Summary
Pathogenic fungi present a special problem in the clinic as the range of drugs that can be used to treat these types of infections is limited. This situation is further complicated by the presence of robust inducible gene networks encoding different proteins that confer tolerance to many available antifungal drugs. The transcriptional control of these multidrug resistance systems in several key fungi will be discussed. Experiments in the non-pathogenic Saccharomyces cerevisiae have provided much of our current understanding of the molecular framework on which fungal multidrug resistance is built. More recent studies on the important pathogenic Candida species, Candida albicans and Candida glabrata, have provided new insights into the organization of the multidrug resistance systems in these organisms. We will compare the circuitry of multidrug resistance networks in these three organisms and suggest that, in addition to the well-accepted drug efflux activities, the regulation of membrane composition by multidrug resistance proteins provides an important contribution to the resistant phenotypes observed.
Introduction
The range of antifungal drug action is limited to five different classes of mechanisms (46). This limited spectrum of drugs is further compromised by the inevitable acquisition of resistance by the pathogenic organisms. Along with selection of organisms that become resistant to a single class of antifungal drug, multidrug resistant fungi are routinely observed in the clinic (34). Genetic changes that produce multidrug resistant fungi lead to major complications in chemotherapy to eliminate these pathogens from a patient. This is a serious issue owing to the high mortality associated with these persistent fungal infections (82).
Candidemia represents the 4th most common nosocomially acquired infection (65). The commonly deployed azole drugs have been relatively effective in controlling the major Candida species (C. albicans) but the extensive use of the drugs has contributed to the rise in incidence of other species such as C. glabrata (70). Both Candida species can develop resistance to azole chemotherapy and, along with this phenotype, a particularly problematic broad range tolerance to antifungal drugs called multidrug resistance or Mdr.
Most of what is currently known concerning the molecular underpinnings of fungal multidrug resistance comes from studies in the nonpathogenic yeast Saccharomyces cerevisiae. We will restrict our discussion of fungal multidrug resistance to S. cerevisiae, C. albicans and C. glabrata since these organisms offer the best delineated genetic pictures of this resistance phenotype. We will differentiate genes and proteins from these different yeasts with the prefixes Sc (S. cerevisiae), Ca (C. albicans) and Cg (C. glabrata). Our goal here is to suggest that while the direct drug efflux activity of multiple transporter proteins under the aegis of multidrug resistance is clearly important in development of this phenotype, changes in overall membrane structure and function are likely to have similarly important contributions. The consideration of transcriptional control mechanisms in these yeasts will be limited to the major regulators of this multigene network, the Zn2Cys6 cluster transcription factors (51). A second class of transcriptional regulatory protein, the basic region-leucine zipper (bZip) family, also plays a role in regulation of multidrug resistance but their contribution is less pronounced. Action of bZip proteins in multidrug resistance has been reviewed elsewhere (4, 60).
Transcriptional control of multidrug resistance
Saccharomyces cerevisiae
Multidrug resistance in fungi involves two classes of transporter proteins consisting of members of the ATP-binding cassette (ABC) or major facilitator superfamily (MFS) groups of transporters. In Saccharomyces cerevisiae the Mdr phenotype is known as pleiotropic drug resistance (PDR) and has principally been attributed to the overexpression of the ABC transporters like ScPdr5, ScYor1 and ScSnq2. The genes encoding these important drug pumps are regulated by transcription factors including ScPdr1, ScPdr3 and ScYrr1 (Figure 1).
Figure 1. Control of multidrug resistance in S. cerevisiae.
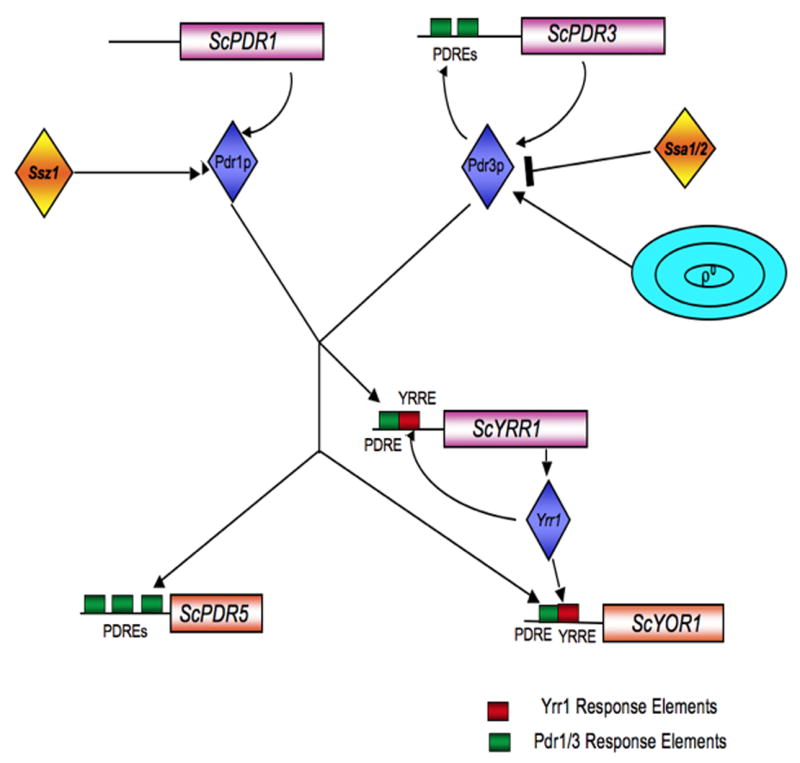
Pathways regulating the expression of multidrug resistance genes are depicted here. Positive interactions are denoted by an arrow while negative effect are indicate by a T-bar. Loss of the mitochondrial DNA gives rise to ρ0 cells that activate ScPdr3 post-translationally. ScYrr1 binds to its target element which is juxtaposed with the Pdr1/3 response elements (PDREs) in some promoters. See the text for details.
ScPdr1 and ScPdr3 are Zn2Cys6 finger transcription factors that regulate the expression of their target genes by binding to a consensus sequence in the promoter region of their target genes called Pdr1/Pdr3 Response Elements or PDREs (36). The PDR3 gene promoter itself contains two PDREs and hence is under autoregulation (16). ScPdr1 and ScPdr3 are 33% identical and share common domain organization with an N-terminal DNA binding domain, a central regulatory domain and C-terminal acidic activation domain (2, 17, 36, 42). Additionally, transcriptional microarray experiments indicate that ScPdr1 and ScPdr3 regulate similar target genes (18). Co-immunoprecipitation experiments have shown that ScPdr1 and ScPdr3 can form both homo or heterodimers in vivo (52). Single point mutations have been isolated in the N- or C-terminal region of the regulatory domain and in the activation domain of both ScPdr1 and ScPdr3 that makes them hyperactive (5, 61, 83). Transactivation experiments revealed that these mutants induced the activation at PDRE-containing promoters.
