Abstract
The unanimous agreement that cellular processes are (largely) governed by interactions between proteins has led to enormous community efforts culminating in overwhelming information relating to these proteins; to the regulation of their interactions, to the way in which they interact and to the function which is determined by these interactions. These data have been organized in databases and servers. However, to make these really useful, it is essential not only to be aware of these, but in particular to have a working knowledge of which tools to use for a given problem; what are the tool advantages and drawbacks; and no less important how to combine these for a particular goal since usually it is not one tool, but some combination of tool-modules that is needed. This is the goal of this review.
Keywords: protein–protein interactions, protein–protein interfaces, binding site prediction, docking, web servers, databases
PROTEIN–PROTEIN INTERACTIONS
Proteins are parts of complex networks or pathways rather than isolated entities. The different levels of complexity of biological systems arise not only from the number of the proteins (genes) of the organism, but also from combinatorial interactions among them as well as from alternative splicing, and chemical and structural alterations of the proteome [1]. Cellular processes are largely governed by different types of interactions between proteins, and the function of a protein can be better understood considering its interactions. The broad recognition of the importance of characterizing all protein interactions in a cell has rendered the development of experimental and computational techniques to detect and predict interacting protein partners. Experimental methods can be divided into two categories:
Screening large scale protein interactions involves high-throughput experiments where each protein encoded in the genome of interest is expressed and exhaustively probed for mutual interactions by assays such as the yeast two hybrid systems (Y2H) [2, 3], protein-fragment complementation assay (PCA) [4, 5], affinity purification [6], phage display libraries [7] indicating physical interactions. Protein/DNA microarrays [8, 9], and ‘synthetic lethals’ (genes whose simultaneous knockout is lethal to the organism [10]) are also important since they provide functional association between proteins.
Screening specific protein interactions where the experiments may be individually designed to identify and validate a small number of specifically targeted interactions [11]. Studies of specific protein interactions can provide structural information, kinetic and dynamic characterization [12]. The methods include X-ray crystallography [10], NMR spectroscopy [10], fluorescence resonance energy transfer (FRET) [13] and surface plasmon resonance (SPR) [14].
In this review, we provide a comprehensive organized list of available (i) web servers and tools to analyze protein–protein interactions and protein–protein interfaces and (ii) large-scale experimental and computational databases. For each section, we present a table listing these, with several discussed in the text. We highlight the merits and shortcomings of these for particular applications and provide two case studies to illustrate how to use a tool-combination toward specific problems. Our goal here is to provide a useful guide.
Protein–protein interaction databases
While most protein–protein interaction (PPI) databases present experimentally verified interactions, some contain computationally predicted interactions and some both. Apart from physical associations, some databases also provide indirect associations (e.g. functional gene links) while others provide interactions at the domain level. All databases grow rapidly as the pace of interaction–detection experiments, genome sequencing and the number of proteins with solved structures increase.
The Database of Interacting Proteins (DIP) is a catalogue of experimentally determined PPIs [15]. Each interacting pair contains accession codes linking to public protein databases. In October 2008, it contained 57 099 interactions between 20 244 proteins for 244 organisms. The Biomolecular Interaction Network Database (BIND) [16], is a collection of records documenting molecular interactions. It includes high-throughput data submissions and hand-curated information gathered from the literature which covers 188 517 interactions. BIND records are created from experimental interactions published in at least one peer-reviewed journal. The Molecular Interactions Database (MINT) [17] focuses on experimentally verified protein interactions with emphasis on proteomes from mammalian organisms. In October 2008, the database contained 111 847 interactions spanning 30 organisms (25 204 mammalian, 7412 C. elegans, 23 375 D. melanogaster, 44 813 Yeast interactions). The Munich Information Center for Protein Sequences (MIPS) [18] is a collection of genome and protein sequence databases. It includes a S. cerevisiae specific protein interaction database containing 15 488 interactions (9103 physical, 6385 genetic, September, 2004), annotated through nine different high throughput analyses. The BioGRID General Repository for Interaction Datasets [19], is a comprehensive compilation of genetic and physical interactions for 22 organisms. It contains 99 104 non-redundant physical and 52 672 non-redundant genetic interactions (October, 2008). Table 1 lists some of the available PPI databases in alphabetical order. The second column gives the source organisms of the PPIs. The third column indicates the interaction detection methods used: high and low throughput experiments (EH, EL), interactions coming from the literature (Lit) and computational prediction (P). The interaction type can be direct (physical) or indirect (functional, genetic), as shown in the sixth column. The database compilation method and the characteristics provided in the table can indicate a measure of confidence in the interactions. By using this table, the most appropriate database for a specific problem can be chosen. For validated protein interactions, curated databases such as BIND, BioGRID, DIP and MINT are useful. In the absence of experimental data or to enrich the dataset with possible protein interactions, predicted protein–protein interaction can be used. Some examples of these databases are PRISM, OPHID and 3D-partner. For a specific organism, for example human, HPRD, HPID and MIPS are favored. If binding site information is needed, PSIbase and DOMINE are useful, since they indicate where the two proteins interact. However, in general, combination of several databases is preferred in protein interaction analyses.
Table 1:
Databases of protein–protein interactions in alphabetical order
| Web server and link | Organism | Detection type | Curation | Structure | Interaction type | Number of interactions (as of December 2008) |
|---|---|---|---|---|---|---|
| BIND [16] http://bind.ca | No species restriction | EH, EL, Lit | Yes | No | Both | 58 266 (as of September 2004) |
| BioGRID [19] http://www.thebiogrid.org/ | No species restriction | EH, EL, Lit | Yes | No | Both | 152 150 |
| DIP [15] http://dip.doe-mbi.ucla.edu/ | No species restriction | EH | Yes | No | Direct | 57 330 |
| DOMINE [123] http://domine.utdallas.edu/cgi-bin/Domine | No species restriction | EL, P | No | Yes | Direct | 20 513 |
| HPID [124] http://wilab.inha.ac.kr/hpid/ | Human | EH, EL, Lit, P | – | No | Both | N/A |
| HPRD [125] http://www.hprd.org/ | Human | EH, EL, Lit | Yes | No | Direct | > 30 000 (as of 2006) |
| IntAct [126] http://www.ebi.ac.uk/intact/site/index.jsf | No species restriction | EL, Lit | Yes | No | Direct | 115 757 |
| MINT [17] http://cbm.bio.uniroma2.it/mint/index.html | No species restriction | EH, EL, Lit | Yes | No | Direct | 111 847 |
| MIPS [18] http://mips.gsf.de/proj/ppi/ | Mammals | EH, EL, Lit | Yes | No | Direct | > 1800 |
| OPHID [127] http://ophid.utoronto.ca/ophidv2.201/ | Human | EH, EL, P | – | No | Direct | 424 066 |
| PRISM [106] | No species restriction | P | – | Yes | Direct | > 100 000 |
| PSIbase [23] http://psibase.kobic.re.kr/ | No species restriction | EL | No | Yes | Direct | N/A |
| STRING [122] http://string.embl.de/ | No species restriction | EH, Lit, P | No | No | Both | > 50 million |
| 3D-partner [128] http://3D-partner.life.nctu.edu.tw | No species restriction | P | – | Yes | Direct | N/A |
The compilation methods and their characteristics are given. The source organism, detection type, whether curated or not, structure availability, interaction type and number of interactions can be found in the columns. See text for notation
INTERFACES ARE THE REGIONS WHERE PROTEINS INTERACT
The region where two protein chains come into contact is the binding site; or for both sides, an interface. In order to identify interface residues and regions that line the protein surfaces, it is essential to know the structures of the proteins. Surface residues are usually determined by calculations of the surface area which is accessible to the solvent [20–22] or distance-based calculations taking into account how close the two proteins are to each other [23–25].
