Abstract
Ubiquitinated endosomal proteins that are deposited into the lumens of multivesicular bodies are either sorted for lysosomal-mediated degradation or secreted as exosomes into the extracellular milieu. The mechanisms that underlie the sorting of cellular cargo proteins are currently unknown. In this study, we show that the COP9 signalosome (CSN)-associated protein CSN5 quantitatively regulated proteins that were sorted into exosomes. Western blot analysis of exosomal proteins indicated that small interfering (si)RNA knockdown of CSN5 results in increased levels of both ubiquitinated and non-ubiquitinated exosomal proteins, including heat shock protein 70, in comparison with exosomes isolated from the supernatants of 293 cells transfected with scrambled siRNA. Furthermore, 293 cells transfected with JAB1/MPN/Mov34 metalloenzyme domain-deleted CSN5 produced exosomes with higher levels of ubiquitinated heat shock protein 70, which did not affect non-ubiquitinated heat shock protein 70 levels. The loss of COP9-associated deubiquitin activity of CSN5 also led to the enhancement of HIV Gag that was sorted into exosomes as well as the promotion of HIV-1 release, suggesting that COP9-associated CSN5 regulates the sorting of a number of exosomal proteins in both a CSN5 JAB1/MPN/Mov34 metalloenzyme domain-dependent and -independent manner. We propose that COP9-associated CSN5 regulates exosomal protein sorting in both a deubiquitinating activity-dependent and -independent manner, which is contrary to the current idea of ubiquitin-dependent sorting of proteins to exosomes.
Cells receive information from the microenvironment and pass the information from the cell surface to internal compartments, from one compartment to another, and even from cell to cell. Exosomes packed with a number of proteins and RNAs deliver messages to the recipient cells and execute their role in cell-cell communication.1,2,3 Therefore, the machinery for sorting exosome molecules determines the composition of exosomes and the subsequent biological effects on recipient cells. The endosomal sorting pathway controls a variety of cellular processes and plays a role in the sorting of ubiquitinated cargo proteins into the lumen of multivesicular bodies (MVBs)4 and viral budding.5,6,7 Ubiquitinated proteins are recognized on the endosomal membrane and sorted, which results in either MVB fusion with the lysosome to degrade its contents,8,9 or releasing the contents into the extracellular environment via exosomal vesicles.10 Released exosomes can be taken up by neighboring cells or distant cells as a means to pass information between cells. The exosome pathway could also be hijacked by certain viruses, such as HIV, to escape host immune recognition.5,6
The mechanism underlying the regulation of a protein to be degraded in the lysosomal pathway or packed into the exosomes for budding out of cells is not clear. Data published recently support the idea that incorporation of proteins into exosomes seems to be a selective and regulated process.10 Ubiquitinated proteins may be deubiquitinated before or after sorting into the MVB and before their actual incorporation into luminal vesicles or exosomes. Ubiquitinated proteins may also escape deubiquitination and end up in exosomes. During this process, deubiquitinating enzymes may work against E3-ligase activity so as to regulate exosomal protein stability and thus its biological activity. Deubiquitinating enzymes might be recruited early in the pathway where they could provide a proofreading mechanism and rescue cargo before commitment to the degradation pathway or exosomal pathway. Therefore, deubiquitination versus ubiquitination of exosomal proteins could be regulated by deubiquitin enzymes that have not been identified.
CSN5 is identical to the fifth component of the COP9 signalosome complex (CSN), which is well conserved in species from yeast to humans, and is involved in a variety of biological responses, such as DNA metabolism, apoptosis, checkpoint control, DNA repair, and cell cycle control in various organisms.11,12,13,14,15,16,17,18,19,20 The CSN core is composed of eight subunits (CSN1-8),20 and associates with different types of proteins including kinases, ligases, and adaptor proteins. CSN is also related structurally and architecturally to the lid subcomplex of the 26S proteasome that mediates degradation of ubiquitin-conjugated proteins.21 Recent reports show that the Rpn11 subunit of the proteasome lid subcomplex possesses ubiquitin isopeptidase activity that is responsible for depolymerization of the ubiquitin chains from substrates before protein degradation.22,23 Likewise, the CSN5 subunit of COP9, which shares sequence similarity to Rpn11, has been shown to have isopeptidase activity that deconjugates NEDD8 from cullin-NEDD8.24 CSN-mutations lead to deficient NEDD8-deconjugating activity and accumulation of NEDD8-conjugated cullins.24,25,26 Thus, CSN could serve as a negative regulator of ubiquitin ligase activity by deconjugating NEDD8 from cullin-NEDD8.
Here, we demonstrate that COP9-associated CSN5 has ubiquitin isopeptidase activity, and regulates ubiquitin-dependent protein sorting into exosomes. Furthermore, experiments with a mutant CSN5 indicate that the CSN5-associated deubiquitin activity contributes to deconjugation of ubiquitin on exosomal proteins. Deletion of the JAB1/MPN/Mov34 metalloenzyme (JAMM) domain of CSN5 leads to the enhancement of ubiquitinated protein, eg, HSP70 and HIV Gag, sorting into exosomes. More ubiquitinated HIV Gag sorted into exosomes also correlates with increased release of HIV-1 from infected cells. Taken together these results indicate that COP9-associated CSN5 deubiquitin activity regulates the sorting of exosomal proteins, and thus enhancement or attenuation of COP9-associated CSN5 deubiquitin activity that might lead to changes in the biological effects exosomes would have on recipient cells. In addition, CSN5 also plays a role in sorting non-ubiquitinated proteins into exosomes in a JAMM domain-independent manner.
Materials and Methods
Cells
The 293 cells with a doxytetracycline inducible siRNA CSN5 were cultured in complete modified Dulbecco medium (Invitrogen, Carlsbad, CA) supplemented with exosome depleted 10% fetal bovine serum as described previously.24 A control 293 cell line was established by transfection of 293 cells with a control vector pGC95 (control scrambled siRNA, 5′-CGTGCAAGGTCAGTACATGTTCAAGAGACATGTACTGACCTTGCACG-3′). To induce siRNA expression, 293 cells were fed with doxytetracycline (1.3 μg/ml in modified Dulbecco medium)24 and analyzed for the presence of CSN5 by Western blot. MDA-MB-231 cells originally derived from a pleural effusion were obtained from the American Type Culture Collection (Manassas, VA) and were routinely cultured in RPMI 1640 supplemented with 10% fetal bovine serum. 4T1 and TS/A mouse mammary carcinoma cell lines were cultured using a method as described previously.27
CSN5 Plasmid DNA Constructs
A pGEX-CSN5 construct was generated by PCR amplification from the pcDNA3-CSN5 template,19 using forward primer 5′-TTGTTGGGATCCATGGCGGCGTCCGGGAGCGGTAT-3′ and reverse primer 5′-CTATTAGAATTCTTAAGAGATGTTAATTTGATT-3′ (BamHI and EcoRI restriction sites are underlined). The pGEX-CSN5 construct was generated by ligating annealed primers in-frame into BamHI-EcoRI digested pGEX-2T vector (Amersham Biosciences). Sequencing of plasmids was performed using an ABI sequencer (Applied Biosystems, Foster City, CA).