Sc PDR5 has three PDREs in its promoter and is strongly responsive to the levels of ScPdr1/Pdr3 activity (37). Hyperactive alleles in either Sc PDR1 or Sc PDR3 lead to massive overexpression of Pdr5p with an associated increase in drug resistance (19, 55). The observation that substitution mutants in Sc PDR1 or Sc PDR3 produce hyperactive forms of these factors is consistent with their protein products normally being under some form of negative regulation. Our understanding of the molecular basis of regulation of ScPdr1 and ScPdr3 is still incomplete but some details are emerging. Cells that have lost their mitochondrial genome (ρ0 cells) strongly induce levels of Sc PDR5 in a ScPdr3-dependent manner (30). Signals from dysfunctional mitochondria were blocked partially by deletion of Sc LGE1 (93). ScLge1 is a nuclear protein involved ubiquitination of histone H2B (32) although this function does not appear to play a role in Sc PDR5 regulation (93). Recent work has identified a second participant in the mitochondrial to nucleus (or retrograde (26)) regulation of Sc PDR5 called ScPsd1. This protein is the mitochondrial phosphatidylserine decarboxylase (converts phosphatidylserine to phosphatidylethanoamine) (7) and loss of Sc PSD1 prevents normal retrograde induction of Sc PDR5 in ρ0 cells. Interestingly, overexpression of ScPsd1 in wild-type ρ+ cells also leads to induction of Sc PDR5 in a ScPdr3p-dependent manner, indicating engagement of the same regulatory circuit under different genetic situations.
Along with the mitochondrial control of ScPdr3 activity, another regulatory input has been identified for both this protein and ScPdr1. These Zn2Cys6-containing transcription factors have been found to be responsive to levels of different Hsp70 proteins produced in S. cerevisiae. S. cerevisiae encodes 14 different Hsp70 proteins that carry out a range of activities throughout the cell (10). Experiments searching for high-copy-number regulators of the Pdr system recovered the Hsp70 protein ScPdr13 (now referred to as ScSsz1) as a positive regulator of Sc PDR5 gene expression (27). Further analyses established that ScSsz1 acted strictly via ScPdr1 and was unable to further elevate the activity of hyperactive forms of this transcription factor (29). The dnaJ partner protein of ScSsz1, called ScZuo1, was also found to stimulate ScPdr1 activity (23). The ScSsz1/Zuo1 complex acts to fold nascent polypeptides on the ribosome but the ability of these factors to stimulate ScPdr1 activity and drug resistance involves an extra-ribosomal role for these chaperone proteins.
Biochemical purification of proteins associating with ScPdr3 led to the identification of the ScSsa1 Hsp70 as a regulator of this transcription factor (81). ScSsa1 (and its close relative ScSsa2) act to repress the transcriptional activating capacity of ScPdr3 in ρ+ cells. Loss of the mitochondrial genome leads to a decrease in the level of Hsp70 protein associating with ScPdr3 with a concomitant rise in ScPdr3-dependent transactivation. No influence of these Hsp70 proteins was found on ScPdr1.
Along with these upstream regulators of ScPdr1/Pdr3 action, more recent work has identified important downstream effectors of these transcription factors. Analysis of proteins capable of binding to a key domain in the RNA polymerase II mediator component ScMed15 (aka ScGal11p) led to the identification of ScPdr1 as the major associated factor (86). The mediator component is a multi-protein complex that allows communication between site-specific transcriptional activators and RNA polymerase II (reviewed in (48)). In this same work, binding experiments were used to provide evidence that ScPdr1/Pdr3 act like xenobiotic receptors with the central region of these factors directly binding to drugs. This binding is predicted to restructure ScPdr1/Pdr3 and allow enhanced recruitment of ScMed15 to target promoters.
A number of other Zn2Cys6-containing transcription factors have been demonstrated to bind to and regulate PDR genes in S. cerevisiae. Sc YRR1 encodes an especially interesting member of this family of less well-studied factors. ScYrr1 was first identified as a determinant of resistance to the cell cycle inhibitor reveromycin A (13). This positive regulator of transcription was found to induce expression of the genes encoding the ABC transporter ScYor1 and ScSnq2 and strikingly the Sc YRR1 gene itself (11, 92). ScYrr1 shares a number of similarities with ScPdr3 as mutations within its central regulatory domain cause it to behave as a hyperactive transcriptional regulator and it is autoregulated (13, 92).
Another feature shared between ScYrr1 and ScPdr3 is the presence of a second factor sharing strong sequence similarity and binding specificity. ScYrm1 likely recognizes the same DNA element as ScYrr1 (50) in a manner similar to the mutual PDRE-binding of ScPdr1 and ScPdr3 (37). ScYrm1 exhibits a complex regulatory relationship with ScYrr1 as ScYrm1 appears to become more active when ScYrr1 is removed from the cell (50). This can also be seen when considering the ScPdr1/Pdr3 pair as some ScPdr1 responses are blunted when ScPdr3 is present and vice versa (see (92) for an example). Clearly, there is more to learn of the functioning of these pairs of related transcription factors.
Candida glabrata
Candida glabrata is a relatively newly recognized fungal pathogen in terms of its clinical importance. The incidence of fungal infections associated with C. glabrata has risen dramatically in recent years, in part due to the low intrinsic and high acquired resistance to azole drugs that are commonly used to treat fungal infections (reviewed in (66)). This attribute has led to intensive examination of the molecular basis for drug resistance in C. glabrata isolates. Work from several labs has provided a consistent link between high azole tolerance and overexpression of ABC transporter-encoding genes in this pathogenic yeast that is closely related to the resistance program described above for S. cerevisiae. Although C. glabrata and S. cerevisiae are close on the genomic evolutionary scale (22) their pathogenicity is quite different. However, examination of loci involved in specifying the multidrug resistance in C. glabrata indicated that extensive similarity existed between this pathogen and S. cerevisiae.
The first identified participant in C. glabrata multidrug resistance was the ABC transporter-encoding gene PDH1 (56) which is now referred to as Cg CDR2 (77). A second ABC transporter protein called CgCdr1 (NCBI Accession: AAF0506) was found (78) and demonstrated by sequence alignment to be more related to ScPdr5 while CgCdr2 (NCBI Accession: AAF64315) more closely aligns with ScPdr15. Both of these ABC transporter proteins are involved in azole tolerance in clinical isolates of C. glabrata.