In order to understand binding principles, properties which distinguish interfaces (or, binding sites) from the rest of the protein surface need to be characterized [20–22, 24–43]. Protein interfaces have greater residue conservation and tend to be planar or well packed depending on the type of interaction. The stability and specificity of the interacting protein pairs relate to the presence of H-bonds, electrostatic interactions, van der Waals forces, salt bridges and hydrophobic attractions. Although rare in protein interfaces, disulphide bonds provide rigidity and stability in the interactions [44]. Residue composition usually differs between transient and obligate complexes: the former relying more on salt bridges and hydrogen bonds, whereas the latter more on hydrophobic interactions [21, 22, 37]. However, recent studies on transient/obligate complexes have shown that obligate interfaces are not more hydrophobic than transient ones due to the presence of water-mediated interactions [45, 46]. Physical binding is further governed by shape and chemical complementarity [37, 39, 47, 48], molecular flexibility and environmental conditions. In agreement with the broadly-accepted notion that binding and folding are similar processes [40–42], structural comparison between highly populated folds and highly populated binding sites illustrates that interface regions are generally structurally similar to cores of globular proteins [49].
Protein–protein interface databases
Protein interfaces have long been studied at both the protein level and the domain level. They have been represented in interface data sets and deposited into databases such as PiBASE [50], InterPare [51], SCOWLP [52], 3DID [53], SCOPPI [54] and PRINT [49]. Table 2 lists some widely used interface databases. The first column lists the databases. The next columns give the attributes of the databases; which domain definition is used, where the dataset is extracted from (interface level, either chains or domains, peptidic or solvent mediated), whether interfaces are classified or not and which thresholds are used in defining the interfaces. Below, we provide some useful attributes of several of the frequently used databases.
Table 2:
Some widely used protein–protein interface databases with their attributes, i.e. domain definition, interface level, classification and extraction methods
| Web server and link | Domain def. | Interface level | Classification | Similarity | Features used in clustering | Extraction method |
|---|---|---|---|---|---|---|
| InterPare [51] | – | Any atom distance at ≤ 5 Å | ||||
| http://interpare.kobic.re.kr/ | SCOP | Domain | No | No | Also, ASA and Voronoi diagram methods are available | |
| PDBSUM [118] http://www.ebi.ac.uk/pdbsum/ | Pfam | Chain | No | – | No | N/A |
| PIBASE [50] http://modbase.compbio.ucsf.edu/pibase/queries.html | SCOP, CATH | Domain | Yes | Structure, topology | Hierarchical clustering, based on inter-residue contacts, ASA, number of interface residues | Any atom distance at ≤ 6.05 Å |
| PPIDB http://ppidb.cs.iastate.edu/ppidb/ | GO | Residue | No | – | No | Both ASA and alpha carbon distance (user defined thresholds) |
| PRINT [49] http://prism.ccbb.ku.edu.tr/interface | SCOP, GO | Chain | Yes | Structure | Iterative hierarchical structural clustering | Sum of van der Waals radii + 0.5 Å |
| ProtCom [129] http://www.ces.clemson.edu/compbio/protcom/ | – | Chain | No | – | No | Sum of van der Waals radii + diameter of water molecule, 2.8 Å |
| SCOPPI [54] http://www.scoppi.org/ | SCOP | Domain | Yes | Sequence & structure | Clustering based on sequence and structure | any atom distance at ≤ 5 Å |
| SCOWLP [52, 56] http://www.scowlp.org/ | SCOP | Domain, peptide, solvent | Yes | Structure | Agglomerative hierarchical clustering | For H-bonds 3.2 Å donor–acceptor distance; for salt bridges 4 Å distance criteria |
| 3DID [53, 55] http://gatealoy.pcb.ub.es/3did/ | Pfam, GO | Domain, peptide | Yes | Structure | Hierarchical clustering by using distance matrices of each interface. | At least 5 contacts (H-bonds, salt bridges, van der Waals interactions) |
PIBASE is a comprehensive database of structurally determined protein interfaces formed between domain pairs. Domain–domain interfaces are generated by distance calculation with a default threshold of 6.05 Å to allow water mediated contacts. The generated domain interfaces are filtered by an accessible surface area (ASA) threshold (300 Å2) and for duplicated domain–domain interactions. The dataset is characterized by geometric, physicochemical and topological properties. PIBASE also provides the contact topology of the domains, polar versus nonpolar ASAs, etc. [50]. InterPare uses three methods to generate domain–domain interfaces which highly overlap [51]: (i) atomic distance calculation, (ii) ASA and (iii) Voronoi Diagram, a computational geometry method. 3DID contains domain–domain interactions annotated with GO functions. One can query the database to retrieve a network of interacting domains. For each domain–domain interaction, the server provides residue pairs with favorable interactions [53]. In the new release of the 3DID, peptide mediated interactions are also available. By defining interaction types, the authors are able to obtain the different interfaces used by a specific domain. Currently, it contains 115 559 domain–domain interfaces classified into 4887 unique interface types. 829 hand-curated domain-peptide interactions enrich the database [55]. SCOPPI is a database of structurally classified protein interfaces. To extract protein interfaces, atomic distances are used and SCOP is used for domain classification. In addition to multiple sequence alignment, SCOPPI applies structural alignment to the SCOP families. The domain–domain binding regions, named faces, are clustered by sequence and structure resulting in approximately 8400 interface types [54]. SCOWLP is a database of detailed domain–domain interfaces enriched with peptidic interfaces and solvent mediated contacts. Solvent mediated contact residues are named ‘wet spots’ [52]. It also classifies binding sites of each domain hierarchically leading to 9334 binding regions over 2561 domain families. 65% of the families in the database contain more than one binding site [56]. PRINT is a database of chain level interfaces extracted from the Protein Data Bank (PDB) [57]. These PPIs are clustered by structural similarity. On February 2006, there were 49512 interfaces which were clustered into 8205 unique interface structures. Interface interaction types are deposited in PRINT and annotated as crystal or biological interfaces, obligate or non-obligate, homo- or hetero-interfaces. For each individual interface, information on GO annotations, SCOP domains, interaction type, residue propensities and structural cluster are given [49]. PPIDB is a periodically updated database which differs from the others by allowing user defined thresholds to extract interface residues.
The physical and chemical properties of protein–protein interfaces
Numerous studies aim to obtain the general patterns of protein binding sites. In a pioneering work by Jones and Thornton [22], 59 different PPIs were divided into four groups: homodimers, enzyme–inhibitor complexes, antibody complexes and hetero-complexes. The complexes were characterized by six properties: size and shape, complementarity, residue interface propensities, hydrophobicity, segmentation, secondary structure and conformational changes. Homodimer interfaces preferred hydrophobic residues and had relatively large surface areas. Heterodimer complexes were less hydrophobic. The interfaces of homodimers, permanent hetero-complexes and enzyme–inhibitor complexes were more complementary than antigen–antibody complexes. In spite of significant differences, a distinct pattern for protein interfaces compared to the rest of the surfaces was not identified [22]. Larsen et al also studied protein recognition mechanisms on a dataset of 136 homodimeric proteins, observing that one-third of the interfaces have a distinguishable large hydrophobic core, polar contacts and water-mediated interactions [35]. Amino acid propensity is another important property which can be used to distinguish interaction types. Ofran and Rost explored six types of PPIs, homodimers versus heterodimers, transient versus permanent dimers and same domain versus different domain interfaces. Using only amino acid composition and residue-contact preferences, 63–100% accuracy was achieved in interaction type prediction [58].
Structural features of interfaces
For strong interactions, good shape complementarity is crucial. The chemical character of protein interfaces is similar to the average protein surface, whereas their packing density is close to the density of the protein core. On average, the accessible surface area of interfaces is 1600 Å2 [37]. Chakrabarti and Janin [28] analyzed 70 protein complexes. They considered small binding regions as single patch and large binding regions as multipatches. Bahadur et al. split interfaces into core and rim. The core region is buried in the interface; the rim contains solvent accessible residues [59]. Interface core regions were found to be similar to the protein interior in residue frequency; rims to protein surface.
