Abstract
Members of the Ets transcription factor family are widely expressed in both the developing and mature mammalian intestine, but their biological functions remain primarily uncharacterized. We used a dominant repressor transgene approach to probe the function of epithelial Ets factors in the homeostasis of the crypt-villus unit, the functional unit of the small intestine. We show that targeted expression in small intestinal epithelium of a fusion protein composed of the Engrailed repressor domain and the Erm DNA-binding domain (En/Erm) results in marked disruption of normal crypt-villus homeostasis, including a cell-autonomous disturbance of epithelial maturation, increased epithelial transit, severe villus dysmorphogenesis, and crypt dysmorphogenesis. The epithelial maturation disturbance is independent of the regulation of TGFβRII levels, in contrast to Ets-mediated epithelial differentiation during development; rather, regulation of Cdx2 expression may play a role. The villus dysmorphogenesis is independent of alterations in the crypt-villus boundary and inappropriate β-catenin activation, and thus appears to represent a new mechanism controlling villus architectural organization. An Analysis of animals mosaic for En/Erm expression suggests that crypt nonautonomous mechanisms underlie the crypt dysmorphogenesis phenotype. Our studies thus uncover novel Ets-regulated pathways of intestinal homeostasis in vivo. Interestingly, the overall En/Erm phenotype of disturbed crypt-villus homeostasis is consistent with recently identified Ets function(s) in the restriction of intestinal epithelial tumorigenesis.
Ets factors comprise a large family of transcription factors found in metazoans.1,2 Numbering as many as 27 in humans, Ets factors are related to each other by a conserved DNA-binding domain (DBD), the Ets domain.1,2,3 The Ets domain is a winged helix-turn-helix structural motif, which binds to a core GGA(A/T) DNA sequence.1,2,3,4 On binding to DNA, Ets factors regulate gene promoter activity directly via intrinsic activation or, less commonly, repression domains, or indirectly through interactions with other transcription factors.1,2 Frequently acting as nuclear effectors of growth factor receptor-mediated signaling pathways, Ets factors control the expression of genes involved in diverse cellular processes, including cell proliferation, apoptosis, differentiation, and cell-cell/cell-matrix interactions.1,2,5,6
Ets factors are widely expressed in a variety of developing and adult mammalian tissues.7 Genetic inactivation of individual Ets factors in the mouse has revealed unique, essential Ets functions in diverse biological processes, including hematopoiesis, immune function, lymph/angiogenesis, neurogenesis/neuromuscular function, spermatogenesis, early embryonic patterning, and development of extraembryonic tissues.1,7,8,9,10,11,12,13,14,15,16 Interestingly, such studies have been remarkable for the absence of phenotypes in a number of tissues with Ets expression, especially those comprising solid organs. The relative paucity of Ets phenotypes in such tissues appears to be attributable to the multiplicity of expression of different Ets factors in the same cell, combined with the ability of co-expressed Ets factors to compensate for each other’s function.4,17,18
Most Ets genes are expressed in the developing and/or mature mammalian intestine, frequently in a tissue compartment-specific and/or temporally dynamic manner.19 However, of the 16 (of 26 total) murine Ets genes inactivated to date, only 1, Elf3/ESE1/ESX/ERT/JEN, has been reported to have an intestinal phenotype.20 As in other systems,21 the absence of phenotypes in other Ets knockout mice is likely attributable to Ets genetic compensation in vivo. The use of genetically manipulated Ets factors with dominant activity has proven an effective way to overcome Ets genetic compensation. Such an approach has been used successfully to uncover and characterize Ets functions in a number of in vivo and in vitro systems, including lung morphogenesis, mammary tumorigenesis, and neuromuscular synapse function in the mouse,21,22,23 neural crest differentiation,24,25 Schwann cell survival,26 and oncogenic cellular transformation.27,28 In the present study, we used the dominant Ets approach to probe the spectrum of functions of Ets transcription factors in the epithelial compartment of the mammalian intestinal crypt-villus unit. Specifically, we used an Ets-dominant repressor, composed of the repressor domain of the Drosophila Engrailed (En) protein fused to the DNA-binding domain of the Ets factor Erm/Etv5, to block endogenous Ets activity in vivo. As shown herein, En/Erm expression in the small intestinal epithelium under control of the well-characterized villin promoter/enhancer reveals Ets functions in multiple aspects of crypt-villus homeostasis, including epithelial maturation, epithelial transit, and complex architectural organization of the crypt-villus unit.
Materials and Methods
DNA Constructs
The expression constructs pSG5-HA/En/Erm, pSG5-HA/ErmDBD, and pSG5-HA/EnRD were derived from the construct pTRE-HEEN (generously provided by John Shannon, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH). pTRE-HEEN contains the murine Erm DNA-binding domain (DBD) (amino acids 364 to 449) and the Drosophila Engrailed repressor domain (EnRD; amino acids 2 to 298). pSG5-HA/ErmDBD and pSG5-HA/EnRD were generated by polymerase chain reaction (PCR) amplification of the ErmDBD and EnRD, respectively, from pTRE-HEEN and subcloning into pTRE-HA (Clontech, Palo Alto, CA), followed by PCR amplification of the HA-tagged inserts and subcloning into pSG5.29 pSG5-HA/En/Erm was generated by PCR amplification of both the EnRD and ErmDBD from pTRE-HEEN and subcloning into pTRE-HA, followed by PCR amplification of the HA-tagged En/Erm fusion and subcloning into pSG5. A seven amino acid (GGGSGGG) spacer was added between the EnRD and ErmDBD of the En/Erm fusion during the first PCR cloning step. All constructs also contained a C-terminal nuclear localization sequence (NLS; PKKKRKV, from the SV40 large T antigen), added during the first PCR amplification step. pSG5-HA/Erm was generated by subcloning a full-length Erm cDNA, amplified from a mouse embryonic brain library by reverse transcriptase (RT)-PCR, into pTRE-HA (Clontech), and then subcloning of the HA-tagged insert into pSG5. pSG5-HA/Ets2 was generated by subcloning a full-length mouse Ets2 cDNA (generously provided by James Hagman, National Jewish Medical and Research Center, Denver, CO) into pCGN2-HA,30 and then subcloning the HA-tagged insert into pSG5. pSG5-HA/Elf3 was generated by subcloning HA-tagged full-length human Elf331 into pSG5. The reporter construct 8x(EBS)-TK-luciferase was generated by subcloning the 8xpal sequence (containing eight copies of the DNA-binding site GCAGGAAGCA from the rat stromelysin promoter) from 8xpal-pBLCAT31 into pA3-TK-luciferase.32 The transgenic construct villin-En/Erm was generated by subcloning the HA-tagged En/Erm fusion (also containing the C-terminal NLS) from pTRE-HA/En/Erm into the p12.4-kb Vill plasmid (generously provided by Deborah Gumucio, University of Michigan, Ann Arbor, MI). All plasmid DNA constructs were confirmed by diagnostic restriction enzyme digestion and, when PCR was used in the cloning process, DNA sequencing.
