Abstract
The ultraviolet B (UVB) waveband within sunlight is an important carcinogen; however, UVA is also likely to be involved. By ascribing mutations to being either UVB or UVA induced, we have previously shown that human skin cancers contain similar numbers of UVB- and UVA-induced mutations, and, importantly, the UVA mutations were at the base of the epidermis of the tumors. To determine whether these mutations occurred in response to UV, we exposed engineered human skin (EHS) to UVA, UVB, or a mixture that resembled sunlight, and then detected mutations by both denaturing high-performance liquid chromatography and DNA sequencing. EHS resembles human skin, modeling differential waveband penetration to the basal, dividing keratinocytes. We administered only four low doses of UV exposure. Both UVA and UVB induced p53 mutations in irradiated EHS, suggesting that sunlight doses that are achievable during normal daily activities are mutagenic. UVA- but not UVB-induced mutations predominated in the basal epidermis that contains dividing keratinocytes and are thought to give rise to skin tumors. These studies indicate that both UVA and UVB at physiological doses are mutagenic to keratinocytes in EHS.
Skin cancers are the most frequent tumors in Caucasians. Ultraviolet (UV) radiation contains three components, UVC (100 to 290 nm), UVB (290 to 320 nm), and UVA (320 to 400 nm). UVB can induce gene mutations,1,2 immune suppression,2,3 and results in the development of skin cancer in animal models.4 Therefore, UVB has been considered to be the dominant carcinogen in sunlight and the chemical and cosmetics industries initially developed sunscreens that protected better from UVB than UVA. However the role of UVA has recently aroused growing concern. UVA comprises at least a 20-fold greater proportion of sunlight than UVB, although UVB photons have more energy than UVA. In addition, initiation of skin tumors presumably requires the penetration of UV photons to the dividing basal/stem cell layer for acute DNA damage to become fixed as heritable genomic mutations,5 and more (>20%) UVA than UVB (<10%) would be expected to reach this basal layer.6 UVA is likely to contribute to skin cancer development. Indeed, UVA is mutagenic in mammalian cells,7,8 human skin tissues,9 and induces skin tumors in animal models.10,11 UVA also causes immunosuppression in humans2 and has been implicated in the development of malignant melanoma in humans.12,13
It is generally accepted that UVB absorbed by two adjacent cytosine or thymine residues in DNA causes the formation of cyclobutane pyrimidine dimers (CPDs), which if unrepaired lead to G:C> A:T transitions,14,15 and pyrimidine (6-4) pyrimidone photoproducts. The mechanistic involvement of UVA in mutagenesis is still unclear. UVA has been shown to induce oxidative DNA damage16 and A:T> C:G transversions have been considered as UVA fingerprints as they were mainly found in UVA- but not UVB-exposed Chinese hamster ovary cells.7 However, recent studies also show that UVA can induce CPDs.17,9 It is important to characterize the waveband and spectra of UV that causes skin cancer in humans to aid development of preventive measures. This cannot be determined directly, and therefore biological factors such as mutations that lead to cancer need to be examined.
Engineered Human Skin (EHS) is an ideal model to investigate UV-induced damage and repair18 and thus is a powerful tool for detection of UV-induced gene mutations. Monolayer cell cultures and mouse epidermis, which are much thinner than human epidermis, do not model UV penetration of intact human skin. Therefore, we used EHS to model the complex organizational structure of human skin, with dividing cells at the base and stratified fully differentiated cells and a cornified layer at the surface. This enables wavebands to interact with target cells in a manner dependent on both depth of penetration of that waveband into human skin, and localization of the target cells within the epidermis. The effects on p53 mutations of UVA, UVB and solar simulated UV (ssUV; mixture of UVA and UVB that mimics sunlight) irradiation at doses to which humans can be realistically exposed were examined. p53 mutations were pre-screened by denaturing high-performance liquid chromatography (dHPLC), and were confirmed by DNA sequencing. We found that as few as four doses of UVA and UVB both induce a similar number of p53 mutations in EHS, but UVA is the dominant mutagen in the basal layer of the epidermis.
Materials and Methods
Reconstruction of EHS
The same strains of cells were used for the whole study. Normal human keratinocytes were isolated from breast skin of a fair-skinned female, age 24, obtained after mammary reduction and used at the first passage for creation of EHS. Normal human dermal fibroblasts were isolated after spreading from mammary skin explants, and cultured in Dulbecco’s Modified Eagles Medium + 10% fetal calf serum.19 Dermal fibroblasts were used at passage 7 for EHS production. Tissue was obtained following institutional ethics committee approval and French Medical Research Council guidelines.
Dermal equivalents were prepared as previously described19 using a collagen-fibroblast mixture containing 106 fibroblasts. After contraction, human keratinocytes were seeded onto this support. The culture was maintained for 7 days in immersed conditions and raised to the air-liquid interface for another 7 days to obtain a complete differentiation process. Before UV exposure, EHS were rinsed twice with PBS to remove culture media components such as phenol red. EHS on grids were then irradiated without medium to avoid any filtering of UV by medium.
UV Sources and Irradiations
The EHS were irradiated with UVA, UVB or ssUV, with 2 different doses per spectra, as previously described20 using a 1000-W Xenon lamp equipped with a dichroic mirror (Oriel, Les Ulis, France) filtered by a UG5, 2-mm thick (Schott, Clichy, France) filter. UVA radiation alone was obtained with a WG335 (3 mm) Schott filter, which removed all of the photons below 320 nm. The UVB spectrum was obtained using a custom made filter obtained by deposition of thin layers on fused silica (MicroModule, Le Plessix Paté, France). The spectra is highly physiological because it only corresponds to the UVB portion contained in the solar spectrum, and has a spectral distribution closely matched to UVB in natural sun exposure. The measured irradiance of the ssUV source obtained with a WG 320 (1.5 mm) Schott filter, complied with the European Cosmetic and Perfumery Association21 ssUV criteria. The spectral irradiances were carefully measured with a spectroradiometer (Macam Photometrics, Livingston, UK) calibrated against traceable standard lamps (National Physics Laboratories, Teddington, UK).
The doses were 12.5 and 6.25 J/cm2 UVA, and 0.1 and 0.05 J/cm2 UVB. The doses of ssUV were 1.4 (1.3 J/cm2 UVA + 0.1 J/cm2 UVB) and 0.75 J/cm2 (0.7 J/cm2 UVA + 0.05 J/cm2 UVB). EHS were exposed 4 times at days 1, 3, 5, and 8 after obtaining a fully differentiated epidermis and the samples were collected at day 11. Each sample was cut into two pieces. One was fixed in 10% formalin for histological analysis and the other was frozen in liquid nitrogen for p53 mutation analysis. A total of 24 EHS were irradiated (3 spectra × 2 doses × 4 repeats). At the same time, four EHS were generated as un-irradiated controls. In total 28 EHS were included in this study.
