Abstract
γ-Aminobutyrate transaminase (GABA-T) catalyses the breakdown of GABA to succinic semialdehyde. In this report, the previously identified Arabidopsis thaliana (L.) Heyhn GABA-T (AtGABA-T) was characterized in more detail. Full-length AtGABA-T contains an N-terminal 36 amino acid long targeting pre-sequence (36 amino acids) that is both sufficient and necessary for targeting the enzyme to mitochondria. Removal of the pre-sequence encoding this N-terminal targeting domain and co-expression of the resulting truncated AtGABA-T cDNA with the GroES/EL molecular chaperone complex in Escherichia coli yielded good recovery of the soluble recombinant proteins. Activity assays indicated that purified recombinant GABA-T has both pyruvate- and glyoxylate-dependent activities, but cannot utilize 2-oxoglutarate as amino acceptor. Kinetic parameters for glyoxylate- and pyruvate-dependent GABA-T activities were similar, with physiologically relevant affinities. Assays of GABA-T activity in cell-free leaf extracts from wild-type Arabidopsis and two knockout mutants in different genetic backgrounds confirmed that the native enzyme possesses both pyruvate- and glyoxylate-dependent activities. The GABA-T transcript was present throughout the plant, but its expression was highest in roots and increased as a function of leaf development. A GABA-T with dual functions suggests the potential for interaction between GABA metabolism and photorespiratory glyoxylate production.
Keywords: Enzyme kinetics, GABA transaminase, gene expression, Glyoxylate, Photorespiration, Recombinant protein, Subcellular localization
Introduction
γ-Aminobutyric acid (GABA) is a ubiquitous four C, non-protein amino acid found in prokaryotes and eukaryotes (Satya Narayan and Nair, 1990; Shelp et al., 1999). It was first identified in potato tubers during the 1950s, and has since been characterized as a neural inhibitor in the central nervous system of mammals. A model for the function of GABA in plants remains elusive, despite evidence linking the metabolite to both stress and signalling (Shelp et al., 1999, 2006; Bouché and Fromm, 2004; Fait et al., 2008).
GABA is produced in the cytosol (Breitkreuz and Shelp, 1995) via the decarboxylation of glutamate in a reaction catalysed by glutamate decarboxylase, a calcium/calmodulin-dependent enzyme (Baum et al., 1993; Ling et al., 1994; Snedden et al., 1995, 1996). GABA is then transaminated to succinic semialdehyde (SSA) via GABA-transaminase (GABA-T) in the mitochondrion (Van Cauwenberghe and Shelp, 1999; Van Cauwenberghe et al., 2002). SSA is oxidized to succinate via SSA dehydrogenase (SSADH) in the mitochondrion (Breitkreuz and Shelp, 1995; Busch and Fromm, 1999), or reduced to γ-hydroxybutyrate by SSA reductase activities in the cytosol and chloroplast (Allan et al., 2003, 2008; Breitkreuz et al., 2003; Fait et al., 2005; Allan and Shelp, 2006; Hoover et al., 2007; Simpson et al., 2008).
GABA-T activity in plants differs from that in most other organisms. GABA-T in mammals, yeast, and Escherichia coli uses 2-oxoglutarate exclusively as an amino donor (André and Jauniaux, 1990; Bartsch et al., 1990; De Biase et al., 1995), whereas both pyruvate- and 2-oxoglutarate-dependent activities occur in plants (Shelp et al., 1995; Van Cauwenberghe and Shelp, 1999). The 2-oxoglutarate-dependent activity from tobacco is highly unstable during purification, and production of a purified protein has not been possible (Van Cauwenberghe and Shelp, 1999). However, the pyruvate-dependent activity has been purified to homogeneity, thereby enabling partial amino acid sequencing and identification of an Arabidopsis (designated hereinafter as AtGABA-T) cDNA (Van Cauwenberghe et al., 2002). Recombinant expression of the cDNA in E. coli confirmed that it encodes a pyruvate-dependent GABA-T that lacks detectable 2-oxoglutarate-dependent activity. Recombinant expression of the protein was minimal and primarily insoluble, making more detailed analysis of the enzyme impracticable (Van Cauwenberghe et al., 2002). Prediction of subcellular localization using PSORT (Nakai and Kanehisa, 1992) suggested that AtGABA-T, like the mammalian enzyme, contains an N-terminal mitochondrial matrix targeting signal (Van Cauwenberghe et al., 2002), which is in agreement with a previous subcellular fractionation study of soybean protoplasts (Breitkreuz and Shelp, 1995).
In this study, a truncated AtGABA-T cDNA lacking the putative N-terminal targeting pre-sequence was co-expressed with the molecular chaperones GroES/EL in E. coli. Substrate specificity and kinetic studies revealed that the recombinant enzyme utilizes only GABA as the amino donor in the production of SSA. As previously observed, the enzyme used pyruvate but not 2-oxoglutarate as an amino acceptor (Van Cauwenberghe et al., 2002). In addition, the enzyme was found to have previously unreported glyoxylate-dependent GABA-T activity. A transient expression system, confirmed that AtGABA-T is localized to mitochondria, and that proper sorting of the enzyme does require an N-terminal targeting pre-sequence. The use of knockout mutants of Arabidopsis confirmed that the native enzyme utilizes both pyruvate and glyoxylate as amino acceptors and suggested that there is no 2-oxoglutarate-dependent GABA-T activity in Arabidopsis.
Materials and methods
Production, purification, and analysis of recombinant Arabidopsis GABA-T
The full-length cDNA of an Arabidopsis thaliana (L.) Heynh GABA-T (GenBank accession no. AF351125), with or without its mitochondrial targeting domain (predicted to be 36 N-terminal amino acids based on the location of the mitochondrial cleavage site) was cloned into the pET-15b expression vector (includes a His6 tag on the N-terminus; Novagen, San Diego, CA, USA) using standard techniques (Sambrook et al., 1989). The no-target GABA-T was amplified for plasmid ligation using primers 5′ GGA ATT CCA TAT GAC TAC TGA GGC AGC ACC TG 3′ and 5′ GCG GGA TCC TCA CTT CTT GTG CTG AGC C 3′.
Full-length and truncated GABA-T was expressed in E. coli BL-21(DE3) Rosetta (pLysS) cells (Novagen) carrying the pREP4-GroESL vector (Dale et al., 1994). Cells were grown at 37 °C and 225 rpm in Luria–Bertani broth containing 50 μg ml−1 ampicillin, 34 μg ml−1 chloramphenicol, and 30 μg ml−1 kanamycin, to an OD600 of 0.6. Cultures were cooled to 25 °C, isopropyl-β-D-thiogalatopyranoside (IPTG) was added to a concentration of 0.25 mM, and the cultures were shaken at 225 rpm and 25 °C for 2 h. Cells were pelleted by centrifugation at 3000 g at 4 °C for 10 min and stored at –80 °C for 1–2 weeks.
Pellets containing recombinant protein were suspended in 10 ml of 50 mM TRIS-HCl (pH 8.2), 1 mM EDTA, 0.5 M NaCl, 0.5 mM phenylmethylsulphonyl fluoride (PMSF), 1 μg ml−1 pepstatin A, 2 μg ml−1 leupeptin, 10 mM imidazole, and 10% glycerol. Lysozyme was added to 1 mg ml−1 and the mixture was incubated on ice with gentle shaking. After 30 min, 6 mM 3-[3-cholamidopropyl)dimethylammonio]-1-propanesulphonate (CHAPS) was added and the mixture was shaken for a further 30 min. Then the mixture was made up to 10 mM MgCl2 and 5 mM ATP, a few DNase crystals added, and incubated at room temperature for 20 min with gentle rocking, followed by centrifugation at 3000 g and 4 °C for 10 min. The supernatant was collected and passed over ProBond nickel resin (Invitrogen, Carlsbad, CA, USA) equilibrated with 50 mM TRIS-HCl (pH 8.2) and 0.5 M NaCl. The nickel resin was washed with 50 mM TRIS-HCl (pH 8.2) containing 20 mM imidazole and 10% glycerol, and the recombinant protein was eluted with 50 mM TRIS-HCl (pH 8.2) containing 500 mM imidazole and 10% glycerol. Pyridoxal-5-phosphate at 2 μg ml−1 was added to all buffers used in the purification of the full or truncated GABA-T. Protein concentration in the eluate was determined using the Bradford assay method (Bradford, 1976). For visual confirmation, 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with 10 μl protein samples and standard protocols, and then the gels were stained with Coomassie brilliant blue R250 (Sambrook et al., 1989; Van Cauwenberghe et al., 2002). Immunoblotting was carried out according to standard protocols (Sambrook et al., 1989) using the HisTag monoclonal antibody (Novagen), the anti-mouse antibody (Sigma-Aldrich, Oakville, ON, Canada), and the alkaline phosphatase conjugate substrate kit (BioRad Laboratories, Hercules, CA, USA) for detection. Molecular weight standards were purchased from Fermentas (Burlington, ON, Canada).