Cell Culture, Transfection, Reporter Assays, and Immunoblotting
HeLa cells were grown as previously described.31 For assays of transcriptional activity, cells were plated in 96-well plates at a density of 4 × 104 cells per well, and were transfected 15 to 18 hours later with 100 ng of the 8xEBS-TK-luciferase reporter plasmid, 1 ng of Renilla-luciferase plasmid, and varying amounts of expression plasmid(s), with the total amount of DNA kept constant by the addition of empty pSG5 expression vector. The cells were harvested 18 to 24 hours later, and luciferase activity was measured as previously described.31 For protein expression analysis, HeLa cells (3 × 106 cells in 200 μl of medium) were mixed with varying amounts of expression plasmid(s), the total amount of DNA being kept constant at 10 μg by the addition of empty pSG5 expression vector. Cells were transfected by electroporation using a Bio-Rad (Hercules, CA) Gene Pulser set at 220 V and 500 μF. Electroporated cells were diluted into 3 ml of medium in 60-mm plates and incubated for 24 hours. Cells were harvested in 0.5 ml of phosphate-buffered saline (PBS)/ethylenediaminetetraacetic acid, pelleted, and lysed in 100 μl of hot (65°C) TEA lysis buffer (55 mmol/L triethanolamine, pH 7.5, 111 mmol/L NaCl, 2.2 mmol/L ethylenediaminetetraacetic acid, and 0.44% sodium dodecyl sulfate) with the complete protease inhibitor cocktail (Roche, Indianapolis, IN). The lysates were vortexed, placed on ice, boiled for 5 minutes, returned to ice, and passed 7 to 10 times through a 27-gauge needle using a 1-cc syringe. Lysate protein concentration was determined by the Bradford assay, using the Bio-Rad Protein Assay reagent. Equal amounts of total extract protein were resolved on a sodium dodecyl sulfate-polyacrylamide gel and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Immunoblotting was performed as previously described.31 Primary antibodies used were anti-HA mouse monoclonal (1:1000; Covance, Princeton, NJ) and anti-tubulin mouse monoclonal (1:1000; Oncogene, Cambridge, MA); secondary antibody used was horseradish peroxidase-conjugated goat anti-mouse IgG (1:5000, Bio-Rad). Detection was performed using the SuperSignal West Pico chemiluminescent substrate kit (Pierce, Rockford, IL).
Transgenic Animals
The villin-En/Erm transgenic construct, excised from the vector backbone by digestion with PmeI, was injected into the pronuclei of fertilized ova from FVB/N mice by the University of Colorado Cancer Center Transgenic/Knockout Core Facility. Transgenic animals were identified by PCR genotyping of tail-biopsy DNA using primers to the Engrailed repressor domain (5′-TGGAGTTTAGCCGGCAACAG-3′ and 5′-TGGCATCGCTCATCTTGGAG-3′); PCR of mouse actin DNA (primers 5′-TATCCTGACCCTGAAGTACC-3′ and 5′-GGTCAGGATCTTCATGAGGT-3′), performed in the same reaction, served as a control. Transgenic animals were maintained in an FVB/N background. Adult transgenic animals were subjected to phenotypic analysis, with littermates or age-matched nontransgenic animals serving as controls.
Histology and Immunohistochemistry
For BrdU-labeling experiments, animals were injected intraperitoneally with 50 mg/kg body weight of BrdU (Sigma, St. Louis, MO) in PBS, 2 or 24 hours before euthanasia. All animals were euthanized using CO2. The small intestine was immediately harvested and cut into three segments of approximately equal length. Fecal contents were gently expelled, the lumen was injected with fixative (4% paraformaldehyde), and the intestine was rolled concentrically and placed in a histology cassette. Tissues were fixed for 24 hours in 4% paraformaldehyde at 4°C, after which the tissues were placed in 70% ethanol, processed further on a standard histology processor, and paraffin-embedded. Sections 4-μm-thick were stained with hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), or processed further for immunohistochemical staining. For immunohistochemical staining, sections were deparaffinized and rehydrated. Antigen retrieval was performed by incubating the slides in 10 mmol/L sodium citrate buffer, pH 6.0, for 1 hour in a Biocare (Walnut Creek, CA) medical decloaker. Endogenous peroxidase activity was blocked by incubation in 3% H2O2 for 10 minutes. For BrdU immunostaining, slides were incubated for 90 minutes in 2 N HCl, washed with ddH2O, incubated for 5 minutes in 0.1 mol/L sodium borate, and washed again with ddH2O, before blockade of peroxidase activity. Immunohistochemical staining was performed using the Vectastain ABC or ImmPRESS kit (Vector Laboratories, Burlingame, CA), and developed using diaminobenzidine (DAKO, Carpinteria, CA, or Sigma). Primary antibodies used were: horseradish peroxidase-conjugated rat anti-HA (1:25, Roche) rabbit anti-iAP (1:500; Abcam, Cambridge, MA), rabbit anti-iFabp (Jeffrey Gordon, Washington University. St. Louis, MO); goat anti-Mcm6 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Ki-67 (1:200; NeoMarkers, Fremont, CA), mouse anti-BrdU (1:40; BD Biosciences, San Jose, CA), rabbit anti-TGFβRII (1:100, Santa Cruz Biotechnology), mouse anti-Cdx2 (1:25, Abcam), and mouse anti-β-catenin (1:500; BD Biosciences/Transduction Laboratories). Biotinylated secondary antibodies used were: goat anti-rabbit IgG and goat anti-mouse IgG (Vector Laboratories), and donkey anti-goat IgG (Santa Cruz Biotechnology). All immunohistochemically stained slides were counterstained with hematoxylin, dehydrated, mounted, and coverslipped.
