Abstract
At least seven isoforms (PsABI3-1 to PsABI3-7) of a putative, pea ABI3-like factor, originated by alternative splicing, have been identified after cDNA cloning. A similar variability had previously only been described for monocot genes. The full-length isoform, PsABI3-1, contains the typical N-terminal acidic domains and C-terminal basic subdomains, B1 to B3. Reverse transcriptase-PCR analysis revealed that the gene is expressed just in seeds, starting at middle embryogenesis; no gene products are observed in embryo axes after 18 h post-imbibition although they are more persistent in cotyledons. The activity of the isoforms was studied by yeast one-hybrid assays. When yeast was transformed with the isoforms fused to the DNA binding domain of Gal4p, only the polypeptides PsABI3-2 and PsABI3-7 failed to complement the activity of Gal4p. Acidic domains A1 and A2 exhibit transactivating activity, but the former requires a small C-terminal extension to be active. Yeast two-hybrid analysis showed that PsABI3 is able to heterodimerize with Arabidopsis thaliana ABI5, thus proving that PsABI3 is functionally active. The minimum requirement for the interaction PsABI3–AtABI5 is the presence of the subdomain B1 with an extension, 81 amino acids long, at their C-terminal side. Finally, a transient onion transformation assay showed that both the active PsABI3-1 and the inactive PsABI3-2 isoforms are localized to nuclei. Considering that the major isoforms remain approximately constant in developing seeds although their relative proportion varied, the possible role of splicing in the regulatory network of ABA signalling is discussed.
Keywords: Abscisic acid, ABA, ABI3, ABI5, alternative splicing, isoforms
Introduction
Abscisic acid (ABA) signalling plays important roles in plants and many of the effects of this phytohormone include changes in gene expression (Busk and Pagès, 1998). In particular, many of the genes involved in seed development and dormancy and in the response to multiple stresses are targets for ABA (for reviews, see Finkelstein et al., 2002, 2008; Xiong et al., 2002; Santos-Mendoza et al., 2008; Suzuki and McCarty, 2008). The mechanisms for ABA signal perception are elusive although other aspects of ABA signalling, such as the involvement of MAP kinase pathways and their cross-talk with other signals (reviewed in Mishra et al., 2006; Hirayama and Shinozaki, 2007) are known.
ABA-regulated genes are characterized by the presence of several cis elements (Marcotte et al., 1989). Among them, the ABA response elements (ABREs) were soon characterized (Guiltinan et al., 1990; Mundy et al., 1990; Skriver et al., 1991). They share a sequence with the highly conserved motif ACGT, acting in co-ordination with other elements, which have mainly been studied in Arabidopsis thaliana. They comprise the proximal RY elements (consensus sequence CATGCA), and some distal sequences, such as the coupling elements (CE), and the sequences binding MYB and MYC-like transcription factors (reviewed in Finkelstein et al., 2002). ABREs bind bZIP-type transcription factors (Guiltinan et al., 1990), and some of them were identified as putative ABA-responsive factors in several species (Oeda et al., 1991; Nakagawa et al., 1996; Nantel and Quatrano, 1996), but until the cloning of rice TRAB1 (Hobo et al., 1999) and Arabidopsis ABI5 (Finkelstein and Lynch, 2000) was achieved, a clear link between bZIP factors and ABA signalling was not established. Since then, many other ABRE-binding bZIP-type factors have been identified (see, for instance, Nieva et al., 2005), and it is known that they are able to heterodimerize with the ABI3-type transcription factors, which may thus be tethered to their target promoters (Finkelstein et al., 2002). The maize VP1 gene (McCarty et al., 1991) is orthologous to Arabidopsis ABI3 and both ABI5 and ABI3 families of ABA-responsive trans-acting factors are conserved in monocots and dicots (Finkelstein et al., 2002, and references herein).
The physical interaction between ABI3 and ABI5-type factors has been demonstrated (Nakamura et al., 2001), although other authors were unable to reproduce those results (López-Molina et al., 2002). The activator domains of the VP1/ABI3 factors reside in the N-terminal region (Hoecker et al., 1995). The VP1/ABI3 family is characterized by the presence of three basic domains in their C-terminal region of their members. The B1 domain is required to interact with ABI5 (Nakamura et al., 2001), the B2 domain is involved in both transactivation and nuclear localization (Marella and Quatrano, 2007) and the RY-binding capacity resides in the B3 domain (Suzuki et al., 1997). The presence of B3 domains is not restricted to the VP1/ABI3 factors. On the contrary, they are found in the members of a superfamily of plant-specific DNA-binding proteins, the AFL (ABI3/FUS3/LEC2) B3 domain factors (for a recent review, see Suzuki and McCarty, 2008).
Recent evidence indicates that the transcripts of genes of the ABI3/VP1 and ABI5 families are subject to alternative splicing and/or unusual post-transcriptional processing in monocots. For instance, wheat and rice VP1 transcripts undergo many mis-splicing events, which often result in truncated polypeptides (McKibbin et al., 2002; Wilkinson et al., 2005; Fan et al., 2007). Apart from the evident relation of this multiple processing with preharvest sprouting, the possibility exists that the developmental regulation of post-transcriptional processing of VP1 transcripts (Fan et al., 2007) plays a distinct functional role. This question needs further clarification, because the two different variants of the OsABI5 factor that result from the alternative splicing of a single gene transcript exhibit different regulatory properties (Zou et al., 2007).
In our laboratory we had characterized a chromosomal protein, p16, which accumulates in the mature seeds of Pisum sativum, and the psp54 gene, which encoded a bicupin precursor of p16 was cloned. The expression of the psp54 gene is regulated, among other factors, by ABA (Castillo et al., 2000, 2002, 2005). These circumstances focused our attention on the mechanisms of ABA-driven gene induction in pea seeds, and we first thought of characterizing some of the ABA-responsive, pea trans-acting factors. A P. sativum DNA sequence sharing homology with A. thaliana ABI3 (AtABI3) was obtained by Nakako and Mori (2002) and deposited in the GenBank database (accession no. AB080195). Nevertheless, no further characterization of either the gene or its products has been described.
In the present paper, the cloning of a P. sativum ABI3-like gene (PsABI3) is described, which gives rise, through alternative splicing events, to seven mRNAs. The three major mRNAs are differentially expressed during seed maturation and germination and two of the polypeptide products display transactivating properties in a yeast one-hybrid system and are able to form heterodimers with A. thaliana ABI5 factor, while the third major isoform is inactive. The possible regulatory properties of the alternative splicing are discussed.