The standard activity assay contained 50 mM N-tris(hydroxymethyl)methyl-4-aminobutanesulphonic acid (TABS; pH 9), 1.5 mM dithiothreitol (DTT), 0.625 mM EDTA, 0.1 mM pyridoxal-5-phosphate, 10% glycerol, and 0.5 μl of purified recombinant protein or 50 μl of desalted crude leaf extract, and was conducted at 30 °C. A discontinuous assay used a final volume of 500 μl and 1 mM amino acceptor; the reaction was initiated by the addition of 1 mM amino donor or water (control), incubated in a water bath for 15 min and 3 h, for determination of the specific activity and substrate specificity or leaf activity, respectively, and then terminated by the addition of ice-cold sulphosalicylic acid to 60 mM (Van Cauwenberghe and Shelp, 1999). The supernatant was neutralized with 1 N NaOH, and the production of specific amino acids was monitored via reverse-phase HPLC as described previously (Allan and Shelp, 2006). A continuous assay used a final volume of 800 μl and contained 125 μM NAD or NADH; the reaction was initiated by the addition of an appropriate amino donor. The influence of pH on enzyme activity was determined at 2 mM pyruvate and 8 mM GABA using 50 mM N-tris[hydroxymethyl]-methyl-2-aminoethane-sulphonic acid (pH 7.5–8.2), 50 mM TABS (pH 8.2–9.5), and 50 mM 3-(cyclohexylamino)-1-propanesulphonic acid (pH 9.5–10.5). Substrates were tested at pH 9 over the following concentration range to determine Km and Vmax values: pyruvate, 0.00625–2 mM; GABA, 0.0125–8 mM; glyoxylate, 0.00625–2 mM; alanine, 0.05–16 mM; and SSA, 0.005–0.5 mM. The following inhibitors of the forward reaction were tested with 2 mM pyruvate and 0.5–4 mM GABA: β-alanine, 0.5–2 mM; ornithine, 0.25–1 mM; glycine, 0.5–4 mM; and vigabatrin, 0.5–2 mM. Reactions that generate SSA and pyruvate were coupled to NAD-dependent SSADH (1.14 U or 50 μg ml−1 SSADH) (see Supplementary Materials and methods and Supplementary Fig. S1 available at JXB online) and NADH-dependent lactate dehydrogenase (LDH; 1 U or 31.25 μg ml−1, hog muscle, Boehringer Mannheim, Burlington, Canada), respectively. The rate of each reaction was monitored as the change in NADH concentration at 340 nm using a Beckman DU640 Spectrophotometer (Mississauga, Canada) equipped with temperature control. All assays were conducted in triplicate. Kinetic parameters were calculated using non-linear least-squares analysis (SigmaPlot2000, version 6.1; Enzyme Kinetics Module, version 1.0; Systat Software Inc., Point Richmond, CA, USA). Inhibition data were fit to appropriate forms of the Michaelis–Menten equation using non-linear least-squares analysis. The inhibition constant, Ki, and the mode of inhibition for each product were confirmed by secondary plots of slopes and intercepts from the double-reciprocal transformation. The equilibrium coefficient was calculated according to Van Bemmelen et al. (1985) using the following equation:
where f and r represent the forward and reverse reactions, respectively.
Expression and activity of native Arabidopsis GABA-T
The Arabidopsis knockout lines used in this study, POP2-3 (CS6387) and GABAT1-1 (Salk_007661) (ecotypes Columbia and Wassilewskija, respectively), have been characterized by Palanivelu et al. (2003) and Miyashita and Good (2008), respectively. All wild-type and knockout Arabidopsis plants were grown in controlled-environment chambers (Ecological Chambers Inc., Model GC8-2H, Winnipeg, Canada) set at 23/19 °C day/night temperatures, a photosynthetic photon flux density of 150 μmol m−2 s−1 at the top of the seedling trays (supplied by cool white fluorescent lighting, Sylvania, Mississauga, Canada), and 65% relative humidity, and supplied as necessary with tap water. For analysis of gene expression, Arabidopsis Columbia plants were grown in a Fox sandy loam (pH 6.5) under a 14 h photoperiod (i.e. long day conditions) and supplied once weekly with a modified quarter-strength nutrient solution (Shelp et al., 1992), and total RNA was isolated using the Qiagen RNeasy kit (Qiagen, Mississauga, Canada). The bottom four leaves of the rosette were harvested for RNA extraction at 2 (young), 4.5 (mature), and 10 (senescent) weeks, whereas root, stem, and flower samples were collected at 6 weeks. For analysis of amino acid composition and in vitro analysis of enzyme activity in mature leaves, plants were grown for 4 weeks in Sunshine professional growth mix (Sun Gro Horticulture Canada, Seba Beach, Alberta) under a 10 h photoperiod (i.e. short day conditions), and not supplied with fertilizer. Amino acid profiles were determined via reverse-phase HPLC as described previously (Allan and Shelp, 2006).
Real-time polymerase chain reaction (PCR) was preformed using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) with a BioRad iCycler (Hercules, CA, USA). The following pairs of gene-specific primers were used to amplify the GABA-T gene (5′ TGG ATC AGA TGC CAA CGA TA 3′ and 5′ GTG GAG CCA TGG TAC GAT TT 3′) and the 18S rRNA gene (5′ TCT GGC TTG CTC TGA TGA TT 3′ and 5′ TCG AAA GTT GAT AGG GCA GA 3′). Total RNA was treated with Turbo DNase (Ambion, Austin, TX, USA). For cDNA synthesis, 300 ng of total RNA were incubated with 1 μl of random hexamer primers, brought up to 15 μl with RNase free water, incubated at 75 °C for 10 min, and chilled on ice. A 2 μl aliquot of 10× reaction buffer, 40 U of RNase inhibitor (Ambion), and 10 nM dNTPs were added and then incubated at 25 °C for 10 min. One hundred units of M-MLV reverse (Ambion) were added to bring the final reaction volume to 20 μl, then the reaction was incubated at 37 °C for 1 h and inactivated at 95 °C for 10 min. Each quantitative PCR used 1× SYBR Green qPCR mix, 0.2 μM forward and reverse primers, and 1 μl of cDNA in a 20 μl volume. All tubes were subjected to 3 min at 95 °C, followed by 40 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. SYBR Green absorbance was detected at 72 °C, and all reactions were conducted in triplicate. GABA-T expression in each sample was normalized to the level of 18S rRNA (Nicot et al., 2005).
Cell-free extracts were prepared by grinding leaf material in 5 vols of ice-cold 50 mM TRIS-HCl buffer (pH 8.2) containing 3 mM DTT, 1.25 mM EDTA, 2.5 mM MgCl2, 10% glycerol, 6 mM CHAPS, 2 μg ml−1 pyridoxal-5-phosphate, 1 mM PMSF, and 2.5 μg ml−1 leupeptin and pepstatin A. The homogenate was incubated on ice for 20 min with gentle rocking and then centrifuged at 3000 g for 10 min at 4 °C, and the supernatant was desalted using a Zebra desalt spin column (Pierce, Rockford, IL, USA). The total protein concentration was determined using the Bradford assay method (Bradford, 1976). GABA-T activity was measured as described above.
Wild-type and knockout GABA-T plants were grown on three types of half-strength MS medium: (i) standard MS medium containing N; (ii) MS medium with 10 mM GABA as the sole N source; and (iii) MS medium containing no N and 10 mM potassium sulphate.
Transient expression and subcellular localization of Arabidopsis GABA-T in tobacco BY-2 cells
Four plant expression plasmids containing AtGABA-T, or a modified version thereof, were constructed for this study including: (i) pUC18/AtGABA-T–GFP, encoding the entire open reading frame (ORF) of AtGABA-T fused to the N-terminus of the green fluorescent protein (GFP); (ii) pRTL2/AtGABA-T-myc, encoding the entire ORF of AtGABA-T fused at its C-terminus to the myc epitope tag (Fritze and Anderson, 2000); (iii) pUC18/1–46-AtGABA-T–GFP, encoding the N-terminal 46 amino acids of AtGABA-T, which represents the protein's predicted mitochondrial matrix targeting pre-sequence fused to the N-terminus of GFP; and (iv) pUC18/1–46ΔAtGABA-T–GFP, encoding an N-terminal truncated version of the fusion protein AtGABA-T–GFP lacking the N-terminal amino acids 1–46 of AtGABA-T. Construction of these four plasmids was carried out by amplifying (via PCR) each of the above-mentioned AtGABA-T sequences with appropriate forward and reverse primers that introduced a 5′ and 3′ NheI site (for primer sequences refer to Supplementary Table S1 at JXB online). The PCR products were subcloned into pCR2.1 (Invitrogen), and the resulting plasmids were digested with NheI. The NheI DNA fragments were then ligated into either NheI-digested pUC18/NheI–GFP, yielding pUC18/AtGABA-T–GFP or pUC18/1–46-AtGABA-T–GFP, or NheI-digested pRTL2/NheI-myc, yielding pRLT2/AtGABA-T-myc or pRLT2/1–46AtΔGABA-T-myc.
The plasmid pUC18/NheI-GFP is a general purpose GFP fusion cassette whereby an in-frame NheI site was introduced at the 5′ end of the GFP ORF using PCR site-directed mutagenesis along with the appropriate (complementary) mutagenic forward (5′ GAC GAC CTG CAG GTC GAC GCT AGC ATG GTG AGC AAG GGC 3′) and reverse (5′ GCC CTT GCT CAC CAG CTA GCC GTC GAC CTG CAG GTC GTC 3′) primers and pUC18/GFP as template DNA (Chiu et al., 1996). pRTL2/NheI-myc, a modified version of the plant expression vector pRTL2/myc-XbaI (Murphy et al., 2003), was constructed in two steps. First, sequences encoding the XbaI site downstream of the myc epitope tag (-EQKLISEEDL-; Fritze and Anderson, 2000) were modified (via PCR mutagenesis) to encode a stop codon. Sequences of the (complementary) mutagenic primers used in this PCR were (forward) 5′ GAA GAT CTG TCT TGA ACT CCG CAA AAA TCA CC 3′) and (reverse) 5′ GGT GAT TTT TGC GGA CTT CAA GAC AGA TCT TC 3′. Secondly, the resulting plasmid pRTL2/myc-stop was modified (via PCR mutagenesis) using the complementary forward (5′ CTA GAA CGC TAG CAT GGA ACA AAA GTT G 3′) and reverse (5′ CAA CTT TTG TTC CAT GCT AGC GTT CTA G 3′) primers to introduce a unique in-frame NheI site immediately upstream of the myc epitope sequence, yielding pRLT2/NheI-myc. pBIN35SB60catE9, encoding the N-terminal pre-sequence of the β-subunit of F1-ATPase from Nicotiana plumaginfolia L. fused to the N-terminus of the bacterial passenger protein chloramphenicol acyltransferase (CAT) and serving as a well-established mitochondrial marker protein, was provided by F Chaumont (University of Lauvain, Place Croix de Sud, Belgium) (Chaumont et al., 1994).
