Abstract
Background
Bet v 1 is an important cause of hay fever in northern Europe. Bet v 1 isoforms from the European white birch (Betula pendula) have been investigated extensively, but the allergenic potency of other birch species is unknown. The presence of Bet v 1 and closely related PR-10 genes in the genome was established by amplification and sequencing of alleles from eight birch species that represent the four subgenera within the genus Betula. Q-TOF LC-MSE was applied to identify which PR-10/Bet v 1 genes are actually expressed in pollen and to determine the relative abundances of individual isoforms in the pollen proteome.
Results
All examined birch species contained several PR-10 genes. In total, 134 unique sequences were recovered. Sequences were attributed to different genes or pseudogenes that were, in turn, ordered into seven subfamilies. Five subfamilies were common to all birch species. Genes of two subfamilies were expressed in pollen, while each birch species expressed a mixture of isoforms with at least four different isoforms. Isoforms that were similar to isoforms with a high IgE-reactivity (Bet v 1a = PR-10.01A01) were abundant in all species except B. lenta, while the hypoallergenic isoform Bet v 1d (= PR-10.01B01) was only found in B. pendula and its closest relatives.
Conclusion
Q-TOF LC-MSE allows efficient screening of Bet v 1 isoforms by determining the presence and relative abundance of these isoforms in pollen. B. pendula contains a Bet v 1-mixture in which isoforms with a high and low IgE-reactivity are both abundant. With the possible exception of B. lenta, isoforms identical or very similar to those with a high IgE-reactivity were found in the pollen proteome of all examined birch species. Consequently, these species are also predicted to be allergenic with regard to Bet v 1 related allergies.
Background
Birch trees grow in the temperate climate zone of the northern hemisphere and release large amounts of pollen during spring. This pollen is a major cause of Type I allergies. The main birch allergen in northern Europe is a pathogenesis-related class 10 (PR-10) protein from the European white birch (Betula pendula) termed Bet v 1 [1,2]. Pollen of other Fagales species contains PR-10 homologues that share epitopes with Bet v 1 [3], as do several fruits, nuts and vegetables [4-7]. An IgE-mediated cross-reaction to these food homologues causes the so-called oral allergy syndrome (OAS) [8,9]. PR-10 proteins constitute the largest group of aeroallergens and are among the four most common food allergens [10].
The genus Betula encompasses over 30 tree and shrub species that are found in diverse habitats in the boreal and temperate climate zone of the Northern Hemisphere. The taxonomy of the Betula genus is debated, as is the number of recognized species. The genus is either divided into three, four or five groups or subgenera [11-13]. B. pendula occurs in Europe and is the only species whose relation to birch pollen allergy has been extensively investigated. Sensitization to birch pollen is also reported across Asia and North America, where B. pendula is not present [14,15]. Other Betula species occur in these areas, but their allergenic potency is unknown. Betula species may vary in their allergenicity as variation in allergenicity has been found among cultivars of apple [16-18], peach and nectarine [19], and among olive trees [20].
PR-10 proteins are present as a multigene family in many higher plants, including Gymnosperms as well as Monocots and Dicots [21-23]. The classification as PR-proteins [24] is based on the induced expression in response to pathogen infections by viruses, bacteria or fungi [25-27], to wounding [28] or to abiotic stress [29,30]. Some members of the PR-10 gene family are constitutively expressed during plant development [31] or expressed in specific tissues [23]. Multiple PR-10 genes have been reported for B. pendula as well [32]. mRNAs of these genes have been detected in various birch tissues, including pollen [1,33,34], roots, leaves [28,30], and in cells that are grown in a liquid medium in the presence of microbial pathogens [27]. PR-10 genes share a high sequence similarity and form a homogeneous group. Homogeneity is believed to be maintained by concerted evolution [35]. Arrangements of PR-10 genes into clusters, such as found for Mal d 1 genes in apple, may facilitate concerted evolution [22].
Several Bet v 1 isoforms have been described for B. pendula [1,32-34,36], including both allergenic and hypoallergenic isoforms [37]. Individual B. pendula trees have the genetic background to produce a mixture of Bet v 1 isoforms with varying IgE-reactivity [32]. The relative abundance of individual isoforms at the protein level will influence the allergenicity of the pollen. Molecular masses and sequences of tryptic peptides from Bet v 1 can be determined by Q-TOF MS/MS [38]. The recently developed Q-TOF LC-MSE method enables peptide identification, but has the additional advantage of being able to determine relative abundances of peptides in a single run [39]. By quantifying isoforms with a known IgE-reactivity [37], the allergenicity of particular birch trees can be predicted. The existence of allergenic and hypoallergenic isoforms indicates that PR-10 isoforms vary in allergenicity, and some PR-10 isoforms do not bind IgE at all. This has already been demonstrated for two truncated Bet v 1 isoforms [33]. Therefore, not all PR-10 isoforms are necessarily isoallergens.
Knowledge on the allergenicity of birch species may facilitate selection and breeding of hypoallergenic birch trees. To investigate the presence and abundance of Bet v 1 isoforms in Betula species that are potential crossing material, we: (I) cloned and sequenced PR-10 alleles from eight representative Betula species to detect PR-10 genes at the genomic level, (II) applied Q-TOF LC-MSE to identify the pollen-expressed Bet v 1 genes, (III) determined relative abundances of isoforms in the pollen proteome, and (IV) compared these isoforms to isoforms with a known IgE-reactivity.
Results
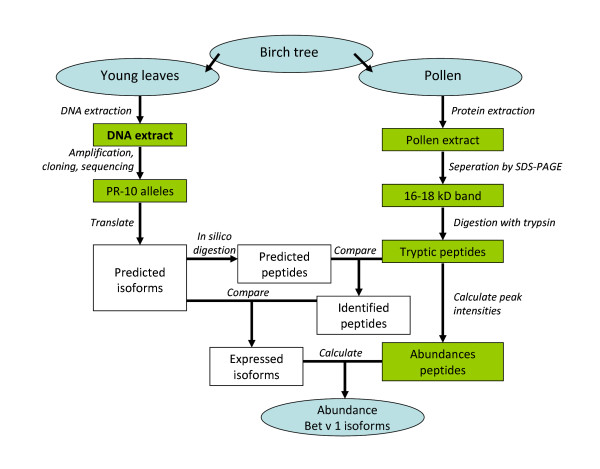
This study encompasses several experimental and analytical steps, involving both genomics and proteomics. All main steps have been summarized in Fig. 1.
Figure 1.
Study workflow diagram. This diagram gives an overview of the experimental steps (green boxes) and analyses (white boxes) performed in this study.
PR-10 subfamilies
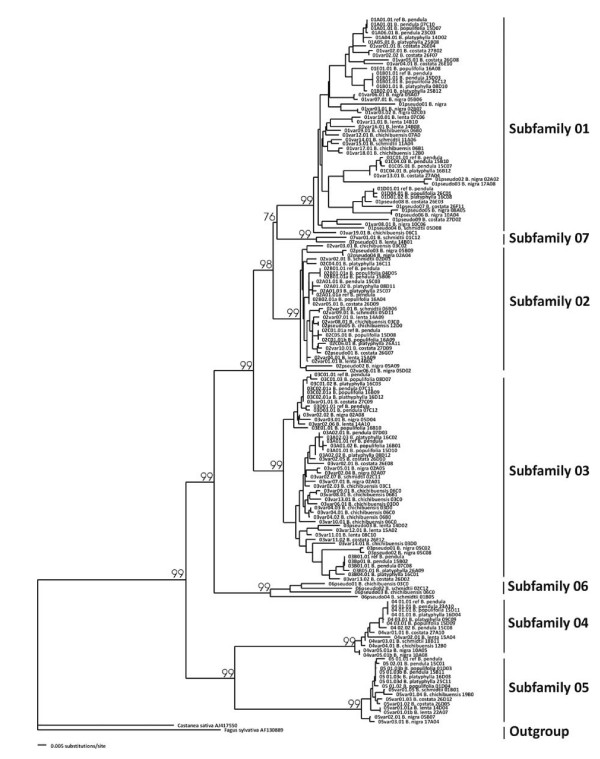
We examined eight Betula species for the presence of PR-10 genes by sequencing 1029 individual clones in both directions (Table 1). Sequences that contained PCR artifacts were excluded by combining information from independent PCRs. The Open Reading Frames (ORF) of the sequences were highly conserved, making the alignment straightforward. The consensus sequence of the exon had 452 positions excluding the 31 bps in the primer regions. 228 out of the 274 variable consensus positions were phylogenetically informative. The sequences grouped into seven well-supported clusters in the Neighbor Joining (NJ) tree (Fig. 2). Five clusters coincided with the division between subfamilies as found in B. pendula [32]. Two new subfamilies (06 and 07) were identified, which occurred only in two species, contrary to the previously described subfamilies 01 to 05 that were found in all species (Table 1). In all sequences, an intron was located between the first and second nucleotide of codon 62. This intron was highly variable in length and composition, which was an additional characteristic for inferring the proper subfamily. Intron sequences were excluded from the phenetic/phylogenetic analysis because introns evolve at a different speed compared to exons.
Table 1.