Materials and methods
Plant materials and growth conditions
Pea (P. sativum, cv. Lincoln) seeds were obtained locally. They were cultured in a greenhouse when adult plants were required to study gene expression in different organs of the plant. When the ungerminated or germinated embryos were needed, seeds were imbibed at 4 °C for 20 h. Germination was then carried out at 28 °C in the dark. Plant materials were collected just post-imbibition or at the appropriate times afterwards, as specified in a HAI (hours after imbibition) scale. Total RNA was obtained from 2 g of plant material, following standard procedures (Ausubel et al. 1995). Genomic DNA was extracted from 2 g young leaves as described by Michaels et al. (1994).
Isolation and sequencing of pea PsABI3 cDNAs and genomic gene
To clone VP1/ABI3-like cDNAs from pea, two primers P1f and P2r (see Supplementary Table S1 at JXB online for the sequences of all the primers used) flanking the initiation and termination translation sites were designed according to the sequence of an ABI3-like cDNA obtained by Nakako and Mori in 2002 from axillary buds of P. sativum (AB080195). Briefly, 2 μg of total RNA from seeds was retrotranscribed by using Superscript II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was properly diluted (5–25-fold), and 1 μl was subjected to PCR under the following conditions: 94 °C for 5 min followed by 30 cycles of amplification (94 °C for 30 s, 54 °C for 30 s, 72 °C for 3 min) and a final step at 72 °C for 7 min. The PCR reaction yielded two major products denoted PsABI3-1 and PsABI3-2, as well as other minor fragments, as detected on agarose gels. The PCR products were cloned into the pGEM-T vector (Promega). The recombinant plasmids were transformed into E. coli DH5α. About 70 clones were analysed after restriction with NcoI and SalI, which do not possess target sites within the Nakako and Mori's sequence.
The genomic gene sequence was obtained with two primer pairs: P1f and P5r; P4f and P2r, using genomic DNA as template. Both sequences were assembled to obtain the full gene sequence. Southern blot analysis was carried out as previously described (Ríos et al., 2007). The probe used for hybridization was the full-length cDNA of PsABI3-1. Sequence alignments were performed with the DNAMAN program (Version 4.03). To search for Arabidopsis DNA sequences the Arabidopsis Information Resource (TAIR) site (www.arabidopsis.org) was used.
Analysis of gene expression
To determine the level of PsABI3 transcripts at different stages of embryo development, seed germination, and in different tissues of plants, total RNA was obtained and RT-PCR reactions were carried out with the primers P1f and P3r to include the first canonical intron. The following conditions were used: 94 °C for 5 min, followed by 34 cycles of amplification (94 °C for 30 s, 56 °C for 30, 72 °C for 2 min) and a final step at 72 °C for 7 min. Simultaneously, a 185 bp actin gene fragment (U81047) was amplified with specific primers Act1f and Act2r (see Supplementary Table S1 at JXB online) as a reference control for the RT-PCR, by using the same PCR conditions but with 30 rounds of amplification instead of 34.
Transactivation analysis in yeast
Full-length sequences corresponding to each isoform were excised by digestion with NcoI and SalI from the pGEM-T vector (Promega). When required, partial sequences were amplified by PCR, using full-length cDNAs as templates. The synthetic primers used contained the restriction sites NcoI and SalI (see Supplementary Table S1 at JXB online). All the inserts were cloned into the pGBK-T7 vector (Clontech), containing the GAL4 DNA binding domain. All constructs were confirmed by sequencing.
The Saccharomyces cerevisiae strain Y190 harbouring LacZ and HIS3 reporter genes were used as an assay system (Clontech). The transformants were selected by growth on SC medium minus Trp (Formedium) at 30 °C for 3–4 d. Subsequently, to test the expression of the LacZ gene, a colony lift filter assays was performed using X-GAL as a substrate (Clontech yeast handbook). The transformants were also grown in SC medium minus His and minus Trp (Formedium), containing 50–100 mM 3-amino-1,2,4-triazole for the growth assay without histidine. The activities of reporters were analysed in 10 independent transformants.
For the yeast two-hybrid system, the S. cerevisiae strain Y190 was transformed with a translational fusion between A. thaliana ABI5 and the activation domain of GAL4 constructed in the pGAD vector (a construction kindly provided by Dr Finkelstein) and the pGBK-T7 vector carrying either the complete or the partial sequences of the different PsABI3 isoforms, fused to the GAL4 DNA binding domain. Transformants were selected on plates containing SC medium lacking tryptophan and leucine (Formedium). The β-galactosidase assays and the ability to grow in SC medium without histidine were performed as described above.
Western blot analysis
Yeast total protein extracts were prepared according to Kushnirov (2000). The proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes (Amersham, Hybond). The membranes were incubated overnight at 4 °C with c-myc mouse monoclonal antibody (Roche) diluted 1:4000. The antigen–antibody complex on the membrane is visualized with a goat anti-mouse (IgG)-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) diluted 1:50 000 and a chemiluminiscent substrate (ECL Advance Western Blotting Detection kit, GE Healthcare) according to the manufacturer's instructions. The membranes were exposed to Fuji Medical X-Ray Film.
Transient expression in onion epidermal cells
PsABI3-1 and PsABI3-2 cDNAs were cloned into the NcoI site of the ppk100 vector containing a double CamV 35S promoter. The cDNAs were amplified by PCR and fused in the 3′ region with the GFP gene. Transformation of onion epidermal monolayer cells using particle bombardment (Bio-Rad) was performed as described previously by Varagona et al. (1992). After 24 h, samples were visualized by Leica TCS SP confocal laser-scanning microscope (Leica, Heidelberg).
Results
Cloning and characterization of putative P. sativum ABI3 genes
To clone and characterize P. sativum ABI3 genes, total RNA was first isolated from mature seeds and the steps described under the Materials and methods were followed. Several PCR products were detected by agarose electrophoresis, which were processed and sequenced as described. More than 70 transformants were analysed after restriction with NcoI and SalI, which do not possess target sites within the Nakako and Mori's sequence. Inserts of different sizes were detected, and two or three inserts of each type were sequenced. In this way, seven putative ABI3-like cDNAs were identified, which are further referred to as PsABI3-1 to PsABI3-7. The sequences of these cDNAs were deposited in GenBank and their accession numbers, together with other properties, are shown in Table 1. The major isoforms are PsABI3-1 and PsABI3-2.
Table 1.
Accession number and properties of the different isoforms of PsABI3
| Name of cDNAs from PsABI3 | Accession number | mRNA length (nt) | Predicted protein lengtha | Predicted protein size (kDa) |
| PsABI3-1 | EU026375 | 2262 | 753 | 83 |
| PsABI3-2 | EU026376 | 1548 | 515 | 56 |
| PsABI3-3 | EU026377 | 2052 | 683 | 75 |
| Nakako and Mori | AB080195 | 2193 | 730 | 80 |
| PsABI3-4 | EU026378 | 907 | 249 | 27 |
| PsABI3-5 | EU026379 | 942 | 313 | 34 |
| PsABI3-6 | EU026380 | 787 | 68 | 7.5 |
| PsABI3-7 | EU026381 | 1386 | 53 | 5.8 |
In number of residues.