BL21 competent E. coli cells (Stratagene) were transformed with the pGEX-CSN5 construct. For large-scale induction of glutathione S-transferase (GST)-CSN5 protein, an overnight culture was diluted 1:100 in 1 L Luria-Bertani broth with appropriate antibiotics and cultured for 3 hours at 37°C. GST fusions were induced with 1 mmol/L isopropyl β-d-1-thiogalactopyranoside for 3 hours at 37°C. The cells were resuspended in 25 ml PBS with 1% Triton X-100 and 0.8 mmol/L phenylmethylsulfonyl fluoride, pulse-sonicated 4 minutes on ice and centrifuged at 10,000 × g for 20 minutes at 4°C. Crude bacterial lysate supernatant was purified using a Glutathione Sepharose 4B slurry (Amersham Biosciences). CSN5 protein purity and integrity after purification were confirmed by Western blotting. Protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL).
Plasmids, including pLEYFP-CSN5, pLEYFP-CSN5JAMMDN expressing CSN5-YFP, YFP-CSN5-JAMM-DN, were produced by subcloning human XhoI-HindIII fragment of pcDNA3-CSN5 containing CSN5 into BglII-HindIII sites of pLEYFP (Clontech) mammalian expression vectors. The expression of fusion proteins of YFP-CSN5 was verified by Western blot analysis for CSN5. The deletion of CSN5 JAMM domain between amino acid positions 53 to 85 in pLEYFP-CSN5 was accomplished using a PCR technique and standard cloning techniques as described previously.28
CSN5 with point mutations at amino acid positions 138 and 140 was generated as described previously29 and both mutations replaced histidines with alanines. The CSN5 gene was cloned into the retroviral packaging vector pMSCVhyg (Clontech, Mountain View, CA). pcDNA3-HIVGag-green fluorescent protein (GFP) was provided by Dr. Stephen J. Gould and described previously.5 All plasmid DNAs described above were used for transfection of 293 cells purchased from ATCC (Manassas, VA). Transfections were performed using the Lipofectamine 2000 method as described previously.19
Western Blot Analysis
Western blot analysis of exosomal proteins was done using a method described previously.27 In brief, exosomes were boiled in SDS sample buffer, and total protein was loaded into each well of an SDS gel for separation by polyacrylamide gel electrophoresis (PAGE). After electrophoresis, the proteins were then stained in the gels using a Colloidal Blue Staining Kit (Invitrogen) or transferred onto nitrocellulose membranes, and the blotted membranes blocked with PBS-Tween 20 (0.25 M/L Tris, pH 7.5, PBS, 150 mmol/L sodium chloride, and 0.2% Tween 20) containing 5% bovine serum albumin. Blots were then probed overnight with 1 μg/ml of a primary antibody, washed three times with PBS-Tween 20 and then probed for 1 hour with the appropriate Alexa Fluor 680 (Molecular Probes) or IRdye 800 (Rockland Immunochemicals) conjugated secondary antibody. After three washes with PBS-Tween 20, blotted proteins were detected and quantified using an Odyssey infrared imaging system (LI-COR, Lincoln, Nebraska).
Exosome Purification
Exosomes were recovered from the supernatant of cells cultured for 36 hours in depleted medium (ie, exosome depleted fetal bovine serum; accomplished by overnight centrifugation at 100,000 × g). Exosomes were purified by differential centrifugation using a previously described method.27 After differential centrifugation, exosomes were sedimented by centrifugation and were resuspended in PBS and re-sedimented at 70,000 × g. Sedimented exosomes were resuspended in 5 ml of 2.6 M/L sucrose, 20 mmol/L Tris-HCl, pH 7.2, and floated into an overlaid linear sucrose gradient (2.0 to 0.25 M/L sucrose, 20 mmol/L Tris-HCl, pH 7.2) in an SW41 tube and centrifuged for 16 hours at 270,000 × g to remove non-membranous protein (complexes). Gradient fractions (1 ml) were collected via the bottom of the tube and washed with PBS by centrifugation at 70,000 × g for 1 hour. Finally, the exosomes (fraction 3) were resuspended in PBS. The morphology of exosomes was examined by electron microscopy using a method described previously.27 The protein content of the exosomes was determined using a BCA protein assay kit (Bio-Rad, Hercules, CA).
Immunoprecipitation and Antibodies
293 Cells were lysed by boiling the cells in 150 μl SDS lysis buffer (1% SDS, 150 mmol/L NaCl, 50 mmol/L Tris, pH 8.0) for 10 minutes at 95°C. The lysate was diluted to match the radioimmunoprecipitation assay (RIPA) buffer concentrations (0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 150 mmol/L NaCl, 50 mmol/L Tris, pH 8.0). After sonication, the lysate was cleared by centrifugation for 10 minutes at 17,000 x g, and 100 μl of diluted primary antibody or a control nonimmune IgG coupled to protein G-agarose beads added to the cleared lysate supernatant. After four washes with washing buffer (Pierce, Rockford, IL), the protein complexes were eluted with 100 μl of 0.1 mol/L glycine (pH 2.8) and collected. The samples were mixed with sample buffer containing 2% β-mercaptoethanol, heated to 95°C, centrifuged at 5000 × g for 5 minutes and the supernatant subjected to 10% SDS-PAGE. The proteins in the gels were either transferred to a nitrocellulose membrane (Bio-Rad) followed by Western blot analysis or colloidal blue-stained for liquid chromatography-tandem mass spectrometry identification using a method described previously.27 The following antibodies were used for immunoprecipitation and Western blots: anti-ubiquitinated protein monoclonal antibody (clone FK2, which recognizes mono- and polyubiquitinylated proteins, but not free ubiquitin, MBL, Woburn, MA), anti-ß-actin (1:5000, Sigma, St. Louis, MO), CSN5, (1:1000; Santa Cruz Biotechnology), rabbit anti-human CD63 (1:500; Santa cruz biotechnology, Santa Cruz, CA), anti-HSP70 (1:1000; Santa cruz biotechnology), anti- CSN1–8 sampler kit (1:500; Boston Biochemicals), anti-GFP antibody (Clontech), and anti-HIV Gag antibody.
In Vitro Deubiquitination Assay
Exosomes were sonicated in deubiquitination buffer (50 mmol/L Tris-HCl, pH 7.2 or pH 8.3, 25 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L dithiotrheitol). The deubiquitination reaction was performed in a 37°C water bath for 30 or 60 minutes, and the reaction stopped in a 4°C ice bath for 10 minutes, followed by addition of 1× SDS sample buffer.