Number of identified PR-10 sequences in nine birch species.
| Species |
Number of sequenced clones |
Subfamily 01 |
Subfamily 02 |
Subfamily 03 |
Subfamily 04 |
Subfamily 05 |
Subfamily 06 |
Subfamily 07 |
Total |
||||||||
| Seqs | Genes | Seqs | Genes | Seqs | Genes | Seqs | Genes | Seqs | Genes | Seqs | Genes | Seqs | Genes | Seqs | Genes | ||
| Subgenus Betulaster: | |||||||||||||||||
| B. nigra | 155 | 10 | 6 | 4 | 3 | 7 | 4 | 2 | 1 | 2 | 1 | - | - | - | - | 25 | 15 |
| Subgenus Neurobetula: | |||||||||||||||||
| B. chichibuensis | 170 | 5 | 4 | 3 | 2 | 10 | 7 | 1 | 1 | 1 | 1 | 2 | 2 | - | - | 22 | 17 |
| B. schmidtii | 184 | 3 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 12 | 10 |
| Subgenus Betulenta: | |||||||||||||||||
| B. lenta | 106 | 3 | 2 | 3 | 2 | 4 | 4 | 1 | 1 | 2 | 1 | - | - | 1 | 1 | 14 | 11 |
| Subgenus Betula: | |||||||||||||||||
| B. costata | 103 | 9 | 8 | 3 | 2 | 5 | 5 | 1 | 1 | 2 | 1 | - | - | - | - | 20 | 17 |
| B. pendula | 102 | 5 | 3 | 2 | 2 | 5 | 4 | 2 | 1 | 2 | 1 | - | - | - | - | 16 | 11 |
| B. plathyphylla | 103 | 6 | 4 | 4 | 3 | 6 | 3 | 2 | 1 | 2 | 1 | - | - | - | - | 20 | 12 |
| B. populifolia | 106 | 4 | 4 | 4 | 2 | 5 | 3 | 2 | 1 | 2 | 1 | - | - | - | - | 17 | 11 |
| B. pendula reference*2 | - | - | 4 | - | 3 | - | 4 | - | 1 | - | 1 | - | - | - | - | - | 13 |
The number of clones sequenced in both directions and the number of identified sequences and genes are shown per species.1 Subfamily 01 to 05 were previously identified [32], while subfamily 06 and 07 are new. Homology to mRNA sequences suggests that subfamily 01 and 02 are expressed in pollen.
*1 Species were diploid (2n) as measured by flow cytometry. The identification of alleles of a single gene is based on the criterion of having > 98.5% sequence similarity, and by allowing maximally two alleles per gene.
*2 Genes identified in B. pendula [32].
Figure 2.
Grouping of PR-10 sequences into subfamilies. Clustering of the PR-10 sequences from eight Betula species in a Neighbor Joining tree with Kimura two-parameter distances. The sequences group into seven subfamilies. Bootstraps percentages on the branches indicate support for these groups.
PR-10 sequences and genes
We recovered 12 to 25 unique PR-10 sequences per species, adding up to 146 sequences in total (Table 1). Out of the 134 unique sequences, over 100 sequences have never been described before. B. pendula, B. plathyphylla and B. populifolia are closely related members of the subgenus Betula and consequently had multiple alleles in common. These species shared one allele with B. costata, which is another member of the subgenus Betula. We applied a predefined cut-off level of 98.5% to attribute all sequences to different genes, while allowing maximally two alleles per gene per species. These criteria coincided in the majority of cases, but several genes of B. chichibuensis in the large cluster in subfamily 03 and of B. lenta in subfamily 02, and the genes 02A/02B and 03C/03D in B. pendula were more than 98.5% similar. Table 1 shows the total number of identified PR-10 genes per species. Out of the 13 genes that have previously been identified in B. pendula (Table 1; Fig. 2), 11 genes were recovered from the newly sequenced B. pendula cultivar 'Youngii'. This study identified no new genes in this cultivar. This indicates that the majority of genes has been recovered by sequencing over 100 clones per species, and that only a small number of genes might be missing in the dataset.
Homologues of the PR-10 genes of B. pendula were identified in B. populifolia and B. plathyphylla. Sequences from these species were labeled according to the procedure described by Gao et al. [22] that was previously used for B. pendula [32]. These labels consist of the subfamily's number, followed by a letter for each distinct gene, then a number for each unique protein variant and an additional number referring to silent mutations. When applicable, an additional letter indicates variations in the intron. The PR-10 genes in B. costata displayed a considerable degree of homology to the genes in B. pendula, but differentiating homologues and paralogues was not always possible. It was not possible to differentiate between homologues and paralogues of the PR-10 genes in B. lenta, B chichibuensis, B. nigra, and B. schmidtii. Rather than developing a separate denomination scheme for each species, we labeled sequences with the PR-10 subfamily number, followed by a number for each unique protein variant and an additional number referring to silent mutations. This facilitates the protein analysis which distinguishes protein variants rather than separate alleles or genes.
The PR-10 gene copy number varied between different birch species. This is caused by evolutionary processes such as duplication, extinction, and recombination. The overall clustering pattern appears to reflect a combination of such events. Genes from the same species tend to group close to each other on several positions in the NJ tree (Fig. 2). Examples are the clusters of highly similar sequences from B. costata in subfamily 01 and from B. chichibuensis in subfamily 03, which either reflect unequal crossing-over, gene conversion or duplication events. The B. populifolia genome harbors two clear examples of unequal crossing-over. Allele 01E01.01 is a recombination between the 01A gene and the 01B gene. The first part matches exactly to allele 01A01.01, while the second part differs by 1 SNP from 01B01.01 with position 267 of the ORF as the point of recombination. Both original genes were also present. Similarly, allele 03E01.01 is a recombination between the 03B gene and the 03D gene. In this case, the recombination probably occurred without gene duplication, since the original 03B gene, as present in B. pendula, was absent.
PR-10 protein predictions
Not all PR-10 alleles will be expressed as a full-sized protein. 112 unique sequences had an intact ORF, while the remaining 22 sequences contain early stop codons or indels in the ORF that result in frame shifts followed by an early stop codon. The latter sequences were denoted as pseudogenes, although it cannot be excluded that these sequences produce truncated proteins. We calculated Ka/Ks ratios within each subfamily. The suspected pseudogenes displayed higher Ka/Ks ratios than the alleles with an intact ORF in the subfamilies 01, 02 and 03 (Table 2). This points to an alleviated selection pressure in the pseudogenes. The other PR-10 subfamilies do not contain sufficient numbers of both genes and pseudogenes to perform this comparison. The majority of sequences had 5' splicing sites of AG:GT and 3' splicing sites of AG:GC, AG:GT or AG:GA, which is in concordance with known motifs for plant introns. Notable exceptions were: an AC:GT (B. schmidtii, 01pseudo04) and an AG:AT (B. nigra, 04var05.01a) 5' splicing site, an AC:GC (B. schmidtii, 01pseudo04) and a TG:GC (B. nigra, 02pseudo04) 3' splicing site, and two deletions (B. costata, 01pseudo05 and 02pseudo01) at the 3' end of the intron. Except for the AG:AT splicing site, all exceptions belonged to sequences that were denoted as pseudogenes, providing additional evidence for these designations.
Table 2.
Sequence conservation within subfamilies of the PR-10 family among eight Betula species.
| Subfamily | 01 | 02 | 03 | 04 | 05 | 06 | 07 |
| Sequences with an intact ORF | |||||||
| n = | 33 | 19 | 39 | 6 | 14 | 0 | 1 |
| Ka/Ks ratio | 0.18 | 0.27 | 0.10 | 0.36 | 0.09 | n. d. | n. d. |
| Range substitutions | 0 – 16 | 0 – 9 | 0 – 8 | 0 – 6 | 0 – 4 | n. d. | n. d. |
| Average # substitutions | 7.0 | 3.1 | 2.8 | 3.3 | 0.9 | n. d. | n. d. |
| Pseudogene sequences | |||||||
| n = | 9 | 5 | 3 | 0 | 0 | 4 | 1 |
| Ka/Ks ratio | 0.38 | 0.30 | 0.20 | n. d. | n. d. | 0.57 | n. d. |
n = number of unique sequences. Ka/Ks ratio = ratio between non-synonymous and synonymous mutations. Range substitutions = minimum and maximum number of amino acid substitutions in pair wise comparisons between sequences of the same subfamilies. n. d. = not determined.
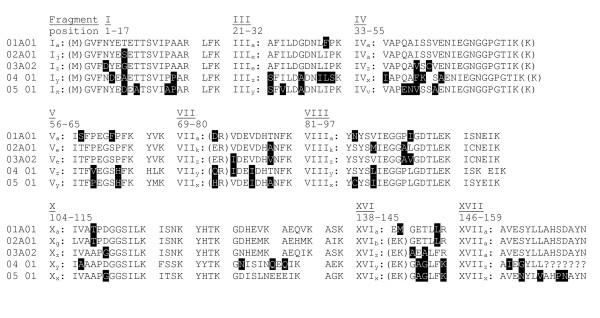
Depending on the subfamily, Ka/Ks ratios ranged from 0.09 to 0.36 for sequences with an intact ORF (Table 2), indicating strong purifying selection. The PR-10 alleles in birch encode a putative protein that consists of 160 amino acids, yielding a relative molecular mass of approximately 17 kDa. The only exception is 01var17.01 in B. chichibuensis, which contains an indel that results in the deletion of two amino acids. The allelic variation is lower at the protein level than at the nucleic acid level, which is consistent with the low Ka/Ks ratios. Hence, the 112 unique genomic sequences encode 80 unique isoforms. The PR-10.05 gene is an extreme example for which only four putative isoforms are predicted, despite the presence of 14 allelic variants. One of these isoforms is predicted in all species except B. nigra. Parts of the PR-10 protein sequences are highly conserved, as is demonstrated in the amino-acid alignment of five PR-10 isoforms (one per subfamily) from B. pendula (Fig. 3). The most prominent region lies between Glu42 and Ile56 and contains only a single amino acid variation among all 80 isoforms. A phosphate-binding loop with the sequence motive GxGGxGx characterizes this region. Additional conserved Glycine residues are present at positions 88, 89, 92, 110 and 111.
