest decreases in down-regulated miRNAs. However, flow-based, liquid-phase profiling is technically demanding with respect to quality consistency in the production of miRNA probes.5 Data obtained by either of these two methods needs to be validated independently by a second technique, such as Northern blot or quantitative real-time polymerase chain reaction, to confirm the miRNA expression profile. The use of these techniques revealed aberrant miRNA expression in numerous tumors when they were compared with their normal counterparts, thereby suggesting that a link does exist between miRNAs and cancer (Table 1).6,7,8,9,10,11,12,13,14
Table 1.
Selected miRNAs Aberrantly Expressed in Tumors
| Selected miRNAs
|
||||
|---|---|---|---|---|
| Organ | Disease type | Up-regulated | Down-regulated | References |
| Liver | HCA and FNH* | 224 | 122a, 422b, 203, 200c | 6 |
| HCC*† | 21, 224, 10b, 221, 222, 20, 18 | 199a, 199b, 200b, 223, 122, 214,145, 150 | 6 | |
| Cholangiocarcinomas* | 21, 23a, 141, 200b, 27a | 6 | ||
| Pancreas | PET* | 23a, 342, 26a, 30d, 26b, 103, 107 | 155, 326, 339, 326 | 6 |
| Insulinomas* | 203, 204, 211, | 6 | ||
| PACC* | 23a, 342, 26a, 30d, 26b, 103, 105 | 155, 326, 339,326 | 6 | |
| Ductal adenocarcinomas*† | 21, 221, 181a, 155, 222, 181b, 107 | 148a, 375 | 6 | |
| Esophagus | ESCC* | 25, 424, 151 | 100, 99, 29c, 140, 205,203, 202 | 6 |
| Stomach | Adenocarcinomas*† | 21, 223, 25, 17-5p, 125b, 181b, 106a, 107, 92, 103, 221, 93, 100, 106b | 136, 218, 212, 96, 339 | 6 |
| Colon | Adenomas* | 21 | 6 | |
| Adenocarcinomas*† | 21, 92, 20a, 106a, 92, 223, 203 | 6 | ||
| Adenocarcinomas stage II* | 145 | 6 | ||
| Hematopoietic tissue | CLL* | 190, 33, 19a, 140, 123, 10b, 92, 188, 154, 217, 101, 196, 134, 141, 132, 192, 16, 15 | 181b, 220 | 7 |
| Ovary | Carcinomas* (serous, clear cell, endometrioid) | 200a, 200c | let-7d, 100, 101, 105, 125a, 125b, 126, 133a, 137, 140, 143, 147, 199a, 199b, 224, 9, 9*, 99a | 8 |
| Breast | Carcinomas* | 155, 21 | 125b, 145, 10b | 9 |
| Lung | NSCLC† | 21, 191, 155, 210 | 126*, 224 | |
| Pituitary gland | Adenoma* | 15, 16 | 10 | |
| Prostate | Carcinomas† | 32, 182, 31, 26a, 200c | 520h, 494, 490, 133a, 1, 218, 220, 128a | 11 |
| Thyroid | Papillary carcinomas* | 221, 221, 146a, 181b | 12, 13 | |
| Anaplastic carcinomas* | 30 days, 125b, 26a, 30a-5p | 14 | ||
HCA, hepatocellular carcinomas; FNH, focal nodular hyperplasia; PET, pancreatic endocrine tumors; PACC, pancreatic acinar cell carcinomas; ESCC, esophageal squamous cell carcinomas; CLL, chronic lymphocytic leukemia; NSCLC, non-small cell lung cancer.
miRNA comparative analysis: nontumor or tumor tissue versus normal tissue.
miRNA comparative analysis: tumor tissue versus adjacent nontumor tissue.
Cause of Abnormal miRNA Expression
MiRNA expression can be altered by several mechanisms in human cancer including chromosomal abnormalities, epigenetic changes, mutations and polymorphisms (SNPs), and defects in the miRNA biogenesis machinery.