Quantification of HIV-1 Released in the Cultured Supernatants of 293 Cells
To analyze the effect of CSN5 on HIV-1 exosomal release, the pNLENG1-ES-IRES proviral DNA was used. NLENG1-ES-IRES represents a recombinant HIV-1 proviral DNA that expresses GFP subsequent to infection and proviral DNA integration. NLENG1-ES-IRES was constructed and has the characteristics of YFP and CFP reporter viruses reported by Levy et al.30 The NLENG1-ES-IRES construct is replication defective since it does not express envelope glycoprotein (gp160). Infectious virus stocks were generated by cotransfecting 293T cells with NLENG1-ES-IRES plus the vesicular stomatitis virus G protein expression plasmid, and used to infect 293 cells stable transfected with either CSN5 pLEYFP-CSN5-JAMMDN or pLEYFP-CSN5. Eight hours postinfection, the cells were washed twice with complete medium to remove the excess virus. Fresh medium was added to each well. Two hours after washing, the residual virus, as determined by detecting p24 using an enzyme-linked immunosorbent assay (ELISA), was at an undetectable level in the supernatants (data not shown). Forty-eight hours postinfection, culture supernatants were collected from each well and analyzed by HIV-1 p24 antigen ELISA (Perkin Elmer). The infected cells were quantified by fluorescence-activated cell sorting analysis for GFP-positive cells using a method as described previously.31
Statistics
Results were expressed as means ± SD, and assessed by one-way analysis of variance with Bonferroni correction.
Results
CSN5 Regulates Exosomal Protein Ubiquitination and Sorting
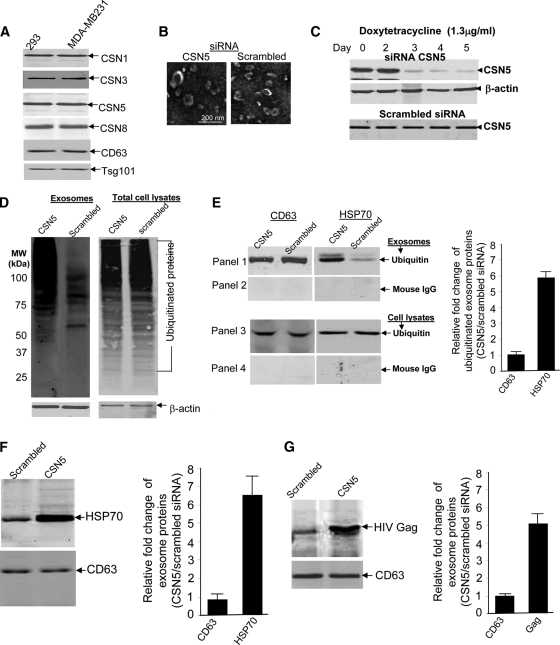
Ubiquitination/deubiquitination of proteins plays an important role in sorting a protein into exosomes via the MVB pathway. Exosomal proteins have been demonstrated to be ubiquitinated.32 To determine whether CSN5, which possesses a potential deubiquitin domain, is recruited into exosomes, exosomes isolated from the supernatants of 293 cells and a human breast tumor cell line (MDA-MB-231) were used for Western blot analysis of proteins using the antibodies indicated in Figure 1A. COP9-associated CSN5 was detected in both human tumor cell lines (Figure 1A) and also the murine breast tumor cell lines 4T-1 and TS/A (data not shown), suggesting that CSN5 may regulate exosomal protein ubiquitination and sorting. To determine whether CSN5 plays a role in regulation of exosomal protein ubiquitination, CSN5 in 293 cells was inducibly knocked down by the addition of doxytetracycline. Exosomes were purified from the supernatants of 293 cells (Figure 1B, a representative image is shown). Morphologically, there are no visible differences in the shapes and sizes of the exosomes as a result of depletion of CSN5 (Figure 1B). Western blot analysis indicated that CSN5 was knocked down dramatically at day 3 after the addition of doxytetracycline (1.3 μg/ml) (Figure 1C, top panel) but not in 293 cells stably transfected with a scrambled siRNA (Figure 1C, bottom panel). Treatment of 293 cells with doxytetracycline alone did not appear to have an effect on β-actin expression (Figure 1C, middle panel). The results of Western blot analysis of ubiquitinated exosomal proteins demonstrated a much higher concentration of ubiquitinated proteins in the exosomes isolated from the supernatants of 293 cells transfected with siRNA CSN5 when compared with control 293 cells (Figure 1D, lanes 1 and 2). Knockdown of CSN5 does not seem to have an effect on ubiquitination of total cellular proteins (Figure 1D, lanes 3 and 4). The increase in signal intensity of ubiquitinated proteins in the exosomes isolated from the supernatants of 293 cells with CSN5 knockdown is not due to protein loading on the gel because similar amounts of β-actin were detected in all test samples (Figure 1D, lanes 1 and 2 and the bottom panel). The difference in intensity of the β-actin signals when comparing exosomes and cell lysates (Figure 1D, bottom panel) suggests that proteins in exosomes are enriched in general (50 μg/lane).
Figure 1.
siRNA knockdown of CSN5 increases ubiquitination of exosomal proteins. A: Exosomes were analyzed by Western blotting using a monoclonal antibody, anti-CSN5, and rabbit polyclonal antibodies to CSN1, 3, and 8, of COP9. Exosome markers, including CD63 and Tsg101, were also analyzed. 293 cells stably transfected with pGC93 (siRNA CSN5 expression vector) or 293 cells transfected with scrambled siRNAs were treated with doxytetracycline (1.3 μg/ml) over a 5-day period. The cells were then used for the following experiments (B through F). In other experiments, 293 cells stably transfected with either pGC93 or scrambled siRNA were transfected with pcDNA3-HIVGag-GFP before being treated with doxytetracycline and subsequently used in experiment G. B: Exosomes were purified from supernatants using sucrose gradients and analyzed by electron microscopy (scale bar = 200 nm). The micrograph is representative of two independent experiments. C: Fifty μg of total cell lysate from 293 cells pretreated with doxytetracycline over 5 days were loaded on 10% SDS gels for PAGE separation. The separated proteins were transferred to a nitrocellulose membrane for Western blotting and probed using antibodies as indicated in Figure 1C. Representative Western blots are shown. D: Fifty μg of protein extracted from exosomes isolated from the supernatants of 293 cells (lanes 1 and 2) or total cell lysates (lanes 3 and 4) were used for ubiquitin Western blot analysis. β-actin detection served as a control for Western blotting analysis. E: Five-day doxytetracycline treated 293 cells (E, panels 3 and 4) or exosomes (E, panels 1 and 2) isolated from the supernatants of 293 cells were lysed using immunoprecipitation lysate buffer. Immunoprecipitation assays were performed using 500 μg aliquots of the total protein lysates and an anti-ubiquitin monoclonal antibody (E, panels 1 and 3) or mouse IgG as a control (E, panels 2 and 4). Precipitated proteins were run in western blots probed with anti-CD63 and anti-HSP70. The presence of HSP70 (F, top panel) and CD63 (F, bottom panel) were determined by Western blot analysis of exosomes purified from the same 293 cells as described above. G: The presence of HIV Gag (top panel) and CD63 (bottom panel) in exosomes purified from 5-day doxytetracycline treated 293 cells were determined by Western blot analysis. Data presented in A–D represent three independent experiments that yielded similar results. The signal intensity of each protein analyzed in E–G was quantified using an Odyssey infrared imaging system (LI-COR). The ratios of signal intensity of each protein in exosomes of CSN5 siRNA: scramble siRNA knockdown 293 cells were calculated and plotted (E–G, bar graphs). The data are presented as means of three independent experiments.