Figure 3.
Alignment of theoretical tryptic peptides of PR-10 proteins in B. pendula 'Youngii'. For clarity, one amino acid sequence is shown per subfamily. Only those fragments that are large enough to be detected by Q-TOF LC-MS/MS are labeled. Variable amino acids are marked in black.
Bet v 1 expression in pollen
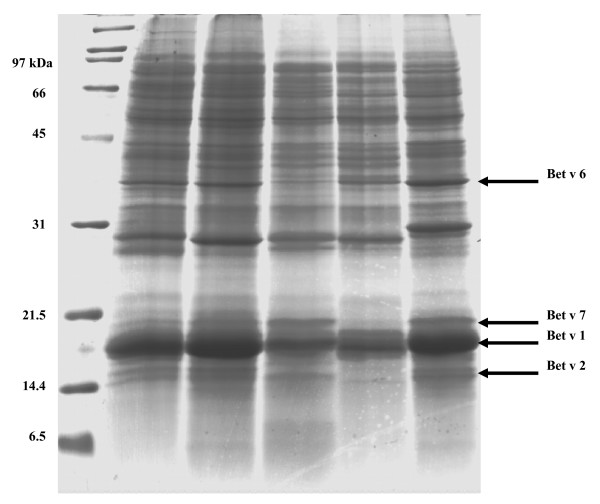
The presence of Bet v 1-like proteins was examined in pollen of B. nigra, B. chichibuensis, B. lenta, B. costata and B. pendula 'Youngii'. Pollen proteins were solubilized in an aqueous buffer and analyzed by SDS-PAGE. Each sample displayed an intense protein band after CBB-staining at the expected molecular mass of Bet v 1, between 16–18 kDa (Fig. 4), while other intense bands were visible at 28 kDa and 35 kDa. No 16–18 kDa band was visible when the pellet that remained after extraction was separated by SDS-PAGE (not shown), indicating the efficiency of the extraction procedure with regard to Bet v 1.
Figure 4.
SDS-PAGE analysis of birch pollen extracts. (Lane 1) B. chichibuensis, (2) B. costata, (3) B. nigra, (4) B. lenta and (5) B. pendula. Bands of allergens that were analyzed and identified with Q-TOF LC-MS/MS are indicated by arrows. (M) LMW size marker proteins.
To establish the identity of the proteins in the 16–18 kDa band, we cut out this band from the lane of B. pendula (Fig. 4) and performed in-gel digestion with trypsin. Q-TOF LC-MS/MS analysis of the tryptic peptides yielded multiple Bet v 1 isoforms (details given below). The bands just above and below the 16–18 kDa band were also sequenced and checked for the presence of Bet v 1. The lower band at 14 kDa contained birch profilin (Bet v 2; GenBank AAA16522; 2 peptides, coverage 24%) and contained no Bet v 1 fragments. The higher band at 19 kDa contained birch cyclophilin (Bet v 7; CAC841116; 3 peptides, coverage 28%) and some minor traces of Bet v 1 (Bet v 1a; CAA33887; 1 peptide, coverage 14%). Bollen et al. [4] detected a band of ~35 kDa when purified Bet v 1 was analyzed by SDS-PAGE, consisting of (dimeric) Bet v 1. We identified the intense band at ~35 kDa in our B. pendula extract as isoflavone reductase (Bet v 6; GenBank AAG22740; 19 peptides, coverage 49%) and detected no Bet v 1 fragments in this band.
Analysis of Bet v 1 isoforms by Q-TOF LC-MSE
The tryptic digests of the 16–18 kDa bands were examined in detail to elucidate the expression of separate Bet v 1 isoforms in pollen. Trypsin cleaves proteins exclusively at the C-terminus of Arginine and Lysine. Fig. 3 shows an example of the fragments I to XVII that are theoretically formed after tryptic digestion of isoforms from the subfamilies 01 to 05. Isoforms of different subfamilies can be discriminated by several fragments on the basis of peptide mass and sequence. The number of discriminating fragments becomes lower for Bet v 1 isoforms within a subfamily. A new mass spectrometric technique called Q-TOF LC-MSE allows simultaneous identification and quantification of peptides (see Method section for details). A distinct feature of the LC-MSE procedure is that information is obtained for all peptides. This contrasts MS/MS, in which a subset of peptides is selected for fragmentation. A software program analyses the data, while using a search database for interpretation of the fragmentation spectra. This database contained the sequence information of all PR-10 isoforms described in this paper and of previously described PR-10 isoforms from B. pendula [32].
The LC-MSE results indicated that PR-10 proteins of subfamily 01 and 02 are expressed in the pollen of the five examined birch species. We found no evidence for the expression of genes from subfamilies 03 to 07 in pollen. For example, we identified 22 Bet v 1 peptide fragments in B. pendula (Table 3), all of which were predicted from the gDNA sequences. Eight detected peptides could distinguish between isoforms of subfamily 01 and 02. The B. pendula genome contains seven genes from subfamily 01 and 02. The expression of four of these (01A, 01B, 01C, 02C) was confirmed (Table 3). Sequence coverage of the expressed isoforms amounted to 71 to 79% (Table 3). Four peptides were specific for isoform 01B01, while one peptide was specific for isoform 02C01. Two peptides were specific for both isoforms of gene 01A, while two others were specific for both isoforms of gene 01C. Isoforms 02A01 and 02B01 could not be separated, so either one or both of them are expressed. Table 3 also shows the peptide fragments that were long enough to be detected in the tryptic digest, but were not observed. Information on absent fragments can be used to exclude expression of particular isoforms, such as isoform 01D01 in B. pendula.
Table 3.
Peptides fragments of PR-10 isoforms in pollen from five Betula species as identified by Q-TOF LC-MSE.
| Species | Fragment | I | III | IV | V | VII | VIII | X | XVI | XVII | Sequence coverage | |
| Isoform | Gene | *2 | ||||||||||
| B. pendula | 01A01 | 1A | A | a | a | a | a | A | a? | a | a | 79% |
| 01A06 | 1A | A | a | a | b | a | A | a? | a | a | 79% | |
| 01B01 | 1B | B | B | a | b | a | C | B | a | a | 79% | |
| 01C04 | 1C | D | a | a | a | a | D | c? | a | a | 71% | |
| 01C05 | 1C | D | a | a | a | a | D | c? | a | a | 71% | |
| 01D01 | 1D | (E) | a | a | (C) | (C) | (E) | a? | a | a | - | |
| 02A01 | 2A | j | e | a | e | k | k | g? | (B*3) | a | 74% | |
| 02B01 | 2B | j | e | a | e | k | k | g? | (c*3) | a | 74% | |
| 02C01 | 2C | j | F | a | e | k | k | a? | (c*3) | a | 74% | |
| 03 *1 | (C), (z) | e | (z), (Y) | e | (Z), (y) | (z), (Y) | (z), (Y) | (z) | a | - | ||
| 04 *1 | (Y) | (Z) | (X), (W) | (Z) | (X) | (X) | (X) | (Y) | (Z), (Y) | - | ||
| 05 *1 | (X) | (Y) | (V) | (Y) | (W) | (W) | (W) | (X) | (X) | - | ||
| B. chichibuensis | 01var09 | 1A | a | a | a | a | a | d | a? | a | a | 79% |
| 01var12 | 1B | C | a | a | a | a | d | a? | a | a | 79% | |
| 01var17 | 1C | a | a | a | a | H | (H) | c? | a | a | - | |
| 01var18 | 1C | a | a | a | a | H | d | c? | a | a | 71% | |
| 01var19 | 1D | (E) | a | a | (C) | (J) | (E) | a? | a | a | - | |
| 02var03 | 2A | j | e | B | e | k | k | a? | (c*3) | a | 74% | |
| 02var08 | 2B | j | F | a | e | k | k | a? | (c*3) | a | 74% | |
| B. costata | 01var01 | 1A | F | a | a | a | a | c | a | a | a | 79% |
| 01var02 | 1B | a | a | a | a | a | c | a | a | a | 79% | |
| 01var04 | 1C | a | (C) | a | b | (E) | c | (D) | a | a | - | |
| 01var05 | 1D | a | a | a | b | a | c | (E) | a | a | 71% | |
| 01var13 | 1E | D | a | a | a | a | D | a | a | a | 79% | |
| Unknown | E | ? | ||||||||||
| 02var05 | 2A | j | (J) | a | e | k | k | a | (c*3) | a | - | |
| 02var10 | 2B | j | G | a | e | k | k | a | (c*3) | a | 74% | |
| Unknown | F | ? | ||||||||||
| B. lenta | 01var10 | 1A | G | A | a | (A) | (F) | d | a | (a*3) | a | 60% |
| 01var11 | 1A | G | A | a | (A) | (G) | d | a | (a*3) | a | 60% | |
| 01var16 | 1B | B | D | a | D | A | d | a | (a*3) | a | 74% | |
| 02var01 | 2A | j | e | a | e | k | k | a | (c*3) | a | 74% | |
| 02var04 | 2B | j | H | a | e | k | k | a | (c*3) | a | 74% | |
| 02var07 | 2C | j | F | a | e | k | k | a | (c*3) | a | 74% | |
| B. nigra | 01var03 | 1A | B | a | a | B | a | F | a | a | a | 79% |
| 01var06 | 1B | a | a | a | a | - | C | a | a | a | 74% | |
| 01var07 | 1C | a | a | a | a | a | C | a | a | a | 79% | |
| 01var08 | 1D | a | a | a | a | a | D | F | a | a | 79% | |
| 02var06 | 2A | J | K | a | e | K | K | a | (c*3) | a | 74% | |
| unknown | F | ? | ||||||||||
Each isoform is displayed on a separate line. When isoforms are encoded by the same gene this is indicated in the third column. Note that gene labels in one species do not correspond to gene labels in other species. Peptide fragments are shown at the top of the table and are labelled with Roman numbers as indicated in Fig. 3. Each variant of these fragments is displayed in the Table by a letter. Bold capital letters indicate that a fragment is unique for the isoforms of a particular gene. Bold italic letters indicate that a fragment is unique for the isoforms of a particular subfamily. Letters displayed between brackets indicate that a particular fragment was predicted, but was absent in the PR-10 mixture. Finally, the last column displays the coverage of the total protein sequence, including the fragments that were too small to be detected (II, VI, IX, XI, XII, XIII, XIV, XV). Fig. 3 displays the representative amino acid sequences of the isoforms 01A01 and 02A01.