Histology and Immunohistochemistry
Samples were fixed in 10% neutral formalin and treated for histology. Paraffin sections were stained with hematoxylin, eosin, and saffron as previously described.19 Immunohistochemical staining of p53 was performed using mouse monoclonal antibody against human p53 (clone D07, Dako, France) and Dako EnVisions+ System Peroxidase (AEC, Dako, France) as described.22 Goat anti-mouse-HRP (Dako, France) was used as secondary antibody.
Frozen sections were used for Ki-67 (M7240, Dako, Denmark) and CPD (H3 clone 4F6, Dako Denmark) immunohistochemical staining. EnVisions G/2 system/AP (K5353, Dako, Denmark) was used for visualization according to manufacturer’s instructions. Both mouse IgG1 (X0931, Dako, Denmark) and omitted primary antibodies were used as negative controls. Adult human skin biopsies, which were collected 24 hours after a 2500 mJ/cm2 ssUV irradiation, were used as positive controls.
DNA Extraction and PCR
DNA was isolated from one or two 8-μm frozen sections (for dHPLC analysis) or 50 laser-captured keratinocytes (for DNA sequencing) using QIAamp DNA Micro Kit (QIAGEN, Valencia, USA). DNA isolated from 50 keratinocytes was amplified by GenomiPhiTM DNA Amplification Kit (Amersham Biosciences, Piscataway, NJ) before PCR. Exons 5 to 9 of p53 were initially amplified in all samples using the primers and cycling parameters described.23,24 Negative controls without DNA were included in each PCR cycle. PCR cycles were performed in a Hybaid Px2 thermal cycler. PCR products were checked by 2% agarose gel electrophoresis before sequencing. Any samples containing potential mutations indicated by sequencing were re-amplified with high fidelity VELOCITY DNA polymerase (Bioline, Boston, USA) and re-sequenced. Cycling parameters were 96°C for 3 minutes, followed by 45 cycles at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, with a final step at 72°C for 10 minutes.
dHPLC Analysis
dHPLC analysis was undertaken on the Transgenomic Wave nucleic Acid Fragment System (Model 3500 HT; Transgenomic, Omaha, NE), controlled by Navigator software (Transgenomic). To enhance heteroduplex formation, untreated PCR products were denatured at 95°C for 5 minutes, followed by gradual reannealing to 25°C over 45 minutes. Eight to twenty-μl products were automatically loaded on a DNASep Cartridge and eluted with linear gradient changes of buffer A and buffer B, with the ratio determined by Navigator software. The temperature for heteroduplex detection was also determined using Navigator software, based on the melting profiles of the amplicons sequence.
Microdissection
The microdissected samples were sequenced only if any variants were identified from whole frozen section(s) by dHPLC. Each of these interesting EHS samples were microdissected to isolate groups of 50 keratinocytes from four regions (two suprabasal and two at the base of the epidermis within two vertical planes, Figure 1A) of one 8-μm cryosection using a Pixcell II laser capture microdissection system (Arcturus Engineering, Mountain View, CA) and procedures we have described previously.25,23
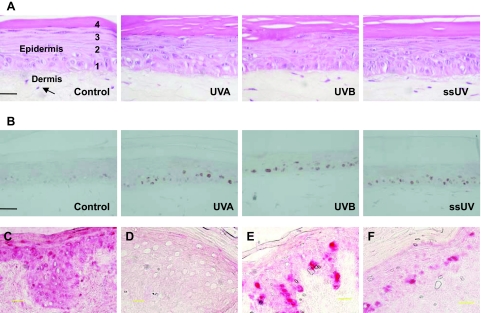
Figure 1.
The regions of microdisseted 50 keratinocytes for DNA sequencing (A). The 50 suprabasal cells (S1 and S2) were directly overlying the 50 basal layer cells (B1 and B2). About 50% of the microdissected basal cells were in contact with the basement membrane, as this region was limited to the bottom two cell layers. Emission spectra obtained from UVA, UVB, and ssUV sources used in this study (B).
DNA Sequencing
Amplicons, which were purified by ExoSAP-IT (Amersham Biosciences), were sequenced in both forward and reverse directions using an Applied Biosystems model 373A. Sequence readouts were checked as previously described.23 The GenBank database accession number is X54156. A mutation was recorded only if detected in both TaqDNA polymerase and VELOCITY DNA polymerase amplified amplicons.
Results
UVA, UVB, and ssUV Sources
Figure 1B shows the emission spectra of the UVA, UVB, and ssUV sources. Using the WG335 schott filter, all photons below 320 nm were cut, which excluded UVB. There is little visible light between 400 and 430 nm, thus the mutations were induced by UVA, but they may not be specific to UVA. The UVB source contains 71% UVB with 26% UVA, mainly below 330 nm. Therefore, UVA within UVB may make a small contribution to UVB induced mutations. The ssUV source contains 88% UVA and 8% UVB.
UVA and UVB Increase p53 Protein Expression
EHS resembles human skin morphologically (Figure 2A). It has an underlying dermis, with the epidermis showing the complete layering and differentiation pattern of normal human skin. Basal, spinous, granular, and keratinised layers were clearly distinguishable in EHS. The epidermis was of similar thickness to normal human adult skin. The underlying dermal equivalent was composed of dermal fibroblasts embedded into a collagen matrix. Overall, the morphology of irradiated EHS was very similar to the un-irradiated control. However, a slight thickening of the stratum corneum was observed in irradiated EHS compared with control, which was more obvious with higher doses of irradiation compared with lower doses. No distinguishable differences in morphology were observed between UVA-, UVB- or ssUV-irradiated samples. UVA, UVB, and ssUV all increased levels of p53 protein as detected by immunostaining (Figure 2B). A large number of p53 positive cells were observed within the basal layer, in addition to staining in the suprabasal compartments. The large number of p53 positive cells in the basal layer may relate to the large number of nuclei also present in this layer compared with suprabasal layers. Sunburn cells were not observed in irradiated EHS, which was expected with these low doses.20 CPD staining could be detected in adult human skin, collected 24 hours after one ssUV exposure (Figure 2C). In contrast, no positive CPD lesions were observed in our EHS samples used for mutation analysis (Figure 2D). This is not surprising as UV-induced photo-lesions are normally repaired with 24 to 48 hours,26 and our EHS were collected 3 days after the final UV exposure. Proliferating Ki-67 keratinoctyes were mainly localized at basal layers, but some Ki-67 positive cells were seen in the suprabasal epidermis (Figure 2, E and F) of our UV irradiated EHS. These data are consistent with previous findings.27 The isotype and omitted antibody controls were negative for staining.