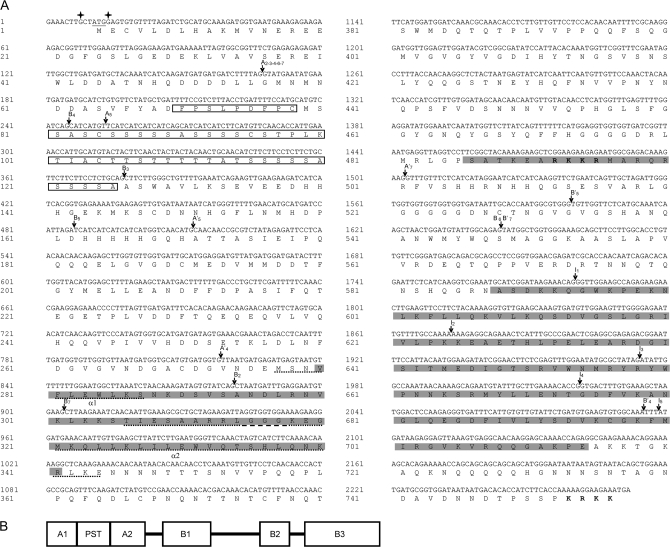
The sequence of the longest cDNA, i.e. PsABI3-1, is given in Fig. 1A, which also shows the predicted amino acid sequence. The context of the initiation codon (GCTATGG) is in accordance with the conserved features of higher plants mRNAs reported by Joshi et al. (1997) in that purines are present at positions −3 and +4. The typical domains of the factors of the ABI3 family are observed in the PsABI3-1 sequence. The N-terminal region of the encoded polypeptide possesses two acidic domains (A1 and A2), separated by a proline-, serine-, and threonine-rich region (PST). The C-terminal moiety contains the three basic domains (B1, B2, and B3) highly conserved in all the orthologues known to date (see Fig. 1B for a schematic representation of the modular structure of PsAIB3-1 polypeptide).
Fig. 1.
Structure of the PsABI3 gene. (A) Nucleotide and deduced amino acid sequence of PsABI3-1. The two purines (G) located at –3 and +4 around the initiation codon are labelled with stars. The location of the five canonical introns is indicated with black arrows marked I with the subscripts 1 to 5. The sites of alternative splicing are identified by black arrows pointing to their 5′ (arrows labelled A) and 3′ (arrows B) sites. When a second splicing process occurs, the arrows are marked A′ and B′. The subscripts 2 to 7 refer to the different isoforms. The basic domains B1, B2, and B3 are shadowed in grey. The two putative amphipathic α-helical within domain B1, α1 and α2 are underlined with dots. Bold letters represent two putative nuclear localization signals in the protein sequence. The PST domain is boxed. (B) Schematic diagram of modular structure of PsABI3-1 polypeptide.
The sequence RKKR, identified as a NLS in VP1 (Marella and Quatrano, 2007) and highly conserved in P. sativum, is found within domain B2. In view of the length of the polypeptide PsABI3-1, the presence of other NLSs cannot be discarded, in accordance with Marella and Quatrano (2007). The last four residues of the PsABI3-1 sequence (KRKK) may well play that role.
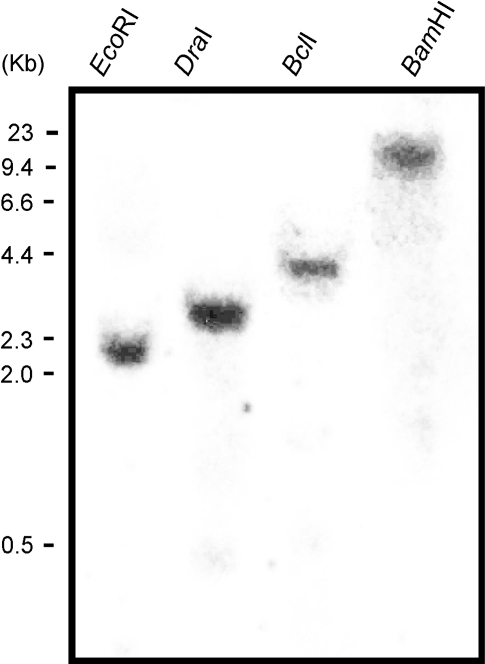
The different isoforms of PsABI3 cDNA may result either from a multigenic family or from an alternative splicing of a single gene. To decide between these possibilities, the Southern analysis was carried out as shown in Fig. 2. There is an EcoRI site at 1741 in cDNA, but the other enzymes used do not cut PsABI3-1. The results of Fig. 2 are compatible with the existence of a single-copy gene, which encodes all the cloned sequences. The presence of a single band after EcoRI digestion may be explained because the 3′ fragment generated would be too small to be detected in the autoradiogram. Nevertheless, the possibility that a second copy of the gene exists cannot be entirely ruled out. This question will be discussed further later on.
Fig. 2.
Southern blot analysis of the PsABI3 gene. Genomic DNA from leaves (30 μg) was digested with restriction enzymes EcoRI, DraI, BclI, and BamHI, blotted and hybridized to a probe containing the PsABI3-1 coding sequence. The migration of size markers is shown at the left.
The multiplicity of isoforms must be explained by an alternative splicing. To find the cause of this alternative splicing, the intron organization of the gene was examined first. To do this, a genomic copy of the gene was obtained by PCR. Two primer pairs were used to this purpose: P1f and P5r; P4f and P2r. Both PCR products were cloned into pGEMT, sequenced, and assembled to obtain the full gene sequence. When the complete genomic sequence was aligned with that of the PsABI3-1 cDNA, the existence of five introns, all of them located within domain B3, can be inferred (Fig. 1A). The lengths of the six exons, from 5′ to 3′, are 1770, 93, 101, 47, 77, and 174 bp, and those of the introns 250, 101, 129, 89, and 82 bp. All five introns are conserved in other members of the VP1/ABI3 family but it can be concluded from the comparison of these lengths with those of the introns of Zea mays VP1 and A. thaliana ABI3 genes (McCarty et al., 1991; Giraudat et al., 1992), that, while the location of the introns is conserved, the size is not.