*1 The isoforms in subfamily 03 to 05 were summarized into a single row and not displayed for the other species, because specific peptides were not detected in any of the species.
*2 Fragments Xa and Xg have exactly the same mass and cannot be distinguished. The peak of peptide Xc overlaps with the first isotope peak of peptide Xa = g because they differ exactly 1 Da in size and have the same charge. As a consequence, Xc cannot be identified separately.
*3 The XVI-peptides are not always detected because of their small size.
Altogether, at least 4 to 6 isoforms were expressed in each of the five examined species. In total, the presence of unique peptides confirmed the expression of 14 isoforms among the five species in total (Table 3). An additional 15 isoforms lacked one or more unique peptides to distinguish them from other isoforms or from each other, but several of these must be expressed. The expression of five isoforms was ruled out, because multiple unique peptides from these variants were lacking from the peptide mixture. Two identified peptides in B. costata and one peptide from B. nigra did not match to any sequence that was recovered from these species. These peptides belong to "unknown isoforms" (Table 3) and this indicates that the sequences that encode these isoforms are missing from the dataset. Finally, conflicting evidence was found for expression of the isoforms 01var10 and 01var11 in B. lenta. Two peptides that were unique for these isoforms were detected, while three peptides that were expected if the isoforms would be expressed were lacking. Expression of an allele that is missing from our dataset is a more likely explanation than the expression of 01var10 or 01var11.
Quantification by Q-TOF LC-MSE
We determined the relative amounts of individual Bet v 1 isoforms in pollen from B. pendula 'Youngii' (Table 4). This information can be deduced from the peak intensities of Bet v 1 peptides in the tryptic digest. Not all identified fragments can be used for quantification, because the peak detection algorithm groups peaks with highly similar masses and retention times together, also when they might belong to different fragments. For example, fragment Ia (1854,91 Da) and VIIa (1854,89 Da) have a retention time that is marginally different, causing a strong overlap in peak area. The relative amounts of two isoforms could be estimated directly: peptide IIIf is unique for isoform 02C01 and comprises 17% of all fragment III-variants, while peptides IIIb and Xb are unique for 01B01 and comprise 18–19% of all fragment III and X-variants. The isoforms 02A01 and 02B01 could not be separated, but together they comprise 13% of the mixture based on fragment IIIe. The relative amounts of the other isoforms were estimated indirectly. Isoform 01A06 and 01B01 share fragment Vb, which comprises 23% of all fragment V-variants. 01A06 is thus estimated to comprise 4–5% of the mixture. The ratio between 01B01 and 01C04 plus 01C05 can be deduced from fragment Ib. 01C04 plus 01C05 are thus estimated to comprise 6% of the mixture. This leaves 40–41% of the total amount of Bet v 1 for isoform 01A01.
Table 4.
Quantification of identified peptides by Q-TOF LC-MSE in the pollen of B. pendula 'Youngii'.
| Fragment | I*1 | III | IV | V | VII | VIII*1 | X *2 | XVII | Direct coverage estimate | Indirect coverage estimate | Subfamily | Direct estimate | |
| Isoform | Gene | ||||||||||||
| 01A01 | 1A | Ia: n.q. | IIIa: 51 | IVa: 100 | Va: 46 | VIIa: 75 | VIIIa: n.q. | Xa+g+c: 82 | XVIIa: 100 | - | 4–41% | 01 | 68–75% |
| 01A06 | 1A | Ia: n.q. | IIIa: 51 | IVa: 100 | Vb: 23 | VIIa: 75 | VIIIa: n.q. | Xa+g+c: 82 | XVIIa: 100 | - | 4–5% | ||
| 01B01 | 1B | Ib: 69 | IIIb: 19 | IVa: 100 | Vb: 23 | VIIa: 75 | VIIIc: n.q. | Xb: 18 | XVIIa: 100 | 18–19% | - | ||
| 01C04/01C05 | 1C | Id: 31 | IIIa: 51 | IVa: 100 | Va: 46 | VIIa: 75 | VIIId: 100 | Xa+g+c: 82 | XVIIa: 100 | - | 6% | ||
| 01D01 | 1D | Ie: 0 | IIIa: 51 | IVa: 100 | Vc: 0 | VIIc: 0 | VIIIe: 0 | Xa+g+c: 82: | XVIIa: 100 | 0% | - | ||
| 02A01/02B01 | 2A | Ia: n.q. | IIIe: 13 | IVa: 100 | Ve: 32 | VIIk: 25 | VIIIk: n.q. | Xa+g+c: 82 | XVIIa: 100 | 13% | - | 02 | 25–32% |
| 02C01 | 2C | Ia: n.q. | IIIf: 17 | IVa: 100 | Ve: 32 | VIIk: 25 | VIIIk: n.q. | Xa+g+c: 82 | XVIIa: 100 | 17% | - | ||
Numbers indicate the relative amount of fragment variants compared to the total amount of homologues fragments. Amounts were averaged over the two duplicates. Note that quantification was not possible for all peptide variants1,2 and that the displayed abundances indicate the relative amounts among those variants that could be quantified. n.q. = not possible to quantify.
* 1 Quantification was not possible for all the peptide variants, because Ia (1854,91 Da) and VIIIa (1854,89 Da), and Ij (1840,89 Da) and VIIIc (1840,88 Da) had a similar mass. Fragment VIIIk overlaps with a keratin peptide.
* 2 Fragments Xa and Xg have exactly the same mass and cannot be distinguished. The peptide peak of Xc overlaps with the first isotope peak of peptide Xa = g because they differ exactly 1Da in size and have the same charge. Xc cannot be identified as a result.
Isoform 01A01 is identical to isoform Bet v 1a, which had the highest IgE-reactivity in several tests performed by Ferreira et al. [37]. Pollen of B. costata, B. nigra and B. chichibuensis contained isoforms that are highly similar to Bet v 1a and differ by only 1–3 amino acids from this isoform. We determined the expression of individual Bet v 1 isoforms in a similar fashion as reported for B. pendula. The Bet v 1a-like isoforms were estimated to comprise 38% (B. chichibuensis), 36–44% (B. nigra) and 36–41% (B. costata) of the total amount of Bet v 1. B. lenta differed from the other species, because the isoform with the highest similarity to Bet v 1a differed by seven amino acids. This isoform was estimated to comprise 12–19% of the total amount of Bet v 1. The expression of subfamily 01 isoforms relative to subfamily 02 isoforms was another major difference between B. lenta and the other species. In B. lenta, subfamily 02 accounted for 74–83% of the total amount of Bet v 1, compared to 25–40% in B. pendula, B. nigra and B. chichibuensis and 49–56% in B. costata.
Discussion
PR-10 gene family organization and evolution
The presence and diversity of Bet v 1 and closely related PR-10 genes in eight birch species was established by amplification and sequencing of more than 100 clones per species. The eight species belong to four different subgenera/groups in the genus Betula [13] and thereby represent a large part of the existing variation within the genus. Each birch species contains PR-10 genes, as could be expected given the broad range of plant species in which PR-10 genes are found [21-23]. The PR-10 genes grouped into subfamilies, as previously reported for B. pendula [32]. Five subfamilies were recovered from all species. Two new subfamilies were identified, but these were each restricted to two species and were mostly composed of pseudogenes.
The PR-10 subfamily has a complex genomic organization. Differentiating between paralogues and homologues was not possible beyond closely related species. One likely explanation is concerted evolution, for which cladistic evidence was found (Fig. 2). Concerted evolution causes genes to evolve as a single unit whose members (occasionally) exchange genetic information through gene conversion or unequal crossing-over [40]. Tandemly arranged genes have increased conversion rates, while such an arrangement is a prerequisite for the occurrence of unequal crossing-over [41]. Most PR-10 genes in apple are arranged in a duplicated cluster [22], thus facilitating the main mechanisms for concerted evolution. We obtained two alleles that appear the direct result of unequal crossing-over between Bet v 1 genes. On a higher taxonomic level, cladistic evidence for concerted evolution is present in the overall gene tree of the PR-10 family [35], as sequence divergence is generally smaller between different genes from the same species than between genes from different species.