Figure 2.
UV radiation does not induce detectable morphological changes in EHS (A). Fibroblasts (arrow) encased in a type I collagen lattice provides dermal support for the epidermis. EHS collected on day 11, 3 days after the final of four UV exposures, were fixed and H&E-stained. Distinct basal (1), spinous (2), granular (3), and cornified (4) layers are recognizable in un-irradiated control, 12.5 J/cm2 UVA-, 0.1 J/cm2 UVB-, and 1.4 J/cm2 ssUV- irradiated EHS. Sections from the same groups of EHS were immunostained for p53 protein (B). Anti-CPD immunostaining in adult human skin collected 24 hours after UV irradiation (C) and in UV-irradiated EHS used for mutation detection (D). Anti-Ki-67 immunostaining in UV-irradiated adult human skin (E) and in UV-irradiated EHS (F). p53 positive cells are stained brown. CPD and Ki-67 positive cells are stained red. Scale bar = 25 μm.
UVA and UVB Induce p53 Mutations
To confirm and extend our earlier report that UVA and UVB contribute to sunlight-induced p53 mutations in human skin cancers,23 we created and irradiated the EHS as described in the Materials and Methods. dHPLC allows automated detection of single base substitutions as well as small insertions and deletions based on heteroduplex formation. The sensitivity of dHPLC has been reported to range from 96 to 100%28,29 and can detect variants at a frequency as low as 2.5% of cells.30 Therefore it was used for screening p53 mutations in exons 5 to 9 from one or two 8-μm thickness frozen sections from each of these EHS samples. DNA sequencing was then used to confirm the variants found by dHPLC. To increase the sensitivity of sequencing, DNA was extracted from regions containing 50 keratinocytes isolated by laser capture microdissection. We also wanted to investigate whether mutations were localized to small regions within the EHS or occurred throughout the sample. Therefore four regions containing 50 microdissected keratinocytes each, two regions within the suprabasal epidermal layers (S1, S2), and two within the basal epidermal layers (B1, B2) in each sample were investigated, respectively. To ensure that any UV-induced mutations were accurately detected, sequencing was performed on samples independently amplified with TaqDNA polymerase and high fidelity VELOCITY DNA polymerase. Only mutations detected in both sets of sequencing were used for data analysis.
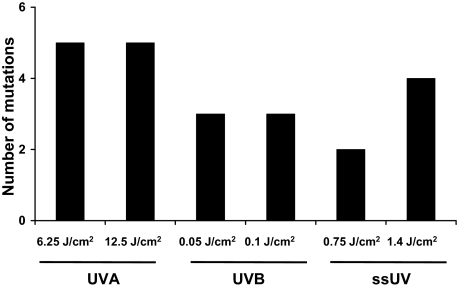
About 80% of variants detected by dHPLC were confirmed as mutations by sequencing on at least one microdissected sample amplification with TaqDNA polymerase. Of these, 63% were independently confirmed following amplified with high fidelity VELOCITY DNA polymerase. There were good concordances between these detection systems. As shown in Table 1, 29 mutations were detected in 15 out of 24 UV-irradiated EHS, and none in un-irradiated EHS. Some of these were repetitive detection of the same mutation in different microdissected regions of the same EHS. There were seven mutations in the 12.5 J/cm2 UVA-irradiated group. A mutation at exon 5, codon 178 was detected in two different microdissected regions in each of samples 7 and 8. Therefore five different mutations were due to this dose of UVA (Figure 3), with two having spread to multiple microdissected regions. Of seven mutations detected in 6.25 J/cm2 UVA-irradiated samples, a mutation at codon 178 was detected in three locations in sample 9. Hence there were five different mutations induced by the lower dose of UVA. Three mutations were found in the 0.1 J/cm2 UVB-irradiated samples and none were detected at more than one region, therefore three mutations were induced by this dose of UVB. Five mutations were found in 0.05 J/cm2 UVB-irradiated samples but identical mutations were detected at two locations in sample 25 (exon 5, codon 148) and sample 26 (exon 5, cordon 156), and therefore the lower dose of UVB induced three mutations. Four mutations were found in 1.4 J/cm2 ssUV-irradiated samples and none were detected at more than one location. Among three mutations detected in 0.75 J/cm2 ssUV-irradiated samples, the same mutation was detected at two locations in sample 20 (exon 5, codon 178). Therefore the lower dose of ssUV induced two mutations. In total, 22 mutations were found to be induced by the three types of radiation tested, UVA, UVB, or ssUV. In another words, about 0.9 mutations were found per UV-irradiated EHS.
Table 1.
Details of Mutations Detected in Exons 5–9 of p53 in EHS Samples
| S-E-L* | Base change | Codon & change | Mt† | CpG‡ | Py-Py |
|---|---|---|---|---|---|
| 12.5 J/cm2 UVA | |||||
| 5-5-B1 | G:C>A:T | 162, ATC to ATT | S | − | + |
| 5-5-B1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 6-6-B1 | G:C>C:G | 202, CGT to CCT | M | + | − |
| 7-5-S1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 7-5-B2 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 8-5-S1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 8-5-B2 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 6.25 J/cm2 UVA | |||||
| 9-5-S1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 9-5-S2 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 9-5-B1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 9-6-S2 | G:C>C:G | 202, CGT to CCT | M | + | − |
| 11-6-B1 | G:C>C:G | 222, CCG to CCC | S | + | − |
| 12-5-B1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 12-6-S2 | G:C>C:G | 222, CCG to CCC | S | + | − |
| 0.1 J/cm2 UVB | |||||
| 24-5-S2 | A:T>C:G | 146, TGG to GGG | M | − | − |
| 24-5-S2 | G:C>C:G | 156, CGC to CGG | S | + | − |
| 24-5-B1 | G:C>A:T | 139, AAG to AAA | S | − | + |
| 0.05 J/cm2 UVB | |||||
| 25-5-S1 | G:C>T:A | 148, GAT to TAT | M | − | + |
| 25-5-S2 | G:C>T:A | 148, GAT to TAT | M | − | + |
| 26-5-S2 | G:C>C:G | 156, CGC to CGG | S | + | − |
| 26-5-S2 | G:C>C:G | 158, CGC to CGG | S | + | − |
| 26-5-B1 | G:C>C:G | 156, CGC to CGG | S | + | − |
| 1.4 J/cm2 ssUV | |||||
| 13-5-S2 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 13-9-B2 | G:C>T:A | 312, ACC to ACA | S | − | + |
| 15-9-B2 | G:C>A:T | 312, ACC to ACA | S | − | + |
| 16-5-S2 | A:T>G:C | 174, AGG to GGG | M | − | + |
| 0.75 J/cm2 ssUV | |||||
| 18-5-S1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 20-5-S1 | A:T>C:G | 178, CAC to CCC | M | − | − |
| 20-5-B2 | A:T>C:G | 178, CAC to CCC | M | − | − |
S-E-L, indicates sample number, exons and layers. There were 4 EHS per group, 5–8 12.5 J/cm2 UVA, 9–12 6.25 J/cm2 UVA, 21–24 0.1 J/cm2 UVB, 25–28 0.05 J/cm2 UVB, 13–16 1.4 J/cm2 ssUV, and 17–20 0.75 J/cm2 ssUV. S1 and S2 are regions 1 and 2 in the suprabasal layers of the epidermis; B1 and B2 are regions 1 and 2 in the basal layers of the epidermis.