More recently, information has been obtained illuminating the identity of a key regulator of the transcription of multidrug resistance gene expression in C. glabrata. Search of the completed C. glabrata genome (22) and direct functional analysis led to the identification of a single homologue of ScPdr1 that exhibited striking sequence similarity with this regulatory protein. This locus was designated Cg PDR1 and has been shown to be a crucial regulator of multidrug resistance in this yeast. Loss of Cg PDR1 from multidrug resistant clinical isolates led to a loss of the high level drug resistance and overproduction of Cg CDR1 seen in these backgrounds (87, 90). DNA sequence determination of Cg PDR1 alleles present in multidrug resistant isolates demonstrated that substitution mutant forms of CgPdr1 were likely to behave as hyperactive transcriptional regulators in a fashion similar to mutants described in Sc PDR1 and PDR3. Analysis of the transcriptome under CgPdr1 control by DNA microarray determined that significant overlap was seen with genes up-regulated by ScPdr1/Pdr3 in S. cerevisiae (89).
While studies on control of CgPdr1 activity are at an early stage, several important findings are already in hand (Figure 2). First, CgPdr1 is likely under negative control as evidenced by the finding of substitution mutations that convert this factor to a hyperactive transcription regulator. Second, CgPdr1 appears to be autoregulated in a manner consistent with that seen for ScPdr3 (87, 90). Third, ρ0 C. glabrata cells are highly drug resistant, induce Cg CDR1 transcription and require CgPdr1 for this regulatory circuit to be completed (87). These data support the view that CgPdr1 may actually be regulated more like ScPdr3 than ScPdr1 even though the sequence similarity is higher between Cg and Sc Pdr1 factors. Most recently, experiments have been carried out to confirm that CgPdr1, like its S. cerevisiae counterparts Pdr1 and Pdr3, directly binds to the CgMed15 mediator component (86). Intriguingly, C. glabrata contains two MED15 homologous genes but only one of these loci is required for CgPdr1-mediated drug resistance.
Figure 2. Control of multidrug resistance in Candida species.
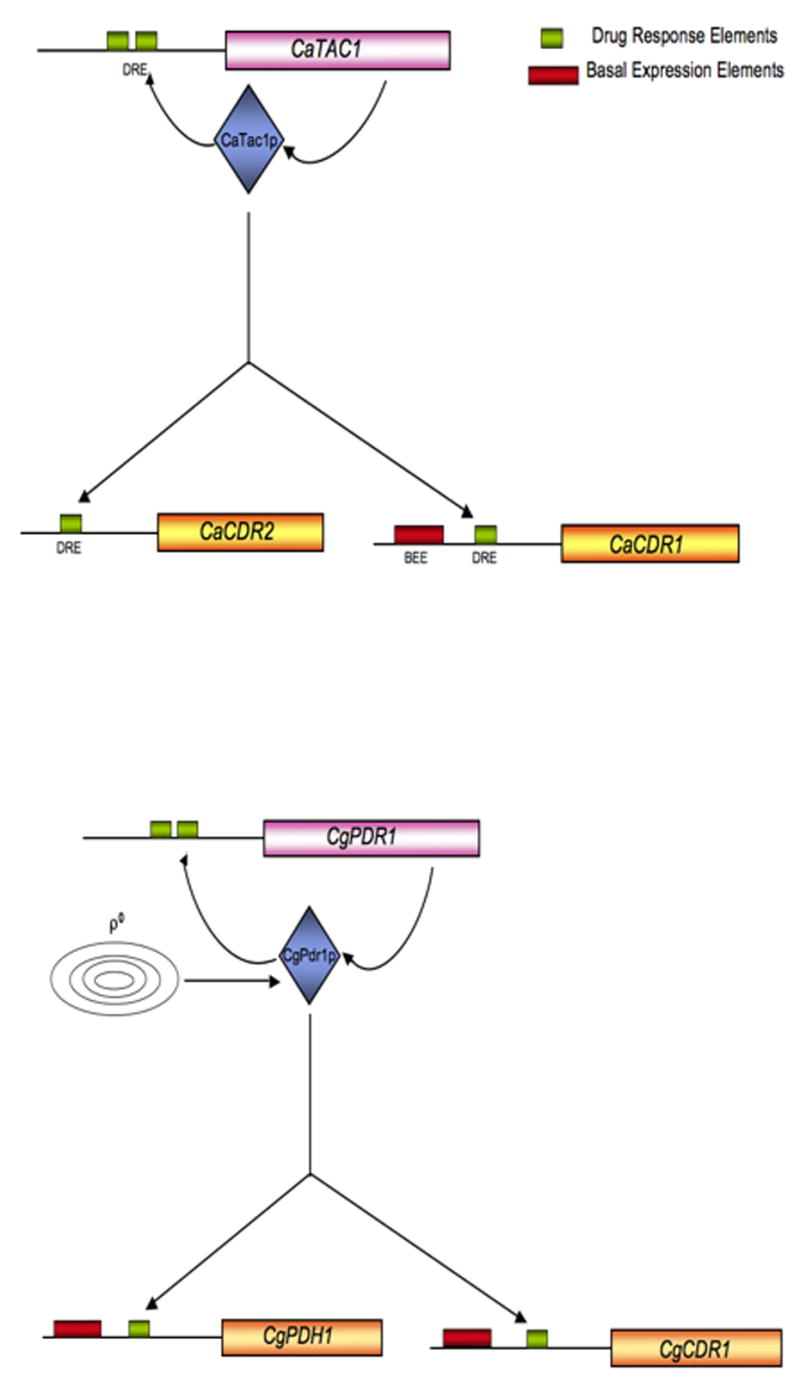
Similar transcriptional control mechanisms in Candida albicans (top) and Candida glabrata (bottom) are shown. No mitochondrial input has yet been described for regulation of multidrug resistance in C. albicans.
Candida albicans
While C. glabrata is increasing in incidence of diagnosed fungal infections, C. albicans still represents the major species associated with candidemias (recently reviewed in (65)). Over the same time period that C. glabrata has risen in detection frequency by roughly a factor of 10, C. albicans association with human disease has remained relatively constant. These changes have been suggested to be due to the relative differences in azole susceptibility of these two Candida species although this conclusion is controversial (80).