Protein surfaces are not flat rather they are filled with pockets, crevices and indentations [60]. Cavities that remain unfilled after the complementary protein associates are unfilled pockets. Complemented pockets are the result of two well fitted protein partners [61], as in the key and lock model. Complemented pockets frequently pre-exist binding. In an analysis of 18 protein complexes with complementary pockets whose unbound structures are known, 16 of the pockets were found to pre-exist in the unbound state [24]. The pockets’ size, shape and functional group distribution are critical in protein engineering and designing new peptides/proteins that will selectively bind to these regions.
Conservation of the interfaces
Valdar and Thornton analyzed six homodimers observing that interface residues are more conserved than the rest of the surface [62]. Caffrey et al. extended this observation on a larger dataset [63]. However, analysis of surface and interface patches did not show a significant difference. Using their Bayesian approach to estimate residue conservation, Tseng and Liang noticed that residue substitution rates in protein cores are significantly different than those in solvent-exposed surfaces of the proteins [64]. Conservation alone is insufficient to reliably predict protein binding sites, but it can be combined with other interface properties. Tools to extract conservation profiles of surface residues can be very helpful for large-scale characterization of functional regions in proteins. ConSurf [65, 66] is a web server, which incorporates several phylogenetic-based algorithms [67] to predict conservation scores of residues for a given protein structure. The SCORECONS web server scores residue conservation in multiple sequence alignment [68]. In the absence of structural data, SCORECONS is preferred. Otherwise, Consurf is very useful given its options and visual interface.
Energetic distribution in the interfaces: hot spots
The energetic contribution of residues in protein interfaces are not distributed uniformly. Some key ‘hot spot’ residues can contribute dominantly to the binding free energy. Experimentally, hot spot residues are identified via Alanine Scanning Mutagenesis; if a residue has a significant drop in binding affinity when mutated to alanine it is labeled a hot residue. Thorn and Bogan [69] deposited hot spots from alanine scanning mutagenesis experiments in the ASEdb database. BID [70] is another database of experimental hot spots, which collects all available experimental data related to hot spots in protein interfaces. However, these databases cover only a small portion when compared to available PPIs. In the absence of experimental data, computational methods can be used for hot spot detection [71]. These computational hot spots are either deposited in large scale databases or the methods are presented as web servers. The hot spot related databases/web servers are listed in a subsection of Table 3. The first column gives the available databases and servers; next columns list the strategy and techniques to identify hot spots, such as experimental or computational alanine scanning, and machine learning, respectively. Some of the servers are based on energy calculation [72–74]. Robetta and FoldX are two examples of energy-based hot spot prediction tools. Robetta mutates side chains to alanine and it repacks the side chains, which are within 5 Å radius sphere of the mutated residue. The rest of the protein remains unchanged. The change in the binding free energy is calculated by an energy function [75]. FoldX mutates systematically side chains between two proteins (or protein–peptide) to alanine and the rest of the complex is relaxed. Then, the change in the binding energy is calculated [76]. These predicted changes in binding energies are used to label the residues as hot spots. Molecular dynamics (MD) studies have also been used [77–79]. MD simulations of 11 protein complexes indicate that anchoring residues in protein interfaces have limited mobility and have a tendency to be hot spots [79]. In a related work, Moreira et al. supported the O-Ring hypothesis, which proposes that hot spots are protected from bulk water by a surrounding rim region by using MD simulations [80]. MD analysis of computational alanine scanning of the human growth hormone receptor complex agrees well with the experimental hot spots [72]. Despite their success, MD-based methods are computationally expensive when compared to other methods. As an alternative, conservation is a property that can be used. Structurally conserved residues and hot spots correlate significantly [81–83]. These hot spots are also buried and tightly packed [82] resulting in densely packed clusters of networked hot spots called ‘hot regions’. Another residue conservation-based method is available in HotSprint. Residue conservation alone is not sufficient to identify hot spots. Some amino acids are preferred as hot spots, such as Arg, Tyr and Trp [27]. HotSprint combines the three properties, conservation, ASA and residue propensity, to detect hot spots of available protein–protein interfaces [84]. KFC predicts computational hot spots by a machine learning approach. It uses structural features such as atomic contacts and H-bonds and gives a binary answer whether a residue is a hot spot or not [85]. Recently, a neural network-based approach using interface features such as sequence profiles, solvent accessibility and evolutionary conservation has been employed in computational hot spot prediction (an adaptation of ISIS) [86]. If we compare these databases/web servers, the experimental databases ASEdb and BID are limited in size. On the other hand, the energy-based methods Robetta and FoldX are appropriate for specific interactions providing accurate estimation of free energy changes. For prediction on large-scale data KFC and Hotsprint are preferred because of their computational effectiveness and comparable performance. In general, all of the databases/servers require protein structure, except ISIS. Although it does not perform better than the computational alanine scanning method [74, 75], in the absence of structural information or binding partner it is useful for hot spots prediction [86].
Table 3:
Web servers/tools for characterization of protein interfaces
| Physical and chemical properties | |||
|---|---|---|---|
| Web server/tool and link | Reported characteristics | Results | Visual interface |
| CASTp [60] http://cast.engr.uic.edu | Pockets & cavities | Residue ASA, Residue Volume | 3D (JMol, Chime) |
| ConSurf [65] http://consurf.tau.ac.il/ | Conservation info based of multiple sequence alignment or protein structure. | Conservation score | Yes (JMol) |
| GALINTER [130] (source code is available upon request) | Protein–protein interfaces | Spatial alignment according to the vector representations of van der Waals interactions and hydrogen bonds based on their geometry | No |
| LIGPLOT [131] http://www.biochem.ucl.ac.uk/bsm/ligplot/ligplot.html | Non-covalent interactions of protein complexes | H-bonds, hydrophobic forces and their strengths, atomic ASA. | 2D |
| MAPPIS [132] http://bioinfo3d.cs.tau.ac.il/MAPPIS/ | Protein–protein binding sites | Common spatial arrangements of physico-chemical properties like H-bond donor, acceptor, aliphatic, aromatic, hydrophobic. | 3D (JMol) |
| MolSurfer [133] http://projects.villa-bosch.de/mcm/software/molsurfer | protein–protein and protein–DNA/RNA interfaces. | Complementarity, hydrophobicity and electrostatic potential. | 3D (WebMol) |
| MultiBind [132] http://bioinfo3d.cs.tau.ac.il/MultiBind/ | Small ligand binding sites | Common spatial arrangements of physico-chemical properties like H-bond donor, acceptor, aliphatic, aromatic, hydrophobic. | 2D |
| PIC [134] http://crick.mbu.iisc.ernet.in/~PIC/ | Non-covalent interactions of protein complexes | hydrogen &disulphide bonds, hydrophobic, ionic & aromatic- aromatic interactions, ASA. | 3D (RasMol, JMol) |
| ProFace [135] http://202.141.148.29/resources/bioinfo/ interface/ | Protein–protein interfaces | Interface area, surface area, fraction of non-polar atoms, non-polar interface area, residue propensity, fraction of buried atoms. | 3D (Rasmol, Chime) |
| ProTherm [136] http://gibk26.bse.kyutech.ac.jp/jouhou/ Protherm/protherm.html | Thermodynamic parameters of wild type and mutant protein. | Gibbs free energy change, enthalpy change, heat capacity change, transition temperature Mutated residue numbers | No |
| PROTORP http://www.bioinformatics.sussex.ac.uk/protorp/ | Non-covalent interactions of protein complexes. | Interface ASA, percentage polar atom in interface and on surface, planarity, eccentricity, secondary structure info, hydrogen and disulfide bonds, salt bridges, gap volume, gap volume index. | No |
| Q-SiteFinder [137] http://bmbpcu36.leeds.ac.uk/qsitefinder/ | Ligand binding site | Site volume, protein volume | 3D (Chime, Mage) |
| ScoreCons [68] http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/valdar/scorecons_server.pl | Conservation info based on multiple sequence alignment. | Conservation score | No |
| SurfNet [138] http://www.biochem.ucl.ac.uk/~roman/surfnet/surfnet.html | Surfaces and void regions. | Computation of gaps, clefts, cavities and binding sites, van der Waals surfaces | 3D (Rasmol, Raster3D, Sybyl) |
| Hot spot databases and prediction servers | Strategy | Availability | Technique |
| ASEdb [27] http://nic.ucsf.edu/asedb/ | Experimental | Database | Alanine scanning |
| BID [70] http://tsailab.tamu.edu/BID/ | Experimental | Database | Collection of several experimental techniques. |
| FoldX [76] http://foldx.crg.es/ | Energy-based | Tool and server | Computational alanine scanning |
| HotSprint [84] http://prism.ccbb.ku.edu.tr/hotsprint/ | Conservation, accessibility, residue propensity | Database | Empirical formula |
| ISIS [95] http://cubic.bioc.columbia.edu/services/isis/ | Sequence-based | Tool | Machine learning |
| KFC [85] http://kfc.mitchell-lab.org/ | Atomic contacts, residue size, H-bond | Server | Machine learning |
| Robetta [74] http://robetta.org/submit.jsp | Energy-based | Server | Computational alanine scanning |
In the first part, the web servers and tools for physical and chemical properties of protein interfaces are listed. The hot spot related databases/web servers are listed in the second part where the hot spot detection strategy, the availability and the used technique are given, respectively.