Mt, mutant, S, indicates silent Mt, M, indicates missense Mt.
+ indicates mutations at CpG or Py-Py sites, − indicates mutations outside.
Figure 3.
UVA and UVB induce p53 mutations in EHS. Similar numbers of mutations are induced by UVA, UVB, and ssUV. Each group had four EHS samples, and total mutations in each group are shown, as detected by sequencing.
Localization of UVA- and UVB-Induced Mutations
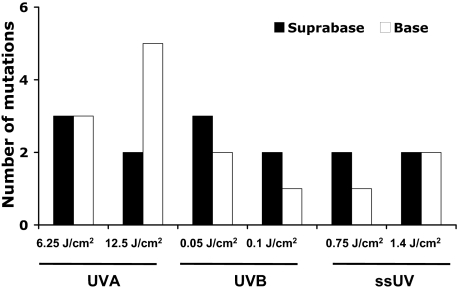
Based on assigning fingerprint mutations to the likelihood of being caused by UVA or UVB, we have previously suggested that UVB-induced DNA damage is confined to the upper epidermal layers of human skin cancers and precancers, whereas UVA-induced DNA damage predominates in the lower layers.23 This stratification is intriguing as the greatest radiation load of both wavebands is at the surface of the epidermis. To confirm this finding, we compared UV-induced mutations in the suprabasal and basal epidermal layers (Figure 4). Note that some mutations were found in both suprabasal and basal epidermis and were therefore counted twice in Figure 4, but only once in Figure 3. Of five mutations induced by the higher dose of UVA, three were found in the basal layer, and two were found in both suprabasal and basal layers. Thus, two and five mutations were in the suprabasal and basal layers, respectively. Among five mutations induced by the lower dose of UVA, two were at the suprabase, two were in the base, and one was in both layers. Therefore, there were three mutations in the base and three at the suprabase. Of the three mutations induced by the higher dose of UVB, two were at the suprabase and one was in the base. Of the three mutations induced by the lower dose of UVB, while one was only at the suprabase, two were in both the basal and suprabasal layers. Therefore, two mutations were in the base and three were at the suprabase. Of the four mutations induced by the higher dose of ssUV, two were in the base only, and two were at the suprabase only, and none were in both layers. Of the two mutations induced by lower dose ssUV, one was at the suprabasal layer only, and one was in both layers. Therefore two mutations were at the suprabase and one in the base. These results show that UVA-induced mutations are predominantly at the basal layer of the epidermis, whereas UVB- and ssUV-induced mutations are evenly distributed to the both suprabasal and basal layers.
Figure 4.
UVA-induced mutations are predominantly at the base of the epidermis. Mutations identified in Figure 3 were divided into whether they were found in suprabasal or basal layers of epidermis.
Types of Mutations Induced by UVA and UVB
Because the amount of UVA energy absorbed by DNA is much lower than UVB,31 its mechanism of action is thought to be different from UVB.32 To examine whether different UV wavebands induce distinct types of mutations in EHS, results from the higher and lower doses of each UV waveband were grouped. Among ten UVA-induced mutations (Figure 5), five were A:T>C:G transversions, which were the most predominant mutations, followed by four G:C>C:G transversions and a G:C>A:T transition. Of the six UVB-induced mutations, the most predominant (three) were G:C>C:G transversions, followed by a A:T>C:G transversion, a G:C>A:T transition, and a G:C>T:A transversion. Among six ssUV-induced mutations, the most predominant were three A:T>G:C transitions. Others include a G:C>A:T transition, a A:T>G:C transition, and a G:C>T:A transversion.
Figure 5.
Types of p53 mutations induced by UV. Mutations from each of the two doses were pooled for each spectrum to give eight EHS per spectrum.
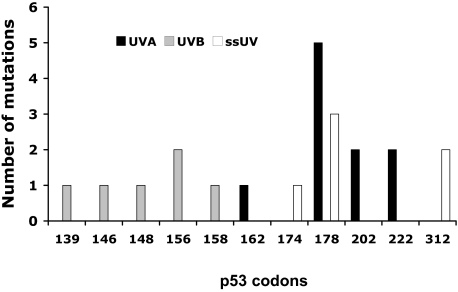
UV-Induced Hotspots in EHS
Hotspots (≥two mutations at the same codon) were found in response to each of the three UV wavebands. As shown in Figure 6, while there were noticeable differences in the mutations’ distribution between each UV waveband, identical hotspots shared across UVA and ssUV were noticed. UVA-induced mutations were found from codons 162 to 222. Five mutations were found at codon 178 and two at 202 and 222. UVB-induced mutations were from codons 139 to 158. Two mutations were found at codon156. The ssUV-induced mutations were found from codons 174 to 312. Three mutations were found at codon 178, corresponding to a UVA hotspot. Two mutations were found at codon 312.
Figure 6.
Distribution of p53 mutations induced by UV. Mutations from each of the two doses were pooled for each spectrum to give eight EHS per spectrum. Mutations were detected by sequencing in microdissected groups of 50 keratinocytes localized at the suprabasal or basal regions of the epidermis.
Other Characterizations of Mutations
Two types of mutations (missense and silent) were found. Of the UVA- and ssUV-induced mutations, 70% and 67% were missense mutations, respectively. In contrast, 67% of UVB-induced mutatons were silent. In addition, 10% of UVA-, 33% of UVB-, and 50% of ssUV-induced mutations were at dipyrimidine sites. Many UVA- and UVB-, but no ssUV-induced mutations were at CpG sites. Most of these mutations were localized to the DNA binding domain (codons 102 to 292), and none were at the nuclear export signal region (316 to 324), or the tetramerization domain (326 to 356).