Irrespective of the differences in azole susceptibility between these two C. albicans and C. glabrata species, C. albicans isolates that exhibit elevated resistance to this important class of antifungal drug are frequently isolated (reviewed in (31)). The first drug resistance gene isolated from C. albicans encoded the major facilitator superfamily (MFS) protein CaMdr1 (24). Later work discovered the ABC transporter-encoding gene Ca CDR1 which was demonstrated to produce a striking homologue of ScPdr5 and complement the drug sensitivity of a pdr5Δ S. cerevisiae strain (69). The same approach of complementation of various S. cerevisiae mutant strain was used to isolate more ABC transporter- as well as transcription factor-encoding genes (79, 85). Use of S. cerevisiae to provide insight into the function of C. albicans membrane proteins has been more successful than the use of this heterologous host to learn about C. albicans gene transcription. Several C. albicans Zn2Cys6 cluster-containing proteins were found to suppress the drug hypersensitivity of a pdr1Δ pdr3Δ double mutant S. cerevisiae strain but surprisingly had no effect on drug resistance in C. albicans (85). The identity of the true C. albicans equivalent of ScPdr1/Pdr3 was unknown until direct experiments in this pathogenic yeast uncovered the Ca TAC1 gene.
The completion of the genomic sequence of C. albicans provided a key starting point in the identification of Ca TAC1. A series of Zn2Cys6 cluster-containing factors were discovered in the region of the mating locus which were then subjected to disruption analysis (8). Although there are more than 70 proteins in the C. albicans genome containing Zn2Cys6 clusters, these factors encoded near the mating locus were deemed of special importance since previous experiments had shown that loss of homozygosity in this region of chromosome V was linked to changes in drug tolerance (74). Disruption of the Ca TAC1 gene produced a strain that was hypersensitive to azole drugs and failed to normally express Ca CDR2 (C. albicans ABC transporter closely related to Ca CDR1). DNA binding experiments using recombinant CaTac1 demonstrated that this protein was able to recognize a previously defined element referred to as the Drug Response Element (DRE) in the promoters of both Ca CDR1/CDR2 (14). The DRE is required for drug induced expression and the high constitutive expression of these ABC transporter-encoding genes that is observed in azole tolerant isolates. Analysis of an epitope-tagged form of CaTac1 have provided evidence that this regulatory protein also controls transcription of its own structural gene (49). Like Sc PDR3 and Cg PDR1, Ca TAC1 is autoregulated.
While strong data indicate that CaTac1 is the functional homologue of ScPdr1/Pdr3, there are important differences between these factors. The sequence similarity shared between CaTac1 and its S. cerevisiae counterparts is dramatically less than that exhibited by CgPdr1 and either ScPdr1 or ScPdr3. The S. cerevisiae protein identified as the closest related polypeptide to CaTac1 is called ScHal9. ScHal9 is thought to be involved in transcriptional control of a salt responsive transporter designated Sc ENA1 (54). Perhaps ScHal9 and CaTac1 share sequence similarity due to these factors sharing more common functional determinants than CaTac1 shares with other drug resistance regulators like ScPdr1/Pdr3 and CgPdr1. We speculate that upstream regulatory signals controlling the function of ScHal9 and CaTac1 may be more closely related than similar signals controlling the drug resistance regulators ScPdr1/Pdr3 and CgPdr1. Further study of CaTac1 and ScHal9 will directly test this suggestion. Finally, the binding site of CaTac1 contains two direct CGG repeats separated by 4 nucleotides (8), an element that is related but distinct from the PDRE (TCCGCGGA (37)). Although CaTac1 controls expression of a highly related suite of genes to those regulated by ScPdr1/Pdr3 and CgPdr1, the molecular basis of this regulation is quite different.
In addition to CaTac1, two other Zn2Cys6-containing transcription factors have been linked to expression of multidrug resistance genes. As mentioned above, the first identified C. albicans gene involved in multidrug resistance was the MFS protein-encoding determinant Ca MDR1. CaMdr1 was shown to be overproduced in a variety of clinical isolates owing to a large elevation in its transcription (35, 72). Only recently has the basis of this transcriptional induction been solved. CaMrr1 was demonstrated to be required for overproduction of Ca MDR1 mRNA in azole resistant strains (58). The CaMrr1 regulatory circuit appears to be distinct from CaTac1 which helps explain previous work in which expression of ABC transporters like CaCdr1/Cdr2 were co-regulated while CaMdr1 was not (see for example (35, 72).).
The second Zn2Cys6-containing factor recently suggested to play a role in multidrug resistance is the regulator of sterol biosynthetic gene expression called CaUpc2 (95). Since azole drugs target sterol biosynthesis, the finding that elevated activity of CaUpc2 increased azole tolerance was expected (reviewed in (1)). However, genomic characterization of CaUpc2 target genes provided the unexpected observation that this factor bound to a number of different multidrug resistance gene promoters including Ca CDR1, CDR2 and MDR1. Transcript levels of Ca CDR1 and MDR1 were also found to be elevated along with CaUpc2 activity. Further study of this interesting link between sterol biosynthesis and multidrug resistance will shed light on the physiological importance of this regulatory connection.
Target genes regulated by multidrug resistance systems
The information described above details a number of important differences in the machinery acting to modulate transcriptional regulation of the suite of genes involved in specifying multidrug resistance. While the regulation of these multidrug resistant genes exhibits striking differences, the genes that compose the target loci share striking similarities. For example, the first known Pdr target gene in S. cerevisiae was the ABC transporter-encoding PDR5 locus. Using this observation as a guide, workers in both Candida species have demonstrated that the Ca and Cg CDR1 genes are major transcriptional targets of the multidrug resistance systems in each of these yeasts. We will discuss the nature of the collection of genes that are commonly regulated in each of these three different yeast upon engagement of their multidrug resistance system.
ABC transporters
As mentioned above, this class of membrane transporters provided the first member of the target genes involved in fungal multidrug resistance, S. cerevisiae PDR5. Both species of Candida express a homologue designated CDR1 (Candida Drug Resistance) that is capable of complementing the strong drug hypersensitivity of a Sc pdr5Δ strain. Coupled with the fact that all three ABC transporter proteins share extremely high sequence similarity, it seem likely that these proteins carry out similar actions in their native environments (recently reviewed in (57).
ScPdr5 is the most extensively analyzed of these related proteins and will be considered as the prototype for the role of this class of factor involved in fungal multidrug resistance. The most widely embraced function of ScPdr5 (and arguably many eukaryotic ABC transporters) is that of a broad specificity, ATP-dependent drug efflux pump. This was convincingly documented (45) in studies demonstrating the wide range of drug phenotypes influenced upon loss of ScPdr5 as well as in a ScPdr5-dependent transport assay now in common use.
However, along with drug efflux activity, experiments from other labs have implicated ScPdr5 in the control of phospholipid asymmetry across the plasma membrane (40, 68). Specifically, evidence has been provided that ScPdr5 can act to control the movement of PE and PC from the inner leaflet of the plasma membrane to the outer leaflet (a process called Flop of phospholipids (reviewed in (67))). Earlier work in mammalian cells demonstrated that mouse Mdr2 exhibited an influence on phospholipid transport (73), indicating that this function is likely a conserved role for ABC transporters.
