Abstract
Proteinases play a critical role in developmental homeostasis and in response to environ-mental stimuli. Our present research reports that a new cysteine protease, NtCP56, is involved in the development of pollen grains in Nicotiana tabacum L. The NtCP56 gene, which encodes a protein of 361 amino acid residues with a calculated molecular mass of 40 kDa, is strongly expressed in anthers. The recombinant NtCP56 showed a high activity towards casein. Kinetic analysis revealed a Km of 2.20 mg ml−1 and Vmax of 11.07 μg ml−1 min−1. The recombinant NtCP56 retained more than 50% of its maximum enzymatic activity from 20 °C to 60 °C with an optimum Tm range of 30–50 °C. The enzyme had a maximum activity at approximately pH 6.5. Suppression of the NtCP56 gene in anti-sense transgenic tobaccos resulted in the sterility of pollen grains. Our data indicated that, as a cysteine protease, NtCP56 might play an important role in pollen development.
Keywords: Cysteine protease, NtCP56, pollen, tobacco
Introduction
Male reproductive processes in flowering plants take place in the anther. In anthers, archesporial cells are thought to divide periclinally to give rise to parietal cells toward the exterior and to sporogenous cells toward the interior (Davis, 1966; Scott et al., 2004). Parietal cells finally differentiate into inner anther walls, including the endothecium, the middle layer and the tapetum, and sporogenous cells undergo several rounds of mitotic division and differentiate to acquire a meiotic cell fate (Nonomura et al., 2007).
Given the significance of male sterility to the agricultural industry, male gametophyte development has always been a major issue in plant studies and a few genes have been identified which control or participate in the development of pollen grains. In Arabidopsis, diverse factors including hormones, transcription factors, and receptors, have been reported to affect male gametophyte development (Sanders et al., 1999; Scott et al., 2004; Mizuno et al., 2007). For instance, a receptor-like protein kinase 2 (RPK 2), is a factor controlling anther development. The RPK2 T-DNA, inserted mutants displayed male sterility due to defects in anther dehiscence and pollen maturation (Mizuno et al., 2007).
More genes participate in the development of pollen grains by controlling tapetum formation and degradation. Two highly similar Arabidopsis LRR (leucine-rich repeat) receptor-like protein kinases, SERK1 and SERK2 (somatic embryogenesis receptor kinase), were shown to be required for tapetum formation in a way similar to that of EMS1/EXS (excess microsporocytes1/extra sporogenous cell) (Albrecht et al., 2005; Colcombet et al., 2005; Li et al., 2006). The Arabidopsis MS1 gene encodes a nuclear protein with a PHD-finger motif which has been shown to participate in the normal formation of the tapetum (Wilson et al., 2001; Ito and Shinozaki, 2002). In rice, an undeveloped tapetum1 (Udt1) gene was also shown to be vital to tapetum differentiation and the formation of microspores (Jung et al., 2005).
Until recently, the rice (Oryza sativa) tapetum degeneration retardation gene (TDR), encoding a putative basic helix–loop–helix protein, was found to be required in the degradation of the tapetum. In the TDR mutant rice, tapetum degeneration was retarded and the middle layer cells persisted, accompanied by aborted pollen development and complete male sterility. It was also verified that the direct targets of TDR were a cysteine protease OsCP1 and its inhibitor (Li et al., 2006).
Cysteine proteases (CPs) have been reported to be the major enzymes responsible for the hydrolysis of most of the storage proteins, hordeins and glutelin, in crops (Lee et al., 2004). CPs also contribute to the regulation of programmed cell death (Solomon et al., 1999). For instance, the expression of SmCP, a S. melongena gene encoding a cysteine proteinase, coincides with several, rather than with specific, events in developmentally regulated programmed cell death in brinjal (Xu and Chye, 1999).
In this study, a new cysteine protease gene isolated from Nicotiana tabacum L. is reported. The NtCP56 gene was strongly expressed in anthers. The recombinant NtCP56 protein showed activity towards casein proteolysis in vitro. In anti-sense NtCP56 transgenic tobacco anthers, the degradation of the tapetum was retarded and pollen development was aborted. These results provide important insights into the crucial role of NtCP56 for pollen development.
Materials and methods
Molecular cloning
The GenBank database was searched using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) with Nicotiana tabacum cysteine protease cDNA (GenBank accession no. AY881010) as query sequence. One Nicotiana tabacum partial mRNA sequence (GenBank accession no. U57825) encoding an endopeptidase-like protein with high similarity to Nicotiana tabacum cysteine protease cDNA was identified. Based on this sequence, five primers (primer 1: 5′-TTGTATGAGAGATGGAGAAGCCAT-3′; primer 2: 5′-AGTTGAAAATGCCCAGCAACTTCC-3′; primer 3: 5′-TGTATGCTCACGAGGATAG-3′; primer 4: 5′-CTGATCTAGAGGTACCGGATCC-3′; and primer 5: 5′-TTTAAACATGAAGAAATTATTACTAC-3′) were designed to amplify Nicotiana tabacum NtCP56 cDNA.