Discussion
This study indicates that UVA as well as UVB is able to induce mutations in p53 in human skin. UVA and UVB induced similar numbers of mutations in EHS, suggesting that both of these UV wavebands contribute to sunlight-induced p53 mutagenesis in human skin. For the higher dose UVA, larger numbers of mutations were found in the basal layer of EHS than in the suprabasal layers. These results are consistent with our previous findings in human skin cancers and precancers,23 where the roles of UVA and UVB were implicated by the types of mutations. However in this study the UV wavebands are known rather than implied. Our previous study was in fully formed tumors while the current study shows that the predominance of UVA-induced mutations in basal cells occurred in response to only four UV exposures.
This study has several strengths that are likely to account for the differences from previous studies. The model is designed to mimic human exposure to sunlight during routine daily activities rather than the higher UV exposures resulting from recreational activities. The dose of ssUV used (1.4 J/cm2) would be delivered by 36 minutes exposure to sunlight in early spring in Sydney, Australia, at midday (0.64 mW cm2).33 Clearly, at more extreme times of year, such as mid summer or winter, shorter or longer exposure times would be required. Therefore, this dose is relatively low and achievable by natural sun exposure. There are a number of complex biological parameters involved in the formation of sunlight-induced mutations in human skin, each of which was mimicked in this study. The exposure spectrum used is a good match to sunlight, not only due to the absence of wavebands lower than are found in sunlight, but the shape of the spectrum. Many irradiation sources are overweight in low wavelength UVB as compared with sunlight. Due to differential wavelength penetration through tissue, our use of EHS that morphologically resembled human skin should result in the dividing basal cells receiving a similar spectrum to basal keratinocytes in humans exposed to sunlight. Monolayer cell cultures or animal models with thinner skin cannot model the UV wavebands likely to reach basal keratinocytes. Cellular interactions and differentiation state have a large impact on many aspects of cell behavior including division and responses to toxic insults such as UV. This has been mimicked by the EHS used in this study where any photodamaged cell would remain localized within the epidermis and subject to regulation via interactions with surrounding cells. The low doses of UV used in this study are also an important consideration because they should be too low to induce apoptosis34,35 or possibly even cell cycle arrest. However, these low doses of UV induced an increase of p53 immunoreactive cells especially at the base of the epidermis, which may indicate damage to p53.22 These considerations may have contributed to our ability to detect mutations in the majority of irradiated EHS despite the small number of irradiations and the low doses. Our data are consistent with a previous study on microdissected sunlight-exposed human skin that found distinct p53 mutations in keratinocytes from different regions of the epidermis at a similar frequency to our study.36 Nevertheless this model indicates that only four exposures to doses of sunlight to which humans can be exposed during normal daily activities are capable of inducing mutations in the dividing basal cells of the epidermis.
UVA-induced mutations at the basal layers of the epidermis may be particularly crucial to skin photocarcinogenesis since this region contains the dividing basal keratinocytes, keratinoctye stem cells, and transient amplifying cells, which are thought to give rise to skin tumors.37 Therefore, UVA may have a more important role in human skin carcinogenesis than has been suggested by photocarcinogenesis experiments in animals,4 or mutation studies in cell monolayers,38 or in psoriasis patients.39 A large percentage of UVA-induced mutations caused amino acid changes, consistent with this. Predominant UVA-induced mutations at the basal layer of the epidermis may be at least partially due to the high intensity of UVA in sunlight and the greater ability of UVA to penetrate to the base of the epidermis. Indeed, thickening of the stratum corneum was not only observed by the UVA- but also the UVB-irradiated EHS samples. Less efficient repair of UVA-induced genetic damage at the base40 would also have this effect. Moreover, it might be possible that mutant p53 increases symmetric replication as wild-type p53 induces asymmetric stem-cell-like division,41 and lead to rapid mutant p53 expansion at the basal epidermis. Chronic UVB exposure drives cells carrying mutant p53 to escape the stem cell compartment42 to suprabasal layers, where the keratinocytes are still able to divide.43 Therefore, the cells harboring DNA damage/mutant p53, and that are dividing at the suprabasal layers, may contribute to UVB- but not UVA-induced mutations at these layers. Moreover, repetitive UV irradiation may result in accumulation of mutations and cell proliferation. Alternatively, UVA-induced photodamage may occur via UVA absorption by an unknown chromophore resulting in production of reactive oxygen species.44 Therefore, localization of the chromophores and anti-oxidant defense mechanisms will also influence the localization of mutations.
Chronic exposure to suberythemal doses of sunlight may result in p53 mutations and contribute to skin cancer development. Suberythemal doses of UV have been shown to induce significant DNA damage including CPDs and (6-4)-photoproducts in human skin of different racial/ethnic groups,45,46 and induces immunosuppression in both humans2 and mice.47 Exposure to about 0.08 minimal erythemal dose with a latency time of 234 to 238 days has been shown to induce squamous cell carcinoma and actinic keratosis in nearly all UV-irradiated mice.48,49 If UV-induced damage is extreme and irreparable, cell death occurs. Low doses of UV such as doses used in the current study may however produce less catastrophic lesions in DNA, so that the cells do not die by apoptosis but instead harbor unrepaired genomic damage that is the first step toward cancer, as observed in this study. The rate of cell proliferation was significantly increased in suprabasal layers in actinic keratosis and skin adjacent to actinic keratosis, and in sun-exposed skin compared with non-sun-exposed skin.50 In addition, it has been shown that epidermal cell proliferation was significantly increased in the progression from normal skin to squamous cell carcinoma.50,51 Thus, UV-induced proliferation of both basal and suprabasal keratinoctyes enables mutations to be established and may contribute to initial process of tumorigenesis. Similarly in our study, both basal and suprabasal keratinocytes in UV-irradiated EHS were Ki-67 positive, indicating that they were dividing. This would enable the fixation of mutations at both of these layers.