Chromosomal Abnormality
MiRNAs often reside in particular genomic regions that are prone to alterations in cancer. These regions could include either a minimal region of loss of heterozygosity, which can harbor a tumor suppressor gene; a minimal region of amplification, which might contain oncogenes; or fragile sites. Fragile sites are preferential sites of sister chromatid exchange, translocation, deletion, amplification or integration of plasmid DNA, and insertion of tumor-associated viruses such as human papilloma virus.15
The high frequency of genomic alterations in miRNA loci was recently confirmed by an extensive study of high-resolution array-based genomic hybridization on 227 human ovarian cancer, breast cancer, and melanomas samples.16 The findings of this study proved that miRNA expression correlated with miRNA copy number, and these data tightly overlap with the miRNA expression data analyzed on a set of breast cancer samples in an independent study.9
Specific examples of miRNAs located in instable genomic regions include the miR-15a/16 cluster, embedded into 13q14, and both miR-143 and miR-145, located at 5q33. In fact, B-cell chronic lymphocytic leukemias and pituitary adenomas, which often harbor 13q14 deletions, showed a decreased expression of miR-15a and miR-16,3,7,10 whereas deletion of the 5q33 region observed in lung cancer seems to contribute to the decreased levels of miR-143 and miR-145 in this tumor.15 Conversely, cluster miR-17-92, located at chromosome 13q31, a region amplified in B-cell lymphomas17 and lung cancers,18 has been found to be overexpressed in these malignancies. Overall, these findings suggest that the location of miRNAs in a genomic region amenable to alterations is not a random event, thereby indicating that the loss or the gain of genomic regions including miRNAs in a specific type of cancer could participate to the cause of this malignancy.
Epigenetic Changes
Recent findings indicate that epigenetic aberrations affect miRNA expression. An extensive analysis of genomic sequences of miRNA genes showed that approximately half of these genes are associated with CpG islands, suggesting that miRNAs can represent candidate targets of the DNA methylation machinery. Analysis of several miRNA-associated CpG islands in five cell lines also indicated that miRNA gene methylation is detectable at high frequencies, both in normal and malignant cells.
Methylation status could possibly explain the deregulated expression of miRNAs in cancer.19 Some examples have been reported by Iorio and colleagues.8 They found that the treatment of a human ovarian cancer cell line (OVCAR3) with the demethylating agent 5-aza-2′deoxycytide (5-AZA-CdR) increased the expression levels of nine miRNAs. Interestingly, three of nine miRNAs, miR-21, miR-203, and miR-205, have been found to be also overexpressed in ovarian carcinomas when compared with their normal counterparts, suggesting that hypomethylation could be the mechanism responsible for their overexpression in vivo.8 Moreover, miR-21 and miR-203 are embedded in a region associated with CpG islands, whereby the DNA methylation machinery could directly affect the expression of these miRNAs.8 Conversely, decreased miR-124a expression was attributed to DNA hypermethylation in colon, breast, and lung carcinomas.20
Mutations and SNPs
Mutations and polymorphisms located in mature miRNA, pre-miRNA, or more likely in adjacent genomics regions can also change miRNA expression by affecting their processing. These events in miRNAs are rarer than in mRNA protein coding genes, because the size of miRNAs and their precursors is small. A few cases have been already reported in the literature, however.
Inherited mutations in the primary transcripts of miR-15a and miR-16–1 have been reported to be responsible for low expression levels in vitro and in vivo. Decreased expression of these miRNAs in familial chronic lymphocytic leukemia and familial breast cancer was also associated with deletion of the normal allele encompassing miR-15a and miR-16–1.21 The importance of this mutation was further supported in a spontaneous mouse model of chronic lymphocytic leukemia.22
MiRNA genomic region can also include SNPs. Hu and colleagues,23 conducted a systematic survey of common pre-miRNA sequences and their surrounding regions and evaluated in detail the association of four selected SNPs in four miRNAs (miR-146a, miR-196a2, miR-499, and miR-149) with the survival of individuals with non-small cell lung cancer. They found that patients with non-small cell lung cancer carrying a variant homozygote of the SNP located in the 3p miRNA region of miR-196a-2 had poor survival, possibly through a mechanism of elevated expression of mature miR-196*.23 These findings suggest that SNPs located in miRNA regions may be prognostic biomarkers of certain malignancies.