Total RNA from anthers of Nicotiana tabacum L., isolated using the Trizol reagent, was used for a reverse-transcription polymerase chain reaction (RT-PCR). The first strand cDNA was synthesized in a volume of 25 μl containing about 2 μg RNA, 4 μl reaction buffer, 1 μl of 10 mM dNTP, 0.5 μl RNase inhibitor, 1 μl adaptor primer (5′-CTGATCTAGAGGTACCGGATCCTTTTTTTTTTTTTTTTT-3′), and 1 μl AMV reverse transcriptase. The transcription reaction was performed at 42 °C for 1 h and terminated at 65 °C for 10 min (Li et al., 2002).
A specific cDNA fragment was first amplified by using primers 1 and 2. To isolate the complete region of this gene, a 3′ race PCR was carried out by using primer 3 and primer 4 according to the instructions of the manufacturer (Takara 3′ full race kit, Japan). The PCR was carried out as follows: 94 °C for 5 min, followed by 25 cycles of 94 °C for 1 min, 56 °C for 1 min, and 72 °C for 2 min. To verify the integrity of the cDNA, sequences of the gene RT-PCR were carried out to amplify the full-length cDNA using primers 4 and 5. The PCR thermal cycles were carried out as follows: 94 °C for 5 min, followed by 25 cycles of 94 °C for 1 min, 45 °C for 1 min, and 72 °C for 2 min. The PCR product was cloned into the pMD18-T vector (Takara, Japan) and sequenced using the chain-terminating method with an ABI automated sequencer (model no. 377). The cDNA from Nicotiana tabacum L. was termed NtCP56 (GenBank accession no. EU429306).
Phylogenetic analysis
A phylogenetic tree between NtCP56 and the other major classes of plant CPs was constructed with MEGA v.2.1 by using the Neighbor–Joining (NJ) method with bootstrapping 500 replicates.
Northern blotting analysis
For the RNA gel blot analysis, 25 μg of RNA was fractionated by gel electrophoresis in a 1.2% formaldehyde agarose gel, then transferred to a Hybond-N+ membrane (Amersham, UK) after UV cross-linking (Yu et al, 2003). The gene-specific DNA probe was prepared by amplifying a 356 bp 5′ cDNA fragment of NtCP56. The probe was labelled using a DIG-primer labelling kit according to the instructions of the manufacturer (Roche, Germany). Membrane-bound DNA was prehybridized for 1 h and the hybridization of probes took place overnight at 42 °C. Two subsequent stringency washes were performed at 25 °C for 10 min and at 65 °C for 30 min. The first washing solution contained 0.1% SDS (w/v), 2× SSC, and the second 0.1% SDS (w/v), 0.5× SSC. Kodak X-Omat AR-5 films were exposed to the blots (Amersham, UK).
Expression, purification, renaturation, and sequencing of recombinant NtCP56
The open reading frame minus the first 60 bp of the cDNA encoding NtCP56 was amplified. The PCR product was digested with KpnI and SacI and subcloned into the plasmid expression vector pET30a (Novagen Inc., Madison, WI. USA). Fresh colonies of BL21 (DE3) bacteria transformed with plasmid pET30a/NtCP56 were incubated overnight at 37 °C. Overnight cultures were diluted to 1:100 in a 1.0 l fermenter and grown to mid-log phase. 0.4 mM IPTG was added and cultures were incubated for 3–5 h following induction. Bacterial pellets were harvested and stored at –70 °C (Smith and Gottesman, 1989).
The resuspended bacteria were disrupted by cold sonication in a Heat Systems-Ultrasonic sonifier cell disruptor at 200 W (120×6 s) after suspension in the buffer; the insoluble protein fractions were isolated by centrifugation for 10 min at 12 000 g. The pellets were washed twice in the same buffer. The insoluble protein pellets were resuspended using 3 ml g−1 bacteria of buffer A (10 mM TRIS-HC1, 8 M urea, 50 mM NaH2PO4 and the pH adjusted to 8.0 using NaOH) and incubated at 28 °C for 1 h.