Analysis of the IARC TP53 Mutation Database (www-p53. iarc.fr) and previous published papers showed that few mutations in our study were the same as those previously seen in actinic keratosis, squamous cell carcinoma or basal cell carcinoma. Some of the mutations found in our study were located on dipyrimidine sites, but were not C to T transitions. In contrast, p53 mutations found in human skin cancer are mainly C to T transitions located at dipyrimidine sites. Prominent hotspots (177, 179, 196, 245, 248, and 278)52 found in human skin cancer are lacking in the current study. However, most of our mutations were within these hotspot regions. Interestingly, mutations at codon 178 appeared frequently in our study as well as in patients with xeroderma pigmentosum with squamous cell carcinoma.52 It is possible that UV exposure may drive mutations at codon 178 to create new mutations in adjacent codons177 and 179, as direct sequence alterations can alter the adjacent base and create transition hotspots.53 p53 mutations have been frequently found in normal human skin, but most of these are lost by epidermal turnover. Only the mutations formed in long residing germinative stem cells,54 or those that give a growth advantage may play critical roles in skin carcinogenesis. Relatively short UV exposures in our experimental model are unlikely to enable selection of clones with a mutation that gives a growth on tumorigenic selective advantage. We therefore expected to see the mutations randomly distributed in p53, not selected for by tumorigenesis. This is exactly what we saw, rather than a predominance of mutations at p53 hotspots in human cancer. Importantly, our study demonstrated that low doses of ssUV with a UVA to UVB ratio that is similar to what is found in sunlight, have the ability to induce p53 mutations. Whether these mutations are within stem cells or proliferating cells, in both basal and suprabasal epidermis is not known, but some of these mutations may modify p53 function as most change amino acids and locate at the DNA binding domain. Despite both UVA and UVB inducing a similar range of mutations, some differences were observed. Furthermore, different mutation hotspots were found between these two UV wavebands, suggesting that the mechanisms of UVA and UVB mutagenesis may differ. As the types of mutations and hotspots induced by ssUV were different to those induced by either UVA or UVB, an interaction between these two UV wavebands within sunlight may occur, contributing to mutagenesis.
A:T>C:G transversions were the most frequent mutational event with UVA exposure but not UVB in our EHS studies. This finding is consistent with A:T>C:G transversions being found in a large proportion of UVA- but not UVB-irradiated Chinese hamster ovary cells.7 UVA mutagenesis has also been suggested to result from oxidative DNA base modifications, such as 7, 8-dihydro-8-oxoguanine.55,56 While a high proportion of G:C>T:A transversions, considered to be the mutagenic hallmark of 7, 8-dihydro-8-oxoguanine, was found in UVA-irradiated rodent cells57,8 and healthy human skin exposed to 40 J/cm2 UVA1 three times a week for 2 weeks,58 none were identified in UVA-irradiated EHS in this study. It is generally accepted that G:C>A:T transitions are UVB fingerprints.14,15 However, Kappes et al38 found that C to T transitions were the predominant type of mutation induced by UVB as well as UVA in cultured primary neonatal human fibroblasts. Formation of CPDs, the precursor of C to T transitions have been found in UVA-irradiated cultured cells and human skin.38,9,59 C to T transitions are present but not predominant in UVA- and UVB-irradiated samples in the current study. G:C>C:G transversions are likely to be formed by reactive oxygen species, and were frequently seen with UVB and UVA in the current study. These lesions were not found in the hrpt gene of UVA- and UVB-irradiated primary human fibroblasts,38 nor in cII57 and lacI transgenes8,60 in UVA-irradiated Big Blue mouse embryonic fibroblasts. In contrast a similar frequency of A:T>G:C transitions were induced by UVA and UVB in the hprt gene of human fibroblasts,38 but only one was found in ssUV-irradiated EHS. Thymine glycol, an oxidized pyrimidine lesion, has been reported to induce A:T>G:C transitions.61 Discrepancies between our data and other studies may be due to a number of factors,62 such as differences in experimental models and design, cell types, UV sources and doses, the detection methods, and relative small numbers of mutations. Nevertheless, both our data and published data suggest that oxidative base damage via different oxidative stress reactions may contribute to UVA, UVB, and ssUV mutagenesis. Thus the inclusion of anti-oxidants in sunscreens may help to prevent sunlight-induced gene mutations. Our use of EHS combined with low doses of spectra that match the respective wavebands of sunlight means that the mutations detected here are likely to be of physiological relevance for human sun exposure.
In conclusion, we have demonstrated three important findings. Firstly, UVA induces mutations in human skin that are predominantly located in the basal layers of the epidermis. This suggests that further UVA protective strategies are required and emphasizes the importance of UVA in causing mutations in humans. Secondly, some similarities were observed between UVA- and UVB-induced mutations. Thirdly, the mutations were induced by low dose of ssUV, demonstrating that sunlight exposures that can be achieved in normal daily activities are mutagenic. The results emphasize the need for greater public awareness that there are no safe UV wavelengths.32
Acknowledgments
We thank François Lejeune for technical assistance in skin reconstruction and François Christiaens for his expertise in monitoring the UV sources.
Footnotes
Address reprint requests to Gary M. Halliday, Dermatology Research Laboratories, Blackburn Building, DO6, The University of Sydney, NSW 2006, Australia. E-mail: garyh@med.usyd.edu.au.
Supported grants, from the NSW Cancer Council to G.M.H, and Cure Cancer Australia Foundation to X.X.H, and Epiderm.