Results
The En/Erm Dominant Repressor Potently Blocks Promoter Activation by Ets Transcription Factors
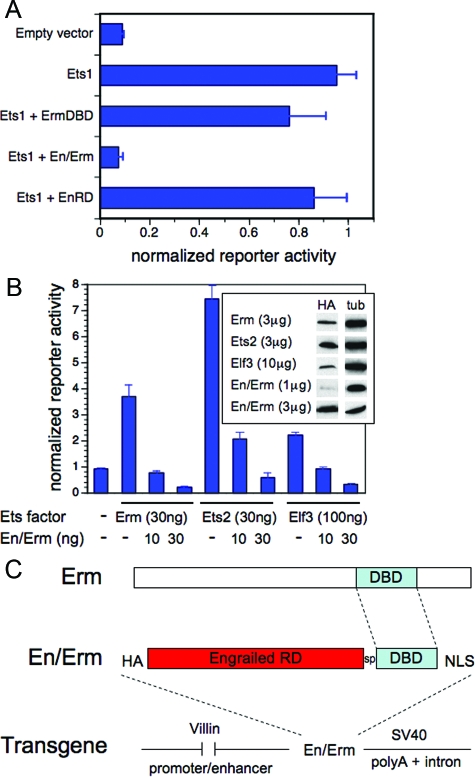
Most Ets transcription factors are expressed in the developing and/or mature mammalian intestine.19 Although 16 (of 26 total) Ets genes have been genetically inactivated in the mouse to date, only 1 (Elf3) has been reported to have an intestinal phenotype.20 The absence of intestinal phenotypes in the other Ets knockout animals is likely attributable to genetic compensation by co-expressed Ets factors. Ets genetic compensation can be overcome by the use of genetically manipulated Ets factors with dominant activity.21,22,23,24,25,26,27,28 We adopted this approach to probe the spectrum of Ets functions in the homeostasis of the intestinal crypt-villus axis in vivo. One possible dominant Ets approach, previously used by some, is the use of the Ets DNA-binding domain alone as a competitive inhibitor.23,25,26,27,28 We considered such an approach, but were concerned that it would rely too heavily on high expression levels, which can be difficult to achieve in transgenic systems. Because most Ets factors function predominantly as transcriptional activators,1,2,4 an alternative approach, used by others, is the use of an Ets dominant repressor, composed of the Ets DNA-binding domain (DBD) and the Drosophila Engrailed repressor domain.21,22,24 The Engrailed repressor domain functions by an active repression mechanism,33 and should thus effect more potent blockade of endogenous Ets promoter activity than the Ets DBD alone at similar expression levels. To test this, we generated the HA epitope-tagged construct En/Erm, composed of the Engrailed repressor domain (EnRD) fused to the amino terminus of the DNA-binding domain of the Ets factor Erm (ErmDBD). As shown in Figure 1A, when tested in a transient co-transfection assay, En/Erm was able to fully block transcriptional activation of an Ets-responsive reporter construct by Ets1. In comparison, an equivalent amount of the Erm DBD alone had a much weaker effect (Figure 1A). Importantly, EnRD alone had little effect on Ets activation (Figure 1A), indicating that the blocking effect of the En/Erm construct requires DNA binding, and is not attributable to nonspecific activity of the En repressor domain alone. These findings are similar to those of Liu and colleagues,21 who, of note, uncovered an Ets pulmonary dysmorphogenesis phenotype using a transgene expressing an Engrailed-ErmDBD fusion, but not the Erm DBD alone. Based on these analyses, we selected En/Erm for blocking Ets-dependent gene expression in vivo.
Figure 1.
Transcriptional blocking activity of the En/Erm dominant repressor in vitro. HeLa cells were transfected with the reporter plasmid 8x(EBS)-TK-luciferase and the indicated plasmid DNA expression constructs. A: Transfected DNA amounts were: Ets1, 50 ng; ErmDBD, 250 ng; En/Erm, 150 ng; EnRD, 50 ng (chosen to normalize for differences in construct expression levels). B: Transfected DNA amounts were as shown. Reporter activity, determined by quantitative luminometry, was normalized to the activity of the co-transfected Renilla-luciferase construct; results are expressed as mean and SD of triplicate transfections. All constructs were expressed from the plasmid pSG5, and all except Ets1 have an N-terminal HA epitope tag. Protein expression (B, inset) was determined by immunoblotting with antibody against HA, and tubulin (tub) as loading control. C: Modular organization of the villin-En/Erm dominant repressor transgene. The transgene consists of the 12.4-kbp villin promoter-enhancer fragment, a HA epitope tag, the Drosophila Engrailed repressor domain (RD), a 7-amino acid (GGGSGGG) spacer (sp), the Erm DNA-binding domain (DBD), a 7-amino acid (PKKKRKV) SV40 large T-antigen nuclear localization sequence (NLS), and a SV40 intron and polyA tail.
Because of the high conservation of the DBD throughout the Ets family,4 we expected that En/Erm would be capable of blocking the activity of a number of different Ets factors. To test this, we examined En/Erm blocking activity against Erm, Ets2, and Elf3, representative members of different subfamilies expressed in the intestinal epithelium.19 Blockade by En/Erm was tested at both low- and high-protein expression levels, relative to Erm, Ets2, and Elf3, in a transient co-transfection assay. As shown in Figure 1B, En/Erm was able to block activation of an Ets-responsive reporter by all three Ets factors, at equivalent (for Ets2) or lower (for Erm and Elf3) relative protein expression levels. Thus, En/Erm has the ability to block the activity of multiple different Ets factors, and should therefore be a good reagent for probing the spectrum of Ets functions in vivo.
Characterization of Villin-En/Erm Transgenic Animals
En/Erm expression was targeted to intestinal epithelium using the well-characterized 12.4-kbp villin promoter.34 This promoter drives gene expression in small intestinal epithelium, and to a lesser extent colonic epithelium, from approximately embryonic day 12.5 on through adulthood, with expression greater in the villi than crypts. The modular organization of the villin-En/Erm transgene is shown in Figure 1C. Multiple transgenic lines were analyzed. Transgene expression was assayed by RT-PCR (not shown) and immunohistochemical staining against the HA epitope tag (Figure 2, A and B). Animals from one stable transgenic line and two independent transgene-positive mosaic founders manifested robust transgene expression detectable by immunohistochemistry (Figure 2B) and similar phenotypes in the small intestine under physiological conditions. These animals were thus further analyzed in detail. The similarity of the phenotypes in animals arising from three independent transgene insertion events confirms that the phenotypes are attributable to En/Erm expression, and not integration site effects. In agreement with previous studies,34 transgene expression was higher in the villi than the crypts (Figure 2B), and greater in the small intestine than the colon (data not shown). The transgene was expressed specifically in the nuclei of epithelial cells (Figure 2B), as expected and required for its dominant repressor effect on Ets-regulated gene expression.