Structure of the PsABI3 transcripts and proteins
Figure 3 shows a scheme of the structure of the different PsABI3 transcripts, deduced from the cDNA sequences. It can be seen that all the transcripts result from processing events starting within the long exon 1. In most of them the 3′ end site also lies within this exon. Three out of the seven transcripts, namely PsABI3-7, PsABI3-5, and PsABI3-4 include an additional splicing event and in the latter case, the 3′ acceptor site is located in exon 5.
Fig. 3.
Structure of cDNAs derived from PsABI3. The Nakako and Mori sequence is depicted in the first row and the numbers on the left of the remaining rows refer to the isoforms found in this work. The left column represents the processed mRNA between the initiation and termination codons as well as the length of the coding region. When applicable, the position of premature stop codons originated by frame shift is indicated. The exon sequences excised from the complete isoform (PsABI3-1) are shown as folded lines. Numbers above the lines stand for the size of the removed or conserved sequences in nt. The arrow heads in Nakako and Mori's sequence indicate the position of insertions relative to PsABI3-1 cDNA. The dinucleotides at the borders of the splicing sites are shown. In both columns, the numbering of the nucleotides is relative to the first translation start site in PSABI3-1.
The larger isoform, PsABI3-1, shows homology with its orthologues, ABI3 from A. thaliana and Phaseolus vulgaris, and VP1 from Zea mays (McCarty et al., 1991; Giraudat et al., 1992; Bobb et al., 1995) and, as mentioned earlier, maintains the typical modular structure of these factors. When the PsABI3-1 sequence is compared with that obtained by Nakako and Mori several differences were found. The most striking one is the absence in the latter of a 78 nt tract, which would code for 26 amino acid residues between subdomains A1 and A2 and includes the short proline-rich sequence FPSLPDFPC (Fig. 1A), which is highly conserved in all the members of the ABI3/VP1 family. Other changes observed when our sequence and that of Nakako and Mori were compared, are two insertions of 6 (ATCATC) and 3 (GGT) nucleotides in the sequence of Nakako and Mori, as well as 19 single nucleotide substitutions that would result in the variation of 9 amino acids in the polypeptidic product. The reason for these differences is not known. Taking into account that the genomic sequence matches that of our cDNA and not that of Nakako and Mori, and in view that, in our hands evidence in support of the presence of this isoform has not been found, we think that the differences may be attributable to the use of another P. sativum variety.
PsABI3-2 has 714 nt fewer than PsABI3-1. Curiously enough, the missing fragment shares with the Nakako and Mori sequence the same 5′ splicing site, but, being larger, affects not only the PST region, but the entire subdomain A2 and part of the B1 domain. The sequence of PsABI3-3 also implies the removal of a fragment with the same GT 5′ border, but with a size intermediate between those of PsABI3-1 and PsABI3-2, as in this case only the PST domain is missing. The DNA sequences missing in isoforms PsABI3-4, PsABI3-6, and PsABI3-7 also start at the same 5′ border, although, as previously mentioned, these forms show additional deletions. One of the sequences missing in isoform PsABI3-4 is exactly the same as that observed in the Nakako and Mori sequence. In the isoforms PsABI3-2, PsABI3-3, PsABI3-6, and PsABI3-7 the borders limiting the missing fragments are GT … AG. These are the splicing consensus borders characteristic of the U2-type introns. The same borders occur in the 5′ missing fragment of PsABI3-4, so it can be assumed that the removal of all these fragments originates from exon skipping using the U2-type spliceosome (Reddy, 2007). Finally, in two of the isoforms, namely PsABI3-4, and PsABI3-5, processing events with atypical splicing donor and acceptor sites occur. These have also been described in the alternative splicing of other plant genes (Gupta et al., 2005). It has to be noted that the 5′ splicing event in PsABI3-5 involves the short direct repeat (TCA)5 and, therefore, the splicing site in isoform 5 is arbitrarily marked in Fig. 1A, because the borders of the removed sequence may be either TC/GA or GC/CA.
The sizes of the putative polypeptides are also given in Table 1, although it is not yet known whether all the mature PsABI3 mRNAs are translated. The processing in PsABI3-4, PsABI3-6, and PsABI3-7 generates premature stop codons resulting in short ORFs.
Finally, a prediction of secondary structure of the putative polypeptides was carried out and it was found that three α-helical amphipathic stretches are possible within domain B1. The second and third segments are interspersed by a three-glycine tract and, perhaps, the whole sequence may form a single helix, referred to as helix 2 (Fig. 1A).
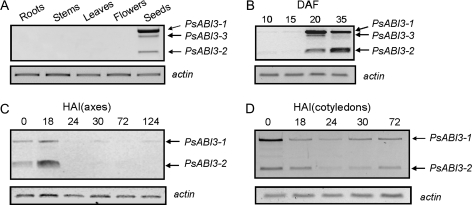
Spatial and temporal patterns of PsABI3 expression
The cloning of PsABI3 cDNAs has been achieved starting from mature seeds. The question as to whether the gene is also expressed in organs or tissues of adult plants immediately arises. The RT-PCR experiments shown in Fig. 4A allowed us to give a negative answer concerning roots, stems, leaves, and flowers and only seeds show a detectable presence of PsABI3 transcripts. The temporal pattern of PsABI3 mRNAs accumulation in developing seeds was investigated next. Although faint bands attributable to PsABI3 transcripts are detectable at early embryogenesis, PsABI3 mRNAs begin to accumulate at middle embryogenesis, 20 DAF, and their level is maintained during the last phases of seed maturation (Fig. 4B). The identity of all the bands was ascertained by sequencing. Just ahead of the band corresponding to PsABI3-3 a faint, fuzzy band is seen at 20 and 35 DAF (Fig. 4B). It was sequenced and it was found that 351 nt were missing, relative to PsABI3-1, from nt 137 onwards. It is thought that it corresponds to a new isoform, which is not included in the list in Table 1 because we did not succeed in obtaining the whole clone.
Fig. 4.
Expression of the PsABI3 gene as determined by RT-PCR. Total RNA was extracted and used for the generation of the corresponding cDNAs. The presence of PsABI3 mRNAs was detected by amplification with primers P1f and P3r (see Table S1 at JXB online), which include the first canonical intron. A 185 bp fragment from actin gene was amplified as an internal control using the primers Act1f and Act2r. The RT-PCR analysis was performed in: (A) different plant organs, (B) during seed development, at different DAF, (C) in embryonic axes, and (D) in cotyledons at different HAI.
On the other hand, the bands of Fig. 4B for 20 DAF and 35 DAF have been integrated by using the ImageJ program, to find that the total amount of transcripts remained roughly constant, although the relative amount of the different isoforms varied (results not shown). The level of transcripts is maintained or even increases in embryonic axes after starting germination, but it drops to negligible amounts when the germination events finish at about 24–30 HAI. Contrarily, mRNAs, though decreasing with the onset of germination, are clearly detectable in cotyledons until 72 HAI (Fig. 4C, D).