Tobacco (Nicotiana tabacum L.) Bright Yellow-2 (BY-2) cells were maintained and prepared for biolistic bombardment as described previously by Banjoko and Trelease (1995). Transient transformations were performed using the Biolistic Particle Delivery System (Bio-Rad) with either 10 μg or 5 μg of plasmid DNA for individual or co-transformations, respectively. Bombarded cells were incubated at 26 °C for ∼20 h in covered Petri dishes to allow transient expression of introduced gene(s) and protein sorting within the cells.
Bombarded BY-2 cells were processed for immunofluorescence microscopy as described by Trelease et al. (1996). Briefly, cells were fixed in formaldehyde and permeabilized with pectolyase Y-23 (Kyowa Chemical Products, Osaka, Japan) and Triton X-100. For experiments designed to demonstrate the mitochondrial import of transiently expressed AtGABA-T-myc, fixed and pecytolyase-treated cells were differentially permeabilized with digitonin (25 μg ml−1) (Sigma Alrich Ltd), rather than Triton X-100, to perforate the plasma membrane, but not organelle membranes (Lee et al., 1997).
Primary and fluorescent dye-conjugated secondary antibodies and sources were as follows: mouse anti-myc (clone 9E10) in hybridoma medium (Princeton University; Monoclonal Antibody Facility, Hybridoma, Princeton, NJ, USA); mouse anti-CAT IgGs (kindly provided by S Subramani, University of California, San Diego, CA, USA); mouse anti-α-tubulin monoclonal antibody (clone DM1A) (Sigma-Aldrich Ltd); rabbit-pea E1β (Luethy et al., 1995); goat anti-mouse Alexa Fluor 488; goat anti-rabbit Alexa Flour 488 (Cedar Lane Laboratories Ltd, Burlington, ON, Canada); and goat anti-rabbit rhodamine red-X (Jackson ImmunoResarch Laboratories Inc., West Grove, PA, USA).
Microscopic visualization of labelled BY-2 cells was performed with a Zeiss Axioskop 2 MOT epifluorescence microscope (Carl Zeiss Inc., Thornwood, NY, USA) with a Zeiss 63× Plan Apochromat oil immersion objective (Carl Zeiss) and a Retiga 1300 charge-coupled device camera (Qimaging, Burnaby, BC, Canada). All images shown were deconvolved and adjusted for brightness and contrast using Northern Eclipse 5.0 software (Empix Imaging Inc., Mississauga, ON, Canada), and then composed into figures using Adobe Photoshop 8.0 (Adobe Systems, San Jose, CA, USA). The images shown are representative of data obtained from viewing several (>50) transformed BY-2 tobacco cells from at least two separate biolistic bombardment experiments.
Results
Production and characterization of recombinant Arabidopsis GABA-T
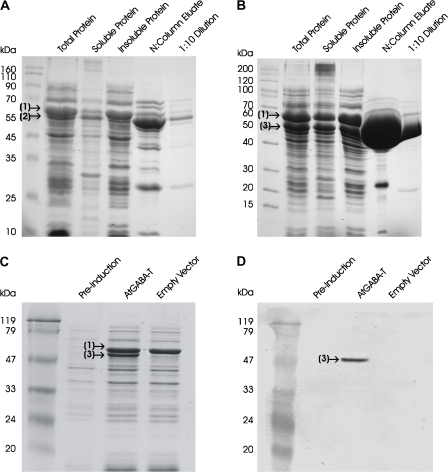
Initial attempts to express the full-length AtGABA-T protein in E. coli were unsuccessful due to the production of insoluble protein aggregates (data not shown). Co-expression of the GroES/GroEL chaperone complex from the pREP4-GroESL plasmid allowed for recovery of a small proportion of the recombinant protein in the soluble fraction (Fig. 1A). Removal of the protein's putative N-terminal mitochondrial targeting pre-sequence at the site of a predicted R-3 mitochondrial cleavage site (36 amino acids) (Sjöling and Glaser, 1998) dramatically improved the proportion of total protein recovered in the soluble fraction (Fig. 1B). Immunoblot analysis of total crude protein, probed with an α-Histag antibody, revealed a single recombinant truncated AtGABA-T band at approximately the predicted molecular mass of 53.4 kDa (Fig. 1C, D). Discontinuous assays revealed that the inability of the enzyme to utilize 2-oxogluarate as an amino acceptor was unchanged by removal of the targeting pre-sequence (Table 1). Furthermore, the mean specific activity (±SE of three biological preparations) for the truncated protein (13.4±0.2 μmol mg−1 protein min−1) was three times that for the full-length protein (4.5±0.2 μmol mg−1 protein min−1), as measured by the continuous assay. Thus, the truncated form of the recombinant enzyme was used for the kinetic experiments described herein.
Fig. 1.
SDS-PAGE analysis of expression and purification of the full-length recombinant AtGABA-T (A) or the truncated recombinant AtGABA-T lacking the mitochondrial targeting domain (B) from E. coli BL21 (DE3) Rosetta pLysS cells co-expressing the GroES/EL chaperone complex. (1) The GroEL subunit of the chaperone complex (60 kDa), (2) the full-length AtGABA-T (57.5 kDa), and (3) the truncated AtGABA-T (53.4 kDa) are indicated on the gels. The right-hand lane on each gel represents the column eluate diluted 10-fold. SDS-PAGE (C) and immunoblot (D) analyses of total crude protein from BL21 (DE3) Rosetta pLysS cells co-expressing the GroES/EL chaperone complex, and either the empty vector or the truncated recombinant AtGABA-T. The immunoblot was probed with a HisTag antibody. (This figure is available in colour at JXB online.)
Table 1.
Pyruvate- and glyoxylate-specific GABA-T activities associated with purified full-length or truncated (lacking the targeting domain) recombinant Arabidopsis protein, and crude cell-free Arabidopsis leaf extract
| Enzyme source | Amino acceptor | GABA-dependent activity |
|
| nmol min−1 mg−1 protein | nmol h−1 | ||
| Recombinant full-length protein | Pyruvate | 3530±90 | 847±21 |
| 2-Oxoglutarate | ND | ND | |
| Recombinant truncated protein | Pyruvate | 7540±360 | 1810±85 |
| 2-Oxoglutarate | ND | ND | |
| Arabidopsis crude extract | Pyruvate | 0.422±0.003 | 54.7±0.4 |
| 2-Oxoglutarate | 0.013±0.003 | 1.8±0.4 | |
Data represent the mean ±SE of triplicate measurements from a typical preparation. Amino acid production was determined via reverse-phase HPLC, and the detection limit for the method was ∼0.02 nmol h−1; ND indicates not detectable.
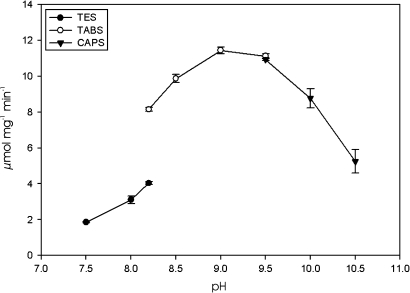
The pH optimum for AtGABA-T in the forward reaction was ∼9.0 (Fig. 2). Preliminary study of the substrate specificity at pH 9 via HPLC analysis of amino acid products revealed that the enzyme utilized glyoxylate, as well as pyruvate, as amino acceptors in the forward reaction, and GABA, but not β-alanine, ornithine, acetylornithine, serine, glycine, asparagine, glutamine, glutamate, valine, leucine, isoleucine, methionine, phenylalanine, histidine, lysine, arginine, aspartate, threonine, tyrosine, tryptophan, proline, or cysteine as amino donors (data not shown). All kinetic parameters for GABA, including specificity constants (i.e. kcat/Km), were similar in the presence of pyruvate and glyoxylate (Table 2, see also Supplementary Fig. S2 at JXB online). The kinetic parameters for pyruvate and glyoxylate were also similar in the presence of GABA. The pyruvate-dependent GABA-T reaction was reversible, whereas the glyoxylate-dependent reaction was not, although glyoxylate could also be used in conjunction with alanine as a substrate in the reverse reaction. The affinity for alanine in the reverse reaction was an order of a magnitude lower than that for GABA in the forward reaction, resulting in specificity constants that were also lower (Table 2; see also Supplementary Fig. S3 at JXB online). In contrast, the affinity and specificity constant for SSA were an order of magnitude higher than the same parameters for the other amino acceptors tested in either the reverse or forward directions. The calculated equilibrium ratio for the reversible pyruvate-dependent GABA-T reaction was 0.68. β-Alanine, ornithine, and vigabatrin were effective competitive inhibitors of pyruvate-dependent GABA-T activity, with Ki values similar to the Km for GABA. Glycine was a slightly less effective inhibitor, with a Ki value in the same range as the Km for alanine (Table 3; see also Supplementary Fig. S4 at JXB online).