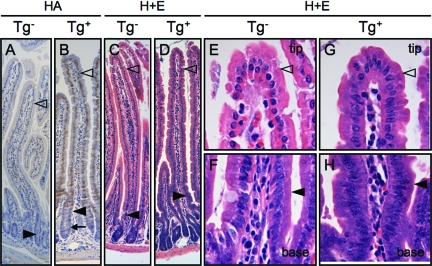
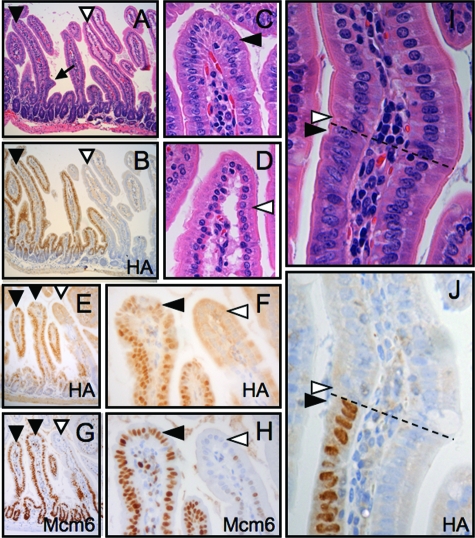
Figure 2.
Transgene expression and impaired enterocyte maturation in En/Erm-expressing small intestine. HA immunostaining of small intestine from control (A) and transgenic (B) animals shows the expression of En/Erm protein specifically in epithelial nuclei of transgenic animals. En/Erm expression was strong in the villus epithelium (B, open arrowhead), and was also observed in the superficial aspects of crypts (B, filled arrowhead), but not the deep aspects of crypts (B, arrow); no immunostaining was observed in control animals (A). H&E-stained sections of small intestine from control (C; detailed views of villus in E and F) and transgenic (D; detailed view of villus in G and H) animals; note morphological resemblance of transgenic enterocytes at the villus tip to enterocytes at the villus base (filled arrowheads, enterocytes at villus base; open arrowheads, enterocytes at villus tip).
En/Erm Expression in Small Intestinal Epithelium Reveals Novel Ets Functions in Villus Epithelial Maturation and Transit
In animals stably expressing immunohistochemistry-detectable En/Erm transgenic protein, the morphology of enterocytes (absorptive epithelial cells) along the length of the small intestine appeared abnormal on H&E-stained histological sections. In enterocytes from nontransgenic animals, a gradual histomorphological change could be seen as the cells progressed from the villus base to the villus tip, characterized by increasing cytoplasmic eosinophilia, and rounding and more basal position of the nucleus (Figure 2, C, E, and F). In contrast, in En/Erm-expressing animals from the stable transgenic line, enterocytes along the length of the villus maintained a morphology that resembled the cells at the base, characterized by darker, more amphophilic cytoplasm, and more elongated and centrally positioned nuclei (Figure 2, D, G, and H). By H&E histomorphology, this phenotype thus suggested impaired enterocyte maturation.
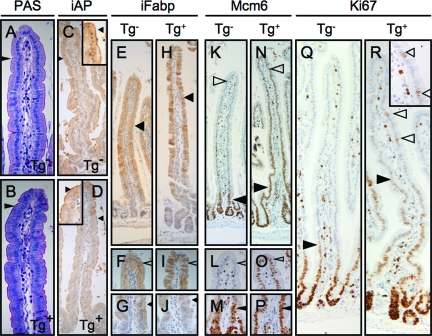
To further characterize this phenotype, we examined the expression of markers of enterocyte differentiation and maturation. The transgenic enterocytes manifested a well-formed PAS-positive glycocalyx, ultrastructurally well-formed microvilli, and expression of intestinal alkaline phosphatase (iAP) and intestinal fatty acid binding protein (iFabp) (Figure 3, A–J; and data not shown); indeed, as in control animals, there appeared to be an appropriate gradient of increased iFabp expression from villus base to tip in the transgenic intestines. Thus, commitment to the enterocyte lineage and some aspects of enterocyte maturation were intact in the transgenic animals. However, in striking contrast to controls, transgenic intestines showed inappropriately persistent expression of Mcm6, a marker of villus epithelial immaturity,35 along the entire villus axis (Figure 3, K–P); furthermore, in the setting of high transgene levels, we observed residual Ki-67 expression in more superficial villus epithelium, not seen in controls (Figure 3, Q and R). Because the expression of these proteins is normally restricted to the less mature cells in the crypt and more basal villus epithelium (Figure 3, K–M, and Q), these findings support the presence of a disturbance of maturation in the transgenic enterocytes. Interestingly, the overall findings of histomorphological immaturity and misexpression of some (eg, Mcm6), but not other (eg, iFabp), enterocyte maturation markers suggest that En/Erm expression results in enterocyte maturation dys-synchrony.
Figure 3.
Characterization of enterocyte maturation disturbance in En/Erm transgenic animals. Histochemical (PAS) and immunohistochemical (iAP, iFabp, Mcm6, and Ki-67) staining of small intestine from control (Tg−: A, C, E--G, K--M, and Q) and transgenic (Tg+: B, D, H--J, N--P, and R) animals. Arrowheads in A and B: PAS-positive glycocalyx on enterocytes; arrowheads in C and D: iAP expression in superficial aspect of enterocytes (insets: detailed views of villus tips); arrowheads in E--J: iFABP expression in enterocyte cytoplasm (F and I: detailed views of villus tips in E and H, respectively; G and J: detailed views of villus bases in E and H, respectively); arrowheads in K--P: Mcm6 expression in enterocyte nuclei at villus base (filled arrowheads) and villus tip (open arrowheads; L and O: detailed views of villus tips in K and N, respectively; M and P: detailed views of villus bases in K and N, respectively; filled arrowheads in Q and R: upper limit of residual Ki-67 immunopositivity in villus enterocytes; open arrowheads in Q and R: solitary ectopic Ki-67 immunopositivity in transgenic superficial villus enterocytes (inset: detailed view).
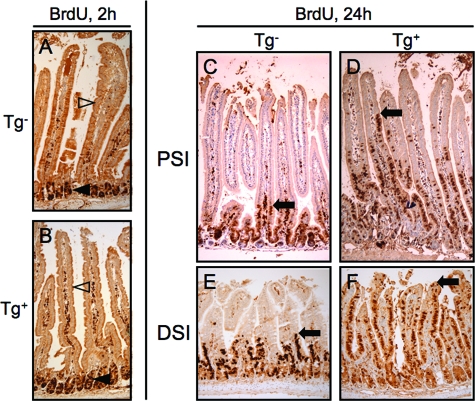
The disturbed maturation of the En/Erm-expressing enterocytes prompted us to examine the intestinal epithelium for proliferative activity, which is normally restricted to the crypt but can inappropriately spread to villi in the context of some genetic manipulations.36 By H&E examination, no mitotic figures were detected in the villus epithelium, and brief (2-hour) in vivo BrdU labeling revealed epithelial proliferative activity appropriately restricted to the crypt compartment, as in controls (Figure 4, A and B). Thus, the immature-appearing En/Erm-expressing enterocytes are postmitotic cells, and the transgenic intestine maintains a normal crypt-villus boundary. We next examined the rate of epithelial transit along the crypt-villus axis because this could in principle account for altered epithelial maturation in the transgenic animals. After cell division in the crypt, intestinal epithelial cells normally migrate in an orderly manner out of the crypt, and from villus base to tip, where they are ultimately shed into the lumen.36 The rate of epithelial transit can be assayed in vivo by a BrdU pulse followed by an extended chase period (eg, 24 hours), which allows postmitotic, BrdU-labeled cells to migrate up the axis.37 As shown in Figure 4, C–F, En/Erm-expressing animals showed much more rapid epithelial migration compared with nontransgenic animals. En/Erm expression thus results in more rapid epithelial transit along the crypt-villus axis.
