Abstract
Cells require actin nucleators to catalyze the de novo assembly of filaments and actin elongation factors to control the rate and extent of polymerization. Nucleation and elongation factors identified to date include Arp2/3 complex, formins, Ena/VASP, and newcomers Spire, Cobl, and Lmod. Here, we discuss recent advances in understanding their activities and mechanisms, and new evidence for their cooperation and interaction in vivo. Earlier models had suggested that different nucleators function independently to assemble distinct actin arrays. However, more recent observations indicate that the construction of most cellular actin networks depends on the activities of multiple actin-assembly promoting factors working in concert.
Introduction
Many cellular processes powered by actin polymerization (e.g. cell motility, endocytosis, and cytokinesis) depend on responsive, rapid bursts of actin filament assembly at specific subcellular locations. Cells typically contain a large pool of actin monomers that is buffered by actin monomer-binding proteins such as thymosin β4 and profilin. These factors suppress spontaneous nucleation of new filaments, yet enable rapid mobilization of monomers for elongation at existing filament ends. This makes nucleation the rate-limiting step in de novo filament formation. Once nucleated, filaments elongate at their fast-growing (barbed) ends at a rate linearly proportional to the concentration of available actin monomers. Elongation at the slower-growing (pointed) ends of filaments may not be physiologically relevant since most actin monomers are bound to proteins that block addition to pointed ends. The extent of filament elongation in vivo is severely limited by the presence of high affinity barbed end capping proteins.
To overcome these barriers to filament nucleation and elongation, cells express actin assembly-promoting factors. First, a variety of actin nucleators with distinct mechanisms respond to cellular signals and regulate the precise timing and location of filament formation. Second, actin elongation factors control the extent of filament growth by protecting barbed ends from capping proteins and influence the rate of actin subunit addition. By employing specific combinations of nucleators and elongation factors, each with distinct mechanisms and modes of regulation, cells gain the versatility required to construct actin networks with specialized architectures and functions.
In this review, we compare the biochemical mechanisms of different actin nucleators and elongation factors, then consider how these activities are used in different combinations to generate cellular actin structures in vivo. Finally, we examine how emerging scaffolding proteins coordinate the spatial and temporal activities of multiple actin assembly factors.
Actin Nucleators
What are the properties of a bona fide actin nucleator? A nucleator can be defined as a factor that stimulates formation of a filament that grows rapidly at its barbed end. In addition, a nucleator should be able to efficiently seed polymerization from a pool of profilin-bound actin monomers (profilin-actin), since this may be the dominant species of available ATP-actin monomers in eukaryotic cells. Spontaneous filament assembly involves sequential formation of highly unstable polymerization intermediates (actin dimers and trimers) that rapidly dissociate, making spontaneous nucleation highly inefficient. In principle, an actin nucleator could use one of three mechanisms to surmount this barrier: (1) structural mimicry of polymerization intermediates, (2) stabilization of spontaneously formed intermediates, or (3) recruitment and alignment of actin monomers to form a polymerization ‘seed’. Nucleators have now been identified that utilize each of these three mechanisms (Figure 1a).
Figure 1. Proposed mechanisms of actin assembly factors.
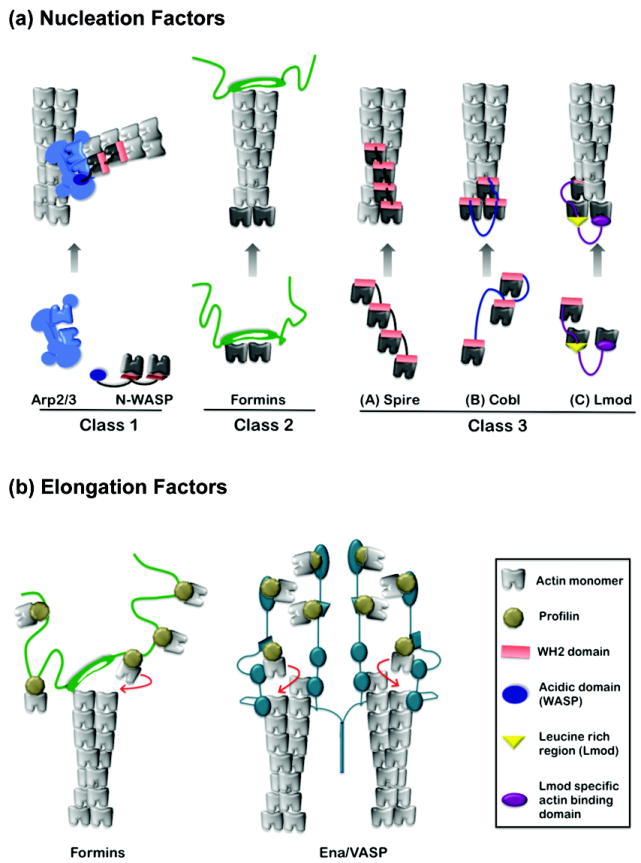