Fig. 2.
Dependence of pyruvate-dependent AtGABA-T activity on pH. Data represent the mean ±SE of triplicate measurements using a typical enzyme preparation; where SE is not shown, it is within the symbol. Three overlapping buffers were used: N-tris[hydroxymethyl]-methyl-2-aminoethane-sulphonic acid (filled circles); N-tris(hydroxymethyl)methyl-4-aminobutanesulphonic acid (open circles); and, 3-(cyclohexylamino)-1-propanesulphonic acid (inverted triangles).
Table 2.
Kinetic parameters for the purified recombinant AtGABA-T
| Varied substrate | Fixed substrate | Km (mM) | Vmax (μmol mg−1 protein min−1) | kcat (s−1) | kcat/Km (s−1 mM−1) |
| Forward | |||||
| GABA | Pyruvate | 0.34±0.02 | 11.9±0.2 | 10.6 | 31 |
| GABA | Glyoxylate | 0.18±0.01 | 7.8±0.1 | 6.9 | 39 |
| Pyruvate | GABA | 0.14±0.01 | 11.9±0.2 | 10.6 | 76 |
| Glyoxylate | GABA | 0.11±0.01 | 10.9±0.3 | 9.7 | 88 |
| Reverse | |||||
| Alanine | SSA | 2.4±0.3 | 17.4±0.6 | 15.4 | 6.4 |
| Alanine | Glyoxylate | 2.2±0.3 | 15.8±0.6 | 14.1 | 6.5 |
| SSA | Alanine | 0.014±0.002 | 12.1±0.5 | 10.8 | 770 |
| Glyoxylate | Alanine | 0.14±0.02 | 13.6±0.4 | 12.1 | 86 |
Parameters were determined from a single typical preparation, and represent the calculated mean ±SE for the best fit of data to the Michaelis–Menten equation by non-linear regression. The forward reaction utilizes GABA and pyruvate or glyoxylate to generate SSA and alanine or glycine. The reverse reaction uses alanine with SSA or glyoxylate to produce pyruvate and GABA or glycine.
Table 3.
Inhibition constants for various inhibitors of pyruvate-dependent AtGABA-T activity
| Inhibitor | Ki (mM) |
| β-Alanine | 0.55±0.13 |
| Ornithine | 0.46±0.12 |
| Vigabatrin | 0.62±0.16 |
| Glycine | 3.70±0.46 |
Parameters were determined from a single typical preparation, and represent the calculated mean ±SE for the best fit of data to the appropriate Michaelis–Menten equation by non-linear regression.
Subcellular localization of Arabidopsis GABA-T
In order to determine the subcellular localization of AtGATA-T, GFP was fused to its C-terminus and the resulting fusion protein (AtGABA-T–GFP) was expressed transiently in tobacco BY-2 suspension-cultured cells serving as a well-characterized in vivo import system (Banjoko and Trelease, 1995; Miao and Jiang, 2007). Figure 3A–C shows that AtGABA-T–GFP expressed in BY-2 cells displayed a punctate fluorescence pattern that was identical to the fluorescence pattern attributable to co-expressed βATPase–CAT, a well-established mitochondrial matrix marker protein (Chaumont et al., 1994). Similar co-localization was observed for expressed AtGABA-T–GFP and immunostained endogenous E1β, a subunit of the pyruvate dehydrogenase complex located in the mitochondrial matrix (Luethy et al., 2001) (Supplementary Fig. S5A–C at JXB online). A C-terminal myc epitope-tagged version of AtGABA-T (AtGABA-T-myc) also co-localized with co-expressed mitochondrial βATPase–CAT in BY-2 cells (Supplementary Fig. S5D–F at JXB online), indicating that the appended GFP moiety in AtGABA-T–GFP did not influence AtGABA-T localization. Figure 3 also shows that the putative N-terminal pre-sequence of AtGABA-T, consisting of residues 1–36 plus 10 immediately adjacent residues, fused to GFP (1–46-AtGABA-T–GFP), exhibited an identical localization to co-expressed mitochondrial βATPase–CAT (Fig. 3D–F). In contrast, when the N-terminal 46 residues were deleted from AtGABA-T–GFP, the resulting truncated fusion protein (1–46ΔAtGABA-T–GFP) accumulated throughout the cytosol and nucleus (Fig. 3G–I).
Fig. 3.
Localization of AtGABA-T–GFP to mitochondria in tobacco BY-2 cells is mediated by its N-terminal 46 amino acid pre-sequence. Co-transiently expressed AtGABA-T–GFP (A) and the mitochondrial marker protein βATPase–CAT (B) co-localize in the same BY-2 cell, as evidenced by the yellow colour in the merged image (C). 1–46-AtGABA-T–GFP (D) co-localizes with co-expressed βATPase–CAT (E) in mitochondria in the same BY-2 cell, as evidenced by the yellow colour in the merged image (F). Expressed 1–46ΔAtGABA-T–GFP localizes throughout the cytosol and nucleus (G) and not to βATPase–CAT-containing mitochondria in the same co-transformed BY-2 cell (H). (I) Differential interference contrast (DIC) image of the same cell shown in (G) and (H). Bar in (A)=10 μm.
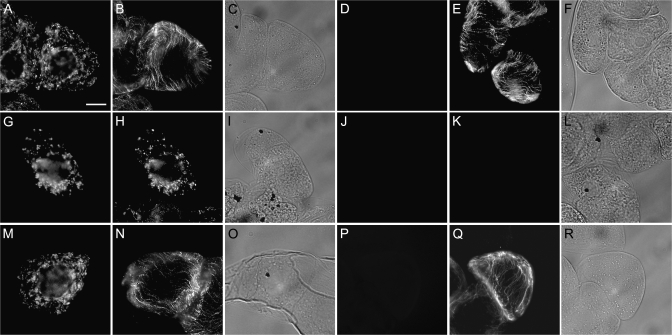
To confirm that AtGABA-T was imported into mitochondria, BY-2 cells transiently expressing AtGABA-T-myc were treated with Triton X-100 or digitonin to permeabilize cellular membranes selectively (Lee et al., 1997). Incubation of cells with Triton X-100 perforates all cellular membranes, including the plasma membrane and organellar membranes, allowing for the immunodetection of both endogenous E1β in the mitochondrial matrix and α-tubulin in the cytosol (Fig. 4A–C). Incubation of cells with digitonin, however, perforates only the plasma membrane, allowing for the immunodection of cytosolic α-tubulin, but not organelle (mitochondrial) membrane-protected E1β (Fig. 4D–F). While transiently expressed AtGABA-T-myc was immunodetected along with endogenous E1β in Triton X-100-permeabilized cells (Fig. 4G–I), neither protein was immunodetected in BY-2 cells permeabilized with digitonin (Fig. 4J–L). Likewise, both expressed AtGABA-T-myc and endogenous tubulin were immunodetected in cells that were permeabilized with Triton X-100 (Fig. 4M–O), whereas only tubulin was immunodetected in digitonin-treated cells (Fig. 3P–R). Taken together, the data presented in Figs 3 and 4 indicate that AtGABA-T is targeted to and imported into mitochondria in BY-2 cells via an N-terminal pre-sequence.
Fig. 4.
Differential permeabilization of AtGABA-T-myc-transformed tobacco BY-2 cells. The presence or absence of immunofluorescence in differential permeabilized cells indicates whether applied antibodies had access to transiently expressed AtGABA-T-myc and endogenous E1β within mitochondria or endogenous α-tubulin in the cytosol. Non-transformed (A–F) or AtGABA-T-myc transiently transformed (G–R) cells were fixed and permeabilized using either Triton X-100 (A–C, G–I, and M-O), which permeabilizes all cellular membranes, or digitonin (D–F, J–L, and P–R), which selectively permeabilizes the plasma membranes, then cells were processed for immunofluorescence microscopy. Immunostaining of endogenous E1β in the matrix of mitochondria (A) and of α-tubulin in cytosolic microtubules (B) in the same non-transformed, Triton X-100-permeabilized cells. Lack of immunostaining of endogenous E1β (D), but the presence of cytosolic tubulin (E), in the same digitonin-permeabilized cells. Immunostaining of expressed AtGABA-T-myc (G and M) and endogenous E1β (H) and tubulin (N) in the same Triton X-100-permeabilized cells. Lack of immunostaining of expressed AtGABA-T-myc (J and P) and endogenous E1β (K), but the presence of cytosolic tubulin (Q), in the same digitonin-permeabilized cells. Differential interference contrast (DIC) images of the corresponding BY-2 cells are shown in (C, F, I, L, O, and R). Bar in (A)=10 μm.