Figure 4.
BrdU immunostaining of small intestine from control (Tg−: A, C, and E) and transgenic (Tg+: B, D, and F) animals pulsed in vivo with BrdU and analyzed 2 hours (A and B) or 24 hours (C--F) later (filled arrowheads: crypt epithelial cells; open arrowheads: villus epithelial cells; arrows: upper limit of epithelial cell transit after BrdU incorporation in the crypt; PSI: proximal small intestine; DSI: distal small intestine).
The identical disturbance of enterocyte maturation was observed in En/Erm-expressing foci of the independent transgenic founders with mosaic expression of the transgene. This was evident by both histomorphological examination (Figure 5, A–D) and Mcm6 immunostaining (Figure 5, E–H). Interestingly, individual mosaic villi in these animals showed an abrupt transition between immature En/Erm-positive cells and directly adjacent En/Erm-negative cells, with the latter consistently exhibiting maturation appropriate for their position along the villus axis (Figure 5, I–J). This indicates that the En/Erm-induced disturbance of enterocyte maturation is cell-autonomous, and thus unlikely to be attributable to global acceleration of epithelial transit alone.
Figure 5.
Characterization of enterocyte maturation disturbance in En/Erm mosaic animals. H&E-stained (A) and HA-immunostained (B) small intestinal focus mosaic for En/Erm expression (filled arrowhead: En/Erm expressing villus; open arrowhead: nonexpressing villus; arrow: incipient villus branch). Detailed views of the tips of the En/Erm expressing (C) and nonexpressing (D) from A (arrowheads: enterocytes). HA (E and F) and Mcm6 (G and H) immunostaining of a small intestinal focus mosaic for En/Erm expression (arrowhead designations are as in A--D). As above, En/Erm expression appears as immunopositivity in epithelial nuclei (filled arrowhead; contrast with absence of nuclear immunopositivity, shown by open arrowheads, in adjacent villus). Detailed view of H&E-stained (I) and HA-immunostained (J) individual villus mosaic for En/Erm expression. Note immature morphology of En/Erm-expressing enterocyte (filled arrowhead), but maturation appropriate for position along villus axis (dashed line) of adjacent nonexpressing enterocyte (open arrowhead).
Enterocytes represent one epithelial lineage in the small intestine, the others being the secretory lineages, composed of goblet cells, Paneth cells, and neuroendocrine cells.36 To determine whether En/Erm expression affected these lineages, we performed quantitative morphometric analysis of secretory epithelial cells. The overall number and localization of goblet cells, neuroendocrine cells, and Paneth cells were not significantly different from nontransgenic controls (data not shown). Thus, En/Erm expression does not affect secretory epithelial cell specification.
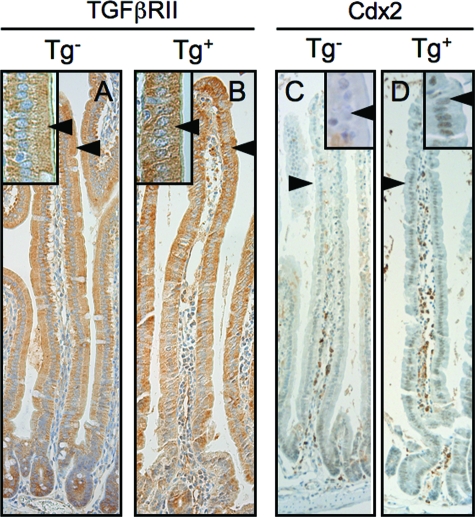
Ets-regulated epithelial expression of transforming growth factor-β type II receptor (TGFβRII) has recently been shown to play an important role in small intestinal epithelial differentiation during embryogenesis.20,38 We thus examined TGFβRII expression in En/Erm-expressing small intestines with disturbed maturation. In contrast to the dysmature embryonic small intestine of Elf3 knockout mice,20 we did not see loss of TGFβRII expression in adult En/Erm-expressing small intestine (Figure 6, A and B). Thus, additional, TGFβRII-independent, Ets-mediated pathways appear to regulate epithelial differentiation in the adult small intestine. Interestingly, in the setting of high-level En/Erm expression, we observed an inappropriate persistence of villus Cdx2 expression in the proximal small intestine (Figure 6, C and D), suggesting a possible role for this important regulator of intestinal epithelial differentiation in the disturbed maturation phenotype.
Figure 6.
Analysis of potential mediators of the disturbed epithelial maturation phenotype. TGFβRII (A and B)- and Cdx2 (C and D)-immunostained small intestine from control (Tg−) and transgenic (Tg+) animals. Note similar level of expression of TGFβRII in control and transgenic animals (arrowheads in A and B), and inappropriate persistence of Cdx2 expression throughout villi of transgenic animals compared with controls (arrowheads in C and D). TGFβRII is cytoplasmic, whereas Cdx2 is nuclear. Insets: Magnified views of villus epithelium.