(a) Three classes of actin nucleators. Nucleator domains are displayed in color, actin subunits used by nucleators to seed polymerization in black, and actin subunits polymerized from nuclei in grey. Class I: N-WASP uses its WH2 domain(s) to recruit actin monomers and its acidic (A) domain to bind to an actin-related protein subunit of Arp2/3 complex. This structure stabilized by N-WASp may mimic an actin trimer. Class II: formins are hypothesized to nucleate actin polymerization by stabilizing spontaneously formed actin dimers and/or trimers. Formins remain associated with the barbed end while permitting addition of actin subunits. Class III: Spire, Cobl and Lmod contain between one and four WH2 domains each, separated by intervening linker sequences of variable length. Their nucleation mechanisms are related, but each may generate an actin nucleus with distinct properties, stabilized by lateral and/or longitudinal contacts between subunits, and in some cases capped at one end. Note, in some respects N-WASp represents a specialized form of Class III nucleator, in which the third actin monomer-binding domain has been replaced with a domain that binds to actin-related proteins. (b) Actin elongation factors. Formins shield barbed end growth from capping proteins by using their dimeric FH2 domains to processively move with the filament end. Adjacent rope-like FH1 domains are used as “arms” to recruit profilin-actin complexes and ‘deliver’ them to the FH2-capped filament end for rapid addition. The elongation mechanism of Ena/VASP is not well understood. However, it tetramerizes, bundles filaments, and may engage multiple barbed ends simultaneously. Its ability to accelerate barbed end elongation could involve a relay or hand-off of actin monomers using multiple actin-binding domains (adapted from model in [19]).
The first nucleator identified, Arp2/3 complex, employs structural mimicry [1,2]. When combined with a ‘nucleation promoting factor’ (NPF), Arp2/3 complex catalyzes polymerization of a new (daughter) filament from the side of an existing (mother) filament at a 70° angle to generate a branched structure. This ‘dendritic’ nucleation activity is used to assemble actin structures such as Listeria comet tails, lamellipodia, focal adhesions, and yeast endocytic patches. The most well understood Arp2/3 complex NPFs are WASp/SCAR/WAVE family proteins, which perform at least two essential roles in nucleation. First, they trigger conformational changes in Arp2/3 complex that bring its actin-related protein subunits (Arp2 and Arp3) into close register, possibly to mimic an actin dimer. Second, they recruit 1-2 actin monomers, which is a critical step in nucleation since Arp2/3 complex alone binds very weakly to monomers.
The second group of nucleators identified, formins, catalyze the formation of linear (unbranched) actin filaments in vitro and assemble diverse actin structures, including stress fibers, cytokinetic actin rings, and actin cables in vivo [3,4]. The mechanism of actin assembly by formins involves high affinity binding of their dimeric donut-shaped FH2 domains to the barbed ends of actin filaments. The FH2 domain lacks detectable affinity for actin monomers, so it has been suggested that formins catalyze polymerization by stabilizing spontaneously formed actin dimers and/or trimers [5]. Although direct experimental evidence for this model is lacking, the idea that an FH2 dimer may stabilize short-pitch associations between two actin subunits is consistent with the co-crystal structure of FH2 bound to actin, which reveals that a single FH2 ‘hemi-dimer’ bridges two actin subunits [6]. In contrast to Arp2/3 complex, which remains associated with the pointed end of the filament it nucleates, the formin FH2 domain remains associated with the barbed end (Figure 1a). After nucleation, the FH2 moves processively with the growing barbed end, allowing rapid insertion of new actin subunits (see below).
The more recently identified nucleators Spire, Cordon bleu (Cobl), and Leiomodin (Lmod) employ yet a third nucleation mechanism that involves actin monomer recruitment to form polymerization seeds. Spire has four tandem actin monomer-binding WASp-homology 2 (WH2) domains separated by short linkers. Electron micrographs supported by hydrodynamic and spectroscopic analyses demonstrate that Spire stably associates with four actin monomers in a prenucleation complex that resembles a short, single-stranded segment of a nascent filament [7,8] (Figure 1a). It has been suggested that Spire remains associated with either the side or the pointed end of a filament after nucleation, allowing free barbed end elongation [7]. However, another group has reported that after nucleation, Spire associates with the barbed end and blocks elongation by profilin-actin [8]. Thus, the post-nucleation effects of Spire have yet to be fully resolved. New insights might be gained by real time microscopy analysis of labeled Spire on single actin filaments. Regardless of the precise mechanism, genetic studies indicate that Spire, along with profilin and the formin Cappuccino, promotes the assembly of cytoplasmic actin meshworks that control cytoplasmic streaming in Drosophila oocytes (see below), and may have roles in membrane trafficking [9].