Nei and Rooney [42] suggested that a combination of recent gene duplications and purifying selection could also explain why tandem gene duplicates appear similar. In their model of birth-and-death evolution of genes, new genes arise due to gene duplications, evolve independently while undergoing purifying selection, and go extinct after becoming non-functional. Pseudogenes are characteristic for this process. The low Ka/Ks ratios clearly point to the occurrence of purifying selection. Pseudogenes are a common feature among the PR-10 genes in birch, since we recovered them from six out of eight species. As much as one-third of the recovered alleles in B. nigra had an interrupted ORF. We did not determine the potential expression of these alleles, since truncated isoforms would have migrated outside the 16–18 kDa band in the SDS-PAGE. None were, however, detected in the 14 kDa band. Basically, all ingredients for the "birth-and-death" model are present, except that independent evolution is questionable due to the presences of duplicates that resulted from unequal crossing-over. Moreover, the clustering of for example the B. chichibuensis alleles (Fig. 2) would suggest an extremely high number of recent duplications. Both processes of "birth-and-death" and concerted evolution may, therefore, be active in the PR-10 gene family. Regardless of the evolutionary processes, its outcome is clear: PR-10 proteins are homogenous as a group and even stronger so within subfamilies. The high homogeneity allowed us to use Q-TOF LC-MSE to quantify the relative expression of separate Bet v 1 isoforms, because large differences in amino acid composition would have distorted the quantification.
Bet v 1 expression
Which PR-10 genes are actually expressed in pollen and are thereby the true Bet v 1 allergens? We used Q-TOF analysis to investigate the expression of Bet v 1 isoforms in pollen of five Betula species. Isoforms from subfamily 01 and 02 were identified in birch pollen, confirming predictions based on mRNA expression [1,33,34]. The single gene in subfamily 05 that was present in all eight birch species, is homologous to ãpr10c, which has a high basal transcription level in roots and a relatively lower basal transcription level in leaves [27,28,43]. Its expression is induced by copper stress [30] and during senescence in leaves [44]. Regarding subfamily 03, the genes PR-10.03C and 03D (= γpr10a and γpr10b) in B. pendula become transcriptionally upregulated upon infection of the leaves with fungal pathogens [27]. Their transcription is induced by wounding or auxin treatment in roots [28,43]. No data have been reported about the expression of the sequenced PR-10 genes in subfamilies 04, 06 and 07.
The pollen-expressed Bet v 1 genes are transcribed during the late stages of anther development [45], but which factors induce transcription is unknown. Bet v 1 is an abundant pollen protein that has been estimated to encompass 10% of the total protein in B. pendula pollen [46]. The Bet v 1 band was the most intense band in the SDS-PAGE gels of birch pollen extracts. Its exact abundance is difficult to estimate due to differences in extraction efficiency between different proteins. However, given the low amount of residual protein in the pellet, our results suggest that the abundance of Bet v 1 is higher than 10% of the total protein content and is likely to exceed 20%. The occurrence of Bet v 1 isoforms in B. pendula has previously been studied in a mixture of pollen from different trees by Swoboda et al. [34]. They analyzed tryptic digests of purified Bet v 1 isoforms by Plasma Desorption Mass Spectrometry (PDMS), a technique that only reveals peptide masses. We examined pollen from individual trees and analyzed the tryptic digests by Q-TOF LC-MSE, which reveals total masses of peptides and the underlying amino acid sequences, based on available sequence information. The ability to determine the peptide sequences yields more accurate information on expression of individual isoforms. We demonstrated that at least 4 to 6 isoforms were expressed in the pollen of one single tree of the birch species B. pendula, B. nigra, B. chichibuensis, B. lenta and B. costata. The actual number is likely to be higher since we could not discriminate each individual isoform due to the high similarity between some isoforms.
Q-TOF LC-MSE has the advantageous ability to simultaneously separate, identify and quantify peptide fragments. A similar strategy has recently been followed by Chassaigne et al. [47]. They identified five peanut-specific peptide ions that were used as specific tags for the peanut allergenic proteins Ara h 1, Ara h 2, and Ara h 3. The relative intensity of the specific peptides even provided information on the processing history of the peanut material. Napoli et al. [48] also used mass spectrometry to analyze an Ole e 1 mixture of multiple isoforms and their post-translational modifications, which could not be separate completely by 2-Dimension gel electrophoresis. A disadvantage of using Q-TOF LC-MSE instead of Q-TOF LC-MSMS in combination with 2D gel electrophoresis and Western blotting – in which allergic sera and specific anti-IgE antibodies are employed – is that our method does not distinguish IgE-binding isoforms from non-IgE-binding isoforms. Therefore, not all described PR-10 isoforms are necessarily true isoallergens.
We included no purification step in the extraction procedure apart from protein separation on SDS-PAGE. This minimizes the chance that certain isoforms are lost during purification, but the Bet v 1 protein band might be contaminated with other pollen proteins with a similar mass. Three peptides of the pollen allergen Bet v 7 were detected in the 16–18 kDa band, but the amount of Bet v 7 was estimated to be less than 2% of the amount of Bet v 1, based on the peak intensities of these peptides. All peptides with high peak intensities could be attributed to Bet v 1 isoforms. Full sequence coverage of Bet v 1 isoforms cannot be achieved by using only trypsin as a protease, as smaller peptides will be lost during peptide extraction from the SDS-PAGE gel. Proteases that cleave at other sites will yield peptides that cover part of the missing protein sequence. Coverage with Q-TOF LC-MSE was 71–79% for the B. pendula isoforms, which is higher than the 57–60% coverage reported for Q-TOF MS/MS [38].
Swoboda et al. [34] estimated that, based on PDMS peak areas of peptides, the relative amount of Bet v 1a (= PR-10.01A01) in the pollen mixture from several B. pendula trees was at least 50% of the total amount of Bet v 1. Ferreira et al. [37] estimated the relative amounts of different Bet v 1 isoforms by NH2-terminal sequencing of purified natural Bet v 1 and reported a ~2:2:1 ratio for isoforms that respectively contain Ser, Thr and Ile at the 7th amino acid position. This would correspond to expression of the isoforms 02A01+02B01+02C01: 01A01+01A06: 01B01+01C04+01C05 in pollen of B. pendula 'Youngii'. When we sum our results in this way, a ratio of 30%:45%:25% is obtained. The similarity between our results and previously obtained estimates suggests that the quantities obtained from B. pendula 'Youngii' are also representative for other B. pendula trees.
Allergenicity
Ferreira et al. [37] distinguished Bet v 1 isoforms with a low, intermediate and high IgE-reactivity. Expression of the isoforms 01B01 (= Bet v 1d, low IgE-reactivity), 02C01 (= Bet v 1c, intermediate IgE-reactivity), 01C04 (= Bet v 1f, intermediate IgE-reactivity) and 01A01 (= Bet v 1a, high IgE-reactivity) in the pollen of B. pendula 'Youngii' was confirmed by identification of unique peptides (Table 3). Isoforms of all three levels of IgE-reactivity were abundant and encompassed 35–38% (high), 22–24% (intermediate) and 18–19% (low) of the total amount of Bet v 1. This leaves 17–22% of the total Bet v 1 for isoforms with an unknown IgE-reactivity. We observed similar quantities in two other B. pendula cultivars as well (results not shown). Since B. pendula is known to be highly allergenic, the presence of isoforms with a high IgE-reactivity is apparently of determining influence on its allergenicity. Interestingly, people that are not yet sensitized to B. pendula pollen come into contact with several abundant isoforms. However, the ability of these isoforms to provoke an IgE-mediated response varies in patients that have become sensitized [49,50]. The factors that cause one isoform to develop into having a high IgE-reactivity and another isoform into having a low IgE-reactivity are currently unknown. The abundance of the isoforms may play a role, but as isoforms with intermediate or low IgE-reactivity are also present in considerable quantities, this is unlikely to be the only factor. Recently, Gao et al. [18], investigated the association of allelic diversity of Mal d 1 and allergenicity in ten pedigree-linked apple cultivars, and found that qualitative as well as quantitative factors were involved.
The opportunities for identifying birch trees that only express hypoallergenic isoforms are limited. The isoforms Bet v 1l and Bet v1d (= 01B01) are currently known as hypoallergenic [37]. The crystal-structure of Bet v 1l has been determined [51] and its allergenicity has recently been tested on a large group of patients [50]. However, none of the examined species contained Bet v 1l, despite thorough examination, and Bet v 1l may represent a sequencing artifact or an unexpressed allele. Only B. pendula, B. populifolia and B. plathyphylla contained 01B01. The most similar isoforms in the other species differed by at least five amino acids. In contrast, the highly allergenic isoform 01A01 is expressed in pollen of B. pendula 'Youngii', while the B. populifolia genome also contains the 01A01 sequence. The other Betula species do not harbor Bet v 1a, but B. chichibuensis, B. costata and B. nigra contain isoforms that differ only by 1–3 amino acids from Bet v 1a. A high similarity between isoforms increases the chance that they share epitopes, although a few amino acid substitutions may influence the allergenicity drastically [52-54]. In all species, these isoforms are abundant. B. lenta forms an exception as the isoform most similar to Bet v 1a has a sequence similarity of 95.5% and encodes a protein that differs by seven amino acids.
Conclusion
We identified 12 to 25 unique PR-10 sequences in each of eight different birch species. Application of Q-TOF LC-MSE revealed that genes from two large subfamilies (01 and 02) were expressed in birch pollen. We showed that Q-TOF LC-MSE allowed fast screening of Bet v 1 isoforms in birch pollen by determining presence and relative abundances of individual isoforms. The pollen of four birch species contained a mixture of Bet v 1 isoforms, with abundant levels of isoforms that were similar to isoforms with a high IgE-reactivity. We predict that the allergenic potency of these species will be high. B. lenta (subgenus Betulenta) lacked isoforms with a high similarity to isoforms with a high IgE-reactivity. This species and related species represent the most promising candidates for further screening of hypoallergenicity by for example skin prick tests or nasal challenges.