From three independent assays, immunoprecipitation of ubiquitinated exosomal proteins followed by Western blot analysis confirmed the presence of HSP70 and CD63 (Figure 1E). Interestingly, knockdown of CSN5 in 293 cells resulted in no changes in the amount of HSP70 from total cell lysates (Figure 1E, third panel from the top) whereas the amount of ubiquitinated HSP70 in the exosomes was remarkably increased (Figure 1E, the top panel, right panel of bar graph). In contrast, the amount of ubiquitinated CD63 sorting into exosomes was not different from the amount present in cell lysates (Figure 1E, lanes 1 and 2). These results suggest that CSN5 selectively regulated the ubiquitination of certain exosomal proteins. The specificity of ubiquitin immunoprecipitation was supported by the fact that no signal was detected for HSP70 or CD63 when a normal mouse IgG was used for immunoprecipitation (Figure 1E, second and fourth panels from the top).
Western blot analysis was done to determine whether CSN5 regulates total HSP70 in addition to potentially playing a role in sorting ubiquitinated HSP70 into exosomes, Western blot analysis of exosomal HSP70 indicated that knockdown of CSN5 results in an increase of HSP70 sorted into the exosomes (Figure 1F, top panel, and right panel of bar graph). The fact that there was no change in the exosomal CD63 signal (Figure 1F, bottle panel) suggests that CSN5 also selectively regulates exosomal protein sorting. Sorting of endogenously expressed HSP70 into exosomes is regulated by CSN5, and in addition siRNA knockdown of CSN5 also results in an increase of the transiently expressed HIV Gag sorted into 293 exosomes (Figure 1G). Therefore, the result of CSN5 regulating HIV Gag sorting into exosomes further suggests that COP9 associated CSN5 may regulate more than HSP70 sorting into exosomes.
Deubiquitination Activity of COP9-Associated CSN5 Regulates HSP70 Ubiquitination
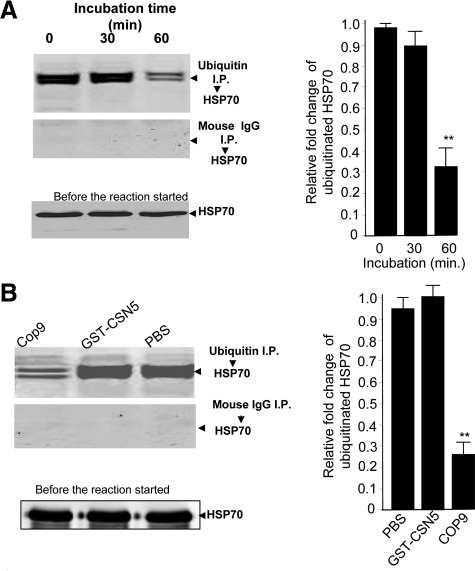
Since knockdown of CSN5 in 293 cells led to an increase of HSP70 sorted into exosomes, and ubiquitination of proteins is considered as a crucial step for sorting into exosomes, we sought to determine whether CSN5 has the deubiquitination activity to regulate HSP70 deubiquitination, as a example. To perform this analysis, sucrose-purified exosomes isolated from the supernatant of 293 cells with CSN5 knockdown were suspended in deubiquitination buffer. Sonicated exosomes were then used in a deubiquitin reaction. The reaction was incubated at 37°C for 0, 30, and 60 minutes and stopped by addition of 1% SDS. Immunoprecipitation of ubiquitin followed by Western blot analysis of HSP70 provided evidence that the addition of recombinant COP9 (Figure 2A, top panel) to the exosomal complex led to significant deubiquitination of exosomal HSP70 (Figure 2A) after a 60 minutes incubation as compared with a 0-minute incubation. The signal intensity of ubiquitinated HSP70 when recombinant CSN5 alone was added was not different from the signal intensity of ubiquitinated HSP70 in a PBS control sample. This finding suggests that the COP9 complex, not CSN5 alone is required for its deubiquitination activity. The reduction of signal intensity of ubiquitinated HSP70 is not due to unequal loading of amounts of HSP70 precipitate used for the deubiquitination assay because similar amounts of HSP70 were detected in all samples before the reaction (Figure 2, A and B, bottom panels). Control samples (Figure 2, A and B, middle panels) did not have detectable signal, indicating the specificity of the anti-HSP70 reaction. In summary, the data presented in Figure 2A, and Figure 1D suggest that exosomal COP9 associated CSN5, by virtue of its deubiquitination activity, plays a role in the regulation of exosomal protein ubiquitination.
Figure 2.
COP9 associated CSN5 regulates the deubiquitination of exosomal proteins. One microgram of a COP9 complex (A, lanes 1, 2, and 3, labeled 0, 30, and 60, respectively) was added to 500 μg of sonicated exosomes isolated from 293 cells transfected with pGC93 vector treated for 5 days with doxytetracycline (1.3 μg/ml). One-third of the mixture was used for Western blot analysis of HSP70 (A, bottom panel), and the remainder used in a deubiquitin assay. A deubiquitin reaction was performed at 37°C for 0, 30, and 60 minutes. The reaction was stopped by addition of 1% SDS sample buffer and subsequently run in an immunoprecipitation assay using agarose beads conjugated to mouse anti-ubiquitin (A, top panel) or mouse IgG antibodies (middle panel), followed by HSP70 Western blot analysis. Five hundred micrograms of 293 cell exosomal protein, as described in (A) were used in a deubiquitin assay in the presence of 1 μg of recombinant COP9 (B, lane 1), 1 μg of GST-CSN5 (B, lane 2), or PBS as a control. One-third of the reacted proteins was used for Western blot analysis of HSP70 (B, bottom panel). The remainder was used for the deubiquitin reaction conducted at 37°C for 1 hour. The deubiquitinated samples were used for ubiquitin (B, top panel) or mouse IgG (B, middle panel) immunoprecipitation, and then Western-blotted and probed with an anti-HSP70 antibody. One representative experiment is presented, out of three independent experiments. The signal intensity of each protein analyzed in A and B was quantified using an Odyssey infrared imaging system (LI-COR). The ratios of signal intensity of ubiquitinated HSP70 at 30 minutes and 60 minutes against 0 minutes after the reaction was terminated were calculated and plotted (A, bar graph). The data are presented as means of three independent experiments. Using a similar method, the ratios of signal intensity of ubiquitinated HSP70 in the 60 minutes reaction containing either GST-CSN5 or COP9 against PBS as a control were calculated and plotted (B, bar graph). The data are presented as means of three independent experiments. **P < 0.0082 (2A); **P < 0.0057 (2B).