Characterization of native Arabidopsis GABA-T
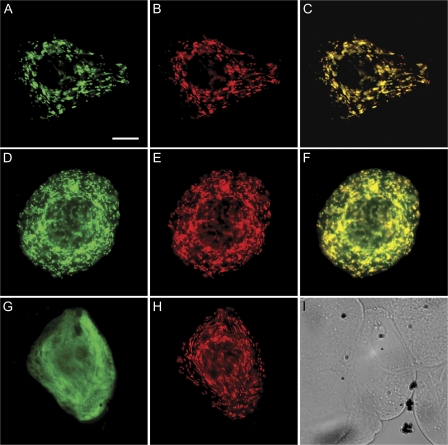
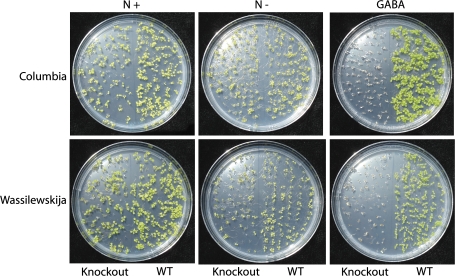
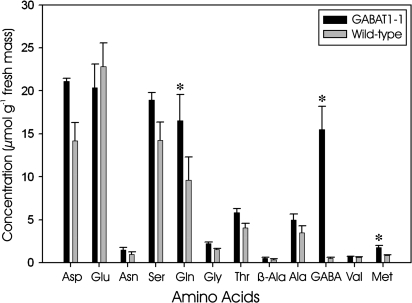
To confirm the presence of glyoxylate-dependent activity in planta, crude cell-free extracts of Arabidopsis leaves from two ecotypes were assayed for GABA-T activity. Both pyruvate- and glyoxylate-dependent GABA-T activities were present in wild-type Arabidopsis, but not in the corresponding knockout mutants (Table 4). Growth of the knockout mutants on minimal medium containing GABA as the sole N source revealed a white, bleached phenotype despite the presumed presence of 2-oxoglutarate-dependent activity (Fig. 5). Indeed, under these conditions, the knockout mutant appeared to exhibit a much shorter life cycle than when grown on minimal medium with no N. The GABAT1-1 knockout mutant also possessed an elevated concentration of leaf GABA, together with smaller but still significant increases in glutamine and methionine (Fig. 6), providing further evidence for the inability of these plants to catabolize GABA. Previous work demonstrated that GABA levels in the POP2-3 mutant are 11-fold higher than those in the wild type (Palanivelu et al., 2003).
Table 4.
Native GABA-T activity in cell-free extracts of two Arabidopsis ecotypes (Columbia and Wassilewskija) and corresponding knockout mutants
| Ecotype/mutant | Amino acceptor | Specific activity (nmol g−1 fresh mass min−1) | Specific activity (nmol m−2 s−1) |
| Columbia | |||
| Wild type | Pyruvate | 13.98±0.37 | 41.99±1.10 |
| GABAT1-1 | Pyruvate | ND | ND |
| Wild type | Glyoxylate | 8.13±0.51 | 24.42±1.53 |
| GABAT1-1 | Glyoxylate | ND | ND |
| Wassilewskija | |||
| Wild-type | Pyruvate | 17.07±0.85 | 51.27±2.55 |
| POP2-3 | Pyruvate | ND | ND |
| Wild-type | Glyoxylate | 7.73±0.25 | 23.21±0.75 |
| POP2-3 | Glyoxylate | ND | ND |
Data represent the mean ±SE of three preparations. Weight was expressed on an area basis by considering 0.018 g of fresh mass to be equivalent to 1 cm2. ND, indicates not detected; the lower limit of detection for the method was 0.25 nmol g−1 fresh mass min−1.
Fig. 5.
Growth of wild-type seedlings and knockout mutants (GABA-T1-1 and POP2-3) of Arabidopsis (ecotypes Columbia and Wassilewskija, respectively) on minimal MS medium containing GABA as the sole source of N (right panels), minimal MS medium with no source of N (centre panels B), and standard MS medium with N (left panels). Similar results were obtained in a replicated experiment.
Fig. 6.
Amino acid profile of mature leaves from Arabidopsis Columbia wild type and the GABA-T knockout (GABA-T1-1). Data represent the mean ±SE of three plants. Significant differences between the wild type and mutants, based on the Student's t-test (P ≤0.05), are indicated with an asterisk.
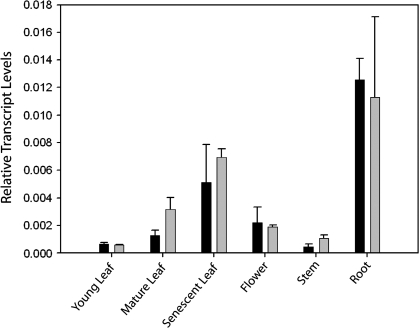
The GABA-T transcript was present in all plant organs analysed, and its relative abundance in those organs was reasonably consistent between experiments (Fig. 7). For leaves, transcript abundance increased with development, although the level in senescent tissue was not as high as that in roots. Young leaves and stems, and mature leaves and flowers displayed similar levels.
Fig. 7.
Relative GABA-T transcript abundance in various organs of wild-type Arabidopsis plants. The results are from two separate experiments (open and filled bars). Data represent the mean ±SE of three plants.
Discussion
Previous research demonstrated production of the full-length AtGABA-T in E. coli using the Xpress™ protein expression system (Invitrogen); however, the expression level in E. coli was low and could only be detected on gels using blotting techniques (Van Cauwenberghe et al., 2002). In the present study, it was possible to increase expression of the full-length AtGABA-T by using the pET15B expression vector (Novagen) with E. coli BL21 (DE3) Rosetta pLysS cells, but the protein formed insoluble aggregates. Efforts to improve the recovery of soluble AtGABA-T (i.e. reducing the temperature during the induction period, lengthening the induction period, lowering the IPTG concentration, and supplementation of the medium with ethanol) were unsuccessful (data not shown). However, co-expression of the GroES/GroEL chaperone complex, as well as removal of the putative mitochondrial targeting pre-sequence, dramatically enhanced the recovery of recombinant AtGABA-T in the soluble form (Fig. 1A, B; Table 1). Increased soluble expression using the chaperone complex is a well-documented phenomenon (Dale et al., 1994; Tibbetts and Appling, 2000; Mitsuda and Iwasaki, 2006). Furthermore, the targeting pre-sequence is probably removed from the mature protein upon localization within the cell (Glaser et al., 1998). Expression of a truncated AtGABA-T lacking the targeting pre-sequence should therefore reflect the mature enzyme in planta and has the added benefit of providing increased levels of expression and solubility in E. coli.
Previous biochemical analysis demonstrated that the recombinant AtGABA-T possesses pyruvate-, but not 2-oxoglutarate-dependent activity (Van Cauwenberghe et al., 2002). In the present report, it was demonstrated that the enzyme catalysed irreversible glyoxylate-dependent, as well as reversible pyruvate-dependent GABA-T activity (Table 2, Supplementary Figs S2, S3 at JXB online). To our knowledge, this is the first report of glyoxylate-dependent GABA-T activity in any species. 2-Oxoglutarate-dependent GABA-T in mammals, fungi, and bacteria is widely documented (Buzenet et al., 1978; Maitre et al., 1978; Der Garabedian et al., 1986; Kumar and Punekar, 1997; Liu et al., 2004), and pyruvate-dependent GABA-T (β-alanine also serves as an amino donor) activity in bacteria and fungi has been reported (Yonaha et al., 1983). Analysis of the amino acid composition in flowers of Arabidopsis GABA-T knockout mutants revealed a 5.7-fold enhancement in β-alanine concentration, together with a 100-fold increase in GABA (Palanivelu et al., 2003). While this could be interpreted as evidence for a promiscuous transaminase that utilizes both GABA and β-alanine, the evidence presented here indicates that the recombinant AtGABA-T does not utilize β-alanine. Indeed, the enzyme utilizes GABA and alanine only as amino donors for the forward and reverse reactions, respectively (Table 2).
The dual specificity of AtGABA-T could be considered analagous to that found with Arabidopsis alanine:glyoxylate (AGT1) and glutamine:glyoxylate (GGT1 and GGT2) aminotransferase. AGT1 exhibits three activities: serine:glyoxylate, serine:pyruvate, and alanine:glyoxylate, with the Km value for alanine (101 mM) being rather high, making its physiological relevance questionable (Liepman and Olsen, 2001). The homologues GGT1 and GGT2 also exhibit substrate promiscuity, catalysing glutamate:glyoxylate, alanine:glyoxylate, glutamate:pyruvate, and alanine:2-oxoglutarate activities (Liepman and Olsen, 2003). Thus, there is a tendency for plant glyoxylate-dependent transaminases to utilize multiple substrates.
The kinetic parameters for pyruvate and glyoxylate were similar (Table 2), indicating that the enzyme could utilize either substrate interchangeably. SSA, the amino acceptor for the reverse reaction, was the most efficient substrate for the enzyme, exhibiting a specificity constant that was an order of magnitude lower than for both pyruvate and glyoxylate. The calculated equilibrium ratio of 0.68 for the pyruvate-dependent reaction indicates that the reverse reaction is slightly favoured. This observation is similar to previous reports for GABA-Ts from rat, Pseudomonas, and mouse, which all favour the reverse direction to varying degrees (see Van Bemmelen et al., 1985). While the reverse reaction is favoured in vitro, two lines of evidence suggest that the enzyme does not generate much, if any, GABA in planta: (i) GABA synthesis and accumulation is highly regulated via stress stimulation/activation of glutamate decarboxylase (Crawford et al., 1994; Snedden et al., 1995, 1996); and (ii) the loss or inhibition of GABA-T activity causes the accumulation of GABA (Fig. 6; Palanivelu et al., 2003; Fait et al., 2005). The affinity of the enzyme for SSA could serve as a safety mechanism to ensure that SSA does not accumulate in the mitochondrion (Lindahl, 1992), a hypothesis that is consistent with the regulation of Arabidopsis SSADH by adenine nucleotides (Busch and Fromm, 1999). The irreversibility of the glyoxylate-dependent reaction is not likely to be due to specific exclusion of glycine by the enzyme, but rather a general transaminase phenomenom related to a high energy requirement for the reverse reaction (Smith, 1985). Indeed, glycine acted as a competitive inhibitor for the reaction, with a Ki value similar to the Km for alanine, indicating that the molecule fits into the active site (Table 3).