Defects in the miRNA Biogenesis Machinery
Despite normal expression levels of pri-miRNA, a number of human primary cancers displayed reduced levels of mature miRNAs. Thomson and colleagues24 explained this difference as a processing defect because of the loss of the RNase III Drosha. Conversely, targeted Drosha activity, directed by the ALL1(MLL) fused gene, appeared to be responsible for miR-191 up-regulation in human acute lymphoblastic leukemia.25 In addition, decreased Dicer endonuclease activity in a proportion of non-small cell lung cancers, correlated with reduced let-7 expression, unfavorable postoperative survival, and poor tumor differentiation status.26 The loss of Dicer likely represents a somatic alteration because Dicer-deficient mice failed to thrive beyond gastrulation because of the lack of multipotent stem cell development.27 Finally, changes in miRNA expression can be attributable also to either the frequent deregulation of transcription factors in cancer or viruses that often integrate within the DNA of tumoral cells.28,29
Significance of the Altered miRNA Expression in Tumors
Oncogenes
High-throughput analyses have reported altered miRNA expression in all tumors investigated to date, suggesting that miRNAs might be implicated in tumorigenesis, likely by regulating oncogene or tumor suppressor genes. The miR-17-92 polycistron represents the first example of miRNA acting as a mammalian oncogene. This cluster is embedded in a human genomic locus, 13q31.3, which is a region that is amplified in several types of lymphoma and solid tumors. Expression of the miR-17-92 cluster in the Eμ-myc transgenic mouse model of B-cell lymphoma accelerated disease onset and progression. In these mice, transcription of the miR-17-92 cluster was directly transactivated by c-Myc, a transcription factor that is frequently overexpressed in cancer cells, through its direct interaction with the putative promoter region.30,31 The miR-17-92 cluster and its paralog, miR-106b-25, seem to be tightly linked to the functions of the E2F family of transcription factors as well. Indeed, members of both clusters target E2F1 and in turn, can be activated by E2F, establishing a negative feedback loop.28
Although these data support a role for the miR-17-92 cluster in promoting tumorigenesis, there is also evidence suggesting that loss-of-function of these miRNAs might be advantageous for cancer cells. Loss-of-heterozygosity at the 13q31.3 locus has been reported in ovarian cancers, breast cancers, and melanomas. Consistent with these observations, introduction of miR-17 into breast cancer cell lines reduced their proliferation.32 This effect was attributable in part to the inhibition of the amplified in breast cancer 1 (AIB1) gene, which encodes a transcriptional co-activator of the estrogen receptor and E2F1. Thus, the same miRNA can have both pro- and anti-tumorigenic activities depending on the targets available in a given cellular context.
The first direct example that a single miRNA has an oncogenic role is miR-155, the overexpression of which has been linked to several types of lymphomas such as Hodgkin and Burkitt lymphoma.33 Recently, Costinean and colleagues34 demonstrated the role of this miRNA in tumorigenesis by producing transgenic mice that specifically overexpress miR-155 in B cells. These transgenic mice developed a preleukemic lymphoproliferative disease that progressed to B-cell leukemia and high-grade lymphoma. A microarray study of the malignant B cells isolated from miR-155 transgenic mice compared with nontransgenic control reported several deregulated protein-coding genes in these cells, potentially identifying direct and indirect targets of miR-155. These findings suggest that miR-155 deregulation could be an early event in oncogenesis, which needs additional genetic alterations for the development of the fully malignant phenotype.