CSN5 JAMM Domain Regulates Exosomal Protein Deubiquitination
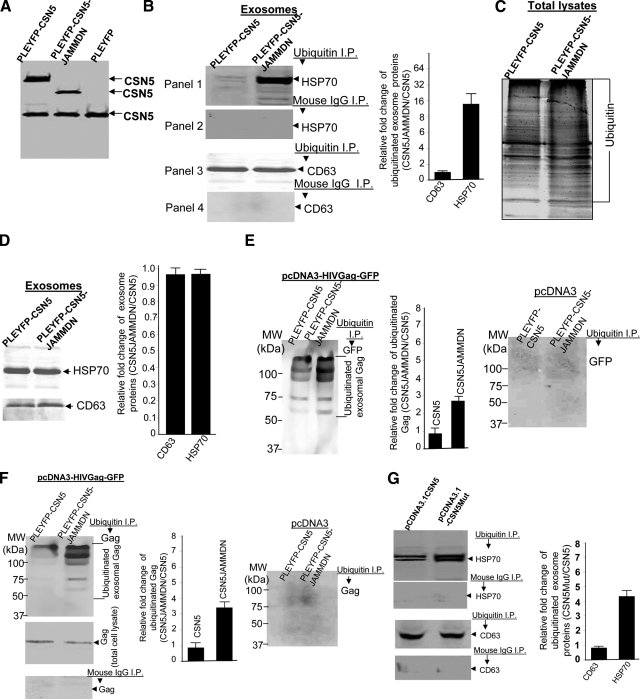
Although CSN5 is involved in removal of NEDD8 from modified proteins as a means to regulate protein activity, the role of CSN5 for the removal of ubiquitin from in vivo ubiquitinated proteins has not been demonstrated in mammalian cells. Given that ubiquitin and NEDD8 have closely related structures,33 we examined whether the JAMM domain of CSN5 plays a role in the ubiquitin isopeptidase of exosomal proteins. 293 cells were stably transfected with the following vectors: pLEYFP-CSN5, pLEYFP-CSN5-JAMMDN, or pLEYFP. Western blot analysis of CSN5 expression in the transfected cell lines indicated that the fusion proteins YFP-CSN5 (Figure 3A, Lane 1) and YFP-CSN5-JAMMDN (Lane 2) could be detected, and that the expression of the fusion protein does not have an effect on the amount of endogenous CSN5 (Figure 3A, bottom protein in each lane). Western blot analysis of immunoprecipitated ubiquitin exosomal proteins indicated that deletion of the JAMM domain of CSN5 led to a significant increase in the ubiquitination of exosomal HSP70, as compared with the exosomes isolated from 293 cells transfected with wild-type pLEYFP-CSN5 (Figure 3B, the top panel, and right panel of bar graph). In contrast, ubiquitinated exosomal CD63 was not affected (Figure 3B, third panel). The signals detected are ubiquitinated-specific as there is no signal detected when a control mouse IgG was used in the assay (Figure 3B, second and fourth panels). The deletion of CSN5 JAMM domain does not appear to have an effect on the ubiquitination of total cellular protein (Figure 3C). Unlike siRNA knockdown of total CSN5 as shown in Figure 1F, deletion of CSN5 JAMM domain does not appear to increase non-ubiquitinated HSP70 sorted into the exosomes (Figure 3D).
Figure 3.
JAMM domain of CSN5 plays a role in the regulation of ubiquitination of exosomal proteins. A: 293 cells were transfected with different plasmid DNAs (10 μg) as indicated in Figure (A). Thirty-six hours after the transfection, the cells (1 × 106) were lysed in lysis buffer and 50 μg of total protein from each lysate were resolved on a 10% SDS gel by PAGE. The proteins were then transferred to a nitrocellulose membrane and the blots were probed with a mouse anti-CSN5 antibody. 293 cells were transfected with pLEYFP-CSN5 or pLEYFP-CSN5-JAMMDN for 36 hours. The exosomes isolated from the supernatants of transfected 293 cells were immunoprecipitated with an anti-ubiquitin monoclonal antibody, followed by Western blot analysis for HSP70 (B, top left panels) and CD63 (B, bottom left panels). In other experiments, 293 cells were lysed and used for Western blot analyses of mono- or polyubiquinated proteins (C) or exosomes were lysed and used for Western blot analysis (D). The exosomes isolated from the supernatants of 293 cells transfected with pLEYFP-CSN5 or pLEYFP-CSN5-JAMMDN PLEYFP plus pcDNA3-HIVGag-GFP (E, left panel; F, top left panel) or plus pcDNA3 (E, right panel, F, right panel) were used for ubiquitin immunoprecipitation, followed by Western blot analysis of GFP (E) or Gag (F). An equal concentration of normal mouse IgG was used as a control for immunoprecipitation, followed by Western blot analysis of Gag (F, bottom left panel). 293 cells were transfected with pcDNA3.1-CSN5 or pcDNA3.1-CSN5Mut for 36 hours. The sucrose purified exosomes were then run in immunoprecipitation assays using an anti-ubiquitin monoclonal antibody (G, panels 1 and 3) or mouse IgG (G, panels 2 and 4). Western blot analysis of HSP70 (G, top left panels) and CD63 (G, bottom left panels) followed. The signal intensity of each protein analyzed in (B, D–F, and G) was quantified using an Odyssey infrared imaging system (LI-COR). The ratios of signal intensity of each protein in exosomes of 293 cells stably transfected with CSN5JAMMDN : CSN5 genes (B, D, E, and F, bar graphs) and CSN5Mut:CSN5 genes (G, bar graph) were calculated and plotted. The data are presented as means of three independent experiments.
Ubiquitination of HIV Gag occurs when HIV hijacks the exosomal pathway.34,35,36,37,38 To examine the role of deubiquitination activity of CSN5 in HIV Gag ubiquitination, ubiquitination of HIV Gag was determined in 293 cells stably transfected with pLEYFP expressing CSN5 with/without the JAMM domain deletion plus pcDNA3-HIVGag-GFP (Figure 3, E and F, left panels) or pcDNA3 (Figure 3, E and F, right panels) as controls. The results of ubiquitin immunoprecipitation followed by Western blot analysis of GFP (Figure 3E,) or HIV Gag (Figure 3F) indicates that more ubiquitinated Gag was detected in exosomes isolated from 293 cells transfected with the JAMM domain deleted CSN5 than cells transfected with wild-type. The signal detected was GFP or Gag specific as no signal was detected when 293 cells were transfected with pcDNA3 instead of pcDNA3-HIVGag-GFP (Figure 3, E and F, right panels). In addition, the identity of each band detected (Figure 3E, left panel) was also confirmed by mass spectrometry. These bands were identified as ubiquitinated HIV Gag (data not shown). The reduction of signal intensity of ubiquitinated HIV Gag in the exosomes isolated from the supernatants of 293 cells transfected with wild-type CSN5 is not due to insufficient expression of HIV Gag-GFP because similar amounts of Gag were detected in both cellular lysates (Figure 3F, bottom/left panel). We also determined the amount of ubiquitinated HIV Gag in exosomes isolated from the supernatants of 293 cells with/without CSN5 knocked down by the addition of doxytetracycline using the identical strategy as described above. siRNA knockdown of CSN5 led to an increase of ubiquitinated HIV Gag in 293 exosomes (data not shown).