An ABC transporter protein called ScYor1 also participates with ScPdr5 in control of phospholipid flop (15, 68) but exhibits a significantly different protein domain structure than ScPdr5. ScYor1 is localized to the plasma membrane and was originally identified as a mediator of resistance to the mitochondrial inhibitor oligomycin (38). ScYor1 was independently identified as a high-copy-number mediator of resistance to aureobasidin A which is an inhibitor of sphingolipid biosynthesis (12). Intriguingly, ScYor1 was also identified as a mediator of resistance to the ceramide synthase inhibitor fumonisin B1 (53). These data suggest that ScYor1 may play a role in controlling levels of endogenous sphingolipid intermediates since both aureobasidin A and fumonisin B1 are thought to act as mimics of natural sphingolipid metabolites.
Sphingolipid and phospholipid homeostasis
Along with ABC transporters, another major category of Pdr target genes are loci encoding proteins involved in biosynthesis of a major plasma membrane lipid called sphingolipids. Sphingolipids make up 30% of the total phospholipids in the S. cerevisiae plasma membrane (64). A number of excellent reviews have detailed the pathways and enzymes involved in the multistep biosynthesis of these important membrane lipids and the reader is urged to examine these for details of this pathway (9, 20, 71). Here we will focus on control of the levels of a key intermediate in sphingolipid biosynthesis, the well-known lipid signaling molecule ceramide. Ceramide is produced by condensation of a very long chain fatty acid with a long chain base (LCB) provided as either dihydrosphingosine or phytosphingosine (PHS) in yeast. Ceramide is then modified by the addition of inositol-phosphate or mannosyl groups to ultimately produce mannosyl-diinositolphosphate ceramide (M(IP)2C) which is the most abundant sphingolipid in S. cerevisiae (21).
Pdr-mediated transcriptional control has been demonstrated to impact sphingolipid biosynthesis at a minimum of three steps. First, the Sc IPT1 gene encoding inositol phosphotransferase was demonstrated to be transcriptionally induced by ScPdr1 and ScPdr3 (28). Second, one component of the multisubunit enzyme ceramide synthase was found to be a Pdr target gene (44). Finally, expression of the putative LCB efflux transporter ScRsb1 is regulated by both ScPdr1 and ScPdr3 (41, 63). Control of intracellular levels of LCBs is critical as inappropriate elevation of these sphingolipid precursors is toxic to the cell (6, 84).
An interesting intersection between function of Pdr target genes and sphingolipid biosynthetic intermediates was found in a genetic screen searching for genes involved in tolerance to PHS (41). Strains lacking the ScPdr5 and Yor1 ABC transporter proteins were observed to be highly resistant to PHS. This PHS resistant phenotype was also shown to be dependent on the presence of the Sc RSB1 and Sc PDR1 genes which led to the suggestion that a regulatory signal may be transduced by ScPdr1 to Sc RSB1 upon imbalance of phospholipid asymmetry in the plasma membrane (41). The induction of Sc RSB1 in a Sc pdr5Δ strain is controversial as this has been seen by one group (41) but not another (63). However, both groups find ScRsb1 to be an important contributor to the elevated PHS resistance exhibited by cells lacking ScPdr5. Irrespective of the exact mechanism underlying the role of ScRsb1 in ScPdr5-regulated PHS tolerance, this finding provides another link between factors involved in multidrug resistance and sphingolipid biosynthesis.
Sc RSB1 homologues referred to as RTA genes have been identified as targets in the multidrug resistance suite of genes in both Candida species. Cg RTA1 and Ca RTA3 are induced under conditions that lead to activation of other multidrug resistance genes (35, 72, 89). A close relative of RTA3 called Ca RTA2 has been directly studied in C. albicans and found to contribute to azole resistance in this pathogen (33). Chromatin immunoprecipitation analyses indicate that Ca RTA3 contains a CaTac1 binding site in its promoter (49). Induction of Rsb1-like proteins is a common theme in regulation of fungal multidrug resistance.
Finally, a common target genes induced during the multidrug resistance response in all these three fungi is the phosphatidylinositol transfer protein (PITP) homologue designated Pdr16. ScPdr16 (or ScSFH3) was first identified as a member of a family of 5 proteins grouped by their similarity to the ScSec14 PITP (reviewed in (59)). ScSec14 is an essential protein that acts to regulate the lipid composition of the late secretory pathway (3). Less is known of the role of ScPdr16 but mutants lacking this protein grow poorly and exhibit sterol handling defects (88). Loss of the PDR16 gene from S. cerevisiae (88), C. albicans (75), or C. glabrata (39) caused an increase in azole sensitivity, again supporting a common role for this factor in all these yeasts.
Transcriptional regulators
ScPdr1 and ScPdr3 are the primary regulators of the Pdr transcriptional response. The effects of these transcription factors are amplified through the control by ScPdr1 and ScPdr3 of genes that specify production of other transcriptional regulatory proteins. The elevated level of expression of these secondarily acting transcription factors is required for the full induction of the multidrug resistance system. Sc PDR3 itself provides a unique example of a transcriptional regulator that is involved both in the primary Pdr response but also is a target gene (16). Sc PDR3 is positively autoregulated and also induced by activation of ScPdr1. This autoregulatory increase in ScPdr3 levels is required for normal activation of the ScPdr3-mediated induction of drug resistance seen in ρ0 cells (30, 94) (see above). Importantly, in C. glabrata, the Cg PDR1 gene is both autoregulated and induced in ρ0 cells in a manner very similar to Sc PDR3 (87, 89). Likewise, Ca TAC1 is autoregulated (49) although no mitochondrial link to control of CaTac1 activity has yet been described.
Another transcription factor that is induced by ScPdr1 and ScPdr3 is the zinc cluster-containing factor ScYrr1 (13). ScYrr1 was first identified through its ability to confer resistance to the cell cycle inhibitor reveromycin A. Later experiments demonstrated that the Sc YRR1 promoter was a target of ScPdr1/Pdr3 regulation as well as positively autoregulated (92). DNA microarray analyses and promoter binding assays (47) demonstrated that ScYrr1 controlled the expression of several genes that did not appear to be direct targets of ScPdr1/Pdr3. Together these data argue that ScYrr1 function is required for cells to express the full spectrum of the multidrug resistant phenotype via activation of a wider range of genes than are regulated by ScPdr1 and ScPdr3. Direct experimentation in C. glabrata suggests that the Pdr1:YRR1 regulatory link is conserved in this pathogenic yeast (76).