Methods
Plant Material
We collected young leaves from eight Betula species (Table 1). A recent phylogenetic analysis identified four groups (subgenera) of species within the genus Betula [13]. Each subgenus is represented by at least one species. Four species from the subgenus Betula were included here to cover the variation within this large group. Plant material was collected from the botanical collections of PPO Boskoop (Boskoop, the Netherlands), the Botanical Garden of Wageningen (Wageningen, the Netherlands) and the Von Gimborn Arboretum (Doorn, the Netherlands). Fresh leaf samples were analyzed by flow cytometry (Plant Cytometry Services, Schijndel, The Netherlands) to estimate the ploidy level. Diploid (B. pendula) and tetraploid (B. pubescens) controls were included. All examined accessions were diploid, thus keeping the number of expected sequences per accession small. During the flowering period of birch in April-May 2004, we collected pollen from the same trees for the species B. nigra, B. chichibuensis, B. lenta, B. costata and B. pendula.
PCR, cloning and sequencing
DNA was extracted using the DNeasy Plant Mini kit (Qiagen) according to the manufacturer's instructions. PR-10 alleles were amplified from genomic DNA with two primer pairs that had been tested and used in previous research on B. pendula [32]. PCR amplification with both primer pairs was performed in 20 μl reactions containing 0.1 mM dNTP, PCR Reaction buffer (Eurogentec), 1.5 mM MgCl2, 0.6 μM forward primer, 0.6 μM reverse primer, 0.5 U Taq polymerase (Goldstar), and 20–80 ng template DNA. PCR reactions started with a heating step at 95°C for 15 minutes, followed by 16–24 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 2 min. A final extension step of 10 min at 72°C was added after the last cycle. To reduce the number of PCR recombination artifacts, we used as few PCR cycles as possible. The minimum number of cycles required to generate sufficient product for cloning was assessed by visual inspection of the amplified products on agarose gel.
PCR products were purified with the MinElute PCR Purification Kit (Qiagen). Purified samples were ligated into the pGEM-T easy Vector (Promega) and established in Escherichia coli Subcloning Efficiency DH5α cells (Invitrogen) according to the manufacturer's instructions. White colonies were picked from agar plates and grown overnight at 37°C in freeze medium. We performed PCR-based screening with vector-specific M13 primers. These PCR products were purified with Sephadex G-50 (Millipore). The DYEnamic™ ET Terminator Cycle Sequencing Kit (Amersham) was used for the sequence reactions. We analyzed sequence products on a 96-capillary system (ABI 3730 × l). The genomic Betula sequences have been submitted to GenBank as EU526132–EU526277.
Phenetic/phylogenetic analysis
Potential PCR artifacts (strand switching and base misincorporation) were excluded by retaining only those sequences that were confirmed in independent PCRs. We included one reference sequence per B. pendula gene in the dataset for comparison with previous results [32]. Nucleotide sequences were aligned using CLUSTAL W with a gap penalty of 10 and a gap extension penalty of 2. We excluded primer traces and introns from further analysis. A Neighbor Joining (NJ) tree was constructed with Kimura two-parameter distances. Gaps were treated as missing characters. Bootstrapping was carried out with 1,000 replicates in PAUP 4.0b10 [55]. The outgroup was composed of PR-10 sequences from Castanea sativa (AJ417550) and Fagus sylvatica (AJ130889), which are two related Fagales species. Parsimony analysis was conducted in PAUP as a heuristic search, while using the following options: 100,000 random additions while holding one tree at each step, TBR branch swapping, the MulTrees option switched on, and ACCTRAN for character optimization. A strict consensus tree was calculated for all of the most parsimonious trees. Branch support was assessed by bootstrap analysis comprising 10,000 replicates consisting of 10 random addition sequences with TBR branch swapping. Both analyses produced highly similar results; therefore, only the NJ analysis is shown.
Protein search database
Nucleotide sequences were aligned codon-by-codon. We analyzed general selection patterns at the molecular level using DnaSp 4.00 [56]. The number of non-synonymous (Ka) and synonymous substitutions (Ks) per site were calculated from pair wise comparisons with incorporation of the Jukes-Cantor correction. Nucleotide data were translated. A Fasta database with the resulting protein sequences was used as a search database in the Q-TOF LC-MSE analysis. As sequence information for the primer region was unavailable, we used the GenBank sequences X15877 (subfamily 01), X77265 (02), X77600 (03), and X77601 (05) to fill these gaps in sequences from the respective subfamilies. The initiating Methionine is removed during PR-10 protein synthesis [30,37] and was therefore removed from the predicted proteins. Protein sequences of birch PR-10 isoforms in GenBank (overview in: Schenk et al., 2006), keratin, trypsin and Bet v 7 (AJ311666) were added to the database.
Protein extraction
Fifty mg of pollen were suspended in 1 ml of 0.05 M Tris-HCl (pH 7.5) following Cadot et al. [57], who found that yield and diversity of the extracted allergens are optimal at pH 7.5 for birch pollen. After incubation under constant shaking at room temperature for 1 hr, the pollen extract was centrifuged at 10.000 rpm for 5 min. The pellet was ground with an Eppendorf-fitting pestle. The extract was then shaken for another hour. The supernatant was collected after centrifugation (10.000 rpm; 5 min) and freeze dried for storage.
SDS-PAGE
The freeze-dried protein extract was redissolved in 0.05 M Tris-HCL (pH 7.5) and analyzed with SDS-PAGE to localize Bet v 1-type proteins. Proteins were separated on a 15% w/v acrylamide SDS-PAGE gel with a 5% w/v stacking gel using the Mini-Protean II gel system (Bio-Rad). After staining with Coomassie BB R-250, the gels were scanned and analyzed by Quantity One (Bio-Rad) scanner software. Relative molecular masses were determined with SDS-PAGE Standards broad range markers (Bio-Rad).
The protein bands at a relative molecular mass of 16–18 kDa were cut out of the SDS-PAGE gel and processed essentially according to Shevchenko [58]. Bands were sliced into 1 mm3-pieces. Bands at 14, 19 and 35 kDa were cut out and analyzed as well. Proteins were reduced with DTT and alkylated with iodoacetamide. Gel pieces were dried under vacuum, and swollen in 0.1 M NaHCO3 that contained sequence-grade porcine trypsin (10 ng/μl, Promega). After digestion at 37°C overnight, peptides were extracted from the gel with 50% v/v acetonitrile, 5% v/v formic acid and dried under vacuum.
Q-TOF LC-MS/MS and Q-TOF LC-MSE
Tryptic digests were analyzed by one-dimensional LC-MS in high-throughput configuration using the Ettan™ MDLC system (GE Healthcare), which was directly connected to a Q-TOF-2 Mass Spectrometer (Waters Corporation, UK). Samples (5 μl) were loaded on 5 mm × 300 μm ID Zorbax™ 300 SB C18 trap columns (Agilent Technologies), and peptides were separated on 100 μm i.d. × 15 cm Chromolith CapRod monolithic C18 capillary columns (Merck) at a flow rate of approximately 1 μl/min. A gradient was applied using two solvents. Solvent A contained an aqueous 0.1% formic acid solution and solvent B contained 84% acetonitrile in 0.1% formic acid. The gradient consisted of isocratic conditions at 5% B for 10 min, a linear gradient to 30% B over 40 min, a linear gradient to 100% B over 10 min, and then a linear gradient back to 5% B over 5 min. MS analyses were performed in positive mode using ESI with a NanoLockSpray source. As lock mass, [Glu1]fibrinopeptide B (1 pmol/μl) (Sigma) was delivered from the syringe pump (Harvard Apparatus, USA) to the reference sprayer of the NanoLockSpray source at a flow rate of 1 μl/min. The lock mass channel was sampled every 10 s.
To identify the 14, 16–18, 19 and 35 kDa bands, the Q-TOF-2 was operating in MS/MS mode for data dependent acquisition. The mass spectrometer was programmed to determine charge states of the eluting peptides, and to switch from MS to MS/MS mode for z ≥ 2 at the appropriate collision energy for Argon gas-mediated CID. Each resulting MS/MS spectrum contained sequence information on a single peptide. Processing and database searching of the MS/MS data set was performed using ProteinLynx Global SERVER (PLGS) v2.3 (Waters Corporation) and the NCBI non-redundant protein database, while taking fixed (carbamidomethylation) and variable (oxidation of Methionine) modifications into account.
After the identification of multiple Bet v 1 isoforms in the 16–18 kDa band, we analyzed the tryptic digest of this band with Q-TOF LC-MSE. The Q-TOF-2 was programmed to alternate between low and elevated levels of collision energy. Collision energy was 5 eV in MS mode and was increased in two steps from 28 to 40 eV in MSE mode. Measuring time in both modes was 0.9 s with an interscan delay of 0.1 s. Unfragmented precursors predominate in low energy mode, while fragmented ions of the precursors are observed in high energy mode. Digests were analyzed in duplicate. MSE data were analyzed according to the procedure described by Silva et al. [39] with the Expression module in PLGS. Different peptide components were detected with an ion detection algorithm, and then clustered by mass and retention time, followed by normalization of the data. The described PR-10 protein search database was used to identify peptides, while taking fixed (carbamidomethylation) and variable (oxidation of Methionine) modifications into account. After processing by PLGS, the so-called Exact Mass and Retention Time (EMRT) table was exported and reclustered using the PACP tool [59] to correct potential misalignments and split peak detection errors. Retention time was normalized and the reclustered EMRT table was further analyzed in Excel.