Finally, although there are no strict rules, properties like binding site size, residue frequency, shape complementarity, conservation, presence of hot spots and hydrophobicity can help in predicting binding sites fairly accurately. Thus, analyses of binding sites and characterization of protein interfaces are useful, and help in improving computational prediction of protein–protein interactions and in designing drugs and inhibitors binding to interrupt these interactions. Table 3 is a comprehensive list of the servers, databases and tools, which characterize the physico-chemical properties of interfaces.
BIOLOGICAL VERSUS CRYSTAL COMPLEXES
Entries in the PDB can have artifacts of crystallization; that is, some of the complexes would not occur in solution or in their physiological states. Determining which contacts are biological and which are not is often difficult, particularly when the oligomeric state of the protein is uncertain or unknown [62]. These crystal packing interactions can cause noise in analyses. A number of studies addressed the problem of distinguishing between biological and crystal packing contacts. Below, some methods are presented to distinguish crystal and biological interfaces.
In the PDB files, BIOMT records, describing how to compute the coordinates of multimers from the explicitly specified single repeating unit, can be used to build quaternary structures. Henrick and Thornton defined crystal packing by assigning a cutoff value (400 Å2) in the buried surface area. Structures passing this requirement are deposited in the PQS server [87]. Crystal structures often have smaller interfaces when compared to biological interfaces. Other studies used different cutoff values to distinguish crystal contacts. Although interface size is the most important feature to identify crystal packing contacts, it is not the sole criterion. There are cases with large interface sizes, yet are biologically irrelevant; examples include the crystal contact in porcine adenylate kinase having an ASA of 2600 Å2, and pancreatic ribonuclease crystals with interface size of 1800 Å2 [88]. Based on the assumption that biological interfaces are more conserved than non-biological interfaces, Valdar and Thornton [89] suggested that crystal interfaces can also be distinguished by residue conservation. They combined both the size and conservation information of the binding sites, achieving an accuracy of 98.3% on their training set. Overall, biological interactions are more conserved and larger than crystal ones [90, 91]. Since the amino acid composition of the protein surface differs from the interface, if the binding site composition is similar to the rest of the protein surface, this interaction is a crystal packing candidate [90]. In the conserved domain interaction approach [92], interacting domain pairs are clustered. If two or more cluster members have similar interface locations, these interfaces possess a conserved binding mode (CBM). Conserved modes are used to distinguish biological from crystal interactions. When tested on interacting globin pairs the accuracy reached 90%. In addition, multiple feature-based approaches are also available; one of them is NOXclass, which is a machine learning-based approach. In NOXclass, six different interface properties are used to distinguish crystal versus biological contacts: interface area, interface area ratio, amino acid composition, a correlation between surface and interface region, gap volume index and conservation score of the interface. NOXclass distinguishes biological and non-biological interfaces with an accuracy of 91.8% based on three parameters (interface area, interface area ratio and area-based amino acid composition) [93]. In a recent method, called DiMoVo, Bernauer et al. used machine learning-based approach with Voronoi tessellation to distinguish biological and crystal complexes [94]. Table 4 lists the available servers/tools that classify PPIs as crystal or biological according to the above properties where the second column describes the strategy of the servers/tools for classification and last column gives information about the results. Among these servers/tools PQS is widely used for distinguishing crystal complexes; however, it is based on ASA and there are counter examples of crystal complexes having large interface size. Another server PreBi is appropriate for homodimers, but the current server is found to be slower than the others. NOXclass with its high accuracy, multiple features and computational effectiveness has been found to be useful. DiMoVo's performance is also compatible with other servers.
Table 4:
Web servers/tools for classification of protein–protein interactions
| Web server/tool and link | Strategy | Result |
|---|---|---|
| CPASS [139] http://bionmr-c1.unl.edu/CPASS_OV/CPASS.htm | Comparison of ligand-defined active sites. | Function assignment |
| DiMoVo [94] http://cgal.inria.fr/DiMoVo/ | Machine Learning-based classification with Voronoi tessellation using interface area, residue and contact frequencies, Voronoi volumes, etc. | Biological versus crystal |
| NOXclass [93] http://noxclass.bioinf.mpi-inf.mpg.de/ | SVM-based classification using interface area, ratio of interface area to surface area, amino-acid composition, correlation between interface and surface, gap volume index, conservation | Percentage whether the complex is obligate, non-obligate or crystal packing. |
| PINS [140] http://pins.ornl.gov/ | Random forest classifier using residue propensity and contacts, interface area, disulphide and H-bonds, conservation, packing density, etc. | Biological versus crystal |
| PISA[141] http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html | Free energy calculation | Biological versus crystal |
| Pita [142] http://www.ebi.ac.uk/thornton-srv/databases/pita/ | Scoring based on crystal symmetry operators. | Biological versus crystal |
| PQS [87] http://www.ebi.ac.uk/msd-srv/pqs/ | Interface size, symmetry operators to form complex | Biological versus crystal |
| PreBi [143] http://pre-s.protein.osaka-u.ac.jp/~prebi/ | Degree of shape and physicochemical complementarity | Biological versus crystal |
| ProFunc [144] http://www.ebi.ac.uk/thornton-srv/databases/ProFunc/ | Sequence and structure comparison | Functional annotation of proteins |
The columns describe the strategy of the servers/tools for the classification and provide information relating to the type of the obtained results, respectively.
BINDING SITE PREDICTION
With no strict quantitative rules to point to the binding mechanism, characteristics of the protein interfaces such as conservation, residue propensity, residue order, geometric and electrostatic complementarity are useful. The combination of multiple protein interface features is successful in discriminating binding sites from the rest of the protein surface. Prediction methods use various features such as structure, sequence and physicochemical properties of protein interfaces [95, 96]. These features can be utilized in different methods such as machine learning-based and empirical scoring functions. Most of these prediction methods use known binding sites for parameterization or training. Prediction also depends on the choice of the training dataset: reliable, diverse and non-redundant gold standard datasets are crucial for a successful training of a prediction method [97]. Table 5 lists several servers to predict binding sites. The first column lists the binding site prediction servers. The second column provides the binding site prediction method. In the third column, the binding site properties that are analyzed for the predictions are given. Below, we provide a brief review of some of these servers.