Within the JAMM domain of the CSN5 protein, amino acids 138 and 140 have been shown to be critical for maintaining deubiquitination of NEDD8 activity. To further identify whether these two amino residues play a role in CSN5-mediated deubiquitination of exosomal HSP70, 293 cells were transfected with wild-type CSN5 or mutant CSN5 with both amino acid mutations. On Western blot analysis of ubiquitinated exosomal HSP70, a stronger signal was detected in the exosomes isolated from the supernatants of 293 cells transfected with the mutations than with a pcDNA3.1CSN5 control vector (Figure 3G). These data suggest that the JAMM domain of CSN5 plays a role in the deubiquitination of exosomal HSP70.
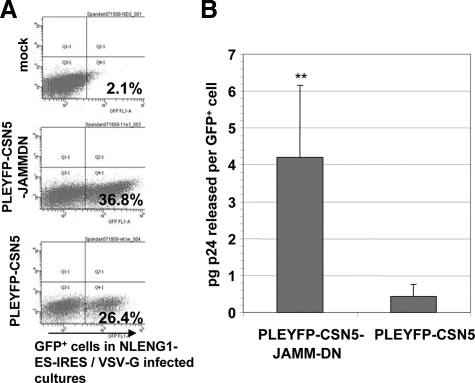
Since HIV-1 Gag ubiquitination has been shown to be important for virus formation, we further determined if the JAMM domain of CSN5 has an effect on the release of HIV-1 through the exosomal pathway, which represents an important pathway of HIV-1 assembly and release in certain cell types.39,40,41,42 ELISA data show that 48 hours after cells are infected with HIV-1, there is a significant increase in the amount of de novo produced virus released from 293 cells transfected with CSN5 JAMM domain deletion in comparison with 293 cells transfected with a wild-type CSN5 (Figure 4).
Figure 4.
293 cells stably transfected with pLEYFP-CSN5-JAMMDN or pLEYFP-CSN5 were infected with vesicular stomatitis virus G pseudotyped HIV-1 NLENG1-ES-IRES. Forty-eight hours later, the cell culture supernatants were analyzed for de novo produced virus by measuring HIV-1 capsid protein using an ELISA for p24. The relative number of HIV-1 infected cells in each culture (JAMMDN mutant versus wild-type) was determined by flow cytometry, measuring NLENG1-ES-IRES+/GFP+ cells (A). The ELISA results are expressed as pg concentration of p24 released per GFP+ cell (B). Data are given as the average values ± SEM obtained for three samples in two independent experiments. **P < 0.001.
Discussion
In this study, we have shown that COP9 associated CSN5 plays a role in exosomal proteins including HSP70 sorting into the exosomes. Our data indicates that siRNA knockout of CSN5 causes a significant increase in both ubiquitinated and non-ubiquitinated proteins including HSP70 detected in the exosomes but not in total cell lysates. Second, we have shown that the JAMM domain of CSN5 has deubiquitin activity that regulates ubiquitinated HSP70 sorting into exosomes. Third, knockdown of CSN5 led to the enhancement of ubiquitinated and total HSP70 sorted into exosomes, supporting the hypothesis that CSN5 is a rate-limiting factor that controls protein sorting into exosomes in a deubiquitin dependent and independent manner. Finally, deletion or point mutation of CSN5 JAMM domain in 293 cells also leads to an enhancement of ubiquitination of other protein, such as HIV Gag, that was sorted into exosomes. This finding suggest that COP9 associated CSN5 regulates other protein sorting into exosomes in addition to HSP70 proteins.
Ubiquitination functions as a sorting signal for endosomes that directs ubiquitination substrates to the interior of MVBs.8,43,44,45 This is the first step that leads to endosomes being transported to lysosomes. An alternative fate of MVBs is their exocytic fusion with the plasma membrane leading to the release of 50- to 90-nm vesicles, called exosomes, into the extracellular milieu. In the present study we observed that COP9-associated CSN5 regulates deubiquitination of exosomal proteins. Although the exact site where exosomal protein sorting is regulated by CSN5-associated COP9 deubiquitin activity is unknown, it is significant that we identified that CSN5-associated with COP9 possesses a deubiquitinase activity for exosomal proteins. Unlike Groisman et al’s finding46 where an artificially polyubiquitinated Cul4A was used as a substrate for in vitro deubiquitination, the proteins we tested, ie, HSP70 and HIV Gag, are derived from exosomes released from the producer cells as a substrate and already demonstrate the deubiquitin activity associated with CSN5. Therefore, our data likely represent what is taking place in vivo during exosome biogenesis. Moreover, since more than just exosomal HSP70 is regulated by CSN5-associated COP9 deubiquitinase activity, identification of other exosomal proteins regulated by the deubiquitinase activity of the CSN5 should provide an important clue to understanding why many pathways regulated by CSN5 are associated with protein degradation. There are still many questions to be addressed. For an example, CSN5 exists as two forms-a free form and a COP9 associated form. We have shown that deubiquitinase activity of CSN5 is associated with COP9. It would be of interest to determine whether the JAMM motif deleted mutant we tested in this study acts as a dominant negative CSN5 to compete with endogenous wild-type CSN5 for inhibition of its deubiquitin activity. If this is the case, then we could theoretically design a specific peptide that is the homologue with CSN5 JAMM domain so as to manipulate the CSN5-associated deubiquitin activity in vivo.
Through this study, we also noticed that unlike ubiquitin patterns of the proteins extracted from total cellular lysates, Western blot analysis of ubiquitinated exosomal proteins, including exosomal HIV Gag, show that they are discrete bands instead of smeared bands. Similar results have been reported by other groups.32 The following factors could contribute to these phenomena although other explanations cannot be excluded. These exosomal proteins may be: 1) dominantly mono-ubiquitinated or polyubiquitinated; 2) initially sorted as polyubiquitinated proteins, but during processing in the exosomal pathway, they may be deubiquitinated so that only certain numbers of ubiquitin moieties remaining attached; or 3) mono-ubiquitinated proteins that are selectively enriched in the exosomes whereas polyubiquitinated exosomal proteins are not enriched and only the mono-ubiquitinated proteins can be detected. Further investigations will need to explore whether COP9 associated CSN5 preferentially regulates mono-ubiquitinated versus polyubiquitinated protein sorting into exosomes. This is of interest since CSN5 associated COP9 has at least two distinct ubiquitin isopeptidase activities: activities that deconjugate ubiquitin and activities that depolymerize polyubiquitin.46
We have shown that COP9 associated CSN5 deubiquitination activity is not restricted to exosomal HSP70, evidenced by the fact that sorting of HIV-ubiquitinated Gag into exosomes is also regulated by CSN5. Knockout of CSN5 resulted in more ubiquitinated Gag being detected in the exosomes, and more p24 released in the supernatants of 293 cells infected with HIV. Ubiquitination of Gag is essential for HIV assembly and budding.35,47,48,49 The amount of p24 released has been used as a marker for virus maturation.40,41 Our result suggests that CSN5 may be a rate-limiting factor for HIV release. One possible mechanism underlying this phenomenon would be that COP9-associated CSN5 deubiquitination activity controls Gag ubiquitination. The ubiquitination of Gag is essential for Gag transport/transfer into the endosomal pathway and subsequently allowing HIV to hijack the exosomal pathway and to bud out of cells. Deubiquitination activity of COP9-associated CSN5 could have an effect on the availability of ubiquitinated HIV Gag in the endosome compartment before HIV budding. CSN5 mediated deubiquitinated HIV Gag would be sorted into a lysosomal pathway for degradation whereas ubiquitinated Gag would be sorted into an exosomal pathway that has been hijacked by HIV for its budding. Therefore, deletion or mutation of CSN5 JAMM domain in 293 cells resulted in more ubiquitinated Gag sorting into exosomes, which, in the end, may result in more viruses being released.