Cobl’s nucleation mechanism is somewhat related to that of Spire, but with key differences. Nucleation by Cobl requires three actin-binding WH2 domains separated by linkers. Deletion of either the first or third WH2 domain greatly diminishes nucleation activity, whereas in Spire, deletion of individual WH2 domains only incrementally reduces activity. Moreover, one of the linkers in Cobl is substantially longer than those found in Spire [10]. Shortening this linker abolishes nucleation, and substituting an unrelated sequence of similar length restores nucleation. Thus, linker length rather than sequence appears to be critical for function. This has led to the suggestion that Cobl stabilizes both short-pitch and long-pitch associations to generate polymerization nuclei (Figure 1a), a model that should be tested through structural analyses. Cobl is highly expressed in the brain, and knock down experiments in primary hippocampal neurons demonstrate that it is required for normal levels of neurite extension and branching. It will be interesting to learn what specific actin structures are assembled by Cobl to promote neurite extension.
The third member of this nucleation group, Lmod, is expressed in cardiac muscle tissue. Nucleation by Lmod depends on a single WH2 domain and two unrelated actin-binding domains similar to those found in tropomodulin (Tmod), a pointed end filament capping protein [11]. Lmod uses this non-uniform array of actin monomer-binding domains to organize 2-3 actin monomers into a polymerization seed, possibly stabilized at its pointed end in a Tmod-like fashion. One of the interesting properties of Lmod is that its nucleation activity is stimulated in vitro by tropomyosin, similar to formins [12], but little else is known about how the activities of Lmod (or Cobl) are regulated.
Actin Elongation Factors
Once nucleated, filaments grow freely at their barbed ends until monomer pools are depleted and/or capping proteins terminate elongation. Because of the high association rate constant for capping proteins, which are abundant in nearly all cell types, filament lengths are severely limited in vivo unless their growing barbed ends are protected or capping protein is locally inactivated. In recent years, it has become evident that cells express proteins that associate and move with the growing barbed ends of filaments, shielding them from capping proteins and controlling the rate of elongation. We refer to the proteins as “actin elongation factors.” Two have been characterized to date, formins and Ena/VASP.
Formins use their dimeric, donut-shaped FH2 domains to crown the barbed end and processively move with the growing filament end [4,13]. This dynamic attachment requires both halves of the formin dimer to be functional, and may involve alternating contacts of the FH2 with the two actin subunits exposed at the barbed end. However, the precise mechanism underlying processive movement remains to be determined. The adjacent FH1 domain appears to be long, unstructured, and possibly rope-like. It contains multiple polyproline tracts that recruit profilin-actin monomers, and by a poorly understood mechanism ‘delivers’ these complexes to the FH2-capped barbed end for rapid addition. This set of interactions can lead to FH1-FH2-dependent acceleration of barbed end elongation, by as much as 5-fold over the rate of elongation at free barbed ends [14] (Figure 1b). Further, different formins accelerate elongation to highly variable extents (1.25 – 5.0 fold). Interestingly, the requirements for formin-mediated actin nucleation and elongation are similar, both requiring two functional halves of an FH2 dimer with intact actin-binding sites. Thus, the same properties of the FH2 dimer that allow it to processively move with an elongating barbed end may be critical for “capture” of polymerization intermediates to nucleate actin assembly.
Enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) is the other known actin elongation factor. Proteins in this family are ubiquitously expressed in mammals, localize to actin-rich zones (e.g. focal adhesions, cell-cell contacts, filopodial tips, and lamellipodia), and contribute to cell motility, axon guidance, cell adhesion, endocytosis, and intracellular pathogen motility [15,16]. Like formins, Ena/VASP proteins bind profilin-actin and multimerize, but form tetramers rather than dimers [17-19]. Unlike formins, they do not appear to nucleate actin assembly under physiological salt conditions in solution [20]. However, this issue may warrant further investigation given the recently reported distinction between Ena/VASP activities in solution versus clustered/immobilized on beads (see below). Similar to formins, Ena/VASP proteins immobilized on beads can protect barbed end growth in the presence of capping proteins [21]; however, this effect requires substantially higher concentrations of Ena/VASP compared to formins, perhaps reflecting differences in barbed end binding affinities and/or rates of dissociation. Live cell imaging experiments show that GFP-VASP remains at the ends of actively growing filopodial tips, which is consistent with either persistent or transient engagement of nascent barbed ends [22].