Table 5:
Servers for binding site prediction
| Web server | Method | Binding site properties |
|---|---|---|
| cons-PPISP [103] http://pipe.scs.fsu.edu/ppisp.html | Neural network | Solvent accessibility and sequence information |
| Firestar [145] http://firedb.bioinfo.cnio.es/Php/FireStar.php | Scoring function | Sequence and structural alignments |
| InterProSurf [146] http://curie.utmb.edu/ | Scoring function | Solvent accessible surface area, propensity of interface residues |
| Meta-PPISP [104] http://pipe.scs.fsu.edu/meta-ppisp.html | Neural network | Combining scores derived from three other servers; ProMate, PINUP, cons-PPISP |
| Patch Finder Plus [147] http://pfp.technion.ac.il/ | Neural network | Surface concavity, surface area, amino acid frequency and composition, hydrogen-bonding potential, sequence conservation |
| PINUP [98] http://sparks.informatics.iupui.edu/PINUP/ | Empirical scoring function | Side chain energy score, conservation score and residue interface propensity |
| PI2PE [148] http://pipe.scs.fsu.edu/ | Neural network | Combining three servers, WESA [149], cons-PPISP [103], DISPLAR [150] |
| PPI-Pred [100] http://bioinformatics.leeds.ac.uk/ppi-pred | Support vector machine | Conservation, electrostatic potential, hydrophobicity, propensity of interface residues, surface shape and solvent accessible surface area |
| PRISM [106] http://prism.ccbb.ku.edu.tr/prism/ | Scoring function | Geometric complementarity, conservation |
| ProMate [99] http://bioportal.weizmann.ac.il/promate/ | Scoring function | Amino-acid propensities, pairwise amino-acid distribution, residue conservation and geometric properties |
| SHARP2 [101] http://www.bioinformatics.sussex.ac.uk/SHARP2/sharp2.html | Patch score calculation | Solvation potential, hydrophobicity, accessible surface area, residue interface propensity, planarity and protrusion |
| SiteEngines [151] http://bioinfo3d.cs.tau.ac.il/SiteEngine/http://bioinfo3d.cs.tau.ac.il/I2I-SiteEngine/ | Scoring function | Structural matching, physico-chemical properties and shapes |
| SPPIDER [102] http://sppider.cchmc.org/ | Neural network | Solvent accessibility |
In the columns, the method and the binding site properties used in the prediction are given, respectively.
ProMate is a scoring function-based approach to identify the location of protein–protein binding sites [98]. The algorithm was trained on a heteromeric transient protein–protein complexes, thus it is most suitable for predicting such interaction interfaces. Amino acid propensities, residue conservation, pairwise amino acid distribution, temperature factors and geometric properties are extracted and optimized to choose the best combination. The location of the interface for about 70% of the proteins was predicted correctly [99]. Another prediction algorithm, PINUP, combines side chain energy score, conservation score and residue interface propensity reaching 44.5% prediction accuracy. Bradford et al. use a support vector machine (SVM) approach combined with surface patch analysis to predict protein–protein binding sites. The SVM is trained on binding site properties (surface shape, conservation, electrostatic potential, hydrophobicity, residue interface propensity and solvent accessible surface area) to distinguish interacting patches from non-interacting patches. PPI-Pred correctly predicts binding site location in 76% of the 180 dataset proteins [100]. Another algorithm that makes use of patch score calculation to predict binding sites is SHARP2. Six binding properties scores (solvation potential, hydrophobicity, accessible surface area, residue interface propensity, planarity and protrusion) are combined for prediction [101].
There are two neural networks-based web servers: SPPIDER, which is based on solvent accessibility [102] and cons-PPISP based on sequence information and solvent accessibility [103]. Zhou and Qin combined the raw scores of cons-PPISP, PINUP and ProMate web servers and built a meta-web server, meta-PPISP with increased accuracy [104]. When the performances of the web servers are compared on two different sets (Enz35 dataset and CAPRI targets), the same rankings are derived (from top to down meta-PPISP, PINUP, ProMate, cons-PPISP, SPIDDER and PPI-Pred). meta-PPISP outperforms other servers [104]. Different from the other servers, PRISM predicts the binding site of two proteins using known template interfaces. It searches for the left and right binding sites of the template interfaces on the two target proteins by structural matching. If one target matches the left partner and the other matches the right of the same interface template, these two targets are predicted to interact through this region [105, 106].
PROTEIN–PROTEIN DOCKING
Docking is the procedure to find the best bound state for given 3-D structures of two (or more) proteins. The docking problem is difficult since there are many potential ways in which proteins can interact, and protein surfaces are flexible. Computational methods were developed to discover the best fit between two proteins and these methods are presented as docking servers/software packages. Several methods ranked well in the CAPRI (http://capri.ebi.ac.uk) community competition. Among these, Z-Dock [107], Gramm-X [108], DOT [109] and ClusPro [110] perform global searches based on fast Fourier transform (FFT) correlation approach. Although the FFT-based initial global search methods are similar, the refinement and filtering steps differ for each server. For example, ClusPro [110] first selects the docked conformations with favorable desolvation and electrostatics properties, and then these filtered structures are clustered using a hierarchical pairwise RMSD algorithm. Clustering is followed by energy minimization step and refinement by SmoothDock [111]. On the other hand, the top predictions of Gramm-X [108] are subjected to the conjugate gradient minimization with a smoothed Lennard-Jones potential. For the resulting minimized predictions, soft Lennard-Jones potential, evolutionary conservation of predicted interface, statistical residue–residue preference, volume of the minimum, empirical binding free energy and atomic contact energy are calculated and used for the application of the SVM filter trained on a benchmark set [112]. The retained predictions are re-scored by a weighted sum of these potential terms. Different from these FFT correlation approaches, PatchDock and SymmDock [113, 114] employ the geometric hashing method as the initial search process. The docked conformations are ranked according to a geometric shape complementarity score. Being computationally faster than other web servers, PatchDock also provides the user an option for submitting potential binding sites of the receptor and ligand. If the binding sites are known, these can yield more accurate results.
In addition to those docking servers that perform global searches, RosettaDock [115] performs a local docking search requiring a reasonable starting conformation. The starting conformation may well be created by any of the servers listed above. The submitted initial docking predictions are refined by a Monte Carlo approach including rigid-body moves and side-chain optimization. A list of selected docking servers/software packages is provided in Table 6. The first column lists the docking servers/softwares. The second and third columns give information about the docking method and filtering/refinement stages, respectively.
Table 6:
Servers for protein–protein docking which ranked well in the CAPRI competition
| Web server/software and link | Docking method | Filtering and refinement |
|---|---|---|
| BDOCK [152] http://www.biotec.tudresden.de/~bhuang/bdock/bdock.html | FFT correlation based on shape complementarity, degree of burial and conservation | Altering the docking solutions with a scoring function |
| ClusPro [110] http://nrc.bu.edu/cluster/ | FFT correlation using DOT [109] | Filtering with empirical potential and clustering, refinement by SmoothDock [111] |
| DOT [109] http://www.sdsc.edu/CCMS/DOT/ | FFT correlation based on electrostatics and shape complementarity | Refinement by energy minimization |
| FireDock [153] http://bioinfo3d.cs.tau.ac.il/FireDock/ | None (refinement server) | Refinement using an energy function |
| GRAMMX [108] http://vakser.bioinformatics.ku.edu/ resources/gramm/grammx | FFT correlation based on shape complementarity, hydrophobicity and smoothed potentials | Clustering and knowledge-based scoring |
| HADDOCK [154] http://www.nmr.chem.uu.nl/haddock/ | Data-driven docking approach based on biochemical and/or biophysical interaction data | None |
| HEX [155] http://www.csd.abdn.ac.uk/hex/ | Spherical polar Fourier correlations | None |
| MolFit [156] http://www.weizmann.ac.il/Chemical_Research_Support//molfit/home.html | FFT correlation based on chemical and shape complementarity | Clustering of the predicted conformations |
| PatchDock [114] http://bioinfo3d.cs.tau.ac.il/PatchDock/ | Geometric hashing and pose-clustering | Ranking according to a geometric shape complementarity score |
| PyDock [157] http://mmb.pcb.ub.es/PyDock/ | FFT based on electrostatics and desolvation energy | Ranking using an energy function |
| RosettaDock [115] http://rosettadock.graylab.jhu.edu/ | Local docking by Monte Carlo search | Ranking using an energy function, clustering |
| ZDOCK [107] http://zlab.bu.edu/zdock/index.shtml | FFT correlation based on shape complementarity, desolvation energy and electrostatics | Refinement by energy minimization |
| 3D-Dock [158] http://www.sbg.bio.ic.ac.uk/docking/ | FFT correlation using FTDOCK [159] | Clustering, refinement of side-chains using Multidock [159] |
Information relating to the docking method and filtering/refinement stages can be found in the columns, respectively.