It is worth noting that the relative level of the different PsABI3 isoforms is time- and tissue-dependent. Figure 4 shows that PsABI3-1, the major isoform in dry seeds, mainly accumulates in cotyledons, while PsABI3-2 represents the most abundant isoform in 18 HAI embryonic axes. Moreover, PsABI3-3 is also detectable in some instances (20 DAF). Although these comments, based on reproducible RT-PCR results, are not entirely reliable from a quantitative point of view, they may imply that the presence of different isoforms of PsABI3 play a functional role, as will be discussed later.
Role of the different isoforms as transcriptional activators
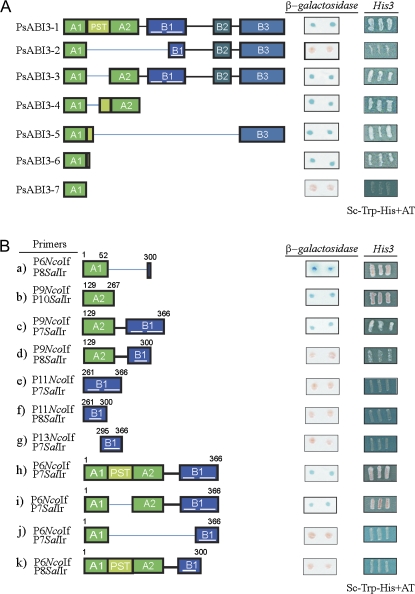
To study the functional ability of the polypeptides encoded by the different PsABI3 mRNAs, a one-hybrid assay was carried out in Saccharomyces cerevisiae. The DNA coding for the binding domain of Gal4p was fused to each of the different PsABI3 cDNAs in the pGBK-T7 vector. These constructs were transformed into yeast and their capability to induce the target genes LacZ or His3 was analysed. Figure 5A shows that, with the exception of PsABI3-2 and PsABI3-7, the remaining polypeptides complement the transactivating activity of Gal4p. The lack of complementation of Gal4p does not result from the absence of the polypeptides, because Western blotting (using an antibody against the c-myc label) showed that the corresponding fusion polypeptides were expressed in yeast (results not shown). The results with the isoform containing the complete sequence, i.e. PsABI3-1 are identical to those obtained with pCL1 vector, which carries the whole Gal4p and was used as positive control. The latter result shows that, in fact, the PsABI3-1 activating domains are functional in yeast. Some of the results with other isoforms are difficult to explain at first sight. For instance, the transactivating activity of PsABI3-3, or PsABI3-4, which lack wholly or in part the PST tract, reveals that this sequence is dispensable for activation. In this line, it might be concluded that the activating subdomain A2 is required for activation (see the results with PsABI3-2, one of the major isoforms). The data obtained with the isoform PsABI3-7, which is a truncated form containing only the subdomain A1, are in accordance with this interpretation, because this isoform is inactive. Nevertheless, the results obtained with isoform PsABI3-6 cast a doubt on the above idea, because a non-PST tail of 15 amino acids added to the C-terminal side of subdomain A1 suffices to restore the transactivating activity. Isoform PsABI3-5 is also active in spite of lacking the activating subdomain A2, but, as in PsABI3-6, it possesses, in addition to the subdomain A1, a tail containing 34 amino acids from the proline-rich sequence as well as the basic domain B3.
Fig. 5.
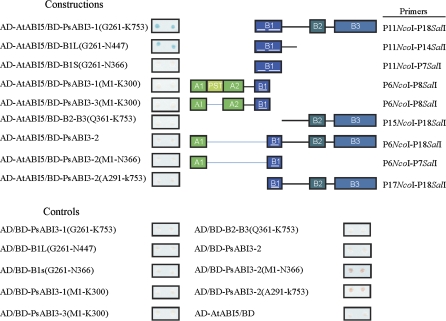
Yeast one-hybrid analysis of the transactivating potential of the different isoforms and artificial constructs. (A) Functional analysis of the full-length PsABI3 isoforms. The β-galactosidase activity was analysed by lift filter assays and the His3 gene expression analysis was performed by growing on SC medium without His. pCL1 and pGBKT7 vectors were used as positive and negative controls, respectively (data not shown). (B) Functional analysis of different constructs derived from PsABI3 isoforms. The primers (see Supplementry Table S1 at JXB online) used for each construct are shown. The numbers over the constructs indicate the position of amino acids in the sequence of PsABI3-1. In both panels, thin blue lines represent the segments missing in every isoform or construct. (This figure is available in colour at JXB online.)
Isoforms PsABI3-1 and PsABI3-2 always appear together (Fig. 4) and in this context the inactivity of the latter is especially interesting. As mentioned above, the polypeptide is actually present in yeast cells, and to explore the causes of its inactivity further, the experiments reported in Fig. 5B were conducted, in which the transactivating activity of several artificial constructs was studied.
It was first shown that, as suspected from the experiments of Fig. 5A, the activating subdomain A1 alone requires a certain C-terminal extension to be active in the absence of other domains. Apart from the data with PsABI3-6 (Fig. 5A), adding 10 amino acids from B1 to A1 restores the activating activity, as shown by the fact that the first 62 amino acids of PsABI3-2 are active (Fig. 5B, a).
The activating subdomain A2 is also active in the absence of other parts of the polypeptide (Fig. 5B, b), and the activity is maintained when A2 is fused to the basic domain B1 (Fig. 5B, c), but not when fused to the first 39 amino acids of this domain, which include the first α-helical segment (Fig. 5B, d). Therefore, the transactivating properties of the basic domain B1 were examined next, to find that neither the whole domain nor its helical segments are able by themselves to activate transcription (Fig. 5B, e–g).
Apart from the constructs a–g mentioned above, the transactivating properties of some artificially truncated forms of the natural isoforms were examined. The deletions of basic domains B2 and B3 (Fig. 5B, h) and even of the PST region (Fig. 5B, i) do not eliminate the transactivating activity of PsABI3-1, but the construct resulting from PsABI3-2 by deletion of the basic domains B2 and B3 is inactive (Fig. 5B, j). By comparing the structure of the isoforms PsABI3-2 and PsABI3-3 it is clear that the only difference is the absence in the former of the activating subdomain A2 and of the first helical segment of B1. It is interesting noting here that the subdomain A2 by itself possesses transactivating activity (Fig. 5B, b) as does the combination of subdomain A2 and the whole basic domain B1 (Fig. 5B, c). Nevertheless, the first helix of B1 neither alone nor in combination with A2 is able to activate transcription in the one-hybrid system (Fig. 5B, d, f). Although some aspects of these results are not easy to explain, a plausible possibility will be discussed later.