Authors' contributions
MFS coordinated the study, performed the analysis and drafted the manuscript. WPCW performed the cloning and sequencing. HHGC performed the SDS-PAGE and Q-TOF LC- MSMS experiments, and participated in drafting the manuscript. AHPA participated in designing the study and in analyzing the Q-TOF LC- MS/MS data. LJWJG participated in the design and coordination of the study. MJMS participated in the design of the study and the analysis of the sequence data. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was partially funded by the Netherlands Proteomics Centre, an Innovative Cluster of the Netherlands Genomics Initiative and partially funded by the Dutch Government (BSIK03009). This research was partially funded by the Allergy Consortium Wageningen (ACW).
Contributor Information
Martijn F Schenk, Email: martijn.schenk@wur.nl.
Jan HG Cordewener, Email: jan.cordewener@wur.nl.
Antoine HP America, Email: twan.america@wur.nl.
Wendy PC van't Westende, Email: wendy.vantwestende@wur.nl.
Marinus JM Smulders, Email: rene.smulders@wur.nl.
Luud JWJ Gilissen, Email: luud.gilissen@wur.nl.
References
- Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, Scheiner O, Breitenbach M. The gene coding for the major birch pollen allergen BetvI, is highly homologous to a pea disease resistance response gene. EMBO Journal. 1989;8:1935–1938. doi: 10.1002/j.1460-2075.1989.tb03597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarolim E, Rumpold H, Endler AT, Ebner H, Breitenbach M, Scheiner O, Kraft D. IgE and IgG antibodies of patients with allergy to birch pollen as tools to define the allergen profile of Betula verrucosa. Allergy. 1989;44:385–395. doi: 10.1111/j.1398-9995.1989.tb04169.x. [DOI] [PubMed] [Google Scholar]
- Niederberger V, Pauli G, Gronlund H, Froschl R, Rumpold H, Kraft D, Valenta R, Spitzauer S. Recombinant birch pollen allergens (rBet v 1 and rBet v 2) contain most of the IgE epitopes present in birch, alder, hornbeam, hazel, and oak pollen: A quantitative IgE inhibition study with sera from different populations. J Allergy Clin Immunol . 1998;102:579–91. doi: 10.1016/S0091-6749(98)70273-8. [DOI] [PubMed] [Google Scholar]
- Bollen MA, Aranzazu G, Cordewener JHG, Wichers HJ, Helsper JPFG, Savelkoul HFJ, Van Boekel MAJS. Purification and characterization of natural Bet v 1 from birch pollen and related allergens from carrot and celery. Mol Nutr Food Res. 2007;51:1527–36. doi: 10.1002/mnfr.200700119. [DOI] [PubMed] [Google Scholar]
- Fritsch R, Bohle B, Vollmann U, Wiedermann U, Jahn-Schmid B, Krebitz M, Breiteneder H, Kraft D, Ebner C. Bet v 1, the major birch pollen allergen, and Mal d 1, the major apple allergen, cross-react at the level of allergen-specific T helper cells. J Allergy Clin Immunol. 1998;102:679–86. doi: 10.1016/S0091-6749(98)70287-8. [DOI] [PubMed] [Google Scholar]
- Karlsson AL, Alm R, Ekstrand B, Fjelkner-Modig S, Schiött A, Bengtsson U, Björk L, Hjernø K, Roepstorff P, Emanuelsson CS. Bet v 1 homologues in strawberry identified as IgE-binding proteins and presumptive allergens. Allergy. 2004;59:1277–1284. doi: 10.1111/j.1398-9995.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- Scheurer S, Son DY, Boehm M, Karamloo F, Franke S, Hoffmann A, Haustein D, Vieths S. Cross-reactivity and epitope analysis of Pru a 1, the major cherry allergen. Molecular Immunology. 1999;36:155–167. doi: 10.1016/S0161-5890(99)00033-4. [DOI] [PubMed] [Google Scholar]
- Bohle B, Radakovics A, Jahn-Schmid B, Hoffmann-Sommergruber K, Fischer GF, Ebner C. Bet v 1, the major birch pollen allergen, initiates sensitization to Api g 1, the major allergen in celery: evidence at the T cell level. European Journal of Immunology. 2003;33:3303–3310. doi: 10.1002/eji.200324321. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Hawranek T, Gruber P, Wopfner N, Mari A. Allergic cross-reactivity: from gene to the clinic. Allergy. 2004;59:243–267. doi: 10.1046/j.1398-9995.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Ebner C. Molecular and biochemical classification of plant-derived food allergens. J Allergy Clin Immunol. 2000;106:27–36. doi: 10.1067/mai.2000.106929. [DOI] [PubMed] [Google Scholar]
- De Jong PC. An introduction to betula: Its morphology, evolution, classification and distribution with a survey of recent work. The IDS Betula Symposium, International Dendrology Soc: 1993; Sussex, UK. 1993.
- Järvinen P, Palmé A, Morales LO, Lännenpää M, Keinänen M, Sopanen T, Lascoux M. Phylogenetic relationships of Betula species (Betulaceae) based on nuclear ADH and chloroplast matK sequences. American Journal of Botany. 2004;91:1834–1845. doi: 10.3732/ajb.91.11.1834. [DOI] [PubMed] [Google Scholar]
- Schenk MF, Thienpont C-N, Koopman WJM, Gilissen LJWJ, Smulders MJM. Phylogenetic relationships in Betula (Betulaceae) based on AFLP markers. pp. 911–924. 4. ISSN 1614-2942.
- Eriksson NE, Holmén A, Möller C, Wihl JÅ. Sensitization according to skin prick testings in atopic patients with asthma or rhinitis at 24 allergy clinica in Northern Europe and Asia. Allergology International. 1998;47:187–196. doi: 10.2332/allergolint.47.187. [DOI] [Google Scholar]
- Abe Y, Kimura S, Kokubo T, Mizumoto K, Uehara M, Katagiri M. Epitope analysis of birch pollen allergen in Japanese subjects. Journal of Clinical Immunology. 1997;17:485–493. doi: 10.1023/A:1027323811296. [DOI] [PubMed] [Google Scholar]
- Bolhaar STHP, Weg WE Van de, Van Ree R, Gonzalez-Macebo E, Zuidmeer L, Bruijnzeel-Koomen CAFM, Fernandez-Rivas M, Jansen J, Hoffmann-Sommergruber K, Knulst AC, et al. In vivo assessment with prick-to-prick testing and double-blind, placebo-controlled food challenge of allergenicity of apple cultivars. J Allergy Clin Immunol. 2005;116:1080–6. doi: 10.1016/j.jaci.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Marzban G, Puehringer H, Dey R, Brynda S, Ma Y, Martinelli A, Zaccarini M, Weg E van der, Housley Z, Kolarich D, et al. Localisation and distribution of the major allergens in apple fruits. Plant Science. 2005;169:387–394. doi: 10.1016/j.plantsci.2005.03.027. [DOI] [Google Scholar]
- Gao Z, Weg EW Van de, Matos CI, Arens P, Bolhaar STHP, Knulst AC, Li Y, Hoffmann-Sommergruber K, Gilissen LJWJ. Assessment of allelic diversity in intron-containing Mal d 1 genes and their association to apple allergenicity. BMC Plant Biol. 2008;8:116. doi: 10.1186/1471-2229-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrazem O, Jimeno L, López-Torrejón G, Herrero M, Espada JL, Sánchez-Monge R, Duffort O, Barber D, Salcedo G. Assessing allergen levels in peach and nectarine cultivars. Ann Allergy Asthma Immunol. 2007;99:42–7. doi: 10.1016/S1081-1206(10)60619-9. [DOI] [PubMed] [Google Scholar]
- Castro AJ, Alche JD, Cuevas J, Romero PJ, Alche V, Rodriguez-Garcia MI. Pollen from different olive tree cultivars contains varying amounts of the major allerge n ole e 1. International Archives of Allergy and Immunology. 2003;131:164–173. doi: 10.1159/000071482. [DOI] [PubMed] [Google Scholar]
- Ekramoddoullah AKM, Yu XS, Sturrock R, Zamani A, Taylor D. Detection and seasonal expression pattern of a pathogenesis-related protein (PR-10) in Douglas-fir (Pseudotsuga menziesii) tissues. Physiologia Plantarum. 2000;110:240–247. doi: 10.1034/j.1399-3054.2000.110214.x. [DOI] [Google Scholar]
- Gao ZS, Weg WE van de, Schaart JG, Schouten HJ, Tran DH, Kodde LP, Meer IM van der, Geest AHM van der, Kodde J, Breiteneder H, et al. Genomic cloning and linkage mapping of the Mal d 1 (PR-10) gene family in apple (Malus domestica) Theoretical And Applied Genetics. 2005;111:171–183. doi: 10.1007/s00122-005-2018-4. [DOI] [PubMed] [Google Scholar]
- Huang JC, Chang FC, Wang CS. Characterization of a lily tapetal transcript that shares sequence similarity with a class of intracellular pathogenesis-related (IPR) proteins. Plant Molecular Biology. 1997;34:681–686. doi: 10.1023/A:1005824306560. [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological and Molecular Plant Pathology. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- Pühringer H, Moll D, Hoffmann-Sommergruber K, Watillon B, Katinger H, Machado MLD. The promoter of an apple Ypr10 gene, encoding the major allergen Mal d 1, is stress- and pathogen-inducible. Plant Science. 2000;152:35–50. doi: 10.1016/S0168-9452(99)00222-8. [DOI] [Google Scholar]