CASE STUDIES
p53-53BP2 Interface
To provide practical examples that reflect the data on protein interactions and interface classification, we illustrate how a database of protein interfaces from the ones explained in the text is used for characterization of the p53-53BP2 protein interface. p53 is one of the major tumor suppressor proteins, which interacts with many other proteins, and 53BP2 is the binding protein partner of p53. We start by querying the PRINT database with this interface, pdbID 1ycs. 1ycs has only one interface, which is formed between chains A (p53) and B (53BP2) [49]. Figure 1 illustrates the complex and highlights the interface between chain A and chain B. The interface region is illustrated in sphere representation. The rest of the protein is drawn as ribbon diagram using VMD [116]. The coordinates of the interface residues can be downloaded from the PRINT database in PDB format. PRINT provides information on this protein interface (1ycsAB) such as interface area (calculated by Naccess [117] as 1500 Å2), gap volume (calculated by Surfnet as 3304 Å3). PRINT also provides the interaction type (using NOXclass). 1ycsAB is found to be a biological interface with a probability of 85% and non-obligate with a probability of 76%. Further, PDBsum [118] indicates that there are nine H-bonds between the chains. HotSprint gives the average conservation score of this interface as six highlighting the computational hot spots, suggesting that this interface is moderately conserved [84].
Figure 1:
An example of protein–protein interfaces (interface region between p53 and its binding protein 53BP2).
Interactions of HRAS
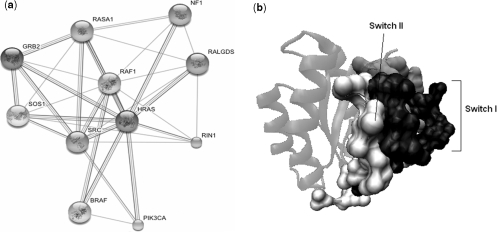
Ras proteins are key regulators of eukaryotic cell growth, functioning in signal transduction pathways. They bind GDP/GTP and have intrinsic GTPase activity. Experimental evidence illustrates the importance of HRAS switch I (residues 25–40) and switch II (residues 57–75) regions in binding to its interaction partners [119–121]. With the goal of characterizing this protein and its network interactions, the interactions of HRAS are obtained using STRING [122], which is a database of known and predicted PPIs. STRING integrates interaction data from high-throughput experiments (DIP, BIND, MINT and GRID databases), co-expression, genomic context and prior knowledge. Its summary network for the experimentally validated interactions is given in Figure 2A.
Figure 2:
(A) HRAS interaction network visualized by STRING [122]. (B) The predicted interacting residues of HRAS (1bkd_R) using SPPIDER and PPI-Pred are displayed as spheres in white and silver color, respectively. The black spheres represent the residues that are common to both web servers. Predictions of SPPIDER and PPI-Pred web servers overlap both Switch I and Switch II regions of HRAS.
In order to locate the binding regions responsible for interactions in the network, several binding site prediction web servers can be used. PPI-Pred [100] provides a list of probable interface residues in the three highest scoring patches. The highest scoring patch successfully overlaps both the switch I and switch II regions of HRAS. The predicted patch is displayed in Figure 2B. The predicted best patch of SHARP2 [101], which uses patch score calculation as the prediction method, corresponds to the switch I region of HRAS. The two web servers which use neural networks in the predictions are SPPIDER [102] and cons-PPISP [103]. Cons-PPISP prediction includes switch I region, whereas SPPIDER includes both switch I and switch II regions of HRAS. The predicted interacting residues with SPPIDER are shown in Figure 2B.
Key Points.
Characterization and prediction of binding sites are crucial to studies of protein interactions. This review provides a comprehensive and organized list of the available databases and web servers of protein binding sites and their characteristics outlining how the tool was constructed, its advantages and drawbacks. In addition, it further provides examples how to use tool-combinations toward particular goals.
These resources can be used to: (i) analyze the physico-chemical properties of interfaces and differentiate between biological complexes and crystal contacts and (ii) predict binding sites in protein structures and of the docked structures of two individual proteins.
In conclusion, a combination of such resources is expected to help biologists explore protein interactions, relate these to cellular processes, and design drugs to target the ‘druggable’ sites.
Acknowledgements
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Biographies
Nurcan Tuncbag is a second-year PhD student at the Computational Sciences and Engineering Program at Koc University, Istanbul. She has been working on protein interactions and interfaces.
Gozde Kar is a first-year PhD student at the Chemical and Biological Engineering Program at Koc University, Istanbul. She has been working on protein interaction networks.
Ozlem Keskin is an associate professor at the Chemical and Biological Engineering Department and the Center for Computational Biology and Bioinformatics at Koc University, Istanbul. Her research focuses on the computational studies of protein interactions and dynamics.
Attila Gursoy is an associate professor at the Computer Engineering Department and the Center for Computational Biology and Bioinformatics at Koc University, Istanbul. His research focuses on the computational studies of protein interactions and parallel computing.
Ruth Nussinov is a professor in the Department of Human Genetics, Medical School, Tel Aviv University, and a Senior Principal Investigator at the SAIC, National Cancer Institute, USA. Her research focuses on computational studies of protein folding, binding and function.
FUNDING
Turkish Academy of Sciences Distinguished Young Investigator Award (to O.K.); the Scientific and Technological Research Council of Turkey Fellowship (to N.T.). Federal funds from the National Cancer Institute; National Institutes of Health; (under contract number N01-CO-12400). Intramural Research Program of the National Institutes of Health; National Cancer Institute; Center for Cancer Research (partial).