References
- Kanjilal S, Pierceall WE, Cummings KK, Kripke ML, Ananthaswamy HN. High frequency of p53 mutations in ultraviolet radiation-induced murine skin tumors: evidence for strand bias and tumor heterogeneity. Cancer Res. 1993;53:2961–2964. [PubMed] [Google Scholar]
- Halliday GM. Inflammation, gene mutation and photoimmunosuppression in response to UVR-induced oxidative damage contributes to photocarcinogenesis. Mutat Res. 2005;571:107–120. doi: 10.1016/j.mrfmmm.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Goettsch W, Garssen J, de Gruijl FR, van Loveren H. UV-B and the immune system. A review with special emphasis on T cell-mediated immunity. Thymus. 1993;21:93–114. [PubMed] [Google Scholar]
- de Gruijl FR, Sterenborg HJ, Forbes PD, Davies RE, Cole C, Kelfkens G, van Weelden H, Slaper H, van der Leun JC. Wavelength dependence of skin cancer induction by ultraviolet irradiation of albino hairless mice. Cancer Res. 1993;53:53–60. [PubMed] [Google Scholar]
- Mukhtar H, Forbes PD, Ananthaswamy HN. Photocarcinogenesis–models and mechanisms. Photodermatol Photoimmunol Photomed. 1999;15:91–95. doi: 10.1111/j.1600-0781.1999.tb00065.x. [DOI] [PubMed] [Google Scholar]
- Bruls WA, Slaper H, van der Leun JC, Berrens L. Transmission of human epidermis and stratum corneum as a function of thickness in the ultraviolet and visible wavelengths. Photochem Photobiol. 1984;40:485–494. doi: 10.1111/j.1751-1097.1984.tb04622.x. [DOI] [PubMed] [Google Scholar]
- Drobetsky EA, Turcotte J, Chateauneuf A. A role for ultraviolet A in solar mutagenesis. Proc Natl Acad Sci USA. 1995;92:2350–2354. doi: 10.1073/pnas.92.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A, Kim SI, Bates SE, Pfeifer GP. Riboflavin activated by ultraviolet A1 irradiation induces oxidative DNA damage-mediated mutations inhibited by vitamin C. Proc Natl Acad Sci USA. 2007;104:5953–5958. doi: 10.1073/pnas.0610534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Woodhead AD, Grist E. Animal model for ultraviolet radiation-induced melanoma: platyfish-swordtail hybrid. Proc Natl Acad Sci USA. 1989;86:8922–8926. doi: 10.1073/pnas.86.22.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastila R, Leszczynski D. Ultraviolet A exposure might increase metastasis of mouse melanoma: a pilot study. Photodermatol Photoimmunol Photomed. 2005;21:183–190. doi: 10.1111/j.1600-0781.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- Moan J, Dahlback A, Setlow RB. Epidemiological support for an hypothesis for melanoma induction indicating a role for UVA radiation. Photochem Photobiol. 1999;70:243–247. [PubMed] [Google Scholar]
- Westerdahl J, Ingvar C, Masback A, Jonsson N, Olsson H. Risk of cutaneous malignant melanoma in relation to use of sunbeds: further evidence for UV-A carcinogenicity. Br J Cancer. 2000;82:1593–1599. doi: 10.1054/bjoc.1999.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Pfeifer GP. Deamination of 5-methylcytosines within cyclobutane pyrimidine dimers is an important component of UVB mutagenesis. J Biol Chem. 2003;278:10314–10321. doi: 10.1074/jbc.M212696200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rosenstein BS, Wang Y, Lebwohl M, Mitchell DM, Wei H. Induction of 8-oxo-7,8-dihydro-2′-deoxyguanosine by ultraviolet radiation in calf thymus DNA and HeLa cells. Photochem Photobiol. 1997;65:119–124. doi: 10.1111/j.1751-1097.1997.tb01886.x. [DOI] [PubMed] [Google Scholar]
- Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernerd F, Asselineau D, Vioux C, Chevallier-Lagente O, Bouadjar B, Sarasin A, Magnaldo T. Clues to epidermal cancer proneness revealed by reconstruction of DNA repair-deficient xeroderma pigmentosum skin in vitro. Proc Natl Acad Sci USA. 2001;98:7817–7822. doi: 10.1073/pnas.141221998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernerd F, Asselineau D. Successive alteration and recovery of epidermal differentiation and morphogenesis after specific UVB-damages in skin reconstructed in vitro. Dev Biol. 1997;183:123–138. doi: 10.1006/dbio.1996.8465. [DOI] [PubMed] [Google Scholar]
- Bernerd F, Vioux C, Lejeune F, Asselineau D. The sun protection factor (SPF) inadequately defines broad spectrum photoprotection: demonstration using skin reconstructed in vitro exposed to UVA, UVB, or UV-solar simulated radiation. Eur J Dermatol. 2003;13:242–249. [PubMed] [Google Scholar]
- COLLPA Brussels: The European Cosmetic Toiletry and Perfumery Association; Sun protection factor test method. 1994:21p. [Google Scholar]
- Seite S, Moyal D, Verdier MP, Hourseau C, Fourtanier A. Accumulated p53 protein and UVA protection level of sunscreens. Photodermatol Photoimmunol Photomed. 2000;16:3–9. doi: 10.1034/j.1600-0781.2000.160103.x. [DOI] [PubMed] [Google Scholar]
- Agar NS, Halliday GM, Barnetson RS, Ananthaswamy HN, Wheeler M, Jones AM. The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations: a role for UVA in human skin carcinogenesis. Proc Natl Acad Sci USA. 2004;101:4954–4959. doi: 10.1073/pnas.0401141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifter M, Jones AM, Walker DM. Epithelial p53 gene expression and mutational analysis, combined with growth fraction assessment, in oral lichen planus. J Oral Pathol Med. 1998;27:318–324. doi: 10.1111/j.1600-0714.1998.tb01963.x. [DOI] [PubMed] [Google Scholar]
- Agar NS, Halliday GM, Barnetson RS, Jones AM. A novel technique for the examination of skin biopsies by laser capture microdissection. J Cutan Pathol. 2003;30:265–270. doi: 10.1046/j.0303-6987.2003.052.x. [DOI] [PubMed] [Google Scholar]
- Tornaletti S, Pfeifer GP. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin cancer. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- Arad S, Konnikov N, Goukassian DA, Gilchrest BA. T-oligos augment UV-induced protective responses in human skin. FASEB J. 2006;20:1895–1897. doi: 10.1096/fj.06-5964fje. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Oefner PJ, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, Upadhyaya M, Sommer SS, McGuffin P. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics. 1998;52:44–49. doi: 10.1006/geno.1998.5411. [DOI] [PubMed] [Google Scholar]
- Keller G, Hartmann A, Mueller J, Hofler H. Denaturing high pressure liquid chromatography (DHPLC) for the analysis of somatic p53 mutations. Lab Invest. 2001;81:1735–1737. doi: 10.1038/labinvest.3780387. [DOI] [PubMed] [Google Scholar]