Interactions between PsABI3 isoforms and AtABI5
It has been mentioned that the interactions between ABI3 and ABI5 are essential for ABA signalling (Santos-Mendoza et al., 2008) and as it has just been shown that PsABI3-2, one of the major ABI3 isoforms in pea, is not able to produce transactivation in the yeast one-hybrid system. To explore the differences in activation between the major PsABI3 isoforms further and to check for their ability to interact with ABI5-like proteins, a yeast two-hybrid analysis was planned to study the heterologous interactions between PsABI3 isoforms and other constructs and AtABI5. To do this, either complete isoforms or constructs derived from PsABI3 were fused to the binding domain of Gal4p and used as bait, while AtABI5 fused to the activating domain of Gal4p was used as prey protein. To study the properties of PsABI3-1, one of the major isoforms able to transactivate in the one-hybrid system, the protein was excised into several fragments, inactive in the yeast one-hybrid system. The results are given in Fig. 6, which also shows the negative controls. Positive interactions were confirmed by the growth of yeast cells in SC medium lacking histidine (data not shown). Figure 6 shows that a fragment containing the three intact B domains is able to interact with AtABI5. In our hands, the minimal fragment showing this interacting ability was one containing the basic domain B1 and an extension of 81 amino acids from the region between B1 and B2 (between Gly261 and Asn447). However, neither the B1 domain alone (Gly261 to Asn366) nor the fragment containing the subdomains A1 and A2 with the interspersed PST region and the first helix of domain B1 are able to interact with AtABI5. The basic domains B2 and B3, by themselves, are not able to interact with the prey protein nor are the isoform PsABI3-2 and the fragments derived from it (Fig. 6). The functional consequences of these results will be discussed below.
Fig. 6.
Yeast two-hybrid analysis of the interaction between several constructs from PsABI3 proteins and AtABI5. The transformants were selected on plates containing SC medium lacking tryptophan and leucine. Positive interactions were confirmed with β-galactosidase filter assays. The thin blue lines represent segments missing in the different isoforms or constructs. The primers (see Supplementary Table S1 at JXB online) used for each construct are shown. Negative controls with empty plasmids are shown. The PsABI3 binding domains in the constructs used as bait were identified by the N- and C-terminal amino acids (one-letter code and sequence number in PsABI3-1). (This figure is available in colour at JXB online.)
Fig. 7.
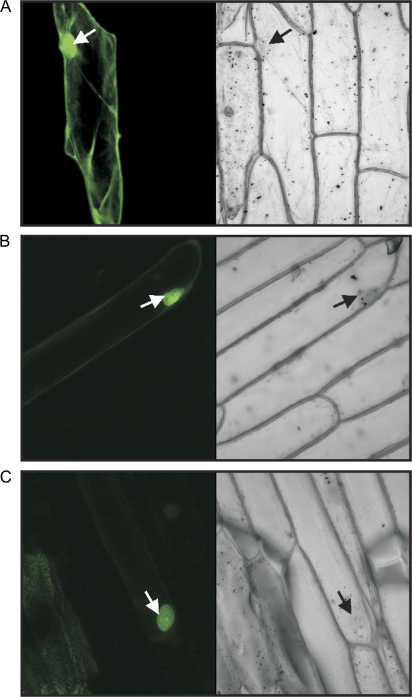
Subcellular localization of PsABI3-1 and PsABI3-2 in onion epidermal cells. Confocal microscopy images of onion cells transformed with (A) GFP alone, (B) construct GFP-PsABI3-1, and (C) construct GFP-PsABI3-2. The fluorescent signal, confined to nuclei in the case of both constructs, is observed in the left panels, while the bright-field images of the same cells is shown in the right panels. Nuclei are arrowed. (This figure is available in colour at JXB online.)
Subcellular localization of PsABI3 isoforms
Our results from yeast one- or two hybrid experiments suggest that isoforms PsABI3-1 and PsABI3-3 are active in plants, while PsABI3-2 is not. Nevertheless, a prerequisite for the activity of a given isoform is its nuclear localization. We have previously commented that a classical NLS occurs within domain B2 and that, perhaps, an additional NLS is located within B3. This would imply that all these three isoforms are localized to nuclei in plant cells. To check the validity of this assumption, a transient onion cell transformation assay was carried out. The results show that both PsABI3-1 and PsABI3-2 are localized to nuclei in plant cells in contrast with the ubiquitous localization of GFP alone. Therefore, the absence of activating properties in PsABI3-2 ought to be explained by the properties of the protein itself and not by the incapacity to enter the nucleus.
In this context, it should be mentioned that the exclusion size of the nuclear pores might allow the small putative polypeptides PsABI3-6, and PsABI3-7 (Table 1) to enter the nucleus by diffusion (Schwechheimer et al., 1998). Anyway, these putative polypeptides would not be capable of binding DNA, as they would lack the B domains.
Discussion
In the present paper we describe the cloning of seven ABI3-like cDNA isoforms from P. sativum, referred to as PsABI3-1 to PsABI3-7. Cloning was achieved by a RT-PCR-based method, using as primers oligonucleotides designed in view of the sequence of an ABI3-like cDNA deposited by Nakako and Mori. However, none of the isoforms found coincide with this sequence. The use of different cultivars might account for this disagreement. Actually, the splicing events may be cultivar-dependent as suggested by some previous data on wheat (McKibbin et al., 2002) and rice (Fan et al., 2007) VP1 genes. Nevertheless, the large number of differences between the Nakako and Mori's sequence and ours suggests that other explanations are possible. For instance, a duplication of a large DNA fragment containing the ancestral gene might have occurred prior to the divergence of both cultivars. If evolutionary changes had not affected the restriction sites, this possibility would be compatible with our Southern analysis results.
Our results show that the different isoforms of PsABI3 described here are originated by alternative splicing. When browsing the Arabidopsis Information Resource (TAIR) site to look for cDNA sequences corresponding to the ABI3 gene, a sequence (AK117245) and an EST (AU238055) were found, which are the product of intron 1 retention, therefore giving an indication that alternative splicing occurs in AtABI3. A stop codon within the retained intron would give rise to a translated polypeptide lacking domain B3. Taken these results together with ours, which represent the first description of the structure and possible role of alternative splicing-generated isoforms of ABI3 in dicots, it can be concluded that the existence of alternative splicing of ABI3-like genes may represent a widespread phenomenon to which more attention should be paid.
Most of the splicing events of ABI3/VP1 transcripts in monocots affect two exons and they often result in frame-shifts and premature termination of the resulting polypeptides. In our case, most of the sequences removed are localized to exon 1 and splicing results in the elimination of defined modules in the putatively translated polypeptides.