References
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Pelletier JN, Arndt KM, Pluckthun A, et al. An in vivo library-versus-library selection of optimized protein–protein interactions. Nat Biotechnol. 1999;17:683–90. doi: 10.1038/10897. [DOI] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–32. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–17. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–68. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–3. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Golemis E. Protein–Protein Interactions. A Molecular Cloning Manual. New York: Cold Spring harbor Press; 2002. [Google Scholar]
- Shoemaker BA, Panchenko AR. Deciphering protein–protein interactions. Part I. Experimental techniques and databases. PLoS Comput Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Marriott G. Analysis of protein interactions using fluorescence technologies. Curr Opin Chem Biol. 2003;7:635–40. doi: 10.1016/j.cbpa.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Karlsson R. SPR for molecular interaction analysis: a review of emerging application areas. J Mol Recognit. 2004;17:151–61. doi: 10.1002/jmr.660. [DOI] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, et al. DIP, the Database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–5. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35(Database issue):D572–4. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Guldener U, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–4. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Marin A, Thornton JM. Protein domain interfaces: characterization and comparison with oligomeric protein interfaces. Protein Eng. 2000;13:77–82. doi: 10.1093/protein/13.2.77. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM. Principles of protein–protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Thornton JM. Analysis of protein–protein interaction sites using surface patches. J Mol Biol. 1997;272:121–32. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- Gong S, Yoon G, Jang I, et al. PSIbase: a database of protein structural interactome map (PSIMAP) Bioinformatics. 2005;21:2541–3. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- Keskin O, Tsai CJ, Wolfson H, et al. A new, structurally nonredundant, diverse data set of protein–protein interfaces and its implications. Protein Sci. 2004;13:1043–55. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Lin SL, Wolfson HJ, et al. A dataset of protein–protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol. 1996;260:604–20. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- Argos P. An investigation of protein subunit and domain interfaces. Protein Eng. 1988;2:101–13. doi: 10.1093/protein/2.2.101. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P, Janin J. Dissecting protein–protein recognition sites. Proteins. 2002;47:334–43. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- Chothia C, Janin J. Principles of protein–protein recognition. Nature. 1975;256:705–8. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone–receptor interface. Science. 1995;267:383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Janin J, Chothia C. The structure of protein–protein recognition sites. J Biol Chem. 1990;265:16027–30. [PubMed] [Google Scholar]
- Keskin O, Ma B, Rogale K, et al. Protein–protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Phys Biol. 2005;2:S24–35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- Keskin O, Nussinov R. Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng Des Sel. 2005;18:11–24. doi: 10.1093/protein/gzh095. [DOI] [PubMed] [Google Scholar]
- Korn AP, Burnett RM. Distribution and complementarity of hydropathy in multisubunit proteins. Proteins. 1991;9:37–55. doi: 10.1002/prot.340090106. [DOI] [PubMed] [Google Scholar]
- Larsen TA, Olson AJ, Goodsell DS. Morphology of protein–protein interfaces. Structure. 1998;6:421–7. doi: 10.1016/s0969-2126(98)00044-6. [DOI] [PubMed] [Google Scholar]
- Lijnzaad P, Berendsen HJ, Argos P. Hydrophobic patches on the surfaces of protein structures. Proteins. 1996;25:389–97. doi: 10.1002/(SICI)1097-0134(199607)25:3<389::AID-PROT10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein–protein recognition sites. J Mol Biol. 1999;285:2177–98. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein–protein interfaces. J Mol Biol. 2002;318:161–77. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein–protein interactions. Curr Opin Struct Biol. 2000;10:153–9. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Lin SL, Wolfson HJ, et al. Studies of protein–protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Nussinov R. Hydrophobic folding units at protein–protein interfaces: implications to protein folding and to protein–protein association. Protein Sci. 1997;6:1426–37. doi: 10.1002/pro.5560060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Xu D, Nussinov R. Structural motifs at protein–protein interfaces: protein cores versus two-state and three-state model complexes. Protein Sci. 1997;6:1793–805. doi: 10.1002/pro.5560060901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Regnier FE. Protein–protein interactions on weak-cation-exchange sorbent surfaces during chromatographic separations. J Chromatogr A. 1998;828:357–64. doi: 10.1016/s0021-9673(98)00641-4. [DOI] [PubMed] [Google Scholar]
- De S, Krishnadev O, Srinivasan N, et al. Interaction preferences across protein–protein interfaces of obligatory and non-obligatory components are different. BMC Struct Biol. 2005;5:15. doi: 10.1186/1472-6807-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Bahadur RP, Chakrabarti P, et al. Hydration of protein–protein interfaces. Proteins. 2005;60:36–45. doi: 10.1002/prot.20478. [DOI] [PubMed] [Google Scholar]
- Teyra J, Pisabarro MT. Characterization of interfacial solvent in protein complexes and contribution of wet spots to the interface description. Proteins. 2007;67:1087–95. doi: 10.1002/prot.21394. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Xu D, Lin SL, Nussinov R. Protein binding versus protein folding: the role of hydrophilic bridges in protein associations. J Mol Biol. 1997;265:68–84. doi: 10.1006/jmbi.1996.0712. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Guney E, et al. Architectures and functional coverage of protein–protein interfaces. J Mol Biol. 2008;381:785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–7. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- Gong S, Park C, Choi H, et al. A protein domain interaction interface database: InterPare. BMC Bioinformatics. 2005;6:207. doi: 10.1186/1471-2105-6-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyra J, Doms A, Schroeder M, et al. SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces. BMC Bioinformatics. 2006;7:104. doi: 10.1186/1471-2105-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33(Database issue):D413–7. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, Henschel A, Kim WK, et al. SCOPPI: a structural classification of protein–protein interfaces. Nucleic Acids Res. 2006;34(Database issue):D310–4. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Panjkovich A, Aloy P. 3did Update: domain–domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37(Database issue):D300–4. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyra J, Paszkowski-Rogacz M, Anders G, et al. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics. 2008;9:9. doi: 10.1186/1471-2105-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y, Rost B. Analysing six types of protein–protein interfaces. J Mol Biol. 2003;325:377–87. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Chakrabarti P, Rodier F, et al. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–19. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web server issue):W116–8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Keskin O, Ma B, et al. Protein–protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J Mol Biol. 2004;344:781–95. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- Valdar WS, Thornton JM. Protein–protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–24. [PubMed] [Google Scholar]
- Caffrey DR, Somaroo S, Hughes JD, et al. Are protein–protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 2004;13:190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Liang J. Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: a Bayesian Monte Carlo approach. Mol Biol Evol. 2006;23:421–36. doi: 10.1093/molbev/msj048. [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–4. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Glaser F, Rosenberg Y, Kessel A, et al. The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins. 2005;58:610–7. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- Pupko T, Bell RE, Mayrose I, et al. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):S71–7. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–41. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001;17:284–5. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- Fischer TB, Arunachalam KV, Bailey D, et al. The binding interface database (BID): a compilation of amino acid hot spots in protein interfaces. Bioinformatics. 2003;19:1453–4. doi: 10.1093/bioinformatics/btg163. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang R, Lai L. Structure-based method for analyzing protein–protein interfaces. J Mol Model. 2004;10:44–54. doi: 10.1007/s00894-003-0168-3. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–87. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein–protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–21. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein–protein interfaces. Sci STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ruiz D, Gohlke H. Targeting protein–protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13:2607–25. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- Huo S, Massova I, Kollman PA. Computational alanine scanning of the 1:1 human growth hormone–receptor complex. J Comput Chem. 2002;23:15–27. doi: 10.1002/jcc.1153. [DOI] [PubMed] [Google Scholar]
- Rajamani D, Thiel S, Vajda S, et al. Anchor residues in protein–protein interactions. Proc Natl Acad Sci U S A. 2004;101:11287–92. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Hot spot occlusion from bulk water: a comprehensive study of the complex between the lysozyme HEL and the antibody FVD1.3. J Phys Chem B. 2007;111:2697–706. doi: 10.1021/jp067096p. [DOI] [PubMed] [Google Scholar]
- Hu Z, Ma B, Wolfson H, et al. Conservation of polar residues as hot spots at protein interfaces. Proteins. 2000;39:331–42. [PubMed] [Google Scholar]