Cell Cycle Regulation
MiRNAs can also contribute to tumorigenesis by modulating the expression of proteins that directly regulate circuitry controlling cellular life and death decisions. MiR-221/222, up-modulated in many tumors, has been reported to target p27Kip1 and p57Kip2 proteins,35,36 two negative cell-cycle regulators that bind to Cdk/cyclin complexes and inhibit the G1/S phase switch. Similarly, members of the miR-17-92 and miR-106b-25 clusters promote cell proliferation by targeting p21Waf/Cip1, which is involved in the same checkpoint.28
Conversely, down-modulated miRNAs in cancer control cell cycle by reducing cyclin and/or Cdk levels. For instance, miR-122, which has been reported to be down-regulated in hepatocellular carcinomas, targets cyclin G1, whose levels are increased in hepatocellular carcinomas and experimental models of hepatocarcinogenesis. Loss of cyclin G1 is associated with a significantly lower tumor incidence after carcinogen treatment.37 Similarly, the mir-15a/16 cluster reduces the levels of cyclin D1, cyclin D3, cyclin E1, and CDK6, suggesting that reduced levels of the miR-16 family trigger an accumulation of multiple cell cycle-promoting genes.38 In addition, these two miRNAs also contribute to apoptosis; they decrease the levels of the apoptotic inhibitor Bcl-2. One report demonstrated that high levels of this protein correlated with low levels of the miR-15a/16 cluster in chronic lymphocytic leukemia.39 Members of the miR-17-92 and miR-106b-25 clusters have also been reported to participate in apoptosis by inhibiting the proapoptotic factor Bim.28,40
Progression and Metastasis
Moreover, a role for miRNAs has been established in the later steps of tumorigenesis, progression, and metastasis. Several miRNAs, such as miR-21 and miR-10b, seem to play an important role. MiR-21 functions as an oncogene and contributes to tumorigenesis, in part through regulation of the tumor suppressor gene tropomyosin 1 (TPM1), a protein involved in cell migration. Recent studies reported that the suppression of miR-21 in metastatic breast cancer cells or malignant hepatocytes significantly reduced invasion and metastasis. Two newly discovered miR-21 targets, programmed cell death 4 (PDCD4) and maspin, have been implicated in these events.41
In another study, Ma and colleagues42 found that the expression of miR-10b was increased in metastatic breast cancer cells compared with healthy or nonmetastatic tumorigenic cells. They also investigated the cause of this up-regulation and identified the transcription factor (Twist) as the inducer of miR-10b overexpression. The authors further validated HOXD10 as a target of miR-10b and showed that a decrease in HOXD10 levels resulted in higher levels of RHOC, which stimulates cancer cell motility.42
MiRNA Applications
miRNAs as Diagnostic and Prognostic Tools
The accumulated data on miRNA expression levels in tumors demonstrate that miRNAs are promising candidates to distinguish different tumors and different subtypes of tumors as well as to predict their clinical behavior. Two large studies have supported the role of miRNAs as either prognostic and/or diagnostic markers. Lu and colleagues analyzed 334 samples, including samples from multiple human cancers. They observed that miRNA expression profile clusters samples by the developmental origin of the tissue. For instance, tissues of epithelial origin cluster separately than tissues of the other origin.5 Moreover, examining the miRNA profile of bone marrow samples from patients with acute lymphoblastic leukemia, they noted that miRNA profile can accurately distinguish tumors reflecting a different mechanism of transformation. Indeed, acute lymphoblastic leukemia samples with different rearrangements, such as mixed lineage leukemia, BCR/ABL, or TEL/AML1, clustered nonrandomly. Tumors of the gastrointestinal tract were also interesting because they all clustered together, reflecting their common derivation from tissue of embryonic endoderm.