Recent efforts to reconcile conflicting data between the behavior of Ena/VASP in solution and on beads have suggested unique requirements for its attachment and protection activity. While bead assays had shown that Ena/VASP can protect barbed end growth in the presence of capping proteins [21], total internal reflection fluorescence (TIRF) microscopy studies monitoring individual actin filaments had suggested that elongation ceases when barbed ends become attached to mouse VASP immobilized on a glass surface [23]. The seemingly conflicting observations left it unclear whether the differences in activity were due to differences in the VASP proteins, purification procedures, and/or assay conditions. These issues appear to have been resolved by a new study, which uses TIRF to directly compare human and Dictyostelium VASP proteins in solution and immobilized on beads for their effects on rate of barbed end elongation, both in the presence and absence of capping protein [24]. Protected elongation was observed with VASP on beads but not in solution, suggesting that the activity requires tethering/clustering of VASP. Thus, Ena/VASP proteins may processively cap and protect filaments in vivo only after their spatial recruitment to cortical foci, e.g. incipient filopodial tips. This study also answers a longstanding question - do Ena/VASP proteins accelerate barbed end elongation? One previous study on mouse VASP showed that it increased rate of filament elongation by only 1.2 fold in solution [23]; however, the new TIRF study shows that human and Dictyostelium VASP accelerate elongation more substantially, by about 2 and 7 fold, respectively [24]. Thus, both VASP and formins can accelerate elongation, although formins require profilin for this activity. An important principle emerges from these collective studies: different formins and Ena/VASP proteins support highly variable rates of elongation. These differences may be tailored in vivo to optimize the assembly of specific actin structures. For example, fast elongation rates may be optimal for filopodial extension compared to cytokinetic actin ring assembly.
What is the mechanism by which VASP protects and accelerates barbed end elongation? The protein has three parts: an N-terminal EVH1 domain that links VASP to key ligands, a central proline-rich region (PRR) that contains at least three separate binding sites for profilin-actin and actin monomers, and a C-terminal EVH2 domain that interacts with F-actin possibly to cap the barbed end. A recent study solved the structures of parts of VASP bound to profilin and actin monomers, and presents an appealing model for how a relay of interactions in the PRR might deliver monomers to the growing barbed end [19]. What is clearly needed next is a more complete structure of the EVH2 domain, in order to understand the basis of VASP association with the barbed end of the filament.
Collaborative assembly of lammelipodia or filopodia
Early observations suggested that Arp2/3 complex and formins may build distinct sets of actin networks in vivo comprised of filaments with different properties: linear versus branched, protected versus unprotected [25-28]. However, new observations are challenging this view, suggesting cross-participation by multiple actin assembly-promoting factors.
One example is in the formation of lamellipodia. These flat sheet-like membrane protrusions contain a dense network of interconnected filaments with their barbed ends generally oriented toward the membrane. The abundance of branched filaments led to over-simplified models suggesting lamellipodia are assembled entirely by Arp2/3 complex without the participation of other actin-assembly promoting factors. However, it has become clear that Ena/VASP and formins contribute to lammelipodial assembly, and recent ultrastructural analyses reveal that lammelipodial actin networks include shorter and longer filaments, and filaments with heterogenous branch angles, which cannot be explained by Arp2/3-nucleated assembly alone [29]. Targeted depletion/relocation of Ena/VASP leads to networks comprised of shorter and more branched filaments, suggesting that Ena/VASP is required for assembly of the longer filaments [21]. These effects likely stem from the ability of Ena/VASP to protect barbed end elongation at the leading edge. Formins may have a similar role in regulating lammelipodial architecture. A recent study found that knock down of mDia2 led to lammelipodial networks comprised of shorter average filament length, whereas mDia2 overexpression produced an excess of long unbranched filaments [30]. Interestingly, there were also fewer lammelipodia per cell after mDia2 knock down, demonstrating that the dendritic nucleation of Arp2/3 complex alone is not sufficient to support normal levels of the flat membrane protrusions. It remains to be determined whether Ena/VASP and formins elongate separate or overlapping sets of barbed ends, and whether Ena/VASP and formins nucleate filaments or instead hijack barbed ends of Arp2/3-nucleated filaments.
A second example of cross-participation is in the formation of filopodia. These finger-like membrane protrusions contain a compact, linear bundle of long, unbranched filaments with their barbed ends oriented toward the filopodial tips. Formins and Ena/VASP localize to filopodial tips, and their genetic requirement for filopodial assembly is undisputed [30-32], but it remains uncertain why filopodial assembly requires the activities of two different actin elongation factors. One possibility is that formins play a more critical role initiating filament assembly, whereas Ena/VASP is more crucial for organizing and controlling the rate of growth of barbed ends. This requires further investigation, which could include testing the combined in vitro effects of Ena/VASP and formins on actin.
It has also been proposed by some groups that Arp2/3 complex makes important contributions to filopodial assembly; however, conflicting data have been reported. One set of studies showed that filopodia are unaffected by knock down of Arp2/3 complex subunits in fibroblasts and melanoma cells or by null mutants of WAVE complex subunits in Dictyostelium [33,34]. Further, a more recent study in carcinoma cells found that knock down of WAVE2 (which is required for lammelipodial formation) increased formin-dependent filopodial assembly, suggesting that Arp2/3 complex activity suppresses filopodial formation [35]. In agreement with this view, another recent study in HeLa cells showed that WAVE2 and Arp2/3 complex directly inhibit filopodial assembly by the formin mDia2 [36]. In this study, knock down of WAVE2 or Arp2/3 complex increased mDia2-dependent filopodia formation, whereas WAVE2 overexpression decreased filopodia. This study suggests that formation of a Dia2-WAVE2-Arp2/3 complex inhibits filopodial extension until EGF-induced activation of Cdc42 locally dissociates the complex, allowing filopodial formation. However, this complex has not been isolated, nor have the activities predicted from this model been demonstrated. Regardless, the collective genetic observations of the studies above indicate that WAVE and Arp2/3 complex have neutral or inhibitory effects on filopodial formation.