The resuspended solution were added to the Ni2+-NTA resin (4 ml) which was equilibrated by buffer A in a 15 ml conical tube and gently shaken at 4 °C for 1 h. Then the bottom cap was removed and the flow-through was collected. The column was washed with 20 column volumes of washing buffer B (10 mM TRIS-HC1, 8 M urea, 50 mM NaH2PO4 and the pH adjusted to 6.3 using HCl) and buffer C (10 mM TRIS-HC1, 8 M urea, 50 mM NaH2PO4 with the pH adjusted to 5.9 using HCl). The over-expressed protein was then eluted in 4 ml fractions in buffer D (10 mM TRIS-HC1, 8 M urea, 50 mM NaH2PO4 and the pH adjusted to 4.5 using HCl) (Handbook for high-level expression and purification of 6× His-tagged proteins, Novagen).
Denatured proteins in a dialysis bag were immersed into a 50-fold volume renaturation buffer (50 mM potassium phosphate, pH 10.7, 5 mM EDTA, 1 mM reduced glutathione, 0.1 mM oxidized glutathione, 6 M/4 M/2 M/0 M urea) (Smith and Gottesman, 1989) and stirred at 4 °C for 3–4 h. Then the buffer was refreshed in turn by a reduction of urea.
Recombinant NtCP56 was processed to a mature form at pH 3.0 with cysteine (Troen et al., 1988). NH2-terminal amino acid sequencing was accomplished by the 2,4-dinitro-l-fluorobenzene (DNFB) method.
Enzyme assays and kinetic studies
Enzyme activity of purified recombinant NtCP56 was routinely assayed with casein (Sigma, USA) as substrate, using the method described by Sasaki et al. (1977) with minor modifications. Assays with up to 1% casein were carried out at 37 °C and pH 7.2, following 30 s activation incubation at 37 °C, pH 3.0, as described by previous reports (Troen et al., 1988). Casein (1 ml of 0.04–1% solution in a phosphate-buffered saline containing 6 mM cysteine) was mixed with recombinant processed protein and the tubes were incubated at 37 °C for 3 min. After incubation, the proteins were precipitated by the addition of 2 ml of a 20% tricarboxylic acid buffer and centrifuged at 12 000 g for 10 min. The absorbance of the clear supernatant was measured by a spectrophotometer at 275 nm. Controls, such as substrate and enzyme blanks, were included to obtain a background absorbance that was subtracted from the enzyme-containing tubes to yield a specific degradation of casein. The following two buffers were used: 100 mM sodium acetate (pH 3.0–5.0) and 100 mM sodium phosphate (pH 6.0–8.0). Kinetic constants were analysed by Matlab software. Leupetin (100 μM), antipain (100 μM), and tosyl-L-lysine chloromethyl ketone (TLCK) (100 μM) were used as inhibitors.
Anti-sense plasmid construction and generation of transgenic plants
The 356 bp fragment of NtCP56 near the 5’ UTR was amplified and a BamHI and XbaI restriction enzyme site was created by PCR. After digestion with BamHI and XbaI restriction enzymes, the fragment was cloned into pBI121 (Clontech, USA) in an anti-sense orientation to create the 35S-anti-NtCP56 fusion construct. The fusion construct plasmid was first introduced into Agrobacterium tumefaciens LBA4404 and verified by using PCR. The empty vector (pBI121) was used as a control. Tobacco plant transformation from leaf explants was carried out as described by Lu et al. (2003).
Histological study
Anthers of control and transgenic tobacco were collected in a series of development stages. Anthers were fixed with FAA (5% formalin, 5% acetic acid, and 70% ethanol, by vol.) for 24 h under negative pressure and dehydrated in a set of increasing solutions (85%, 95%, and 100% by vol.), then decolourized in solutions of 50% dimethylbenzene and 50% ethanol (v/v) and dimethylbenzene and finally embedded in paraffin. Paraffin-embedded anthers were cut to a thickness of 10 μm and stained with safranine and a fast green solution.
Results
Isolation and characterization of NtCP56
A 1289 bp cDNA was cloned from Nicotiana tabacum L. anthers. The cDNA contained a 1083 bp open reading frame (ORF), 7 bp of 5′ untranslated region (UTR), and a 199 bp 3′ UTR. The ORF encodes a peptide of 361 residues which has a predicted molecular mass of 43 kDa (Fig. 1). This cDNA shows 36–83% similarity with other KDEL-tailed plant cysteine proteinases, including the Solanum lycopersicum KDEL-tailed cysteine endopeptidase, Ricinus communis Cys-EP (Schmid et al., 1998), Phaseolus vulgaris EP-C1 (Tanaka et al., 1991), Glycine max CysP1 (Ling et al., 2003), and Vigna mungo sulphydryl endopeptidase SH-EP (Akasofu et al., 1989). Then the cDNA is named as NtCP56. A phylogenetic analysis was performed using the MEGA v.2.1 programme (Fig. 2). The NtCP56 protein is located in the group which contains KDEL-tailed family members. Given this analysis, the NtCP56 protein can be classified as belonging to the subfamily C1A of papain-like cysteine proteinases (MEROPS peptidase database, http://merops. sanger.ac.uk; Rawlings et al., 2008).