Another interesting observation was that miRNAs correlate with cellular differentiation stages. In the same report, they explored also an experimental model in which they treated myeloid leukemia cell line HL-60 with all-trans retinoic acid, a potent inducer of neutrophilic differentiation. MiRNA profiling revealed that the induction of many miRNAs was coincident with the different stages of differentiation. In a different study, Volinia and colleagues,43 by hierarchical clustering analysis of 540 samples including 363 from six of the most frequent solid tumors types (breast, colon, lung, pancreas, stomach, and prostate) and 177 normal controls, showed that a unique miRNA signature enabled the tumor samples to be grouped on the basis of their tissue of origin. Several other miRNA profiling studies have been performed on various cancer types highlighting also the prognostic role of these small RNAs in large set of samples. For example, Murakami and colleagues44 reported that miR-222, miR-106a, and miR-17-92 clusters have been associated with the degree of the differentiation of hepatocellular carcinomas; whereas high expression of miR-21 was associated with poor survival of patients with incidence of pancreas endocrine tumors45 or colon adenocarcinomas.46 This suggests a role for specific miRNAs for identifying disease progression.
On these bases, miRNA profiling has acquired importance in resolving one of the most demanding issues in cancer diagnostics—the origin of metastasis of unknown primary tumor. In a recent study, a miRNA-based tissue classifier was constructed to identify the tissue of origin of metastatic tumors.47 Three-hundred thirty-three formalin-fixed, paraffin-embedded samples, including 205 primary tumors and 131 metastatic tumors, were analyzed on a custom microarray platform and used for this analysis. Briefly, the researchers built the classification algorithm as a branched binary tree: in each node of the tree, classification proceeds to one of two possible branches, grouping together tissues with underlying similarities. The decision at each node is based on level expression of only 48 miRNAs with potential specificity in tissue differentiation and embryogenesis. This allows each cancer type to be assigned to one of two possible branches of the tree. Primary tumors and metastasis from the same tissue were grouped together because no significant differences were observed in miRNA expression. This classifier emerges as more accurate than those using mRNA expression profiling and represents a remarkable advance for determining the origin of metastatic cancer of unknown primary origin.
miRNAs as Therapeutic Tools
Because of the significance of miRNAs in cancer, the management of miRNAs with altered expression in cancer should be considered as a therapeutic strategy. Several reports have demonstrated that miRNAs can be responsible for drug resistance as well.28,48 Thus, miRNAs or anti-microRNA (anti-miRNA) may be used in therapeutics either individually or in combination with other treatments that have lost efficacy. The delivery modality of these molecules critically affects the success of the experimental approach. Hence, several tools have been developed to enable selective targeting of miRNA pathways, which include tools that can either reconstitute reduced miRNAs or reduce overexpressed miRNAs. Recently, technologies have also been described that permit selective protection from miRNAs by limiting accessibility to miRNA binding sites.
Re-Expression of miRNAs
Restoring miRNA expression in diseases in which expression is consistently reduced could be a novel therapeutic approach. Several techniques have been developed to deliver miRNAs in in vitro systems. One of these techniques involves synthetic small RNAs called miRNA mimetics, which contain the exact sequence of the endogenous miRNA. MiRNA mimetics are delivered as perfect complementary duplexes to improve RISC loading of miRNA. Moreover, miRNA mimetics can be modified to have enhanced efficiency by increasing the affinity for a specific target and by reducing other unwanted miRNA effects.49 Another approach to increase expression of specific miRNAs involves introduction of DNA vectors that contain artificial miRNA precursor sequences for miRNA expression. In this case, the mechanism of the silencing of the mRNA target seems to be through mRNA degradation and not translational repression.50 Alternatively, viral vectors might also be used to deliver miRNAs in in vitro systems. Although the devices described above have strong potential to be used in therapy, further studies are necessary to demonstrate that these methods can really operate efficiently by delivering miRNAs in in vivo models.