In contrast, correlative light and electron microscopy studies on migrating melanoma cells and neurons have suggested that the linear filaments in filopodia stem directly from branched filaments in the lamellipodia [30,37,38]. This has led to the ‘convergent elongation’ model, which suggests that filaments nucleated by Arp2/3 complex are captured at their barbed ends and organized by Ena/VASP and/or formins. The model is supported by genetic observations showing that partial knock down of different Arp2/3 complex subunits leads to a 2-3 fold reduction in number of visible filopodia on the cell surface and a reduction in frequency of filopodial initiation [38]. Further support comes from studies showing that a purified mixture of actin, Arp2/3 complex, WASP-VCA, and fascin is sufficient to assemble densely branched networks from which Arp2/3-dependent filopodial-like bundles protrude [39]. We note that additional roles for Arp2/3 complex in this process could exist. In particular, filaments nucleated by formins (and/or Ena/VASP) at filopodial tips might extend their pointed ends into the lammelipodia and be captured by Arp2/3 complex, linking them to the sides of filaments and stabilizing the filopodia. This mechanism would invert the roles of Arp2/3 complex and formins (in nucleation and capture), providing the reciprocal mechanism to convergent elongation. If both mechanisms were at work, this would lead to bidirectional growth and capture.
The reasons for the conflicting genetic results between the two sets of studies above are not yet clear. Different cell lines and/or conditions for adhesion were used in these studies [33,38], which might affect the dependence of filopodial stability on Arp2/3 complex activity. It has also become apparent that there are different types of filopodial extensions, which may have different molecular requirements for their assembly [40]. This underscores the point that new approaches are needed to better define the coordinated series of events involved in assembling complex actin structures. For instance, new insights might be gained by combining high resolution localization of proteins in cellular actin structures with more acute genetic or pharmacological disruption of their functions.
Coordination of actin assembly by multivalent scaffolding proteins
The observations above raise a key question - how are the activities of multiple actin assembly-promoting factors spatially and temporally coordinated to orchestrate the assembly of specific actin structures? This is achieved at least in part by multi-domain scaffolding proteins that can bind and/or regulate two or more actin assembly factors. Three such factors are IQGAP1, IRSp53, and DIP/WISH (Figure 2).
Figure 2. Physical complexes that spatially and temporally coordinate multiple actin assembly factors.
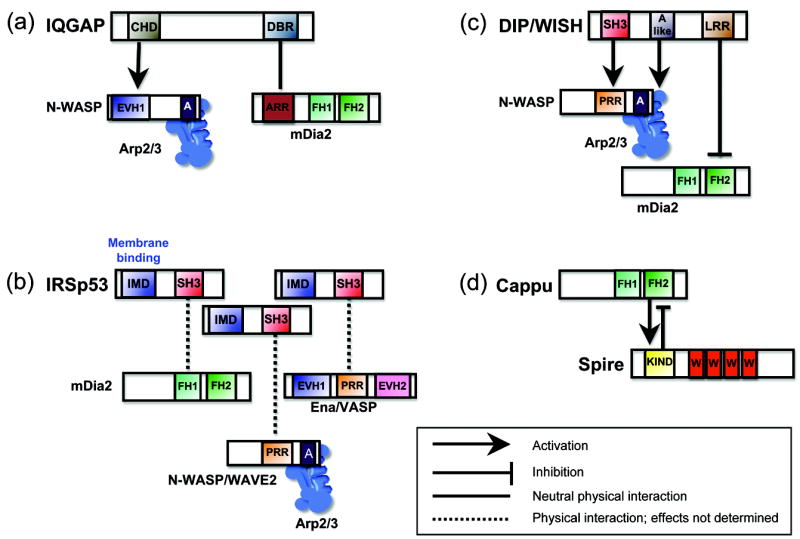
(a) IQGAP uses its calponin homology domain (CHD) to associate with the EVH1 domain of N-WASP, and its dia-binding region (DBR) to associate with the ankrin-rich repeats (ARR) of mDia2. (b) IRSp53 uses its IMD domain to bind and induce curvature of membranes and its SH3 domain to interact with the proline-rich region (PRR) of N-WASP and WAVE2, the PRR of Ena/VASP, and the proline-rich FH1 domain of mDia2. (c) DIP/WISH uses its SH3 domain to bind the PRR of N-WASP and an acidic (A-like) domain to bind Arp2/3 complex, leading to N-WASP-Arp2/3 activation, and its leucine-rich repeat (LRR) domain to directly inhibit FH2 domain of mDia2. (d) Spire uses its KIND domain to inhibit the FH2 domain of Cappuccino, while this same interaction enhances Spire activity.