- Keskin O, Ma B, Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345:1281–94. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- Ma B, Elkayam T, Wolfson H, et al. Protein–protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–7. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney E, Tuncbag N, Keskin O, et al. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008;36(Database issue):D662–6. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell SJ, LeGault L, Mitchell JC. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36(Web server issue):265–9. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y, Rost B. Protein–protein interaction hotspots carved into sequences. PLoS Comput Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–61. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- Janin J. Specific versus non-specific contacts in protein crystals. Nat Struct Biol. 1997;4:973–4. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- Valdar WS, Thornton JM. Conservation helps to identify biologically relevant crystal contacts. J Mol Biol. 2001;313:399–416. doi: 10.1006/jmbi.2001.5034. [DOI] [PubMed] [Google Scholar]
- Carugo O, Argos P. Protein–protein crystal-packing contacts. Protein Sci. 1997;6:2261–3. doi: 10.1002/pro.5560061021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J, Rodier F. Protein–protein interaction at crystal contacts. Proteins. 1995;23:580–7. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci. 2006;15:352–61. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Domingues FS, Sommer I, et al. NOXclass: prediction of protein–protein interaction types. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernauer J, Bahadur RP, Rodier F, et al. DiMoVo: a Voronoi tessellation-based method for discriminating crystallographic and biological protein–protein interactions. Bioinformatics. 2008;24:652–8. doi: 10.1093/bioinformatics/btn022. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Rost B. ISIS: interaction sites identified from sequence. Bioinformatics. 2007;23:13–6. doi: 10.1093/bioinformatics/btl303. [DOI] [PubMed] [Google Scholar]
- Res I, Mihalek I, Lichtarge O. An evolution based classifier for prediction of protein interfaces without using protein structures. Bioinformatics. 2005;21:2496–501. doi: 10.1093/bioinformatics/bti340. [DOI] [PubMed] [Google Scholar]
- Keskin O, Tuncbag N, Gursoy A. Characterization and prediction of protein interfaces to infer protein–protein interaction networks. Curr Pharm Biotechnol. 2008;9:67–76. doi: 10.2174/138920108783955191. [DOI] [PubMed] [Google Scholar]
- Liang S, Zhang C, Liu S, et al. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res. 2006;34:3698–707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvirth H, Raz R, Schreiber G. ProMate: a structure based prediction program to identify the location of protein–protein binding sites. J Mol Biol. 2004;338:181–99. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- Bradford JR, Westhead DR. Improved prediction of protein–protein binding sites using a support vector machines approach. Bioinformatics. 2005;21:1487–94. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Jones S. SHARP2: protein–protein interaction predictions using patch analysis. Bioinformatics. 2006;22:1794–5. doi: 10.1093/bioinformatics/btl171. [DOI] [PubMed] [Google Scholar]
- Porollo A, Meller J. Prediction-based fingerprints of protein–protein interactions. Proteins. 2007;66:630–45. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins. 2001;44:336–43. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–9. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- Aytuna AS, Gursoy A, Keskin O. Prediction of protein–protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics. 2005;21:2850–5. doi: 10.1093/bioinformatics/bti443. [DOI] [PubMed] [Google Scholar]
- Ogmen U, Keskin O, Aytuna AS, et al. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33(Web Server issue):W331–6. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–94. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34(Web Server issue):W310–4. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Roberts VA, Pique ME, et al. Protein docking using continuum electrostatics and geometric fit. Protein Eng. 2001;14:105–13. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, et al. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32(Web server issue):W96–9. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho CJ. Modeling side-chains using molecular dynamics improve recognition of binding region in CAPRI targets. Proteins. 2005;60:245–51. doi: 10.1002/prot.20565. [DOI] [PubMed] [Google Scholar]
- Chen R, Mintseris J, Janin J, et al. A protein–protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, et al. Geometry-based flexible and symmetric protein docking. Proteins. 2005;60:224–31. doi: 10.1002/prot.20562. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web server issue):W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ. The RosettaDock server for local protein–protein docking. Nucleic Acids Res. 2008;36(Web Server issue):233–8. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. London: In. Department of Biochemistry and Molecular Biology, University College; 1993. NACCESS. [Google Scholar]
- Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–2. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BE, Yang SS, Boriack-Sjodin PA, et al. Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J Biol Chem. 2001;276:27629–37. doi: 10.1074/jbc.M101727200. [DOI] [PubMed] [Google Scholar]
- Hiwasa T, Kasama M, Nakadai T, et al. Loss of Raf-1-binding activity of v-Ha-Ras by the deletion of amino acid residues 64–72 and 143–151. Cell Signal. 1996;8:393–6. doi: 10.1016/0898-6568(96)00078-2. [DOI] [PubMed] [Google Scholar]
- Spoerner M, Herrmann C, Vetter IR, et al. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA. 2001;98:4944–9. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, et al. STRING 7 – recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35(Database issue):D358–62. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari B, Tasneem A, Przytycka TM, et al. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2008;36(Database issue):D656–61. doi: 10.1093/nar/gkm761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Park B, Kim H, et al. HPID: the human protein interaction database. Bioinformatics. 2004;20:2466–70. doi: 10.1093/bioinformatics/bth253. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32(Database issue):D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Lewington C, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32(Database issue):D452–5. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–82. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Chen YC, Lo YS, Hsu WC, et al. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007;35(Web server issue):W561–7. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrotas PJ, Alexov E. PROTCOM: searchable database of protein complexes enhanced with domain–domain structures. Nucleic Acids Res. 2007;35(Database issue):D575–9. doi: 10.1093/nar/gkl768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sommer I, Lengauer T, et al. Alignment of non-covalent interactions at protein–protein interfaces. PLoS ONE. 2008;3:e1926. doi: 10.1371/journal.pone.0001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Shulman-Peleg A, Shatsky M, Nussinov R, et al. MultiBind and MAPPIS: webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 2008;36(Web server issue):W260–4. doi: 10.1093/nar/gkn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC, Walther D. MolSurfer: two-dimensional maps for navigating three-dimensional structures of proteins. Trends Biochem Sci. 1999;24:285–7. doi: 10.1016/s0968-0004(99)01412-7. [DOI] [PubMed] [Google Scholar]
- Tina KG, Bhadra R, Srinivasan N. PIC: Protein interactions calculator. Nucleic Acids Res. 2007;35(Web server issue):W473–6. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RP, Bahadur RP, Pal A, et al. ProFace: a server for the analysis of the physicochemical features of protein–protein interfaces. BMC Struct Biol. 2006;6:11. doi: 10.1186/1472-6807-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MD, Bava KA, Gromiha MM, et al. ProTherm and ProNIT: thermodynamic databases for proteins and protein–nucleic acid interactions. Nucleic Acids Res. 2006;34(Database issue):D204–6. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein–ligand binding sites. Bioinformatics. 2005;21:1908–16. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. 307–28. [DOI] [PubMed] [Google Scholar]
- Powers R, Copeland JC, Germer K, et al. Comparison of protein active site structures for functional annotation of proteins and drug design. Proteins. 2006;65:124–35. doi: 10.1002/prot.21092. [DOI] [PubMed] [Google Scholar]
- Bordner AJ, Gorin AA. Comprehensive inventory of protein complexes in the Protein Data Bank from consistent classification of interfaces. BMC Bioinformatics. 2008;9:234. doi: 10.1186/1471-2105-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. J Appl Cryst. 2003;36:1116–22. [Google Scholar]
- Tsuchiya Y, Kinoshita K, Ito N, et al. PreBI: prediction of biological interfaces of proteins in crystals. Nucleic Acids Res. 2006;34(Web server issue):W320–4. doi: 10.1093/nar/gkl267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33(Web server issue):W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G, Valencia A, Tress ML. Firestar – prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res. 2007;35(Web server issue):W573–7. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi SS, Schein CH, Oezguen N, et al. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–99. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski EW, Gregoret LM, Mandel-Gutfreund Y. Annotating nucleic acid-binding function based on protein structure. J Mol Biol. 2003;326:1065–79. doi: 10.1016/s0022-2836(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Tjong H, Qin S, Zhou HX. PI2PE: protein interface/interior prediction engine. Nucleic Acids Res. 2007;35(Web server issue):W357–62. doi: 10.1093/nar/gkm231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–99. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–77. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–33. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Schroeder M. Using protein binding site prediction to improve protein docking. Gene. 2008;422(1–2):14–21. doi: 10.1016/j.gene.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Ritchie DW, Kemp GJ. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–94. [PubMed] [Google Scholar]
- Heifetz A, Katchalski-Katzir E, Eisenstein M. Electrostatics in protein–protein docking. Protein Sci. 2002;11:571–87. doi: 10.1110/ps.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TM, Blundell TL, Fernandez-Recio J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein–protein docking. Proteins. 2007;68:503–15. doi: 10.1002/prot.21419. [DOI] [PubMed] [Google Scholar]
- Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–20. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- Jackson RM, Gabb HA, Sternberg MJ. Rapid refinement of protein interfaces incorporating solvation: application to the docking problem. J Mol Biol. 1998;276:265–85. doi: 10.1006/jmbi.1997.1519. [DOI] [PubMed] [Google Scholar]