A more recently characterized factor that has been found to be inducible by ScPdr1 and ScPdr3 is ScRpn4 (62). ScRpn4 controls the expression of a variety of genes involved in specifying proteasome function (91). While the contribution of proteasome function to multidrug resistance is still under study, evidence has been recently obtained that Sc RPN4 regulation by ScPdr1 and ScYrr1 may be conserved in C. glabrata (76) although further experimentation will be required to confirm this suggestion.
Other target genes
While the genes discussed above represent major classes of genes common to the multidrug resistance system in these different yeast, clearly this is only a first look into the catalog of all relevant loci influencing this phenotype. For example, C. albicans appears to have a pronounced link between expression of multidrug resistance and oxidative stress tolerance genes (35, 72). C. glabrata appears to regulate a much wider range of genes than S. cerevisiae as assessed by DNA microarray but this may reflect the different technology used in and more recent execution of the C. glabrata experiments (89).
Target genes are very similar but their upstream signals vary
While different regulatory systems are employed by these three different yeasts to control expression of their multidrug resistance systems, there are important commonalities to the spectrum of genes induced in each organism. ABC transporters, sphingolipid and phospholipid homeostasis proteins represent conserved targets of the different multidrug resistance systems. Mammalian multidrug resistant cells are often observed to have changes in their sphingolipid profiles that appear associated with the resistance phenotype (reviewed in (25)). This linkage of changes in levels of expression of membrane transporters and the lipid composition of the plasma membrane in which they function seems to be a conserved feature of the multidrug resistance phenotype within all eukaryotic organisms to date.
We hypothesize that the physiological function of the multidrug resistance system might be to regulate both the lipid composition of the plasma membrane as well as to control permeability of that membrane to various substances. If the ABC transporters can act to both efflux drugs and regulate the lipid bilayer of the plasma membrane, then their activity could be amplified through regulation of other proteins embedded in this membrane (Figure 3). For example, loss of ScPdr5 may have direct phenotypic effects on drug resistance (i.e.-fluconazole sensitivity) but pdr5Δ strains also exhibit increased resistance to other compounds such as PHS (41) and oligomycin (36, 43). While the preponderance of evidence supports the idea that ScPdr5 (and its Candida homologues) acts as a drug efflux pump, this ABC transporter also controls the phospholipid asymmetry of the plasma membrane (40, 68). These observations suggest the possibility that ScPdr5 might actually be a multifunctional protein in terms of its contribution to multidrug resistance. Its direct action would be to efflux a broad range of drugs but its indirect contribution would be to control the phospholipid distribution across the plasma membrane that could regulate the function of other transporter proteins.
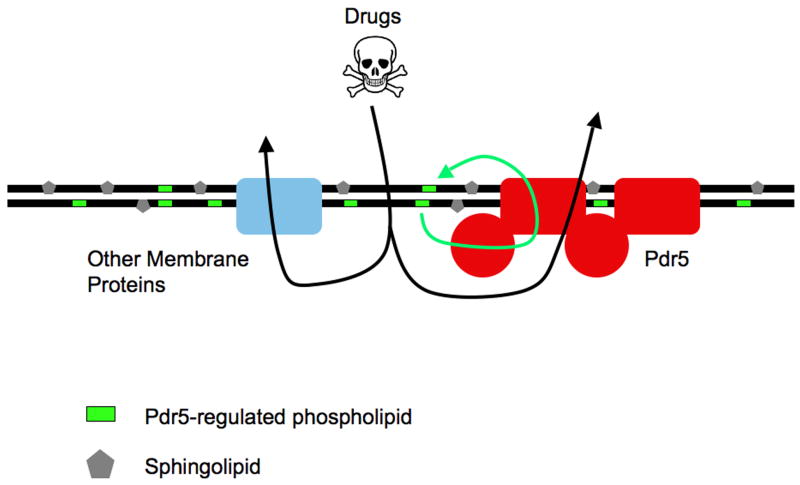
Figure 3. Regulation of multidrug resistance at the plasma membrane.
S. cerevisiae is used as the prototype for the various mechanisms that may act to control drug entry into fungal cells. Drugs can enter the cell in many cases by diffusion and are removed by the direct action of multidrug transporters like Pdr5 but also other membrane transporter proteins (blue box). ScPdr5 has at least one other activity which is to control phospholipid distribution across the plasma membrane. Sphingolipid distribution is also asymmetric across the plasma membrane with the majority of these lipids located in the outer leaflet. Control of the lipid composition of the plasma membrane is likely to influence the activity of embedded proteins.
The view of these ABC transporter proteins as both regulators of the plasma membrane environment and drug pumps is attractive in terms of explaining the broad specificity of the membrane transporters. If the activity of other membrane proteins can be stimulated through the changes in the lipid bilayer, then the apparent effect of a multidrug transporter may actually be due to the stimulated action of other proteins.
Finally, the conservation of the linked changes in sphingolipid, phospholipids and ABC transporter levels in fungal (and possibly mammalian) multidrug resistance suggests that the regulation of these functions is coordinately controlled in all these organisms. All these fungi induce a common spectrum of genes but do so in response to different signals. S. cerevisiae and C. glabrata share the most overlap but even these organisms have striking differences in their regulation of multidrug resistance genes as described above. C. albicans has the same basic architecture for regulation (transcriptional regulation by a Zn2Cys6 cluster protein) but a different DNA binding site is used by CaTac1 and activity of CaTac1 has not yet been described to share any upstream signals with ScPdr1/Pdr3 or CgPdr1. We speculate that the normal multidrug response in all these fungi requires changes both in plasma membrane composition and also the ABC transporters that function in this environment. These mutual changes could elicit synergistic responses in resistance as changes in ABC transporters could both pump drugs and influence the membrane bilayer while changes in the membrane bilayer could also affect transporter function and drug movement across this membrane barrier. Triggering the induction of multidrug resistance seems to be linked to different signals in organisms as closely related as yeasts but ultimately results in similar changes in the cell biology of the target. Understanding the physiology underlying these changes represents a major challenge to the field.