IQGAP1 is a large (350 kDa) effector of Rac1 and Cdc42 required for lamellipodial assembly [41], and uses its calponin homology domain (CHD) to directly bind the EVH1 domain of N-WASP and stimulate Arp2/3-dependent actin assembly [42]. It also binds tightly to mDia1 using a distinct domain and is required for mDia1 localization to the leading edge, but interestingly has no in vitro effects (with or without RhoA) on mDia1 activity [43]. It is not yet clear whether IQGAP1 can bind N-WASP and mDia1 simultaneously, or whether N-WASP binding to IQGAP1 might influence mDia1 activity.
IRSp53 is a 53 kDa BAR-related membrane-binding protein that oligomerizes and induces curvature of membranes to help promote filopodial protrusion [44,45,46]. It is also an effector of Cdc42, and through its SH3 domain binds to Ena/VASP, mDia2, WAVE2 and N-WASP. Thus, IRSp53 may concatamerize on membranes to spatially organize these actin regulators into filopodial tip complexes and coordinate membrane deformation with actin assembly. Consistent with this model, IRSp53 induces filopodia in an N-WASP- and Ena/VASP-dependent manner [46].
DIP/WISH is a 90 kDa protein that has an SH3 domain and binds to both N-WASP and Arp2/3 complex to stimulate Arp2/3-dependent actin assembly [47,48]. It also has a Leucine-rich repeat (LRR) domain, through which it potently inhibits the formin FH2 domain to suppress mDia2-dependent filopodial assembly [49]. Consistent with these activities, knock down of DIP/WISH impairs lamellipodial assembly [48], and over expression of DIP/WISH impairs filopodial formation [49]. Moreover, DIP/WISH localizes to both filopodial tips and the lammelipodial cortex. It is interesting that both IRSp53 and DIP/WISH localize to filopodial tips, and appear to have antagonistic roles in filopodial assembly. It remains uncertain whether these two proteins perform their functions in the same or competing complexes.
Direct interactions between formins and Spire
Actin nucleator cross-regulation can also be direct, as demonstrated by the interactions between Spire and the formin Cappuccino. This topic has been extensively reviewed elsewhere [50,51], so it is only commented on briefly here. Genetically, Cappuccino, Spire and profilin are each required for the formation of a cytoplasmic actin network in oocytes that affects events during development [52]. The precise role of the actin meshwork is not yet fully understood, but it is required for proper regulation of cytoplasmic streaming, microtubule orientation, and cell polarity. Interestingly, loss of the actin network in cappuccino mutants cannot be rescued by overexpression of Spire, but loss of the network in spire mutants can be partially rescued by expression of an activated Cappuccino. Thus, formins may have the central role in the assembly of these networks, enhanced by Spire. In either case, the genetic data available make a strong case that Spire and Cappuccino synergize in vivo to assemble these actin networks, and a possible mechanism for the synergy arose from the observation that they directly interact [53].
Defining the biochemical effects of Spire-Cappuccino interactions on actin assembly has proven to be more difficult. One group reported that Spire KIND domain inhibits Cappuccino-mediated actin assembly, while this interaction activates Spire [54] (Figure 2). A different study concluded that the KIND domain has no effect on Cappuccino activity [53]. Importantly, neither study explains how such effects (inhibitory or neutral) can lead to the genetic observation that Spire is required for Cappuccino-dependent actin meshwork formation. A third study offers a clear biochemical correlation with the in vivo observations, showing that Spire and Cappuccino synergize in a biomimetic in vitro motility assay [8]. However, the mechanism used to explain the observed synergy in this study involves Spire capping barbed ends, which contradicts earlier reports suggesting that Spire caps pointed ends [7]. Clearly, more work is needed to resolve the mechanism of this interaction. It may be informative to use real-time microscopy on individual actin filaments and labeled Spire and Cappuccino to determine if they act sequentially or simultaneously, and at which end of the filament.
Homeostatic cross-talk
The activities of actin assembly-promoting factors can also be balanced by homeostatic mechanisms. One recent study in S. cerevisiae showed that the lethality caused by an overactive formin construct could be rescued by overexpression of yeast WASp or actin monomer binding proteins [55]. The implication of these data is that formins and WASp-Arp2/3 complex compete for a limited pool of actin monomers in cells, and formin hyper-activity can be rescued by up-regulation of WASp-Arp2/3 because it restores the balance of assembly activities between these two nucleator systems. Thus, one important mechanism for maintaining proper homeostasis of actin regulation in cells is the balanced expression and activation of different nucleators. Further evidence for homeostatic mechanisms is provided by a recent study showing that combined knock down of N-WASP and WAVE2 leads to increased RhoA activity, which induces mDia1-dependent formation of stress fibers and filopodia [35]. These data suggest that normal expression of N-WASP and WAVE2 represses RhoA, and therefore inhibits mDia1. Consistent with these observations, another recent study reported that knock down of Arp2/3 complex subunits in B35 neuroblastoma cells led to increased RhoA activity, which in turn caused excessive formation of actin bundles and focal adhesions [38]. Although it is not yet clear how the expression of N-WASP/WAVE2-Arp2/3 machinery alters RhoA activity levels, these observations provide examples of how nucleator cross-talk can be achieved in the absence of physical interactions. Together, these studies point to another mechanism for cross-talk between actin assembly factors, complementary to the mechanisms based on physical associations described in the sections above. These findings stress the importance of considering homeostatic effects when interpreting all phenotypes arising from genetic depletion of an actin regulator.