Fig. 1.
Nucleotide and deduced amino acid sequence of NtCP56. The signal peptide (M1-S20) of NtCP56 is shown in bold. The catalytic triad Cys, His, and Asn and also the Glu active site residue are circled. The GCNGG motif is double-underlined. ERFNIN motif (E53-N72) of NtCP56 is shown in a rectangular box and the KDEL (K358-L361) motif is single underlined. The arrow indicates the site of auto-hydrolysis. Numbers on the left and right margins represent nucleotide and deduced amino acid sequences, respectively.
Fig. 2.
Phylogenetic tree showing relationships between NtCP56 and other plant CPs. Bootstrap values from a sample of 500 replicates are shown on each branch using the following sequences NtCP (Nicotiana tabacum, GenBank accession no. AAW78660); SlCP (Solanum lycopersicum, GenBank accession no. ABV22590); GmCP (Glycine max, GenBank accession no. BAC77523); PvEP-C1 (Phaseolus vulgaris, GenBank accession no. CAA40073); VmCP (Vigna mungo, GenBank accession no. P12412); HaCP (Helianthus annuus, GenBank accession no. BAC75924); AtCP (Arabidopsis thaliana, GenBank accession no. NP_568722); HemCP (Hemerocallis hybrid cultivar, GenBank accession no. P43156); IhCP (Iris×hollandica, GenBank accession no. AAR92155); SaCP (Sandersonia aurantiaca, GenBank accession no. AAD28477); OsCP (Oryza sativa, GenBank accession no. AAD20453); TrCP (Trifolium repens, GenBank accession no. AAP32196); MtCP (Medicago truncatula, GenBank accession no. AAQ63885); LjCP (Lotus japonicus, GenBank accession no. BAF56430); DcCP (Daucus carota, GenBank accession no. BAD29956); AdCP (Actinidia deliciosa, GenBank accession no. ABQ10200); HaCP (Helianthus annuus, GenBank accession no. BAC75923); BoCP (Brassica oleracea, GenBank accession no. AAL60580); ZmCP (Zea mays, GenBank accession no. NP_001104879).
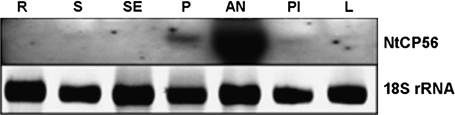
The expression of the NtCP56 gene in different tissues of tobacco is determined using Northern blotting (Fig. 3). The result showed that the NtCP56 gene is strongly expressed in anthers and weakly expressed in petal, pistil, stem, and leaf.
Fig. 3.
Northern hybridization showing NtCP56 expression in the tissue of Nicotiana tabacum L. NtCP56 mRNA abundance was analysed by using the 5’ end of NtCP56 cDNA as a probe. Each lane was loaded with 20 μg total RNA. R, root; S, stem; SE, sepal; P, petal; AN, anther; PI, pistil; L, leaf. To ensure equal sample abundance on gels 18S rRNA was used to monitor loading equivalence.
Expression, purification, and sequencing of recombinant NtCP56 protein
The plasmid which encodes the full-length NtCP56 minus the signal peptide was efficiently expressed in E. coli. Three hours after induction with IPTG, recombinant NtCP56 represented approximately 20% of the total bacterial protein. The molecular weight of the purified recombinant NtCP56 was estimated to be approximately 43 kDa according to the SDS-PAGE analysis (Fig. 4, Lane 3). This 43 kDa protein did not have any determinable activity (data not shown). After renaturation, the purified protein could be processed into a 33 kDa mature form at pH 3.0 (Fig. 4, Lane 4). The NH2-terminal amino acid sequence of processed recombinant NtCP56 was HEDSVPP which, with four additional NH2-terminal amino acids (HEDS), was compared with the predicted autocatalysis.
Fig. 4.
Purification of recombinant NtCP56 and processed protein. Protein samples were subjected to SDS-polyacrylamide gel electrophoresis under reducing conditions and stained with Coomassie Brilliant Blue R-250. Lane M, molecular mass markers with the sizes shown on the left in kDa. Lane 1, induced BL21 (DE3)+PET30a whole cell lysate. Lane 2, induced BL21 (DE3)+pET30a/NtCP56 whole cell lysate. Lane 3, 43 kDa purified recombinant NtCP56. Lane 4, 33 kDa processed mature recombinant NtCP56.