The most frequent splice site in our case is GT-AG, characteristic target of plant U2-type spliceosome (Reddy, 2007). In other instances, however, non-canonical dinucleotides are used for splicing (see Fig. 3, isoforms PsABI3-4 and PsABI3-5). Similar non-U2 or non-U12-type splicing events have been described for other plant genes (Li et al., 2006). The 5′ splicing event in PsABI3-5 involves the short direct repeat (TCA)5. Short repeats either direct or inverted have been found in association with splicing (Fan et al., 2007; Utsugi et al., 2008) and it has been suggested that they may be a cause of missplicing. Intron retention events, the most common cause of alternative splicing in A. thaliana and Oryza sativa (Wang and Brendel, 2006) have not been detected, but exon skipping, which is less common in plants than in humans (Reddy, 2007), occurs in several instances. Finally, three of the isoforms, namely PsABI3-4, PsABI3-5, and PsABI3-7, result from two splicing events. It has to be remarked that the list of plant genes undergoing alternative splicing is continuously increasing with the result that our knowledge of the complexity of the plant transcriptome becomes more and more extensive.
Of course, not all the splicing possibilities ought to occur actually in vivo, but it should be stressed that, in our case, the different isoforms have been identified by oligodT primer-driven RT-PCR, starting from plant RNA. So the existence of the seven transcript isoforms in the plant transcriptome under normal development conditions can be taken for granted and the existence of additional isoforms cannot be discarded. Moreover, at least three out of the seven transcripts are present in an amount high enough to be easily detected by agarose electrophoresis (Fig. 4).
The structure of PsABI3-1, the largest isoform, matches that of the homologous proteins from other plants, in which two acidic, activator domains are present in the N-terminal region, while the C-terminal moiety comprises three basic domains. These are involved in interactions with DNA and other factors, especially with ABI5 and other bZIP-type transcription factors. In our case, both acidic domains exhibit transactivating activity. Domain A2, by itself, is able to transactivate in a one-hybrid system (Fig. 5B, construct b), while domain A1 requires a small C-terminal extension to be active (see PsABI3-6 in Fig. 5A and construct a in Fig. 5B). Therefore, it is tempting to speculate that both A1 and A2 domains are redundant in their activating activity. On the other hand, two-hybrid systems show that the minimal region of PsABI3 required to interact with ABI5 is the complete B1 domain plus an extension at its C-terminal side (Fig. 6, construct b). Other results given in Fig. 5 are difficult to interpret. For instance, the cause by which the A2 domain either alone or fused to the complete B1 subdomain exhibit transactivating activity while the A2 domain fused to the first helix of the B1 subdomain does not, is not clear. Probably, aberrant tertiary structures are formed in some of the artificial constructs that prevent polypeptides from playing the proper biological function and this may explain as well the absence of a trans-activating capacity. Moreover, it has to be kept in mind that the structural changes provoked by the absence of some amino acid tracts may affect the stability of the resulting polypeptides and/or alter their capacity to bind other molecules. For instance, the removal of the PST domain, although it does not abolish the transactivating activity of the PsABI3 isoforms, may result in the absence of post-translational modifications (e.g. phosphorylations), that may be important for the structure and/or function of the protein. Similarly, the presence of an incomplete B1 domain in isoform PsABI3-2 may result in an aberrant tertiary structure causing inactivity.
With respect to the natural isoforms, five out of the seven found are active (Fig. 5). Isoforms PsABI3-4 and PsABI3-6 show transactivating activity in the yeast one-hybrid system (Fig. 5A) but they lack the DNA binding domain, as well as the B1 domain essential for the heterodimerization with ABI5-type factors (Nakamura et al., 2001). Therefore, these isoforms, by themselves, would not display activating properties. Nevertheless, they can play a role in vivo, because, through their intact domain A1, they may be able to compete with the active isoforms PsABI3-1 and PsABI3-3 for the recruitment of other factors. In view of the inactivity of PsABI3-2, we wondered what the structural requirements for the functionality of PsABI3 factors are. The importance of the domain B1 and of its neighbour sequences in both transactivation and interaction with other proteins, such as ABI5, seems to emerge from the results in Figs 5 and 6.
The PsABI3 gene is only expressed during embryogenesis and their products accumulate in the dry seeds to diminish along germination. This is a logical issue, because of the involvement of ABA signalling in seed maturation (Finkelstein et al., 2002). It is worth noting that the relative amount of the major isoforms, namely PsABI3-1, PsABI3-2, and PsABI3-3, changes during development. PsABI3-3, which is abundant at middle embryogenesis (20 DAF), is virtually absent at other moments of seed maturation and germination. At these moments only PsABI3-1 and PsABI3-2 are visible in agarose gels (Fig. 4). As mentioned above, it seems that the relative amount of the isoforms may change without changing the total transcriptional rate of the gene. This might point to a double regulatory circuit for the amount of the different isoforms, in which the regulation of splicing would add to transcriptional control.
The existence of multiple splice variants of PsABI3 immediately poses the question as to whether alternative splicing plays a biological role. Similar questions have been often raised, and in some instances the answer seems to be affirmative (for a recent review, see Reddy, 2007). In the present case, although it is not yet known if the different mRNAs are translated in vivo, the fact that the amount of the major isoforms changes in a precise, probably regulated, way during development, together with the circumstance that one of these isoforms is inactive in transactivation, suggest a putative functional role. The inactive isoform PsABI3-2 has an intact B3 subdomain required to bind the RY element (Suzuki et al., 1997) and, in consequence, it may compete with the active isoforms PsABI3-1 and PsABI3-3 in their binding to the cis promoter elements. Nevertheless, being PsABI3-2 inactive in transactivation, their binding to DNA would be unproductive. In this way, the inactive isoforms would play a role somewhat similar to that of the repressing VAL proteins (Suzuki and McCarty, 2008).
Our results open an interesting question, namely the nature of the mechanisms that regulate the expression of seed storage proteins in dicots of agricultural interest, in which ABA plays a pivotal role. Our data show that the expression of PsABI3 correlates with that of the genes coding the main storage proteins (legumin and vicilin) and other factors involved in seed maturation and desiccation tolerance. RY is the core element of the legumin box and it is present in many genes coding for those proteins. This suggests that PsABI3 and other B3 transcriptional factors are involved in the regulation of their expression. These mechanisms are well established in Arabidopsis (see the excellent reviews of Santos-Mendoza et al., 2008; Suzuki and McCarty, 2008), but they cannot be extrapolated to other species. It has been demonstrated, within the limitations of a two-hybrid assay, the existence of a physical interaction between PsABI3 and AtABI5, but the whole scenario of genetic interactions with LEC1-, LEC2- and FUS3-like genes remains to be studied in the absence of a thorough study of these genes in P. sativuum.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. Sequence of primers designed for PCR reactions.