Perspectives
A new concept coming into focus is that cells express a wide variety of actin assembly-promoting factors, and that many of them associate in functional complexes to build actin structures. There are undoubtedly more actin nucleators and elongation factors to discover, possibly with new and unique mechanisms to add to the complexity of actin regulation. A question that arises is why so many? One answer is functional diversity, to match the immense diversity in architecture of actin structures observed in vivo. A second answer is that the construction of even a single, highly complex actin network requires the signature activities of multiple actin assembly factors working in concert. Future investigations are likely to reveal many more cellular processes that depend on cooperation and cross-talk among actin assembly-promoting factors. The emerging complexity of these systems emphasizes that gaining a solid understanding of the cellular functions of any individual actin regulatory protein will require studying its effects in the context of the multi-protein complexes it forms in vivo.
Acknowledgments
We apologize to those authors whose important work we were unable to cite due to space restrictions. We thank James Moseley and Avital Rodal for critical reading of the review. Work in the authors’ laboratory is supported in part by grants from the National Institutes of Health (GM063691 and GM083137).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 3.Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 5.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 6.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 7.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- •8.Bosch M, Le KH, Bugyi B, Correia JJ, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018.Intensive biochemical characterization of Spire is presented, revealing that the four WH2 domains bind actin monomers cooperatively and may also bind barbed ends of filaments Using a biomemetic motility assay, it is shown that Spire and Cappuccino synergize to assemble actin.
- 9.Kerkhoff E. Cellular functions of the Spir actin-nucleation factors. Trends Cell Biol. 2006;16:477–483. doi: 10.1016/j.tcb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- ••10.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030.Identification of the brain enriched protein Cordon-bleu (Cobl) as an actin nucleator that employs three tandem WH2 domains Due to an extended linker between the second and third WH2 domains, it is proposed that Cobl forms an actin trimer stabilized by lateral and longitudinal contacts In vivo, Cobl is shown to be required for proper neuronal morphology and process outgrowth.
- ••11.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313.Leiomodin (Lmod) is identified as an actin nucleator that uses an array of different actin binding domains to stabilize a nucleus Tropomyosin is shown to enhance Lmod activity and mediate its localization to pointed ends of filaments Lmod knock down disrupts sarcomere assembly in rat cardiomyocytes, suggesting that Lmod nucleates tropomyosin-coated filaments in heart muscle.
- •12.Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46:8146–8155. doi: 10.1021/bi700686p.This study shows that tropomyosin binds to the formin FH2 domain and enables FH2-capped barbed ends to elongate as fast as free barbed ends However, it remains unclear whether tropomyosin can further stimulate formin-mediated elongation in the presence of profilin.
- 13.Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 15.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 16.Trichet L, Sykes C, Plastino J. Relaxing the actin cytoskeleton for adhesion and movement with Ena/VASP. J Cell Biol. 2008;181:19–25. doi: 10.1083/jcb.200710168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci U S A. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••19.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874.Structures are solved for regions of VASP bound to profilin-actin and actin monomers The authors propose a model for elongation involving a relay of monomer-binding interactions that “hands off” subunits to the barbed end.
- 20.Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- ••22.Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990.FRAP analysis is used to show that Ena/VASP is a stabile component of filopodial tip complexes It is also demonstrated that Ena/VASP is required for filopodia assembly in the absence of capping protein, indicating that it makes functional contributions beyond protecting barbed ends from capping protein.
- •23.Pasic L, Kotova T, Schafer DA. Ena/VASP proteins capture actin filament barbed ends. J Biol Chem. 2008;283:9814–9819. doi: 10.1074/jbc.M710475200.TIRF microscopy is used to show that Ena/VASP captures single filament barbed ends and protects them from capping proteins In addition, a modest profilin-dependent increase in rate of elongation is observed for Ena/VASP in solution.
- ••24.Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211.This study reveals that VASP proteins can greatly accelerate elongation of actin filaments, and that different isoforms of VASP support different rates of elongation Further, these activities occur specifically when VASP is immobilized versus free in solution, suggesting that clustering of VASP molecules may be required for their ability to protect and accelerate barbed end elongation.
- 25.Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 27.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- 28.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- ••29.Koestler SA, Auinger S, Vinzenz M, Rottner K, Small JV. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat Cell Biol. 2008;10:306–313. doi: 10.1038/ncb1692.Using correlative live-cell imaging and electron microscopy, the authors observe that actin filaments in lammelipodia, and those that extend into filopodia, have a wide angular distribution, which is inconsistent with proposed Arp2/3 dendritic nucleation models for lamelliopodial formation.