In contrast, siRNA knockdown of CSN5 leads to an increase of both ubiquitinated HSP70 and non-ubiquitinated HSP70. Our findings indicate that deletion of the CSN5 JAMM domain primarily has an effect on the sorting of ubiquitinated HSP70, suggesting that CSN5 regulates not only ubiquitinated proteins but total protein sorted into exosomes as well. We do not know which CSN5 domain(s) regulate total protein sorting into exosomes.
Caution should be exercised in drawing conclusions on the role of CSN5 in terms of deubiquitination of exosomal proteins. CSN5 could directly or indirectly regulate the deubiquitination of exosomal proteins. CSN5 containing the deubiquitin domain with a point-mutation or the CSN5 with a deletion of this domain could result in a reduction of deubiquitination of exosomal proteins. Therefore, CSN5 could act as a deubiquitin enzyme to drive this reaction. Also plausible is that a deubiquitin enzyme associated with CSN5 or other subunits of COP9 may initiate the reaction, and CSN5 regulates the activity of its associated deubiquitin enzyme since COP9-associated CSN5 interacts with many proteins and regulates a variety of signaling pathways. Both possibilities could co-exist. Mass spectrometry analysis of COP9-associated deubiquitin enzymes would be one avenue for determining if the latter possibility is the case. Additionally, siRNA knockdown of the COP9-associated deubiquitin enzyme would provide conclusive evidence that the latter possibility occurs.
It is conceivable that regulation of exosomal components, including exosomal proteins in and out of an exosome, is critical for determining the biological effects on recipient cells under physiological and pathophysiological conditions. Our results point out that the composition of exosomal proteins, such as HSP70 and HIV Gag, can be regulated by manipulating the expression of CSN5 and COP9/CSN5 associated deubiquitinase activity. CSN5 is upregulated in the tumor tissues including breast, colon, pancreatic, and lung carcinomas.50,51,52,53,54,55,56,57,58,59 CSN5 overexpression has been correlated with a poor cancer prognosis. Our findings and other groups’ published results indicate that exosomes released from tumor cells cause immune suppression27,31,60,61; while other data suggest that tumor exosomes stimulate an immune response.62 This discrepancy in exosomal protein activity and biological function could be due to the level of expressed CSN5 in particular tumor cell lines used in each study. This possibility is further validated by our HIV production data, suggesting that CSN5 negatively regulates the production of HIV particles. Interestingly, recently published data report that CSN5 overexpression is negatively correlated with hepatitis B virus infection in hepatocellular carcinoma patients.50 In summary, besides CSN5 being an important factor in several signaling pathways, it may also regulate exosomal protein sorting and may have other effects on exosome mediated cell-to-cell communication, tumor immune suppression, and in the pathogenesis of viral infections (including HIV).
The results presented in this study raise several questions and prompt new avenues of investigation. Future studies should focus on the unique nature of COP9-associated CSN5 deubiquitination activity, the role of COP9-associated CSN5 machinery responsible for driving exosomal protein sorting, how this system functions in relation to ESCRT components and how COP9-associated CSN5 selectively targets certain cargoes. Answers to these issues will assist in clarifying the diverse functions of COP9.
Acknowledgments
We are deeply indebted to Dr. Raymond J. Deshaies of California Institute of Technology for providing 293 cells stably transfected with pGC93 vector expressing siRNA CSN5, Dr. Stephen J. Gould of Johns Hopkins University for providing pcDNA3-HIVGag-GFP, and Dr. Joseph R. Nevins of Duke University Medical Center for providing pCDNA3.1CSN5 and pCDNA3.1-CSN5mut vectors. We thank Dr. A. Jerald Ainsworth for editorial assistance.
Footnotes
Address reprint requests to Dr. Huang-Ge Zhang, University of Alabama at Birmingham, 306 Shelby Building, 1825 University Boulevard, Birmingham, AL 35294-0007. E-mail: Huang-Ge.Zhang@ccc.uab.edu.
Supported in part by grants from the NIH (RO1CA116092, RO1CA107181, R01AT004294), services of the Mucosal HIV and Immunobiology Center/Genetically Defined Mucrobe Core (DK64400), and the Center for AIDs Research/Virology Core (P30AI027767), a grant from the Susan G. Komen Breast Cancer Foundation, and by Birmingham VAMC Merit Review Grants to J.C.K. and H.-G.Z.