Acknowledgments
Our work on multidrug resistance has been funded by NIH grants GM49825 and GM75120. We dedicate this review to the memory of our colleague Dr. Ronald Butow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akins RA. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol. 2005;43:285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]
- 2.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 3.Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer BE, Wolfger H, Kuchler K. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim Biophys Acta. 1999;1461:217–236. doi: 10.1016/s0005-2736(99)00160-1. [DOI] [PubMed] [Google Scholar]
- 5.Carvajal E, van den Hazel HB, Cybularz-Kolaczkowska A, Balzi E, Goffeau A. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol Gen Genet. 1997;256:406–415. doi: 10.1007/s004380050584. [DOI] [PubMed] [Google Scholar]
- 6.Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem. 2000;275:17229–32. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- 7.Clancey CJ, Chang SC, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–90. [PubMed] [Google Scholar]
- 8.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell. 2004;3:1639–52. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation and function. Biochim Biophys Acta. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig EA, Gambill BD, Nelson RJ. Heat shock proteins: molecular chaperones of protein biogenesis. Micro Revs. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Z, Hirata D, Miyakawa T. Functional analysis of the promoter of the yeast SNQ2 gene encoding a multidrug resistance transporter that confers the resistance to 4-nitroquinoline-N-oxide. Biosci Biotechnol Biochem. 1999;63:162–167. doi: 10.1271/bbb.63.162. [DOI] [PubMed] [Google Scholar]
- 12.Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The mutidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 13.Cui Z, Shiraki T, Hirata D, Miyakawa T. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline-N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol Microbiol. 1998;29:1307–1315. doi: 10.1046/j.1365-2958.1998.01027.x. [DOI] [PubMed] [Google Scholar]
- 14.de Micheli M, Bille J, Schueller C, Sanglard D. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol Microbiol. 2002;43:1197–214. doi: 10.1046/j.1365-2958.2002.02814.x. [DOI] [PubMed] [Google Scholar]
- 15.Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 16.Delahodde A, Delaveau T, Jacq C. Positive autoregulation of the yeast transcription factor Pdr3p, involved in the control of the drug resistance phenomenon. Mol Cell Biol. 1995;15:4043–4051. doi: 10.1128/mcb.15.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 18.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 19.Dexter D, Moye-Rowley WS, Wu AL, Golin J. Mutations in the yeast PDR3, PDR4, PDR7 and PDR9 pleiotropic drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- 21.Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- 22.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich J, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszu IR, Lemaire M, Lesur I, Ma L, Muller H, Nicaud J, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard G, Straub M, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet J. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 23.Eisenman HC, Craig EA. Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol Microbiol. 2004;53:335–44. doi: 10.1111/j.1365-2958.2004.04134.x. [DOI] [PubMed] [Google Scholar]
- 24.Fling ME, Kopf J, Tamarkin A, Gorman JA, Smith HA, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 25.Gouaze-Andersson V, Cabot MC. Glycosphingolipids and drug resistance. Biochim Biophys Acta. 2006;1758:2096–2103. doi: 10.1016/j.bbamem.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Gulshan K, Schmidt J, Shahi P, Moye-Rowley WS. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol Cell Biol. 2008;28:5851–5864. doi: 10.1128/MCB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallstrom TC, Katzmann DJ, Torres RJ, Sharp WJ, Moye-Rowley WS. Regulation of transcription factor Pdr1p function by a Hsp70 protein in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallstrom TC, Lambert L, Schorling S, Balzi E, Goffeau A, Moye-Rowley WS. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:23674–23680. doi: 10.1074/jbc.M101568200. [DOI] [PubMed] [Google Scholar]
- 29.Hallstrom TC, Moye-Rowley WS. Hyperactive forms of the Pdr1p transcription factor fail to respond to positive regulation by the Hsp70 protein Pdr13p. Mol Microbiol. 2000;36:402–413. doi: 10.1046/j.1365-2958.2000.01858.x. [DOI] [PubMed] [Google Scholar]
- 30.Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisae. J Biol Chem. 2000;275:37347–37356. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- 31.Hazen KC, Baron EJ, Colombo AL, Girmenia C, Sanchez-Sousa A, del Palacio A, de Bedout C, Gibbs DL. Comparison of the susceptibilities of Candida spp. to fluconazole and voriconazole in a 4-year global evaluation using disk diffusion. J Clin Microbiol. 2003;41:5623–32. doi: 10.1128/JCM.41.12.5623-5632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–6. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 33.Jia XM, Ma ZP, Jia Y, Gao PH, Zhang JD, Wang Y, Xu YG, Wang L, Cao YY, Cao YB, Zhang LX, Jiang YY. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem Biophys Res Commun. 2008;373:631–636. doi: 10.1016/j.bbrc.2008.06.093. [DOI] [PubMed] [Google Scholar]
- 34.Kanafani Z, Perfect J. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis. 2008;46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 35.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother. 2004;48:3064–3079. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzmann DJ, Burnett PE, Golin J, Mahe Y, Moye-Rowley WS. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzmann DJ, Hallstrom TC, Mahe Y, Moye-Rowley WS. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 38.Katzmann DJ, Hallstrom TC, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley WS. Expression of an ATP-binding cassette transporter encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur R, Castano I, Cormack BP. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob Agents Chemother. 2004;48:1600–13. doi: 10.1128/AAC.48.5.1600-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kean LS, Grant AM, Angeletti C, Mahé Y, Kuchler K, Fuller RS, Nichols JW. Plasma membrane translocation of fluorescent-labeled phosphatidylethanolamine is controlled by transcription regulators, PDR1 and PDR3. J Cell Biol. 1997;138:255–270. doi: 10.1083/jcb.138.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kihara A, Igarashi Y. Cross talk between sphingolipids and glycerophospholipids in the establishment of plasma membrane asymmetry. Mol Biol Cell. 2004;15:4949–59. doi: 10.1091/mbc.E04-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolaczkowska A, Kolaczkowski M, Delahodde A, Goffeau A. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol Genet Genomics. 2002;267:96–106. doi: 10.1007/s00438-002-0642-0. [DOI] [PubMed] [Google Scholar]
- 43.Kolaczkowska A, Kolaczkowski M, Goffeau A, Moye-Rowley WS. Compensatory activation of the mutidrug transporters Pdr5p, Snq2p and Yor1p by Pdr1p in Saccharomyces cerevisiae. FEBS Lett. 2008;582:977–983. doi: 10.1016/j.febslet.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolaczkowski M, Kolaczkowska A, Gaigg B, Schneiter R, Moye-Rowley WS. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:880–92. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowski A, Soumillion JP, Konings WN, Goffeau A. Anticancer drugs, ionophoric peptides and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- 46.Kontoyiannis DP, Lewis RE. Antifungal drug resistance of pathogenic fungi. Lancet. 2002;359:1135–44. doi: 10.1016/S0140-6736(02)08162-X. [DOI] [PubMed] [Google Scholar]