En/Erm Expression in Small Intestinal Epithelium Results in Villus and Crypt Architectural Disorder
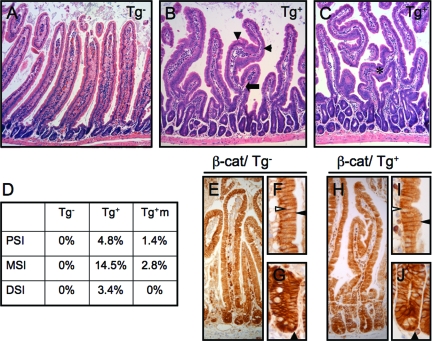
Small intestinal villi maintain an exquisitely ordered architecture under normal homeostasis, characterized by even spacing, unidirectional (radial) growth, relatively constant height for a given intestinal segment, and absence of branching. Compared with villi from nontransgenic animals (Figure 7A), villi in En/Erm-expressing animals showed marked architectural disorganization (Figure 7, B and C), including branch formation (arrows), mid-villus changes in the direction of growth (arrowheads), and villus bridging. The degree of villus dysmorphogenesis correlated with the level of En/Erm expression in individual animals. On average, as many as 14.5% of villi showed branching in a given intestinal segment (Figure 7D). Villus branching was also observed in the independent transgenic animals mosaic for En/Erm expression (Figure 7D). Recently, ectopic activation of β-catenin in villus epithelium, either directly or secondary to blockade of Hedgehog signaling, has been shown to result in marked disruption of villus morphogenesis during intestinal development.39,40 However, we did not observe ectopic nuclear translocation, indicative of activation, of β-catenin in the dysmorphogenic En/Erm-expressing intestine (Figure 7, E–J; in control experiments, the same antibody was able to robustly detect inappropriate activation of β-catenin in the setting of epithelial neoplasia (Supplemental Figure S1 available at http://ajp.amjpathol.org). These findings suggest that the En/Erm-induced villus dysmorphogenesis occurs independently of inappropriate β-catenin activation, and appears to uncover a novel pathway regulating normal villus architecture.
Figure 7.
Villus architectural dysmorphogenesis in En/Erm-expressing small intestine. A–C: H&E-stained sections of small intestine from control (Tg−) and transgenic (Tg+) animals (arrow: villus branching; arrowheads: villus turns; asterisk: villus bridging). D: Quantitation of villus branching in control (Tg−, n = 8) and transgenic (Tg+, n = 3) animals, and transgene-expressing foci in mosaic animals (Tg+m, n = 2), in proximal (PSI), mid (MSI), and distal (DSI) small intestine. β-Catenin immunostaining in control (E–G) and transgenic (H–J) animals. Note nuclear staining (filled arrowheads, G and J) limited to epithelial cells at bases of crypts, and membranous (filled arrowheads, F and I) but no nuclear (open arrowheads, F and I) staining in villi, in both transgenic and control animals.
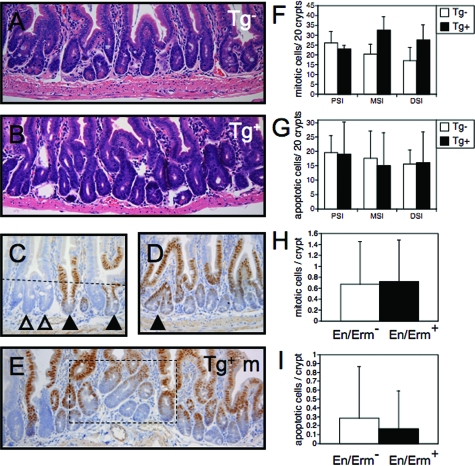
Like the villi, the small intestinal crypts maintain an orderly architecture, with relatively constant spacing, orientation, and size of crypts along the length of the intestine. In contrast to nontransgenic animals (Figure 8A), the crypts in En/Erm-expressing animals showed greater variation in number, size, and orientation (Figure 8B). As in the case of the villus dysmorphogenesis, the crypt phenotype correlated with the level of En/Erm expression in individual animals. By morphometric analysis, we did not observe statistically significant differences in crypt epithelial proliferation or apoptosis, possible mechanisms of altered crypt homeostasis (Figure 8, F and G). There appeared to be an overall trend toward increased crypt number and cellularity in En/Erm-expressing animals, but these differences did not reach statistical significance (data not shown). Interestingly, in the mosaic lines, similar crypt changes were observed, but were dependent on the size of the En/Erm-expressing focus. In areas with highly mosaic expression (ie, extensive intermixing of En/Erm-positive and -negative crypts), there was little effect of En/Erm on individual crypt size and morphology (Figure 8C). However, in larger foci of En/Erm protein expression, alterations in the crypt compartment were observed, including crypt branching and disorder of orientation (Figure 8, D and E). Morphometric analysis of crypt epithelial proliferation and apoptosis in the mosaic animals did not reveal statistically significant differences between En/Erm-expressing and nonexpressing foci (Figure 8, H and I). Thus, the alterations in crypt homeostasis in areas of En/Erm expression appear to involve crypt nonautonomous mechanisms, independent of significant effects on crypt epithelial proliferation or apoptosis.
Figure 8.
Crypt alterations in En/Erm-expressing small intestine. A and B: H&E-stained small intestine from control (Tg−) and transgenic (Tg+) animals. Note crypt disorder, including increased variation in crypt position and size in transgenic animals (B) relative to controls (A). C–E: HA immunostaining of small intestine in mosaic transgenic animals (open arrowheads: non-expressing crypts; black arrowheads: En/Erm-expressing crypts; dashed line in E: crypt-villus boundary; dashed box in G: crypt dysmorphogenesis in a large En/Erm-expressing mosaic focus. Crypt mitotic (F) and apoptotic (G) activity in proximal (PSI), mid (MSI), and distal (DSI) small intestine of control (Tg−, n = 7 and 8, respectively) and transgenic (Tg+, n = 3 for each) animals. Average mitotic (H) and apoptotic (I) cells per En/Erm nonexpressing (En/Erm−, white bars) and expressing (En/Erm+, black bars) crypts in mosaic animals (49 total expressing and nonexpressing crypts along the entire small intestine were scored for each; values are expressed as average and SD). Mitotic and apoptotic cells were scored by their characteristic morphology on H&E-stained sections, in well-oriented, fully-visualized crypts; none of the comparisons between experimental and control groups yielded statistically significant differences (P < 0.05).
Discussion
Ets Factors and Intestinal Epithelial Maturation and Transit
Our studies using targeted expression of the Ets dominant repressor En/Erm in the intestine reveal multiple novel roles for epithelial Ets transcription factors in small intestinal crypt-villus homeostasis. First, En/Erm expression leads to disturbed maturation of the enterocyte epithelial lineage. This phenotype is interesting, and to our knowledge unique, in that it shows features of dys-synchrony, whereby some maturation steps, namely accumulation of cytoplasmic iFabp, proceed normally, whereas others, namely extinction of nuclear Mcm6, do not. Indeed, this phenotype suggests that enterocyte maturation in the adult intestine is controlled by multiple parallel and independent pathways.