Vigabatrin was an effective competitive inhibitor of GABA-T activity (Table 3, Supplementary Fig. S4 at JXB online) as one would expect given its effectiveness as an inhibitor in mammalian systems (De Biase et al., 1991) and its ability to reduce the accumulation of reactive oxygen intermediates in SSADH-deficient Arabidopsis plants (Fait et al., 2005). Both β-alanine and ornithine also competitively inhibited GABA-T activity (Table 3), with Ki values similar to that for vigabatrin, strongly suggesting that all three compounds inhibit the enzyme because of their structural similarity to GABA.
A mitochondrial localization for GABA-T was previously indicated by cell fractionation studies (Breitkruez and Shelp, 1995) and mitochondrial proteome analysis (Sweetlove et al., 2002). This localization was confirmed in the present study by the transient expression of the full-length AtGABA-T cDNA in tobacco BY-2 cells. Herein, AtGABA-T co-localized with the co-expressed mitochondrial marker proteins βATPase–CAT (Chaumont et al., 1994) or E1β (Luethy et al., 2001), regardless of whether it was appended to the GFP or myc epitope tag (Fig. 3; Supplementary Fig. S5 at JXB online). Differential permeabilization of cellular membranes using Triton-X100 and digitonin also confirmed that AtGABA-T was localized inside the mitochondrion rather than associated with the outer membrane (Fig. 4). In addition, when the predicted AtGABA-T N-terminal pre-sequence (i.e. 36 amino acid residues plus 10 immediately adjacent amino acids) was appended to GFP, the fusion protein (1–46-AtGABA-T–GFP) sorted to mitochondria, whereas the corresponding truncated version of AtGABA-T (1–46ΔAtGABA-T–GFP), which lacks the N-terminal targeting pre-sequence, was localized to the cytosol and nucleoplasm (Fig. 3). These latter observations are consistent with previous reports of untargeted GFP in the nucleus and cytosol of plant cells (Köhler et al., 1997), and that the loss of AtGABA-T's mitochondria targeting ability was due to the removal of its N-terminal pre-sequence.
The absence of both pyruvate- and glyoxylate-dependent GABA-T activities in knockout mutants, and the inability of these mutants to grow on GABA as the sole N source, established that the protein under consideration catalyses GABA-T activity in planta (Table 4; Fig. 5). This indicates that Arabidopsis contains only one pyruvate-dependent GABA-T and does not contain a cytosolic form as predicted for rice (Ansari et al., 2005), tomato (AY240230), and pepper (AAC78480). These results, in conjunction with the accumulation of GABA in the knockouts (Fig. 6; Palanivelu et al. 2003), also suggest that a separate 2-oxoglutarate-dependent GABA-T is not present in seedlings, even though cell-free Arabidopsis leaf extracts displayed a low level of 2-oxoglutarate-dependent activity (Table 1), similar to that previously found in tobacco (Van Cauwenberghe and Shelp, 1999). The most likely explanation is that plants lack a second GABA-T enzyme and that the detectable 2-oxoglutarate-dependent activity is actually a combination of pyruvate-dependent GABA-T and alanine:2-oxoglutarate aminotransferase activity (Barbosa, 2002). This hypothesis is consistent with the instability of, and difficulty in purifying, the 2-oxoglutarate-dependent GABA-T from tobacco, which could not actually be separated from the more abundant pyruvate-dependent activity (Van Cauwenberghe and Shelp, 1999), the failure to identify a 2-oxoglutarate-dependent GABA-T in Arabidopsis based on homology (Barbosa, 2002), and the prevention of SSA and γ-hydroxybutyrate accumulation in the ssadh knockout by complementation with pyruvate-dependent GABA-T knockouts (Ludewig et al., 2008). In most cases, 2-oxoglutarate-dependent GABA-T activity in crude cell-free extracts is reported as lower than pyruvate-dependent activity; indeed, it is often over an order of magnitude lower (Table 1; also see references in Van Cauwenburghe and Shelp, 1999). These results may be due to the operation of pyruvate-dependent GABA-T in insufficiently desalted extracts, thereby allowing the coupled reaction to occur. The one exception is a recent paper by Akihiro et al. (2008), which reported that 2-oxoglutarate-dependent GABA-T activity in tomato fruit is three orders of magnitude higher than pyruvate-dependent activity. There are two reasons to treat those results with caution: (i) the pyruvate-dependent activity was assayed with 10 mM pyruvate, which is well above levels known to inhibit the enzyme (Van Cauwenberghe and Shelp, 1999); and (ii) the two activities were assayed under different conditions, with 2-oxogluarate-dependent activity being monitored after a 12 h incubation.
AtGABA-T knockout mutants of Arabidopsis grown with GABA as the sole N source developed a white bleached phenotype that was not observed in seedlings grown without N (Fig. 5). This might be attributed to the promotion of plant growth by GABA, followed by premature death due to the plant's inability to catabolize GABA. While further experimentation is required to test this hypothesis, it does fit with the recent literature suggesting a role for GABA in signalling (see Bouché et al., 2003; Shelp et al., 2006). GABA receptors have not yet been identified in plants; however, the presence of GABA-binding sites on pollen and somatic protoplast membranes has been demonstrated (Yu et al., 2006).
AtGABA-T knockout mutants of Arabidopsis display only one obvious physical phenotype, reduced seed production under self-fertilization, implying that the role of this enzyme is restricted to flower tissue (Palanivelu et al., 2003). More recently, it has been demonstrated that AtGABA-T knockouts also fail to respond to treatment with the C6-volatile E-2-hexanol (Mirabella et al., 2008), although the role of GABA in C6-volatile signalling is unknown. Real-time PCR analysis of the AtGABA-T transcript in Arabidopsis revealed that the gene was expressed in all organs tested, including roots, stem, leaves, and flowers (Fig. 7). In leaves, there was a clear trend of increasing expression with development, a pattern similar to that found in rice (Ansari et al., 2005). Roots contained the highest level of AtGABA-T transcript, though the physiological purpose for that level is not yet known. These results are in agreement with a previous study of AtGABA-T expression in all tissues except those for roots, which were found to have the same level as stem and mature leaf tissue (Miyashita and Good, 2008). Differences in AtGABA-T expression in roots between the two studies might be attributed to the growth conditions (semi-hydroponics versus soil). Nevertheless, the expression of AtGABA-T throughout the Arabidopsis plant indicates that it has a function(s) beyond pollen tube guidance in flowers (Palanivelu et al., 2003).
Identification of glyoxylate-dependent GABA-T activity raises the question of its role in plants. The mitochondrion is not traditionally associated with glyoxylate transamination. However, a glycolate dehydrogenase exists in Arabidopsis mitochondria, which resembles the photorespiratory pathway employed in green algae (Bari et al., 2004; Stabenau and Winkler, 2005). This enzyme could provide a source of glyoxylate for GABA-T, providing the basis for a link between GABA metabolism and photorespiration. Theoretically, these two enzymes could allow the glycolate to glycine reactions of photorespiration to occur in mitochondria. It is noteworthy that knockouts of both glycolate dehydrogenase (Bari et al., 2004) and GABA-T (this study) fail to exhibit typical photorespiration phenotypes when grown under atmospheric conditions. Despite the lack of phenotype, Niessen et al. (2007) demonstrated that glycolate dehydrogenase knockouts have a reduced photorespiration capacity. Sharkey (1988) estimated, based on Rubisco kinetics, that the rate of photorespiration at 210 μmol mol−1 CO2 is 8 μmol m−2 s−1, which compares with a GABA-T activity with 1 mM substrate and optimum pH of 40–50 nmol m−2 s−1 (equivalent to 17–20 μmol mg−1 protein min−1) in Arabidopsis crude cell-free extracts (Table 4). Thus, the GABA-T activity is at least two orders of magnitude lower than the expected rate of photorespiration, indicating that the theoretical contribution of GABA-T to photorespiratory flux would be small. Unfortunately, the glycolate dehydrogenase activity extracted from Arabidopsis is reported on a crude protein basis only (310 mmol mg−1 protein min−1; Niessen et al., 2007), which is extremely high compared with reports of algal glycolate dehydrogenase, which are in the nmol mg−1 min−1 range (Stabenau and Winkler, 2005).
Photorespiration has been proposed to act as a mechanism for stress protection in plants, preventing the over-reduction of the photosynthetic electron chain and photoinhibition, and providing metabolites such as glycine for other processes such as the synthesis of glutathione (Kozaki and Takebo, 1996; Wingler et al., 2000; Mullineaux and Rausch, 2005, and references therein). A mitochondrial version of the photorespiratory pathway would bypass the H2O2 production associated with the glycolate oxidase reaction, and reduce generation of additional reactive oxygen species. The accumulation of GABA during stress is a well documented phenomenom (Bown and Shelp, 1989; Satya Narayan and Nair, 1990; Bown and Shelp, 1997; Shelp et al., 1999; Kinnersley and Turano, 2000) and would provide ample amino donor for the transamination of glyoxylate to glycine during stress. While a model in which GABA metabolism contributes to photorespiration is very appealing, the lack of a photorespiration phenotype for both glycolate dehydrogenase and AtGABA-T knockout mutants suggests caution. Any contribution of AtGABA-T to photorespiration is likely to occur under stress when CO2 availability is restricted. Perhaps AtGABA-T functions to prevent the cellular accumulation of glyoxylate, a metabolite that is highly reactive and known to inhibit Rubisco activity (Campbell and Ogren, 1990; Hausler et al., 1996). Future work utilizing 15N-labelled GABA to determine the fate of GABA-N in planta under conditions designed to manipulate photorespiration may provide a better understanding of the relationship between the two pathways.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. Synthetic oligonucleotide primers used to amplify GABA-T sequences containing NheI restrictions sites.