Anti-miRNAs
Antisense oligonucleotides that bind directly to miRNAs and block their activity are generally named anti-miRNAs. They work by stoichiometric interaction with mature miRNAs, either titrating them from biologically active pools of mature miRNAs or binding to miRNA precursors and inhibiting the biogenesis of mature miRNAs. Early reports describe 2′O-methyl RNA oligonucleotides as efficient anti-miRNAs because the 2′O-methyl group increases the stability of the anti-miRNA molecules. Subsequently, several chemical modifications have been carried to these oligonucleotides to improve their efficacy.51 In vitro experimental procedures demonstrated a powerful inhibitory activity of 2′-O-methoxyethyl oligonucleotides (2′-MOEs), in which every third nucleotide was substituted with a locked nucleic acid (LNA) residue; and of the 2′-fluoro-oligonucleotide with a phosphorothioate backbone.52
The first example of an in vivo knockdown of miRNA has been achieved by using the antagomiR to probe the liver-specific miR-122.53 The antagomiR is a 2′O-methylated oligonucleotide with a cholesterol molecule linked at its 5′ end that improves the cellular uptake of this molecule. However, in two similar studies, Esau and colleagues54 and Elmen and colleagues55 reported that unconjugated 2′-MOE anti-miR-122 and LNA-antimiR-122 also effectively antagonize the liver-expressed miR-122 in nonhuman primates.
Ebert and colleagues56 describe another approach to inhibit the function of specific miRNAs in cells. They used synthetic mRNAs that contained multiple binding sites for an endogenous miRNA. These synthetic mRNAs, or miRNA sponges, bind up the miRNA and prevent its association with its endogenous targets. In culture cells they were at least as effective as LNA anti-miRNAs. Anti-miRNA therapy holds great promise, but care must be taken to confirm that it does not deregulate genes that are not an intended point of therapy.
Target Protectors
A miRNA:mRNA pair usually possesses perfect complementarity between nucleotides 2 and 8 from the 5′ end of the miRNA (seed region). Target protectors (TPs) are small oligonucleotides with perfect complementary to the seed region and to 5′ and 3′ flanking sequences in the 3′ UTR of specific miRNA target genes. Thus, TPs prevent miRNA access to those sites. These specialized oligonucleotides have recently been reported to interfere with miR-430-mediated repression of specific 3′ UTRs in zebrafish.57 Therefore, TPs are advantageous in cases in which the mRNA targets of miRNAs are known. In the years to come, as more and more targets of miRNAs are validated, this technology will become very useful to affect only those miRNA target genes intended as point of therapy.
Conclusion
miRNAs participate in keeping the balance of genes regulating networks that determine the cells’ fate. Deregulation of miRNAs, which is a frequent outcome in human cancer, seriously weakens this balance, thereby contributing to oncogenesis and cancer progression. Several studies have reported that miRNAs affect the expression of genes and pathways involved in cancer pathogenesis from initiation to metastasis disease. However, more research is needed to validate these findings, and murine models genetically modified by deleting or overexpressing putative tumor-suppressor or oncogenic miRNAs could help to identify miRNAs regulating cancer-related pathways. Therapeutics can take advantage of this new knowledge as can diagnostics and prognostics: clinical applications exploiting our understandings of miRNA function will be the next great challenge in cancer research.
Footnotes
Address reprint requests to Carlo M. Croce, M.D., Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 460 West, 12th Ave., Columbus, OH 43210. E-mail: carlo.croce@osumc.edu.
Carlo M. Croce delivered the Keynote Lecture, entitled “Role of MicroRNA Genes in Cancer,” on April 5, 2008 at the annual meeting of the American Society for Investigative Pathology in San Diego, CA.
References
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Visone R, Petrocca F, Croce CM. Micro-RNAs in gastrointestinal and liver disease. Gastroenterology. 2008;135:1866–1869. doi: 10.1053/j.gastro.2008.10.074. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16–1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, Liu CG, Petrocca F, Troncone G, Calin GA, Scarpa A, Colato C, Tallini G, Santoro M, Croce CM, Fusco A. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, Toro JR, Zenger VE, Metcalf RA, Marti GE. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Canaani E, Croce CM. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci USA. 2007;104:10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Motsch N, Pfuhl T, Mrazek J, Barth S, Grasser FA. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- Tsuda N, Ishiyama S, Li Y, Ioannides CG, Abbruzzese JL, Chang DZ. Synthetic microRNA designed to target glioma-associated antigen 1 transcription factor inhibits division and induces late apoptosis in pancreatic tumor cells. Clin Cancer Res. 2006;12:6557–6564. doi: 10.1158/1078-0432.CCR-06-0588. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]