The Ets transcription factor Elf3 has recently been shown to be an important regulator of intestinal epithelial differentiation during embryonic development,20,38 raising the possibility that the En/Erm phenotype is attributable to blockade of Elf3 in the adult intestine. However, although superficially similar (impaired epithelial differentiation/maturation), the En/Erm phenotype differs from the Elf3−/− phenotype in several respects. We do not observe the microvillus dysmorphogenesis or decreased goblet cell numbers seen in Elf3−/− embryonic intestine. Further, in contrast to Elf3−/− embryonic intestine, we do not observe loss of TGFβRII expression in association with the villin-En/Erm disturbed epithelial maturation phenotype. Interestingly, epithelial-specific deletion of TGFβRII in the mouse small intestine is reported to be free of phenotypic changes under conditions of homeostasis.41 Together, these findings suggest that signaling via TGFβRII, although apparently important in the developing intestine, may have a less important role in epithelial differentiation in the adult intestine. Interestingly, our studies suggest a possible role for Cdx2 in Ets-mediated epithelial maturation in the adult intestine. Cdx2 is known to be able to promote intestinal epithelial differentiation.36 However, in the proximal adult small intestine, Cdx2 is expressed in a diminishing gradient along the villus axis (Figure 6C),42 opposite to the gradient of epithelial maturation. Moreover, we observed inappropriately persistent Cdx2 expression in the setting of En/Erm-induced epithelial immaturity. Together, these findings suggest that Cdx2 may function as a negative regulator of intestinal epithelial maturation, and that the role of Ets factors in adult intestinal epithelial maturation may involve down-regulation of Cdx2 expression. Whether the En/Erm epithelial dysmaturation phenotype uncovers TGFβRII-independent activity of Elf3, and/or the activity of other Ets factor(s) remains to be determined.
The increased intestinal epithelial transit in villin-En/Erm animals is the first demonstration of a role for Ets factors in epithelial movement along the crypt-villus axis, an important parameter of intestinal homeostasis. Given the villus-predominant pattern of En/Erm expression and the lack of a crypt-autonomous En/Erm phenotype in the transgenic animals, we think that the increased transit phenotype uncovers a specific function of Ets factors in the control of epithelial movement along the crypt-villus axis, rather than a nonspecific effect of increased crypt epithelial production. Little is currently known about mechanisms specifically controlling epithelial transit in the intestine. E-cadherin has been shown to be one important regulator of this process.43 However, by immunohistochemical staining, we did not observe alterations in E-cadherin expression or localization in villin-En/Erm animals with increased transit (data not shown). Thus, Ets factors appear to regulate intestinal epithelial transit independently, or alternatively downstream, of E-cadherin. The apparent absence of a compensatory increase in epithelial production in response to the accelerated transit in the transgenic animals is somewhat surprising. We suspect that this reflects our inability to detect small changes in proliferation and/or apoptosis in our analyses; indeed, although not statistically significant, there appeared to be a trend toward increased crypt epithelial proliferation in the mid-small intestine of transgenic animals, where phenotypes were also generally most pronounced (Figure 8F). Interestingly, it is worthy of mention that alterations in villus epithelial transit, apparently unaccompanied by changes in crypt epithelial production, have been reported in the setting of other genetic manipulations of the intestine.37,44
Ets Factors and Architectural Organization of the Crypt-Villus Axis
The crypt-villus unit of the small intestine forms relatively late in development,36 and from here on maintains an exquisitely organized architecture: individual units are precisely spaced, and the separate identity, size (depth of crypts and height of villi), and orientation (perpendicular to the length of the intestine) of the crypt and villus components are precisely maintained. Opposing activities of Wnt and Hh signaling, as well as Ephrin signaling, itself regulated by the Wnt pathway, normally restrict epithelial proliferation to the crypt compartment, and differentiation to the villus compartment.36,40 Disruption of the Wnt and Hh pathways, directly or indirectly resulting in ectopic activation of β-catenin, can give rise to marked alterations in villus architecture and loss of the normal crypt-villus boundary.39,40 Interestingly, En/Erm expression causes villus dysmorphogenesis of similar severity, but independent of alterations in the crypt-villus boundary or inappropriate β-catenin activation. Little is currently known about mechanisms specifically regulating parameters of villus morphology, including villus height, direction, and absence of branching. Our studies identify Ets factors as novel regulators of villus morphology in the mature adult small intestine. Our analyses further suggest that Ets factors may regulate crypt number, position, and/or size, by crypt nonautonomous mechanisms possibly involving epithelial-stromal communication pathways.
Summary and Perspectives
Our studies using the Ets dominant repressor En/Erm identify Ets factors as important regulators of epithelial maturation, epithelial transit, and crypt-villus architecture in the adult small intestine. Little is currently known mechanistically about these important parameters of intestinal homeostasis. Examination of candidate mechanisms, namely TGFβRII expression, E-cadherin expression, and β-catenin localization, suggests that these do not contribute to the observed phenotypes. Thus, the mechanisms by which Ets factors regulate adult intestinal homeostasis appear to represent novel pathways. Unraveling of these pathways will in part require precise knowledge of both segmental and subcompartmental Ets expression in the adult intestine. Unfortunately, such information is not currently available, but existing data from whole tissue expression analysis18 allow one to speculate as to the relevant Ets factor(s). Elf3 appears to be the most highly expressed Ets factor in the adult intestine,18 and the En/Erm phenotype is dependent on robust transgene expression. Given the demonstrated role of Elf3 in epithelial differentiation in the developing intestine,20,38 we suspect that the En/Erm phenotype may be attributable to blockade, probably incomplete, of Elf3 and simultaneous blockade of other highly expressed Ets factor(s) in the small intestine, such as Ehf/ESE-3 and/or Ets2.18
What does the En/Erm phenotype reveal about possible Ets roles in intestinal pathology? The overall phenotype—increased cell transit, impaired differentiation, and crypt-villus disorder—is that of a disturbed and overactive crypt-villus axis. Ets factors are generally overexpressed in intestinal epithelial neoplasms, and appear to be tumor promoting.19 Surprisingly, however, in the only genetic study of Ets function in intestinal epithelial neoplasia published thus far, Ets2, at physiological or near-physiological gene dosage, was shown to restrict rather than enhance tumor multiplicity.45 Thus, in contrast to the generally tumor-promoting effects of overexpressed Ets factors in established tumors, the maintenance of crypt-villus homeostasis by physiological levels of at least some epithelial Ets factors may have an overall effect of restricting tumor initiation and/or early promotion. Further studies will be required to determine to what extent this duality of function reflects inherent properties of different Ets factors, promoter context-dependent transcriptional activities (activation versus repression) of individual Ets, and/or gain-of-function type phenomena (ie, promoter effects via lower affinity sites) at supraphysiological expression levels. Finally, the En/Erm phenotype of disturbed crypt-villus homeostasis also suggests possible roles for Ets factors in the control of epithelial regeneration/repair after injury.