Supplementary Material
Acknowledgments
We wish to thank Dr R Finkelstein for kindly providing the plasmid carrying AtABI5. We are very indebted to Dr Gabino Ríos for his valuable comments on the manuscript. This work was supported in part by grant BFU2007-63120 from the Spanish Ministry of Education and Science. AG was a recipient of a predoctoral grant BES-2005-10301 from the Spanish Ministry of Education and Science.
Glossary
Abbreviations
- ABA
abscisic acid
- ABRE
ABA-response element
- bZIP
basic leucine zipper domain
- DAF
days after fertilization
- GFP
green fluorescent protein
- HAI
hours after imbibition
- NLS
nuclear localization signal
- ORF
open reading frame
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PST
proline-, serine-, threonine-rich
- RT-PCR
reverse transcriptase-PCR
- SC
synthetic complete medium
- SDS
sodium dodecyl sulphate
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Deidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley and Sons; 1995. [Google Scholar]
- Bobb AJ, Eiben HG, Bustos MM. PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. The Plant Journal. 1995;8:331–343. doi: 10.1046/j.1365-313x.1995.08030331.x. [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- Castillo J, Genovés A, Franco L, Rodrigo MI. A multifunctional bicupin serves as precursor for a chromosomal protein of Pisum sativum seeds. Journal of Experimental Botany. 2005;56:3159–3169. doi: 10.1093/jxb/eri313. [DOI] [PubMed] [Google Scholar]
- Castillo J, Rodrigo MI, Márquez JA, Zúñiga A, Franco L. A pea nuclear protein that is induced by dehydration belongs to the vicilin superfamily. European Journal of Biochemistry. 2000;267:2156–2165. doi: 10.1046/j.1432-1327.2000.01229.x. [DOI] [PubMed] [Google Scholar]
- Castillo J, Zúñiga A, Franco L, Rodrigo MI. A chromatin-associated protein from pea seeds preferentially binds histones H3 and H4. European Journal of Biochemistry. 2002;269:4641–4648. doi: 10.1046/j.1432-1033.2002.03164.x. [DOI] [PubMed] [Google Scholar]
- Fan J, Niu X, Wang Y, Ren G, Zhuo T, Yang Y, Lu B-R, Liu Y. Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa) Journal of Experimental Botany. 2007;58:3811–3817. doi: 10.1093/jxb/erm231. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(Supplement):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Review of Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Gupta S, Wang BB, Stryker GA, Zanetti ME, Lal SK. Two novel arginine/serine (SR) proteins in maize are differentially spliced and utilize non-canonical splice sites. Biochimica et Biophysica Acta. 2005;1728:105–114. doi: 10.1016/j.bbaexp.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends in Plant Science. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proceedings of the National Academy of Sciences, USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes and Development. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Molecular Biology. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Li J, Li X, Guo L, Lu F, Feng X, He K, Wei L, Chen Z, Qu LJ, Gu H. A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. Journal of Experimental Botany. 2006;57:1263–1273. doi: 10.1093/jxb/erj094. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proceedings of the National Academy of Sciences, USA. 2002;99:10203–10208. doi: 10.1073/pnas.152318599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte WR, Jr, Russell SH, Quatrano RS. Abscisic acid-responsive sequences from the Em gene of wheat. The Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella HH, Quatrano RS. The B2 domain of VIVIPAROUS1 is bi-functional and regulates nuclear localization and transactivation. Planta. 2007;225:863–872. doi: 10.1007/s00425-006-0398-6. [DOI] [PubMed] [Google Scholar]
- Michaels SD, John MC, Amasino RM. Removal of polysaccharides from plant DNA by ethanol precipitation. Biotechniques. 1994;17:274–276. [PubMed] [Google Scholar]
- Mishra NS, Tuteja R, Tuteja N. Signaling through MAP kinase networks in plants. Archives of Biochemistry and Biophysics. 2006;452:55–68. doi: 10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua N-H. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proceedings of the National Academy of Sciences, USA. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Ohmiya K, Hattori T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. The Plant Journal. 1996;9:217–227. doi: 10.1046/j.1365-313x.1996.09020217.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR. Physical interactions between ABA response loci of Arabidopsis. The Plant Journal. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Nantel A, Quatrano RS. Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. Journal of Biological Chemistry. 1996;271:31296–31305. doi: 10.1074/jbc.271.49.31296. [DOI] [PubMed] [Google Scholar]
- Nieva C, Busk PK, Domínguez-Puigjaner E, Lumbreras V, Testillano PS, Risueño MC, Pagès M. Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Molecular Biology. 2005;58:899–914. doi: 10.1007/s11103-005-8407-x. [DOI] [PubMed] [Google Scholar]
- Oeda K, Salinas J, Chua N-H. A tobacco bZip transcription activator (TAF-1) binds to a G-box-like motif conserved in plant genes. EMBO Journal. 1991;10:1793–1802. doi: 10.1002/j.1460-2075.1991.tb07704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annual Review of Plant Biology. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- Ríos G, Gagete AP, Castillo J, Berbel A, Franco L, Rodrigo MI. Abscisic acid and desiccation-dependent expression of a novel putative SNF5-type chromatin-remodeling gene in Pisum sativum. Plant Physiology and Biochemistry. 2007;45:427–435. doi: 10.1016/j.plaphy.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. The Plant Journal. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Zourelidou M, Bevan MW. Plant transcription factor studies. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:127–150. doi: 10.1146/annurev.arplant.49.1.127. [DOI] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J. Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proceedings of the National Academy of Sciences, USA. 1991;88:7266–7270. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has cooperative DNA binding activity. The Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Current Opinion in Plant Biology. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Utsugi S, Nakamura S, Noda K, Maekawa M. Structural and functional properties of Viviparous1 genes in dormant wheat. Genes and Genetic Systems. 2008;83:153–166. doi: 10.1266/ggs.83.153. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. The Plant Cell. 1992;4:1213–1227. doi: 10.1105/tpc.4.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V. Genome wide comparative analysis of alternative splicing in plants. Proceedings of the National Academy of Sciences. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, Lenton J, Holdsworth M. Transcripts of Vp-1 homoeologues are alternatively spliced within Triticeae tribe. Euphytica. 2005;143:243–246. [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. The Plant Cell. 2002;14(Supplement):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F. Characterization of alternative splicing products of bZIP transcripton factors OsABI5. Biochemical and Biophysical Research Communications. 2007;360:307–313. doi: 10.1016/j.bbrc.2007.05.226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.