- Robert N, Ferran J, Breda C, Coutos-Thevenot P, Boulay M, Buffard D, Esnault R. Molecular characterization of the incompatible interaction of Vitis vinifera leaves with Pseudomonas syringae pv. pisi: Expression of genes coding for stilbene synthase and class 10 PR protein. European Journal of Plant Pathology. 2001;107:249–261. doi: 10.1023/A:1011241001383. [DOI] [Google Scholar]
- Swoboda I, Scheiner O, Heberle-Bors E, Vicente O. cDNA cloning and characterization of three genes in the Bet v 1 gene family that encode pathogenesis-related proteins. Plant Cell and Environment. 1995;18:865–874. doi: 10.1111/j.1365-3040.1995.tb00595.x. [DOI] [Google Scholar]
- Poupard P, Strullu DG, Simoneau P. Two members of the Bet v 1 gene family encoding birch pathogenesis-related proteins display different patterns of root expression and wound-inducibility. Australian Journal of Plant Physiology. 1998;25:459–464. [Google Scholar]
- Moons A, Prinsen E, Bauw G, Van Montagu M. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell. 1997;9:2243–2259. doi: 10.1105/tpc.9.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utriainen M, Kokko H, Auriola S, Sarrazin O, Karenlampi S. PR-10 protein is induced by copper stress in roots and leaves of a Cu/Zn tolerant clone of birch, Betula pendula. Plant Cell and Environment. 1998;21:821–828. doi: 10.1046/j.1365-3040.1998.00326.x. [DOI] [Google Scholar]
- Walter MH, Liu JW, Wünn J, Hess D. Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental, dark-induced and exogenous-stimulus-dependent expression. European Journal of Biochemistry. 1996;239:281–293. doi: 10.1111/j.1432-1033.1996.0281u.x. [DOI] [PubMed] [Google Scholar]
- Schenk MF, Gilissen LJWJ, Esselink GD, Smulders MJM. Seven different genes encode a diverse mixture of isoforms of Bet v I, the major birch pollen allergen. BMC Genomics. 2006;7:168. doi: 10.1186/1471-2164-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl-Hajek R, Radauer C, O'Riordain G, Hoffmann-Sommergruber K, Leberl K, Scheiner O, Breiteneder H. New Bet v 1 isoforms including a naturally occurring truncated form of the protein derived from Austrian birch pollen. Molecular Immunology. 1999;36:639–645. doi: 10.1016/S0161-5890(99)00078-4. [DOI] [PubMed] [Google Scholar]
- Swoboda I, Jilek A, Ferreira F, Engel E, Hoffmann-Sommergruber K, Scheiner O, Kraft D, Breiteneder H, Pittenauer E, Schmid E, et al. Isoforms of Bet v 1, the major birch pollen allergen, analyzed by liquid-chromatography, mass-spectrometry, and cDNA cloning. Journal of Biological Chemistry. 1995;270:2607–2613. doi: 10.1074/jbc.270.6.2607. [DOI] [PubMed] [Google Scholar]
- Wen J, Vanek-Krebitz M, Hoffmann-Sommergruber K, Scheiner O, Breiteneder H. The potential of Betv1 homologues, a nuclear multigene family, as phylogenetic markers in flowering plants. Mol Phylogenet Evol 1997 Dec;8(3):317-33. 1997;8:317–33. doi: 10.1006/mpev.1997.0447. [DOI] [PubMed] [Google Scholar]
- Hoffmann-Sommergruber K, Vanek-Krebitz M, Radauer C, Wen J, Ferreira F, Scheiner O, Breiteneder H. Genomic characterization of members of the Bet v 1 family: genes coding for allergens and pathogenesis-related proteins share intron positions. Gene. 1997;197:91–100. doi: 10.1016/S0378-1119(97)00246-1. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Hirtenlehner K, Jilek A, Godnik-Cvar J, Breiteneder H, Grimm R, Hoffmann-Sommergruber K, Scheiner O, Kraft D, Breitenbach M, et al. Dissection of immunoglobulin E and T lymphocyte reactivity of isoforms of the major birch pollen allergen Bet v 1: Potential use of hypoallergenic isoforms for immunotherapy. Journal of Experimental Medicine. 1996;183:599–609. doi: 10.1084/jem.183.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsper J, Gilissen L, van Ree R, America AHP, Cordewener JHG, Bosch D. Quadrupole time-of-flight mass spectrometry: A method to study the actual expression of allergen isoforms identified by PCR cloning. J Allergy Clin Immunol. 2002;110:131–8. doi: 10.1067/mai.2002.125599. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Analytical Chemistry. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- Liao DQ. Concerted evolution: Molecular mechanism and biological implications. American Journal of Human Genetics. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Evolution of gene families. Gene. 2000;259:45–52. doi: 10.1016/S0378-1119(00)00428-5. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupard P, Brunel N, Leduc N, Viémont JD, Strullu D-G, Simoneau P. Expression of a Bet v 1 homologue gene encoding a PR 10 protein in birch roots: induction by auxin and localization of the transcripts by in situ hybridization. Australian Journal of Plant Physiology. 2001;28:57–63. [Google Scholar]
- Valjakka M, Luomala EM, Kangasjärvi J, Vapaavuori E. Expression of photosynthesis- and senescence-related genes during leaf development and senescence in silver birch (Betula pendula) seedlings. Physiologia Plantarum. 1999;106:302–310. doi: 10.1034/j.1399-3054.1999.106307.x. [DOI] [Google Scholar]
- Swoboda I, Dang TCH, Heberle-Bors E, Vicente O. Expression of Bet v 1, the major birch pollen allergen, during anther development – An in situ hybridization study. Protoplasma. 1995;187:103–110. doi: 10.1007/BF01280237. [DOI] [Google Scholar]
- Larsen JN. Isoallergens – significance in allergen exposure and response. ACI News. 1995;7:141–160. [Google Scholar]
- Chassaigne H, Nørgaard JV, Van Hengel AJ. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS) J Agric Food Chem. 2007;55:4461–73. doi: 10.1021/jf063630e. [DOI] [PubMed] [Google Scholar]
- Napoli A, Aiello D, Di Donna L, Moschidis P, Sindona G. Vegetable proteomics: the detection of Ole e 1 isoallergens by peptide matching of MALDI MS/MS spectra of underivatized and dansylated glycopeptides. Journal of proteome research. 2008;7:2723–2732. doi: 10.1021/pr700855u. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Hirthenlehner K, Briza P, Breiteneder H, Scheiner O, Kraft D, Breitenbach M, Ebner C. Isoforms of atopic allergens with reduced allergenicity but conserved T cell antigenicity: Possible use for specific immunotherapy. Int Arch Allergy Immunol. 1997;113:125–7. doi: 10.1159/000237524. [DOI] [PubMed] [Google Scholar]
- Wagner S, Radauer C, Bublin M, Hoffmann-Sommergruber K, Kopp T, Greisenegger EK, Vogel L, Vieths S, Scheiner O, Breiteneder H. Naturally occurring hypoallergenic Bet v 1 isoforms fail to induce IgE responses in individuals with birch pollen allergy. J Allergy Clin Immunol. 2008;121:246–52. doi: 10.1016/j.jaci.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. Journal of Molecular Biology. 2003;325:123–133. doi: 10.1016/S0022-2836(02)01197-X. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Ebner C, Kramer B, Casari G, Briza P, Kungl AJ, Grimm R, Jahn-Schmid B, Breiteneder H, Kraft D, et al. Modulation of IgE reactivity of allergens by site-directed mutagenesis: potential use of hypoallergenic variants for immunotherapy. Faseb Journal. 1998;12:231–242. doi: 10.1096/fasebj.12.2.231. [DOI] [PubMed] [Google Scholar]
- Son DY, Scheurer S, Hoffmann A, Haustein D, Vieths S. Pollen-related food allergy: cloning and immunological analysis of isoforms and mutants of Mal d 1, the major apple allergen, and Bet v 1, the major birch pollen allergen. European Journal of Nutrition. 1999;38:201–215. doi: 10.1007/s003940050063. [DOI] [PubMed] [Google Scholar]
- Neudecker P, Lehmann K, Nerkamp J, Haase T, Wangorsch A, Fötisch K, Hoffmann S, Rösch P, Vieths S, Scheurer S. Mutational epitope analysis of Pru av 1 and Api g 1, the major allergens of cherry (Prunus avium) and celery (Apium graveolens): correlating IgE reactivity with three-dimensional structure. Biochemical Journal. 2003;376:97–107. doi: 10.1042/BJ20031057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sinauer Associates, Sunderland, Massachusetts; 2003. [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Cadot P, Lejoly M, Vanhoeyveld EW, Stevens EAM. Influence of the pH of the extraction medium on the composition of birch (Betula verrucosa) pollen extracts. Allergy. 1995;50:431–437. doi: 10.1111/j.1398-9995.1995.tb01173.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- De Groot JCW, Fiers MWEJ, Van Ham RCHJ, America AHP. Post alignment clustering procedure for comparative quantitative proteomics LC-MS Data. Proteomics. 2008;8:32–36. doi: 10.1002/pmic.200700707. [DOI] [PubMed] [Google Scholar]