Materials and methods. Production and purification of recombinant E. coli succinic semialdehyde dehydrogenase.
Figure S1. SDS-PAGE analysis of expression and purification of the recombinant E. coli K-12 SSADH from BL21 (DE3) Rosetta pLysS cells co-expressing the GroES/EL chaperone complex.
Figure S2. Kinetic characterization of AtGABA-T activity in the forward direction.
Figure S3. Kinetic characterization of AtGABA-T activity in the reverse direction.
Figure S4. Inhibition of pyruvate-dependent AtGABA-T activity by (A) β-Ala, (B) Orn, (C) vigabatrin, and (D) Gly.
Figure S5. Localization of AtGABA-T–GFP and AtGABA-T-myc to mitochondria in tobacco BY-2 cells.
Supplementary Material
Acknowledgments
This research was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to RTM and BJS, and the Ontario Ministry of Agriculture and Food to BJS, and postgraduate awards from the Ontario Graduate Scholarships Program to SMC, RD, and PKD. The authors thank F Chaumont (University of Louvain) for pBIN35Sb60catE9, Jan Miernyk (University of Missouri-Columbia) for rabbit pea-E1β antibodies, and Andrew Hanson (University of Florida) for suggesting use of the GroES/EL chaperone. The authors would also thank the Arabidopsis Biological Resource Center for providing us with POP2-3 seeds, and Yo Miyashita (University of Alberta) for providing GABAT1-1 seeds, which were originally obtained from the Arabidopsis Biological Resource Center.
Glossary
Abbreviations
- BY-2
Bright Yellow-2
- CAT
chloramphenicol acyltransferase
- GABA
γ-aminobutyric acid
- GABA-T
γ-aminobutyrate transaminase
- GFP
green fluorescent protein
- ORF
open reading frame
- PCR
polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- SSA
succinic semialdehyde
- SSADH
succinic semialdehyde dehydrogenase
References
- Akihiro T, Koike S, Tani R, et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant and Cell Physiology. 2008;49:1378–1389. doi: 10.1093/pcp/pcn113. [DOI] [PubMed] [Google Scholar]
- Allan WL, Peiris C, Bown AW, Shelp BJ. Gamma-hyroxybutyrate accumulates in green tea and soybean sprouts in response to oxygen deficiency. Canadian Journal of Plant Science. 2003;83:951–953. [Google Scholar]
- Allan WL, Shelp BJ. Fluctuations of γ-aminobutyrate, γ-hydroxybutyrate and related amino acids in Arabidopsis leaves as a function of the light–dark cycle, leaf age and N stress. Canadian Journal of Botany. 2006;84:1339–1346. [Google Scholar]
- Allan WL, Simpson JP, Clark SM, Shelp BJ. γ-Hydroxybutyrate accumulation in Arabidopsis and tobacco plants is a general response to abiotic stress: putative regulation by redox balance and glyoxylate reductase isoforms. Journal of Experimental Botany. 2008;59:2555–2564. doi: 10.1093/jxb/ern122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André B, Jauniaux J-C. Nucleotide sequence of the yeast UGA1 gene encoding GABA transaminase. Nucleic Acids Research. 1990;18:3046. doi: 10.1093/nar/18.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari IM, Lee R, Chen SG. A novel senescence-associated gene encoding γ-aminobutyric acid (GABA):pyruvate transaminase is unregulated during rice leaf senescence. Physiologia Plantarum. 2005;123:1–8. [Google Scholar]
- Banjoko A, Trelease RN. Development and application of an in vivo plant peroxisome import system. Plant Physiology. 1995;107:1201–1208. doi: 10.1104/pp.107.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa JMF. Auburn: Auburn University; 2002. Physiological, biochemical and molecular aspects of γ-aminobutyric acid (GABA): a stress-responsive non-protein amino acid. PhD thesis. [Google Scholar]
- Bari R, Kebeish R, Kalamajka R, Rademacher T, Peterhansel C. A glycolate dehydrogenase in the mitochondria of Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:623–630. doi: 10.1093/jxb/erh079. [DOI] [PubMed] [Google Scholar]
- Bartsch K, von Johnn-Marteville A, Schulz A. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase (gabT) and characterization of the succinic semialdehyde dehydrogenase (gabD) Journal of Bacteriology. 1990;172:7035–7042. doi: 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain. Journal of Biological Chemistry. 1993;268:19610–19617. [PubMed] [Google Scholar]
- Bouché N, Lacombe B, Fromm H. GABA signaling: a conserved and ubiquitous mechanism. Trends in Cell Biology. 2003;13:607–610. doi: 10.1016/j.tcb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Bouché N, Fromm H. GABA in plants: just a metabolite? Trends in Plant Science. 2004;9:110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Bown AW, Shelp BJ. The metabolism and physiological roles of 4-aminobutyric acid. Biochemistry (Life Science Advances) 1989;8:21–25. [Google Scholar]
- Bown AW, Shelp BJ. The metabolism and functions of γ-aminobutyric acid. Plant Physiology. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breitkreuz KE, Allan WA, Van Cauwenberghe OR, Jakobs C, Talibi D, André B, Shelp BJ. A novel γ-hyrdoxybutyrate dehydrogenase. Identification and expression of an Arabidopsis cDNA and potential role under oxygen deficiency. Journal of Biological Chemistry. 2003;278:41552–41556. doi: 10.1074/jbc.M305717200. [DOI] [PubMed] [Google Scholar]
- Breitkreuz KE, Shelp BJ. Subcellular compartmentation of the 4-aminobutyrate shunt in protoplasts from developing soybean cotyledons. Plant Physiology. 1995;108:99–103. doi: 10.1104/pp.108.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KB, Fromm H. Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiology. 1999;121:589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzenet AM, Fages C, Bloch-Tardy M, Gonnard P. Purification and properties of 4-aminobutyrate 2-ketoglutarate aminotransferase from pig liver. Biochimica et Biophysica Acta. 1978;522:400–411. doi: 10.1016/0005-2744(78)90073-6. [DOI] [PubMed] [Google Scholar]
- Campbell WJ, Ogren WL. Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynthesis Research. 1990;23:257–268. doi: 10.1007/BF00034856. [DOI] [PubMed] [Google Scholar]
- Chaumont F, de Castro Silva Filho M, Thomas D, Leterme S, Boutry M. Truncated presequences of mitochondrial F1-ATPase β subunit from Nicotiana plumbaginifolia CAT and GUS proteins into mitochondria of transgenic tobacco. Plant Molecular Biology. 1994;24:631–641. doi: 10.1007/BF00023559. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Current Biology. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Crawford LA, Bown AW, Breitkreuz KE, Guinel FC. The synthesis of γ-aminobutyric acid in response to treatments reducing cytosolic pH. Plant Physiology. 1994;104:865–871. doi: 10.1104/pp.104.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale GE, Schonfield H-J, Langen H, Stieger M. Increased solubility of trimethoprim-resistant type S1 DHFR from Staphylococcus aureus in Escherichia coli cells overproducing the chaperonins GroEL and GroES. Protein Engineering. 1994;7:925–931. doi: 10.1093/protein/7.7.925. [DOI] [PubMed] [Google Scholar]
- De Biase D, Barre D, Bossa F, Pucci P, John RA. Chemistry of the inactivation of 4-aminobutyrate aminotransferase by the antiepileptic drug vigabatrin. Journal of Biological Chemistry. 1991;266:20056–20061. [PubMed] [Google Scholar]
- De Biase D, Barra D, Simmaco M, John RA, Bossa F. Primary structure and tissue distribution of human 4-aminobutyrate aminotransferase. European Journal of Biochemistry. 1995;227:476–480. doi: 10.1111/j.1432-1033.1995.tb20412.x. [DOI] [PubMed] [Google Scholar]
- Der Garabedian PA, Lotti AM, Vermeersch J. 4-Aminobutyrate:2-oxoglutarate aminotransferase from Candida: purification and properties. European Journal of Biochemistry. 1986;156:589–596. doi: 10.1111/j.1432-1033.1986.tb09618.x. [DOI] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. Highway or byway: the metabolic role of the GABA shunt in plants. Trends in Plant Science. 2008;13:14–19. doi: 10.1016/j.tplants.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Fait A, Yellin A, Fromm H. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Letters. 2005;579:415–420. doi: 10.1016/j.febslet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Anderson TR. Epitope tagging: general method for tracking recombinant proteins. Methods in Enzymology. 2000;327:3–16. doi: 10.1016/s0076-6879(00)27263-7. [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjöling S, Tanudji M, Whelan J. Mitochondrial protein import in plants. Signals, sorting, targeting and regulation. Plant Molecular Biology. 1998;38:311–338. doi: 10.1023/a:1006020208140. [DOI] [PubMed] [Google Scholar]
- Hausler RE, Bailey KJ, Lea PJ, Leegood RC. Control of photosynthesis in barley mutants with reduced activities of glutamine synthetase and glutamate synthase. III. Aspects of glyoxylate metabolism and effects of glyoxylate on the activation state of ribulose-1,5-bisphosphate carboxylase-oxygenase. Planta. 1996;200:388–396. [Google Scholar]
- Hoover GJ, Van Cauwenberghe OR, Breitkreuz KE, Clark SM, Merrill AR, Shelp BJ. Characteristics of an Arabidopsis glyoxylate reductase: general biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Canadian Journal of Botany. 2007;85:883–895. [Google Scholar]