- Yu B, Sawyer NA, Caramins M, Yuan ZG, Saunderson RB, Pamphlett R, Richmond DR, Jeremy RW, Trent RJ. Denaturing high performance liquid chromatography: high throughput mutation screening in familial hypertrophic cardiomyopathy and SNP genotyping in motor neurone disease. J Clin Pathol. 2005;58:479–485. doi: 10.1136/jcp.2004.021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB. The wavelengths in sunlight effective in producing skin cancer: a theoretical analysis. Proc Natl Acad Sci USA. 1974;71:3363–3366. doi: 10.1073/pnas.71.9.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. Revisiting the photochemistry of solar UVA in human skin. Proc Natl Acad Sci USA. 2006;103:13567–13568. doi: 10.1073/pnas.0605833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, Rana S. Waveband and dose dependency of sunlight-induced immunomodulation and cellular changes. Photochem Photobiol. 2008;84:35–46. doi: 10.1111/j.1751-1097.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Latonen L, Taya Y, Laiho M. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene. 2001;20:6784–6793. doi: 10.1038/sj.onc.1204883. [DOI] [PubMed] [Google Scholar]
- Cotton J, Spandau DF. Ultraviolet B-radiation dose influences the induction of apoptosis and p53 in human keratinocytes. Radiat Res. 1997;147:148–155. [PubMed] [Google Scholar]
- Ling G, Persson A, Berne B, Uhlen M, Lundeberg J, Ponten F. Persistent p53 mutations in single cells from normal human skin. Am J Pathol. 2001;159:1247–1253. doi: 10.1016/S0002-9440(10)62511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ. Keratinocyte stem cells: targets for cutaneous carcinogens. J Clin Invest. 2000;106:3–8. doi: 10.1172/JCI10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006;126:667–675. doi: 10.1038/sj.jid.5700093. [DOI] [PubMed] [Google Scholar]
- Wolf P, Kreimer-Erlacher H, Seidl H, Back B, Soyer HP, Kerl H. The ultraviolet fingerprint dominates the mutational spectrum of the p53 and Ha-ras genes in psoralen + ultraviolet A keratoses from psoriasis patients. J Invest Dermatol. 2004;122:190–200. doi: 10.1046/j.0022-202X.2004.22118.x. [DOI] [PubMed] [Google Scholar]
- Javeri A, Huang XX, Bernerd F, Mason RS, Halliday GM. Human 8-oxoguanine-DNA glycosylase 1 protein and gene are expressed more abundantly in the superficial than basal layer of human epidermis. DNA Repair (Amst) 2008;7:1542–1550. doi: 10.1016/j.dnarep.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Sherley JL, Stadler PB, Johnson DR. Expression of the wild-type p53 antioncogene induces guanine nucleotide-dependent stem cell division kinetics. Proc Natl Acad Sci USA. 1995;92:136–140. doi: 10.1073/pnas.92.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Remenyik E, Zelterman D, Brash DE, Wikonkal NM. Escaping the stem cell compartment: sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc Natl Acad Sci USA. 2001;98:13948–13953. doi: 10.1073/pnas.241353198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, Lyons JG. Inflammatory doses of UV may not be necessary for skin carcinogenesis. Photochem Photobiol. 2007;83:1–12. doi: 10.1111/j.1751-1097.2007.00247.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takahashi K, Zmudzka BZ, Kornhauser A, Miller SA, Tadokoro T, Berens W, Beer JZ, Hearing VJ. Human skin responses to UV radiation: pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. FASEB J. 2006;20:1486–1488. doi: 10.1096/fj.06-5725fje. [DOI] [PubMed] [Google Scholar]
- Tadokoro T, Kobayashi N, Zmudzka BZ, Ito S, Wakamatsu K, Yamaguchi Y, Korossy KS, Miller SA, Beer JZ, Hearing VJ. UV-induced DNA damage and melanin content in human skin differing in racial/ethnic origin. FASEB J. 2003;17:1177–1179. doi: 10.1096/fj.02-0865fje. [DOI] [PubMed] [Google Scholar]
- Byrne SN, Spinks N, Halliday GM. Ultraviolet a irradiation of C57BL/6 mice suppresses systemic contact hypersensitivity or enhances secondary immunity depending on dose. J Invest Dermatol. 2002;119:858–864. doi: 10.1046/j.1523-1747.2002.00261.x. [DOI] [PubMed] [Google Scholar]
- Berg RJ, de Vries A, van Steeg H, de Gruijl FR. Relative susceptibilities of XPA knockout mice and their heterozygous and wild-type littermates to UVB-induced skin cancer. Cancer Res. 1997;57:581–584. [PubMed] [Google Scholar]
- Berg RJ, Rebel H, van der Horst GT, van Kranen HJ, Mullenders LH, van Vloten WA, de Gruijl FR. Impact of global genome repair versus transcription-coupled repair on ultraviolet carcinogenesis in hairless mice. Cancer Res. 2000;60:2858–2863. [PubMed] [Google Scholar]
- Einspahr J, Alberts DS, Aickin M, Welch K, Bozzo P, Levine N, Grogan T. Evaluation of proliferating cell nuclear antigen as a surrogate end point biomarker in actinic keratosis and adjacent, normal-appearing, and non-sun-exposed human skin samples. Cancer Epidemiol Biomarkers Prev. 1996;5:343–348. [PubMed] [Google Scholar]
- Einspahr JG, Alberts DS, Warneke JA, Bozzo P, Basye J, Grogan TM, Nelson MA, Bowden GT. Relationship of p53 mutations to epidermal cell proliferation and apoptosis in human UV-induced skin carcinogenesis. Neoplasia. 1999;1:468–475. doi: 10.1038/sj.neo.7900061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Hum Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
- Miller JH. Mutagenic specificity of ultraviolet light. J Mol Biol. 1985;182:45–65. doi: 10.1016/0022-2836(85)90026-9. [DOI] [PubMed] [Google Scholar]
- de Gruijl FR, Rebel H. Early events in UV carcinogenesis–DNA damage, target cells and mutant p53 foci. Photochem Photobiol. 2008;84:382–387. doi: 10.1111/j.1751-1097.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- Darr D, Fridovich I. Free radicals in cutaneous biology. J Invest Dermatol. 1994;102:671–675. doi: 10.1111/1523-1747.ep12374036. [DOI] [PubMed] [Google Scholar]
- Stary A, Sarasin A. Ultraviolet A- and singlet oxygen-induced mutation spectra. Methods Enzymol. 2000;319:153–165. doi: 10.1016/s0076-6879(00)19017-2. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Synold TW, Xi B, Pfeifer GP. G-to-T transversions and small tandem base deletions are the hallmark of mutations induced by ultraviolet a radiation in mammalian cells. Biochemistry. 2004;43:8169–8177. doi: 10.1021/bi049761v. [DOI] [PubMed] [Google Scholar]
- Persson AE, Edstrom DW, Backvall H, Lundeberg J, Ponten F, Ros AM, Williams C. The mutagenic effect of ultraviolet-A1 on human skin demonstrated by sequencing the p53 gene in single keratinocytes. Photodermatol Photoimmunol Photomed. 2002;18:287–293. doi: 10.1034/j.1600-0781.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- Courdavault S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat Res. 2004;556:135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Kim SI, Pfeifer GP, Besaratinia A. Mutagenicity of ultraviolet A radiation in the lacI transgene in Big Blue mouse embryonic fibroblasts. Mutat Res. 2007;617:71–78. doi: 10.1016/j.mrfmmm.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann JM, Basu AK, Loechler EL. Mutagenic specificity of alkylated and oxidized DNA bases as determined by site-specific mutagenesis. Ann Ist Super Sanita. 1989;25:155–161. [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]