References
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex. ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilfelder U, Frank O, Sparacio S, Schon U, Bosch V, Seifarth W, Leib-Mosch C. The potential of retroviral vectors to cotransfer human endogenous retroviruses (HERVs) from human packaging cell lines. Gene. 2007;390:175–179. doi: 10.1016/j.gene.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Raposo G, Marsh M. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 2004;12:310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Callige M, Kieffer I, Richard-Foy H. CSN5/Jab1 is involved in ligand-dependent degradation of estrogen receptor {alpha} by the proteasome. Mol Cell Biol. 2005;25:4349–4358. doi: 10.1128/MCB.25.11.4349-4358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YS, Hao J, Bonin C, Morikawa Y, Cserjesi P. JAB1 enhances HAND2 transcriptional activity by regulating HAND2 DNA binding. J Neurosci Res. 2004;76:613–622. doi: 10.1002/jnr.20105. [DOI] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK. CSN5/Jab1 mutations affect axis formation in the Drosophila oocyte by activating a meiotic checkpoint. Development. 2002;129:5053–5064. doi: 10.1242/dev.129.21.5053. [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, Segal D, Chamovitz DA. COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: effects on Cactus. Dorsal and hematopoiesis. Genes Cells. 2007;12:183–195. doi: 10.1111/j.1365-2443.2007.01049.x. [DOI] [PubMed] [Google Scholar]
- Kim JV, Latouche JB, Riviere I, Sadelain M. The ABCs of artificial antigen presentation. Nature Biotechnol. 2004;22:403–410. doi: 10.1038/nbt955. [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Hanck T, Stricker R, Reiser G. Jab1, a novel protease-activated receptor-2 (PAR-2)-interacting protein, is involved in PAR-2-induced activation of activator protein-1. J Biol Chem. 2006;281:7927–7936. doi: 10.1074/jbc.M510784200. [DOI] [PubMed] [Google Scholar]
- Oh W, Lee EW, Sung YH, Yang MR, Ghim J, Lee HW, Song J. Jab1 induces the cytoplasmic localization and degradation of p53 in coordination with Hdm2. J Biol Chem. 2006;281:17457–17465. doi: 10.1074/jbc.M601857200. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kato JY, Tatsumi E, Takahashi T, Matsuo Y, Yoneda-Kato N. The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood. 2005;105:775–783. doi: 10.1182/blood-2004-04-1242. [DOI] [PubMed] [Google Scholar]
- Wang J, Li C, Liu Y, Mei W, Yu S, Liu C, Zhang L, Cao X, Kimberly RP, Grizzle W, Zhang HG. JAB1 determines the response of rheumatoid arthritis synovial fibroblasts to tumor necrosis factor-alpha. Am J Pathol. 2006;169:889–902. doi: 10.2353/ajpath.2006.051161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1–10. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54. doi: 10.1016/j.bbamcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, Zhang HG. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- Zhang HG, Hanson LA. Deletion of thymidine kinase gene attenuates channel catfish herpesvirus while maintaining infectivity. Virology. 1995;209:658–663. doi: 10.1006/viro.1995.1300. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev. 2006;20:613–623. doi: 10.1101/gad.1345006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA. 2004;101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang HG. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood Cells Mol Dis. 2005;35:398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281:23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- Jager S, Gottwein E, Krausslich HG. Ubiquitination of human immunodeficiency virus type 1 Gag is highly dependent on Gag membrane association. J Virol. 2007;81:9193–9201. doi: 10.1128/JVI.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger PP, Schubert U. The ubiquitin-proteasome system in HIV replication: potential targets for antiretroviral therapy. Expert Rev Anti Infect Ther. 2005;3:61–79. doi: 10.1586/14787210.3.1.61. [DOI] [PubMed] [Google Scholar]
- Mazze FM, Degreve L. The role of viral and cellular proteins in the budding of human immunodeficiency virus. Acta Virol. 2006;50:75–85. [PubMed] [Google Scholar]
- Ott DE, Coren LV, Chertova EN, Gagliardi TD, Schubert U. Ubiquitination of HIV-1 and MuLV Gag. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- Cunningham AL, Dwyer DE, Dowton DN. Viral markers in HIV infection and AIDS. J Acquir Immune Defic Syndr. 1993;6:S32–S35. [PubMed] [Google Scholar]
- Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saag MS. Use of virologic markers in clinical practice. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:S3–S13. doi: 10.1097/00042560-199701001-00002. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell. 2004;15:468–480. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007;18:707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Bultmann A, Eberle J, Haas J. Ubiquitination of the human immunodeficiency virus type 1 env glycoprotein. J Virol. 2000;74:5373–5376. doi: 10.1128/jvi.74.11.5373-5376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan J, Oliaro J, Domigan N, Potter H, Aitken G, Allardyce R, Roake J. Immune response to an 18-kilodalton outer membrane antigen identifies lipoprotein 20 as a Helicobacter pylori vaccine candidate. Infect Immun. 2000;68:3337–3343. doi: 10.1128/iai.68.6.3337-3343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, 2nd, Yoshinaka Y, Oroszlan S, Arthur LO, Henderson LE. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MC, Huang CC, Chang HC, Hu TH, Hung WC. Overexpression of Jab1 in hepatocellular carcinoma and its inhibition by peroxisome proliferator-activated receptor{gamma} ligands in vitro and in vivo. Clin Cancer Res. 2008;14:4045–4052. doi: 10.1158/1078-0432.CCR-07-5040. [DOI] [PubMed] [Google Scholar]
- Wang J, Barnes RO, West NR, Olson M, Chu JE, Watson PH. Jab1 is a target of EGFR signaling in ERalpha-negative breast cancer. Breast Cancer Res. 2008;10:R51. doi: 10.1186/bcr2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MC, Chai CY, Hou MF, Chang HC, Chen WT, Hung WC. Jab1 is overexpressed in human breast cancer and is a downstream target for HER-2/neu. Mod Pathol. 2008;21:609–616. doi: 10.1038/modpathol.2008.23. [DOI] [PubMed] [Google Scholar]
- Hsu MC, Chang HC, Hung WC. HER-2/neu transcriptionally activates Jab1 expression via the AKT/beta-catenin pathway in breast cancer cells. Endocr Relat Cancer. 2007;14:655–667. doi: 10.1677/ERC-07-0077. [DOI] [PubMed] [Google Scholar]
- Berg JP, Zhou Q, Breuhahn K, Schirmacher P, Patil MA, Chen X, Schafer N, Holler TT, Fischer HP, Buttner R, Gutgemann I. Inverse expression of Jun activation domain binding protein 1 and cell cycle inhibitor p27Kip1: influence on proliferation in hepatocellular carcinoma. Hum Pathol. 2007;38:1621–1627. doi: 10.1016/j.humpath.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Osoegawa A, Yoshino I, Kometani T, Yamaguchi M, Kameyama T, Yohena T, Maehara Y. Overexpression of Jun activation domain-binding protein 1 in nonsmall cell lung cancer and its significance in p27 expression and clinical features. Cancer. 2006;107:154–161. doi: 10.1002/cncr.21961. [DOI] [PubMed] [Google Scholar]
- Harada K, Kawashima Y, Yoshida H, Sato M. High expression of Jun activation domain-binding protein 1 (Jab1) is a strong prognostic marker in oral squamous cell carcinoma patients treated by UFT in combination with radiation. Anticancer Res. 2006;26:1615–1619. [PubMed] [Google Scholar]
- Sui L, Dong Y, Watanabe Y, Yamaguchi F, Sugimoto K, Tokuda M. Clinical significance of Skp2 expression, alone and combined with Jab1 and p27 in epithelial ovarian tumors. Oncol Rep. 2006;15:765–771. [PubMed] [Google Scholar]
- Dong Y, Sui L, Watanabe Y, Yamaguchi F, Hatano N, Tokuda M. Prognostic significance of Jab1 expression in laryngeal squamous cell carcinomas. Clin Cancer Res. 2005;11:259–266. [PubMed] [Google Scholar]
- Fukumoto A, Ikeda N, Sho M, Tomoda K, Kanehiro H, Hisanaga M, Tsurui Y, Tsutsumi M, Kato JY, Nakajima Y. Prognostic significance of localized p27Kip1 and potential role of Jab1/CSN5 in pancreatic cancer. Oncol Rep. 2004;11:277–284. [PubMed] [Google Scholar]
- Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176:1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]