Biochemical characterization of processed recombinant NtCP56
The steady-state kinetics of the enzyme was studied in assays with various concentrations of casein as substrates. Processed recombinant NtCP56 obeyed the Michaelis–Menten-type of kinetics towards casein. The Km (2.20±0.04 mg ml−1) and Vmax (11.07±0.23 μg·ml−1 min−1) values for the recombinant enzyme are comparable to those reported for papain-like proteins from different species (Table I). The activity of the recombinant NtCP56 is pH-dependent. The highest activity was observed at pH 6.5, close to the optimal pH 7.0 of commercial papain (Hoover and Kokes, 1946). The enzyme still showed 53% of its maximum activity between pH 3.5 to pH 7.5, but only 28% at pH 8.5 (Fig. 5A), which was similar to the results of a previous report (Smith and Gottesman, 1989). The optimum temperature for NtCP56 activity was at 30 °C. The processed recombinant NtCP56 retained more than 50% of its maximum enzymatic activity between 20–60 °C (Fig. 5B).
Table 1.
Enzymatic properties of recombinant NtCP56
| Recombinant NtCP56 | Papaina | Fruit bromelainb | |
| Km (mg ml−1) | 2.20±0.04 | 1.4709 | 0.7 |
| Vmax (μg ml−1 min−1) | 11.07±0.23 | 5.5252 | 134.2 |
| Kcat (min−1) | 1.025 | ||
| Kcat/Km (ml mg−1 min−1) | 0.46 | ||
| Inhibitor action (%) | |||
| None | |||
| Leupetin (100 μM) | 45.76% | ||
| Antipain (100 μM) | 18.60% | ||
| Tosyl-L-lysine chloromethyl ketone | 11.02% | ||
| (TLCK) (100 μM) |
Fig. 5.
Effect of pH and Tm on the enzyme activity. (A) pH stability of processed recombinant NtCP56. (B) Thermal stability of processed recombinant NtCP56.
Phenotypic analyses of anti-NtCP56 transgenic tobacco
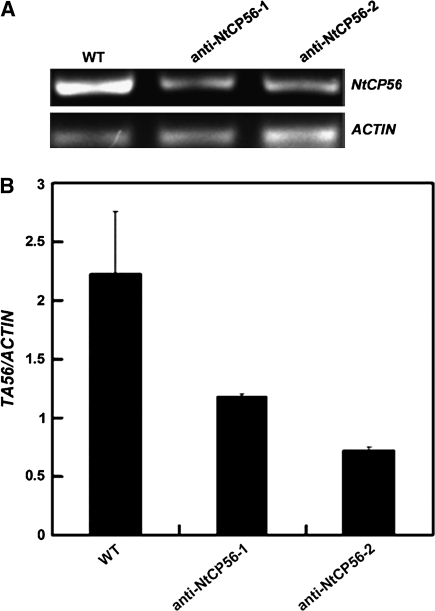
In order to discover the function of NtCP56 in anther development, anti-NtCP56 transgenic tobaccos were obtained. In order to ascertain whether or not the NtCP56 gene plays a crucial role in the growth and development of tobacco plants, the root length and plant height of transgenic and control tobacco plants were measured each week. All 14 transgenic plants are phenotypically indistinguishable from the control plants under normal growing conditions. Compared with the control, the mRNA level of NtCP56 was reduced by 45–67% (Fig. 6A, B).
Fig. 6.
Relative expression level of NtCP56 mRNA in control, anti-NtCP56-1 and anti-NtCP56-2 tobaccos. (A) Results of the semi quantity RT-PCR by 1% agarose. (B) Quantification of the result of the semi quantity RT-PCR.
Anther development was concisely divided into six stages based on a previous classification of anther development (Feng et al., 2001). During the microspore mother cell (MMC) stage, there was no detectable morphological difference in anthers between the control and anti-sense transgenic plants. Normal microsporocytes, an epidermis, an endothecium, a middle layer, and a tapetum were found both in controls and in transformation anthers (Fig. 7A, B). Up to the tetrad stage, there was still no obvious difference in anther cellular morphology between the control and anti-sense transformations. During this period, both in the control and in transgenic anthers, microsporocytes underwent meiosis to form tetrads of four haploid microspores. The tapetal cells had began to differentiate and their cytoplasm became deeply stained (Fig. 7C, D) (Li et al., 2006). Subsequently, the transgenic anthers had detectable morphological abnormalities. At the young microspore stage, in control anthers, microspores were released from tetrads and dispersed independently in locules (Fig. 7E). Partial tapetal cells were degenerated and the tapetum became thin (Fig. 7M). By contrast, in transformation anthers the tapetum remained relatively thick (Fig. 7N). Next, from the young pollen stage to the pollen mitosis stage, in control anthers, the uninucleate pollen developed to dianucleate pollen through mitotic divisions and dispersed in locules (Fig. 7G, I). Meanwhile, almost all tapetal cells had differentiated and degenerated (Fig. 7O). However, in the anti-NtCP56 transgenic anthers, abnormal microspores collapsed and remained undeveloped following the release from tetrads (Fig. 7H, J). Partial tapetum was still retained during this period (Fig. 7P). At the mature pollen stage, control pollen grains were full of starch, lipids, and nutrients (Fig. 7K) and the tapetum was fully degenerated (Fig. 7Q). By contrast, in transformation anthers, many pollen grains had become abnormal and did not mature (Fig. 7L, R). Large numbers of stagnated, deformed, and aggregated pollen grains were observed, and particularly in the corners of the locules (Fig. 7T). Most of the pollen grains did not stain densely, which suggested immaturity. In addition, from the young pollen stage, the cells of the endothecium gradually became fibrotic in control anthers. By contrast, in the transformation of anti-NtCP56, the endothecium cells remained loosely arrayed and did not have clear fibrotic cell walls.