- 47.Le Crom S, Devaux F, Marc P, Zhang X, Moye-Rowley WS, Jacq C. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol Cell Biol. 2002;22:2642–2649. doi: 10.1128/MCB.22.8.2642-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis BA, Reinberg D. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J Cell Sci. 2003;116:3667–75. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 49.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhauser J, Nantel A, Raymond M, Rogers PD. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell. 2007 doi: 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucau-Danila A, Delaveau T, Lelandais G, Devaux F, Jacq C. Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J Biol Chem. 2003;278:52641–52650. doi: 10.1074/jbc.M309580200. [DOI] [PubMed] [Google Scholar]
- 51.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamnun YM, Pandjaitan R, Mahe Y, Delahodde A, Kuchler K. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol Microbiol. 2002;46:1429–40. doi: 10.1046/j.1365-2958.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- 53.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae: an enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 54.Mendizabal I, Rios G, Mulet JM, Serrano R, de Larrinoa IF. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998;425:323–8. doi: 10.1016/s0014-5793(98)00249-x. [DOI] [PubMed] [Google Scholar]
- 55.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 56.Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer DJ, Ward DJ, Marsden K, Bennett JE. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob Agents Chemother. 1998;42:1695–1701. doi: 10.1128/aac.42.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monk BC, Goffeau A. Outwitting multidrug resistance to antifungals. Science. 2008;321:367–369. doi: 10.1126/science.1159746. [DOI] [PubMed] [Google Scholar]
- 58.Morschhauser J, Barker KS, Liu TT, Blab-Warmuth J, Homayouni R, Rogers PD. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mousley CJ, Tyeryar KR, Ryan MM, Bankaitis VA. Sec14p-like proteins regulate phosphoinositide homeostasis and intracellular protein and lipid trafficking in yeast. Biochem Soc Trans. 2006;34:21–49. doi: 10.1042/BST0340346. [DOI] [PubMed] [Google Scholar]
- 60.Moye-Rowley WS. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog Nucleic Acids Res Mol Biol. 2003;73:251–279. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- 61.Nourani A, Papajova D, Delahodde A, Jacq C, Subik J. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol Gen Genet. 1997;256:397–405. doi: 10.1007/s004380050583. [DOI] [PubMed] [Google Scholar]
- 62.Owsianik G, Balzi E, Ghislain M. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol Microbiol. 2002;43:1295–1308. doi: 10.1046/j.1365-2958.2002.02823.x. [DOI] [PubMed] [Google Scholar]
- 63.Panwar SL, Moye-Rowley WS. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J Biol Chem. 2006;281:6376–84. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- 64.Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfaller MA, Diekema DJ. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J Clin Microbiol. 2004;42:4419–31. doi: 10.1128/JCM.42.10.4419-4431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pomorski T, Holthuis JC, Herrmann A, van Meer G. Tracking down lipid flippases and their biological functions. J Cell Sci. 2004;117:805–13. doi: 10.1242/jcs.01055. [DOI] [PubMed] [Google Scholar]
- 68.Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–54. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prasad R, Dewergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 70.Richardson MD. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother. 2005;56 1:i5–i11. doi: 10.1093/jac/dki218. [DOI] [PubMed] [Google Scholar]
- 71.Riezman H. Organization and functions of sphingolipid biosynthesis in yeast. Biochem Soc Trans. 2006;34:367–369. doi: 10.1042/BST0340367. [DOI] [PubMed] [Google Scholar]
- 72.Rogers PD, Barker KS. Genome-wide expression profile anlaysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother. 2003;47:1220–1227. doi: 10.1128/AAC.47.4.1220-1227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruetz S, Gros P. Phosphatidylcholine translocase: a physiological role for the mdr2 gene. Cell. 1994;77:1071–81. doi: 10.1016/0092-8674(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 74.Rustad TR, Stevens DA, Pfaller MA, White TC. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology. 2002;148:1061–1072. doi: 10.1099/00221287-148-4-1061. [DOI] [PubMed] [Google Scholar]
- 75.Saidane S, Weber S, De Deken X, St-Germain G, Raymond M. PDR16-mediated azole resistance in Candida albicans. Mol Microbiol. 2006;60:1546–1562. doi: 10.1111/j.1365-2958.2006.05196.x. [DOI] [PubMed] [Google Scholar]
- 76.Salin H, Fardeau V, Piccini E, Lelandais G, Tanty V, Lemoine S, Jacq C, Devaux F. Structure and properties of transcriptional networks driving selenite stress response in yeasts. BMC Genomics. 2008;9:333. doi: 10.1186/1471-2164-9-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanglard D, Ischer F, Bille J. Role of ATP-binding cassette transporter gene in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob Agents Chemother. 2001;45:1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother. 1999;43:2753–2765. doi: 10.1128/aac.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 80.Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2002;2:73–85. doi: 10.1016/s1473-3099(02)00181-0. [DOI] [PubMed] [Google Scholar]
- 81.Shahi P, Gulshan K, Moye-Rowley WS. Negative transcriptional regulation of multidrug resistance gene expression by an Hsp70 protein. J Biol Chem. 2007;282:26822–31. doi: 10.1074/jbc.M704772200. [DOI] [PubMed] [Google Scholar]
- 82.Shao PL, Huang LM, Hsueh PR. Recent advances and challenges in the treatment of invasive fungal infections. Int J Antimicrob Agents. 2007;30:487–495. doi: 10.1016/j.ijantimicag.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 83.Simonics T, Kozovska Z, Michalkova-Papajova D, Delahodde A, Jacq C, Subik J. Isolation and molecular characterization of the carboxy-terminal pdr3 mutants in Saccharomyces cerevisiae. Curr Genet. 2000;38:248–255. doi: 10.1007/s002940000164. [DOI] [PubMed] [Google Scholar]
- 84.Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2829–2824. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- 85.Talibi D, Raymond M. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J Bacteriol. 1999;181:231–240. doi: 10.1128/jb.181.1.231-240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li D, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Naar AM. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 87.Tsai HF, Krol AA, Sarti KE, Bennett JE. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother. 2006;50:1384–92. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 89.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol. 2006;61:704–22. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 90.Vermitsky JP, Edlind TD. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother. 2004;48:3773–81. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–61. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X, Cui Z, Miyakawa T, Moye-Rowley WS. Cross-talk between transcriptional regulators of multidrug resistance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:8812–8819. doi: 10.1074/jbc.M010686200. [DOI] [PubMed] [Google Scholar]
- 93.Zhang X, Kolaczkowska A, Devaux F, Panwar SL, Hallstrom TC, Jacq C, Moye-Rowley WS. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J Biol Chem. 2005;280:2759–70. doi: 10.1074/jbc.M408333200. [DOI] [PubMed] [Google Scholar]
- 94.Zhang X, Moye-Rowley WS. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the Fo component of the mitochondrial ATPase. J Biol Chem. 2001;276:47844–47852. doi: 10.1074/jbc.M106285200. [DOI] [PubMed] [Google Scholar]
- 95.Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, Raymond M. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell. 2008;7:836–847. doi: 10.1128/EC.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]