Supplementary Material
Acknowledgments
We thank Melissa Gonzalez for generating the 8x(EBS)-TK-luciferase reporter construct; the University of Colorado Cancer Center Transgenic/Knockout Core Facility for generation of villin-En/Erm transgenic mice; Jana Polzer, Janet Lieber, Michael Medina, and Sara Garza-Williams for assistance with mouse histology; Janet Lieber and Gary Mierau for assistance with electron microscopy; Heide Ford and Erica McCoy for assistance with HA immunostaining; John Shannon (Cincinnati Children’s Hospital Medical Center), Deborah Gumucio (University of Michigan), Jeffrey Gordon (Washington University), and James Hagman (National Jewish Medical and Research Center) for reagents; and Heide Ford and Sean Colgan for critical reading of the manuscript.
Footnotes
Address reprint requests to Paul Jedlicka, Department of Pathology, University of Colorado Denver, Anschutz Medical Center, PO Box 6511, MS 8104, Aurora CO 80045. E-mail: paul.jedlicka@ucdenver.edu.
Supported by the Cancer League of Colorado, The Children’s Hospital/University of Colorado Denver Research Institute, and the National Institutes of Health (K08 DK074557 to P.J. and R01 DK37667 and DK46868 to A.G.H.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Present address of L.S.: Department of Genetics and Development, Columbia University, New York, NY.
References
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ET: S-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Hartmann A, Duval DL, Bradford AP. ETS transcription factors in endocrine systems. Trends Endocrinol Metab. 2007;18:150–158. doi: 10.1016/j.tem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, Chandrashekar V, Hofmann MC, Hess RA, Murphy KM. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–2308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Rossant J. Ets2 is necessary in trophoblast for normal embryonic anteroposterior axis development. Development. 2006;133:1059–1068. doi: 10.1242/dev.02277. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Huber RM, Ladle DR, Murphy K, Arber S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron. 2007;55:726–740. doi: 10.1016/j.neuron.2007.07.028. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- O'Leary DA, Noakes PG, Lavidis NA, Kola I, Hertzog PJ, Ristevski S. Targeting of the ETS factor GABPalpha disrupts neuromuscular junction synaptic function. Mol Cell Biol. 2007;27:3470–3480. doi: 10.1128/MCB.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol Dis. 2004;32:143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D, Lapinskas EJ, Visvader J, Lindeman GJ, Thomas R, Ormandy CJ, Hertzog PJ, Kola I, Pritchard MA. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Gutierrez-Hartmann A. Ets transcription factors in intestinal morphogenesis, homeostasis and disease. Histol Histopathol. 2008;23:1417–1424. doi: 10.14670/hh-23.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev Biol. 2003;261:10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol. 2001;11:1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
- de Kerchove D'Exaerde A, Cartaud J, Ravel-Chapuis A, Seroz T, Pasteau F, Angus LM, Jasmin BJ, Changeux JP, Schaeffer L. Expression of mutant Ets protein at the neuromuscular synapse causes alterations in morphology and gene expression. EMBO Rep. 2002;3:1075–1081. doi: 10.1093/embo-reports/kvf220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratore C, Brugnoli G, Lee HY, Suter U, Sommer L. The role of the Ets domain transcription factor Erm in modulating differentiation of neural crest stem cells. Dev Biol. 2002;250:168–180. doi: 10.1006/dbio.2002.0795. [DOI] [PubMed] [Google Scholar]
- Théveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS ONE. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson DB, Langner K, Namini SS, Jessen KR, Mirsky R. Beta-neuregulin and autocrine mediated survival of Schwann cells requires activity of Ets family transcription factors. Mol Cell Neurosci. 2002;20:154–167. doi: 10.1006/mcne.2002.1109. [DOI] [PubMed] [Google Scholar]
- Langer SJ, Bortner DM, Roussel MF, Sherr CJ, Ostrowski MC. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Maira SM, Sobieszczuk P, Wasylyk B. Reversion of Ras transformed cells by Ets transdominant mutants. Oncogene. 1994;9:3665–3673. [PubMed] [Google Scholar]
- Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DF, Lewis SR, Haugen BR, James RA, McDermott MT, Wood WM, Ridgway EC. Pit-1 and GATA-2 interact and functionally cooperate to activate the thyrotropin beta-subunit promoter. J Biol Chem. 1997;272:24339–24347. doi: 10.1074/jbc.272.39.24339. [DOI] [PubMed] [Google Scholar]
- Eckel KL, Tentler JJ, Cappetta GJ, Diamond SE, Gutierrez-Hartmann A. The epithelial-specific ETS transcription factor ESX/ESE-1/Elf-3 modulates breast cancer-associated gene expression. DNA Cell Biol. 2003;22:79–94. doi: 10.1089/104454903321515896. [DOI] [PubMed] [Google Scholar]
- Ng L, Forrest D, Haugen BR, Wood WM, Curran T. N-terminal variants of thyroid hormone receptor beta: differential function and potential contribution to syndrome of resistance to thyroid hormone. Mol Endocrinol. 1995;9:1202–1213. doi: 10.1210/mend.9.9.7491112. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, O'Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- Hauck AL, Swanson KS, Kenis PJ, Leckband DE, Gaskins HR, Schook LB. Twists and turns in the development and maintenance of the mammalian small intestine epithelium. Birth Defects Res C Embryo Today. 2005;75:58–71. doi: 10.1002/bdrc.20032. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Gordon JI. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development. 2000;127:2629–2642. doi: 10.1242/dev.127.12.2629. [DOI] [PubMed] [Google Scholar]
- Flentjar N, Chu PY, Ng AY, Johnstone CN, Heath JK, Ernst M, Hertzog PJ, Pritchard MA. TGF-betaRII rescues development of small intestinal epithelial cells in Elf3-deficient mice. Gastroenterology. 2007;132:1410–1419. doi: 10.1053/j.gastro.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Kim BM, Mao J, Taketo MM, Shivdasani RA. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 2007;133:529–538. doi: 10.1053/j.gastro.2007.04.072. [DOI] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132:279–289. doi: 10.1242/dev.01576. [DOI] [PubMed] [Google Scholar]
- Muñoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, Moses HL, Grady WM. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- Hermiston ML, Wong MH, Gordon JI. Forced expression of E-cadherin in the mouse intestinal epithelium slows cell migration and provides evidence for nonautonomous regulation of cell fate in a self-renewing system. Genes Dev. 1996;10:985–996. doi: 10.1101/gad.10.8.985. [DOI] [PubMed] [Google Scholar]
- Wong MH, Rubinfeld B, Gordon JI. Effects of forced expression of an NH2-terminal truncated beta-catenin on mouse intestinal epithelial homeostasis. J Cell Biol. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.