Fig. 7.
Lengthways section comparison of the anther development of the control and the anti-NtCP56 transgenic tobaccos. Six stages of anther development in the control and the corresponding stages of development in the anti-NtCP56 transformation were compared. Control sections are shown in (A), (C), (E), (G), (I), (K), (M), (O), (Q), (S), and (B), (D), (F), (H), (J), (L), (N), (P), (R), and (T) show corresponding anti-NtCP56 sections. (S) and (T) show the locules corner sections of control and anti-NtCP56 transgenic tobaccos in the last stage. Tds, tetrads; PG, pollen grains; Ms, microspore; Tp, tapetum; Ep, epidermis; En, endothecium.; ML, middle layer. (A, B) The MMC stage; (C, D) The TDS stage; (E, F, M, and N) The young microspore stage; (G, H, O, P) The single nuclear pollen stage; (I, J) The dinuclear pollen stage; (K, L, Q, R, S, T) The mature pollen stage. (Bars=50 μm).
In order to validate the sterility of transformation pollen grains further, the germination rate of pollen grains in vitro were determined. The germination rate of pollen grains is 41.3% of transgenic lines (102 in 247) and 5.3% of control lines (17 in 318). The germination rate of control lines is about eight times higher than that of transgenic lines. This result showed most transgenic pollen grains were abnormal in development.
These results suggested that the suppression of NtCP56 resulted in defects in pollen grain development.
Discussion
In this study, a gene from the tobacco anther, which is coded for a cysteine protease named NtCP56 has been isolated and characterized. Given the alignment to other plant papain-like cysteine proteases, the sequence of NtCP56 was identified as a signal peptide that would be processed prior to enzyme activation. Like other papain-like cysteine proteinases, three highly conserved catalytic residues Cys150, His286, and Asn307 constitute the catalytic triad of cysteine proteases (Guerrero et al., 1998). Another conserved residue, Gln144, is involved in maintaining an active enzyme conformation (Kamphuis et al., 1985). NtCP56 protein also contains a conserved non-contiguous ERFNIN motif (EX3RX3FX2NX3I/VX3N) which is typical for cysteine proteases in the cathepsin L and H-like proteinases but not in cathepsin B-like proteinases (Wiederanders, 2003). A GCNGG motif was identified in NtCP56. With the exception of the central Asn (N) residue, this GCNGG motif is invariant in all ERFNIN proteinases and also in the cathepsin B-like proteinases (Beyene et al., 2006). In addition, a C-terminal tetrapeptide KDEL is present. It has been suggested that this is an endoplasmic reticulum (ER) retention signal (Fig. 1) (Guerrero et al., 1998).
Foreign proteins overexpressed in E. coli frequently formed insoluble aggregates or inclusion bodies in the bacterial cytoplasm (Marston, 1986). When plasmid pET30a-NtCP56 was expressed in E. coli, the recombinant proteins quickly formed inclusion bodies and could only be solubilized in strong detergents such as 8 M urea. Denatured soluble protein was eluted at pH 4.5 after purification by Ni-NTA resin. Renaturation was carried out at neutral or alkaline pH to ensure recovery of the pro-enzyme in order not to be autocatalysed (Gal and Gottesman, 1986). This might be a result of the difference between the maturation conditions in vitro and in the plant (Vernet, 1990).