- ••30.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317.Loss-of-function and gain-of-function approaches are used to show that mDia2 influences filopodial, and more unexpectedly, lamellipodial actin architecture Correlative light and electron microscopy suggests that filopodia form by convergence of lamellipodial filaments into bundles that extend into filopodia.
- 31.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 32.Block J, Stradal TE, Hanisch J, Geffers R, Kostler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- 33.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE-and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci U S A. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••35.Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123.This study shows that knock down of WAVE2 but not N-WASP impairs lamellipodia formation Further, the combined knock down of N-WASP and WAVE2 increases levels of active RhoA, which in turn activates formin-dependent filopodial assembly.
- ••36.Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–857. doi: 10.1038/ncb1745.This paper demonstrates that mDia2 binds to WAVE2, and forms complexes with WAVE2 and Arp2/3 in HeLa cell extracts Further genetic observations lead the authors to propose formation of an mDia2-WAVE2-Arp2/3 complex that inhibits mDia2-induced filopodia while promoting Arp2/3-dependent ruffling.
- 37.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •38.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964.This paper shows that depletion of Arp2/3 complex in neuronal cells impairs both lammelipodial and filopodial protrusion Ultrastructural analysis suggests that the actin filaments in filopodia originate at Arp2/3 complex-generated branch points in the lamellipodia, lending support to the convergent elongation model.
- ••39.Ideses Y, Brill-Karniely Y, Haviv L, Ben-Shaul A, Bernheim-Groswasser A. Arp2/3 branched actin network mediates filopodia-like bundles formation in vitro. PLoS ONE. 2008;3:e3297. doi: 10.1371/journal.pone.0003297.This in vitro study powerfully demonstrates that purified mixtures of actin, Arp2/3 complex, WASp-VCA and fascin are sufficient to assemble dense, branched actin networks from which filopodial-like bundles of filaments protrude.
- 40.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 41.Brandt DT, Grosse R. Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep. 2007;8:1019–1023. doi: 10.1038/sj.embor.7401089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••42.Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282:426–435. doi: 10.1074/jbc.M607711200.This paper shows that IQGAP1 directly activates N-WASP to stimulate Arp2/3-mediated actin assembly in vitro, and is required for N-WASp co-localization with Arp2/3 complex at the leading edge of cells.
- ••43.Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071.IQGAP1 is identified as a novel Dia1 binding partner, and is shown to be required for Dia1 localization to the leading edge of migrating cells and for Dia1-dependent phagocytosis However, IQGAP1 shows no effect on Dia1 activity.
- ••44.Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176.The authors show that the IMD domains of IRSp53 and MIM directly induce curvature of membranes in vitro, and that this membrane-deforming activity of IRSp53 is required for filopodia formation in vivo.
- 45.Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- ••46.Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–20472. doi: 10.1074/jbc.M710185200.This paper shows that the IMD domain of IRSp53 induces membrane protrusions and filopodial formation in vivo. Further, IRSp53-induced filopodial assembly requires Ena/VASP and N-WASP, suggesting that IRSp53 coordinates membrane and actin dynamics to induce filopodial protrusions.
- 47.Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T. A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J Cell Biol. 2001;152:471–482. doi: 10.1083/jcb.152.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim JK, Song WK. Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem. 2006;281:617–625. doi: 10.1074/jbc.M504450200. [DOI] [PubMed] [Google Scholar]
- ••49.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024.This paper identifies DIP/WISH as a direct inhibitor of the mDia2 FH2 domain and shows that DIP/WISH suppresses filopodia formation in vivo.
- 50.Quinlan ME, Kerkhoff E. Actin nucleation: bacteria get in-Spired. Nat Cell Biol. 2008;10:13–15. doi: 10.1038/ncb0108-13. [DOI] [PubMed] [Google Scholar]
- 51.Renault L, Bugyi B, Carlier MF. Spire and Cordon-bleu: multifunctional regulators of actin dynamics. Trends Cell Biol. 2008;18:494–504. doi: 10.1016/j.tcb.2008.07.008. [DOI] [PubMed] [Google Scholar]
- ••52.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13:539–553. doi: 10.1016/j.devcel.2007.09.003.Using rapid fixation techniques, the authors discover a cytoplasmic actin meshwork in Drosophila oocytes that requires Spire, Cappuccino and profilin for its formation.
- 53.Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196.This paper shows that the Spire KIND domain binds directly to the FH2 domain of Cappucciono The interaction inhibits Cappucciono-mediated actin assembly, while stimulating the activity of Spire.
- •55.Gao L, Bretscher A. Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol Biol Cell. 2008;19:1474–1484. doi: 10.1091/mbc.E07-05-0520.This study shows that the lethality caused by overexpression of an active formin construct in yeast can be suppressed by overexpression of WASp/Las17 or actin monomer binding proteins This suggests that formins and Arp2/3 complex are cross-regulated via competition for the same actin monomer pool.