- Kinnersley AM, Turano FJ. Gamma aminobutyric acid (GABA) and plant responses to stress. Critical Reviews of Plant Science. 2000;19:479–509. [Google Scholar]
- Köhler RH, Zipfel WR, Webb WW, Hanson MR. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. The Plant Journal. 1997;11:613–621. doi: 10.1046/j.1365-313x.1997.11030613.x. [DOI] [PubMed] [Google Scholar]
- Kozaki A, Takebo G. Photorespiration protects C3 plants from photooxidation. Nature. 1996;384:557–560. [Google Scholar]
- Kumar S, Punekar NS. The metabolism of 4-aminobutyrate (GABA) in fungi. Mycological Research. 1997;4:403–409. [Google Scholar]
- Lee MS, Mullen RT, Trelease RN. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. The Plant Cell. 1997;9:185–197. doi: 10.1105/tpc.9.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ. Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. The Plant Journal. 2001;25:487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ. Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiology. 2003;131:215–237. doi: 10.1104/pp.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl R. Aldehyde dehydrogenases and their roles in carcinogenesis. Critical Reviews in Biochemistry and Molecular Biology. 1992;27:283–335. doi: 10.3109/10409239209082565. [DOI] [PubMed] [Google Scholar]
- Ling V, Snedden WA, Shelp BJ, Assman SM. Analysis of a soluble calmodulin binding protein from fava bean roots: identification of glutamate decarboxylase as a calmodulin activated enzyme. The Plant Cell. 1994;6:1135–1143. doi: 10.1105/tpc.6.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Peterson PE, Carter RJ, Zhou X, Langston JA, Fisher AJ, Toney MD. Crystal structure of unbound and aminooxyacetate-bound Escherichia coli γ-aminobutyrate aminotransferase. Biochemistry. 2004;34:10896–10905. doi: 10.1021/bi049218e. [DOI] [PubMed] [Google Scholar]
- Ludewig F, Hüser A, Fromm H, Beauclair L, Bouché N. Mutants of GABA transaminase (POP2) suppress the severe phenotype of succinic semialdehyde dehydrogenase (ssadh) mutants in Arabidopsis. 2008 doi: 10.1371/journal.pone.0003383. PLoS ONE3, e3383. doi:10.1371/journal.pone.0003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy MH, David NR, Elthon TE, Miernyk JA, Randall DD. Characterization of a monoclonal antibody directed against the E1α subunit of plant mitochondrial pyruvate dehydrogenase. Journal of Plant Physiology. 1995;145:443–449. [Google Scholar]
- Luethy MH, Gemel J, Johnston ML, Mooney BP, Miernyk JA, Randall DD. Developmental expression of the mitochondrial pyruvate dehydrogenase complex in pea (Pisum sativum) seedlings. Physiologia Plantarum. 2001;112:559–566. doi: 10.1034/j.1399-3054.2001.1120414.x. [DOI] [PubMed] [Google Scholar]
- Maitre M, Ciesielski L, Cash C, Mandel P. Comparison of the structural characteristics of the 4-aminobutyrate:2-oxoglutarate transaminase from rat and humna brain, and of their affinites for certain inhibitors. Biochimica et Biophysica Acta. 1978;522:385–399. doi: 10.1016/0005-2744(78)90072-4. [DOI] [PubMed] [Google Scholar]
- Miao YS, Jiang L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols. 2007;2:2348–2353. doi: 10.1038/nprot.2007.360. [DOI] [PubMed] [Google Scholar]
- Mirabella R, Rauwerda H, Struys EA, Jakobs C, Triantaphylides C, Haring MA, Schurrink RC. The Arabidopsis her1 mutant implicates GABA in E-2hexenal responsivness. The Plant Journal. 2008;52:197–213. doi: 10.1111/j.1365-313X.2007.03323.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda M, Iwasaki M. Improvement in the expression of CYP2B6 by co-expression with molecular chaperones GroES/EL in Escherichia coli. Protein Expression and Purification. 2006;46:401–405. doi: 10.1016/j.pep.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. Contribution of the GABA shunt in hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant and Cell Physiology. 2008;49:92–102. doi: 10.1093/pcp/pcm171. [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynthesis Research. 2005;86:459–474. doi: 10.1007/s11120-005-8811-8. [DOI] [PubMed] [Google Scholar]
- Murphy MA, Phillipson BA, Baker A, Mullen RT. Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiology. 2003;133:813–828. doi: 10.1104/pp.103.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, le Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Niessen M, Thiruveedhi K, Rosenkranz R, Kebeish R, Hirsch HJ, Kreuzaler F, Peterhansel C. Mitochondrial glycolate oxidation contributes to photorespiration in higher plants. Journal of Experimental Botany. 2007;58:2709–2715. doi: 10.1093/jxb/erm131. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell. 2003;114:47–59. doi: 10.1016/s0092-8674(03)00479-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Satya Narayan V, Nair PM. Metabolism, enzymology and possible roles of 4-aminobutyrate in higher plants. Phytochemistry. 1990;29:367–375. [Google Scholar]
- Sharkey TD. Estimating the rate of photorespiration in leaves. Physiologia Plantarum. 1988;73:147–152. [Google Scholar]
- Shelp BJ, Bown AW, Faure D. Extracellular gamma-aminobutyrate mediates communication between plants and other organisms. Plant Physiology. 2006;142:1350–1352. doi: 10.1104/pp.106.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends in Plant Science. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Penner R, Zhu Z. Broccoli (Brassica oleracea var. italica) cultivar response to boron deficiency. Canadian Journal of Plant Science. 1992;72:883–888. [Google Scholar]
- Shelp BJ, Walton CS, Snedden WA, Tuin LG, Oresnik IJ, Layzell DB. GABA shunt in developing soybean seeds is associated with hypoxia. Physiologia Plantarum. 1995;94:219–228. [Google Scholar]
- Simpson JP, Di Leo R, Dhanoa PK, Allan WL, Makhmoudova A, Clark SM, Hoover GJ, Mullen RT, Shelp BJ. Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. Journal of Experimental Botany. 2008;59:2545–2554. doi: 10.1093/jxb/ern123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöling S, Glaser E. Mitochondrial targeting peptides in plants. Trends in Plant Science. 1998;3:136–140. [Google Scholar]
- Smith IK. Aminotransferases utilizing glyoxylate. In: Christen P, Metzler DE, editors. Transaminases. New York: Wiley; 1985. pp. 390–396. [Google Scholar]
- Snedden WA, Arazi T, Fromm H, Shelp BJ. Calcium/calmodulin activation of soybean glutamate decarboxylase. Plant Physiology. 1995;108:543–549. doi: 10.1104/pp.108.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H. Activation of petunia glutamate decarboxylase by calcium/calmodulin or a monoclonal antibody which recognizes the calmodulin binding domain. Journal of Biological Chemistry. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- Stabenau H, Winkler U. Glycolate metabolism in green algae. Physiologia Plantarum. 2005;123:235–245. [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH. The impact of oxidative stress on Arabidopsis mitochondria. The Plant Journal. 2003;32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Tibbetts AS, Applings DR. Characterization of two 5-aminoimidazole-4-carboxaminde ribonucleotide transformylase/inosine monophosphate cyclohydrolase isozymes from Saccharomyces cerevisiae. Journal of Biological Chemistry. 2000;275:20920–20927. doi: 10.1074/jbc.M909851199. [DOI] [PubMed] [Google Scholar]
- Trelease RN, Xie W, Lee MS, Mullen RT. Rat liver catalase is sorted to peroxisomes by its C-terminal tripeptide Ala-Asn-Leu, not by the internal Ser-Lys-Leu motif. European Journal of Cell Biology. 1996;71:248–258. [PubMed] [Google Scholar]
- Van Bemmelen FJ, Schouten MJ, Fekkes D, Bruinvels J. Succinic semialdehyde as a substrate for the formation of γ-aminobutyric acid. Journal of Neurochemistry. 1985;45:1471–1474. doi: 10.1111/j.1471-4159.1985.tb07214.x. [DOI] [PubMed] [Google Scholar]
- Van Cauwenberghe OR, Makhmoudova A, McLean MD, Clark S, Shelp BJ. Plant pyruvate-dependent gamma-aminobutyrate transaminase: identification of an Arabidopsis cDNA and its expression in Escherichia coli. Canadian Journal of Botany. 2002;80:933–941. [Google Scholar]
- Van Cauwenberghe OR, Shelp BJ. Biochemical characterization of partially purified GABA:pyruvate transaminase from Nicotiana tabacum. Phytochemistry. 1999;52:575–581. [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. Photorespiration: metabolic pathways and their role in stress protection. Philosophical Transactions of the Royal Society B: Biological Sciences. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha K, Suziki K, Minei H, Toyama S. Distribution of ω-amino-acid:pyruvate transaminase and aminobutyrate:α-ketoglutarate transaminase in microorganisms. Agricultural and Biological Chemistry. 1983;47:2257–2265. [Google Scholar]
- Yu G, Liang J, He Z, Sun M. Quantum dot-mediated detection of γ-aminobutyric acid binding sites on the surface of living protoplasts in tobacco. Chemistry and Biology. 2006;13:723–731. doi: 10.1016/j.chembiol.2006.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.