The NtCP56 gene is strongly expressed in anthers. This result is analogous to OsCP1 reported in 2004, where its promoter was highly active in the anther, but not in other flower organs and vegetative organs of rice (Lee, 2004). This is different with most homologous CPs which have been reported and participated in processing and degradation of seed storage proteins (Shimada et al., 1994; Toyooka et al., 2000), fruit ripening (Alonso and Granell, 1995) and also in legume nodule development (Naito et al., 2000). But it was similar to OsCP1 isolated from rice, which is involved in pollen development (Lee et al., 2004). It has been suggested that cysteine proteases have separate roles in different tissues (Lee et al., 2004). RD21 (Yamada et al., 2001), a cysteine protease that belongs to the papain family in Arabidopsis, has a role in the degradation of cellular proteins during leaf senescence, rather than in the degradation of seed storage protein after seed germination. Thus, it is speculated that NtCP56 is a novel cysteine protease which might be involved in anther development.
It has been demonstrated that the protease activity increased during anther development (DeGuzman and Riggs, 2000). This enhanced activity can be correlated with morphological and biochemical events during late micro-sporogenesis which require proteolytic enzymes. Pollen development is a dynamic process that involves protein synthesis required for the development (Lee et al., 2004). In addition, the formation and degradation process of the tapetum which supplies nutritional sources to developing pollen was also vital to pollen development (Ku et al., 2003).
An anti-sense approach was used to analyse the role of NtCP56 in pollen development. Until the tetrad stage, the anther development of transgenic plants was normal. However, after that, development of both tapetum and microspores was disrupted in anti-NtCP56 anthers. It suggested that NtCP56, as a cysteine protease, may participate in new protein synthesis for pollen development or aid in the degenerative process of the tapetum. It is either activating hydrolytic enzymes that degrade cellular macromolecules, or by functioning as a hydrolytic enzyme, or both (Koltunow et al., 1990). At present, the mechanism of NtCP56 during pollen development remains to be explained.
Acknowledgments
Financial source: National Natural Science Foundation of China (Grant No.30070612). This work was supported by grant from the National Basic Research Program of China (2009CB119104). The authors also thank Jing-chuan Du, Xu Wang (The Chinese Academy of Agriculture Sciences), Nan Qiao and members of the Botany laboratory for support during data analysis.
References
- Akasofu H, Yamauchi D, Mitsuhashi W, Minamikawa T. Nucleotide sequence of cDNA for sulfhydryl-endopeptidase (SHEP) from cotyledons of germinating Vigna mungo seeds. Nucleic Acids Research. 1989;17:6733. doi: 10.1093/nar/17.16.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. The Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Granell A. A putative vacuolar processing protease is regulated by ethylene and also during fruit ripening in citrus fruit. Plant Physiology. 1995;109:541–547. doi: 10.1104/pp.109.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene G, Foyer CH, Kunert KJ. Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco (Nicotiana tabacum L.) leaves. Journal of Experimental Botany. 2006;57:1431–1443. doi: 10.1093/jxb/erj123. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. The Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TA. Floral structure and stamens in Bombax ceiba L. Journal of Genetics. 1966;59:294–299. [Google Scholar]
- DeGuzman R, Riggs CD. A survey of proteinases active during meiotic development. Planta. 2000;210:921–924. doi: 10.1007/s004250050698. [DOI] [PubMed] [Google Scholar]
- Feng JH, Lu YG, Liu XD, Xu XB. Pollen development and its stages in rice (Oryza sativa L.) Chinese Journal of Rice Science. 2001;15:21–28. [Google Scholar]
- Gal S, Gottesman MM. The major excreted protein of transformed fibroblasts is an activable acid-protease. Journal of Biological Chemistry. 1986;261:1760–1765. [PubMed] [Google Scholar]
- Guerrero C, Calle M, Reid MS, Valpuesta V. Analysis of the expression of two thioprotease genes from day lily (Hemerocallis spp.) during flower senescence. Plant Molecular Biology. 1998;36:565–571. doi: 10.1023/a:1005952005739. [DOI] [PubMed] [Google Scholar]
- Hoover SR, Kokes ELC. Effect of pH upon proteolysis by papain. Journal of Biological Chemistry. 1946:199–207. [PubMed] [Google Scholar]
- Ito T, Shinozaki K. The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant and Cell Physiology. 2002;43:1285–1292. doi: 10.1093/pcp/pcf154. [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. The Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis IG, Drenth J, Baker EN. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence from cathepsins B and H, and stem bromelain. Journal of Molecular Biology. 1985;182:317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. The Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S, Yoon H, Suh HS, Chung YY. Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta. 2003;217:559–565. doi: 10.1007/s00425-003-1030-7. [DOI] [PubMed] [Google Scholar]
- Lee S, Jung KH, An G, Chung YY. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Molecular Biology. 2004;54:755–765. doi: 10.1023/B:PLAN.0000040904.15329.29. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. The Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QZ, Li XG, Bai SN, Lu WL, Zhang XS. Isolation of HAG1 and its regulation by plant hormones during in vitro floral organogenesis in Hyacinthus orientalis L. Planta. 2002;215:533–540. doi: 10.1007/s00425-002-0796-3. [DOI] [PubMed] [Google Scholar]
- Ling J, Kojima T, Shiraiwa M, Takahara H. Cloning of two cysteine proteinases genes: CysP1 and CysP2, from soybean cotyledons by cDNA representational difference analysis. Biochimica et Biophysica Acta. 2003;1627:129–139. doi: 10.1016/s0167-4781(03)00082-4. [DOI] [PubMed] [Google Scholar]
- Lu H, Zeng QY, Zhao YL, Jiang XN. Xylem-specific expression of a GRP1.8 promoter::4CL gene construct in transgenic tobacco. Plant Growth Regulation. 2003;41:279–286. [Google Scholar]
- Marston FA. The purification of eukaryotic polypeptides synthesized in Escherichia coli. Biochemical Journal. 1986;240:1–12. doi: 10.1042/bj2400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Shinozaki KY. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. The Plant Journal. 2007;50:751–766. doi: 10.1111/j.1365-313X.2007.03083.x. [DOI] [PubMed] [Google Scholar]
- Naito Y, Fujie M, Usami S, Murooka Y, Yamada T. The involvement of cysteine proteinase in the nodule development in Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei. Plant Physiology. 2000;124:1087–1096. doi: 10.1104/pp.124.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. The Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. MEROPS, the peptidase database. Nucleic Acids Research. 2004;32:160–164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction. 1999;11:297–322. [Google Scholar]
- Sasaki M, Minakata K, Yamamoto H, Niwa M, Kato T, Ito N. A new serum component which specifically inhibits thiol proteinases. Biochemical and Biophysical Research Communications. 1977;76:917–924. doi: 10.1016/0006-291x(77)91589-3. [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Kalousek F, Gietl C. A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta. 1998;206:466–475. doi: 10.1007/s004250050423. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. The Plant Cell. 2004;16:46–60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme of soybean that converts proproteins to the corresponding mature forms. Plant and Cell Physiology. 1994;35:713–718. doi: 10.1093/oxfordjournals.pcp.a078648. [DOI] [PubMed] [Google Scholar]
- Smith SM, Gottesman MM. Activity and deletion analysis of recombinant human cathepsin L expressed in Escherichia coli. Journal of Biological Chemistry. 1989;264:20487–20495. [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne B, Menachem E, Levinea A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. The Plant Cell. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yamauchi D, Minamikawa T. Nucleotide sequence for an endopeptidase (EP-C1) from pods of maturing Phaseolus vulgaris fruits. Plant Molecular Biology. 1991;16:1083–1084. doi: 10.1007/BF00016081. [DOI] [PubMed] [Google Scholar]
- Toyooka K, Okamoto T, Minamikawa T. Mass transport of a proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. Journal of Cell Biology. 2000;148:453–563. doi: 10.1083/jcb.148.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troen BT, Ascherman D, Atlas D Gottesman MM. Cloning and expression of the gene for the major excreted protein of transformed mouse fibroblasts. Journal of Biological Chemistry. 1988;263:254–261. [PubMed] [Google Scholar]
- Varnier AL, Mazeyrat-Gourbeyre F, Sangwan RS, Clement C. Programmed cell death progressively models the development of anther sporophytic tissues from the tapetum and is triggered in pollen grains during maturation. Journal of Structural Biology. 2005;152:118–128. doi: 10.1016/j.jsb.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Vernet T, Tessier DC, Richardson C, Laliberte F, Khouri HE, Bell AW, Storer AC, Thomas DY. Secretion of functional papain precursor from insect cells. Journal of Biological Chemistry. 1990;265:16661–16666. [PubMed] [Google Scholar]
- Wiederanders B. Structure–function relationships in class CA1 cysteine peptidase propeptides. Acta Biochimica Polonica. 2003;50:691–713. [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. The Plant Journal. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- Xiao GP. Ultrasonic extraction technology of papain and its enzymatic properties. Journal of Fujian Agriculture and Forestry University. 2005;34:318–323. [Google Scholar]
- Xu FX, Chye ML. Expression of cysteine proteinase during developmental events associated with programmed cell death in brinjal. The Plant Journal. 1999;17:321–327. doi: 10.1046/j.1365-313x.1999.00370.x. [DOI] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiology. 2001;127:1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shena WH. The tobacco A-type cyclin, Nicta; CYCA3;2, at the nexus of cell division and differentiation. The Plant Cell. 2003;15:2763–2777. doi: 10.1105/tpc.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WL, Wu W, Li CM, Wang XJ. Kinetic study of fruit bromelain. Journal of China Agricultural University. 1999;4:11–13. [Google Scholar]