Abstract
SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) genes have been demonstrated to play a role in somatic embryogenesis in several plant species. As more is learnt about these genes, the view of their role in plant development has broadened. The Medicago truncatula MtSERK1 gene has been associated with somatic embryogenesis and in vitro root formation. In order to study the role of MtSERK1 in development further, the MtSERK1 promoter sequence has been isolated and cloned into a promoter–GUS analysis vector. SERK1 promoter-driven GUS expression was studied in A. tumefaciens-transformed cultures and regenerated plants, in A. rhizogenes-transformed root clones, and in nodulation. In embryogenic cultures, GUS staining is detected after 2 d of culture at the edge of the explant and around vascular tissue. Expression at the explant edge intensifies over subsequent days and then is lost from the edge as callus formation moves inward. MtSERK1 expression appears to be associated with new callus formation. When somatic embryos form, GUS staining occurs throughout embryo development. Zygotic embryos show expression until the heart stage. The in planta studies reveal a number of interesting expression patterns. There appear to be three types. (i) Expression associated with the primary meristems of the root and shoot and the newly formed meristems of the lateral roots and nodule. (ii) Expression at the junction between one type of tissue or organ and another. (iii) Expression associated with the vascular tissue procambial cells. The data led us to conclude that MtSERK1 expression is associated with developmental change, possibly reflecting cellular reprogramming.
Keywords: 2HA, Development, lateral roots, Medicago truncatula, nodulation, SERK, somatic embryogenesis
Introduction
SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) genes encode leucine-rich repeat receptor-like kinases (LRR-RLKs), and the first SERK gene identified was reported in carrot (Daucus carota) suspension cultures where it was specifically expressed in cells which developed into somatic embryos (Schmidt et al., 1997). SERK genes have since been linked to somatic embryogenesis (SE) in a number of species including Dactylis glomerata (Somleva et al., 2000), Arabidopsis thaliana (Hecht et al., 2001), Medicago truncatula (Nolan et al., 2003), sunflower (Helianthus annuus) (Thomas et al., 2004), Ocotea catharinensis (Santa-Catarina et al., 2004), Citrus unshiu (Shimada et al., 2005), and Theobroma cacao (de Oliveira Santos et al., 2005). SERK genes have also been described in relation to apomixis in Hieracium (Tucker et al., 2003) and Poa pratensis (Albertini et al., 2005) as well as zygotic embryogenesis in carrot, Arabidopsis, and wheat (Triticum aestivum) (Schmidt et al., 1997; Hecht et al., 2001; Singla et al., 2008). The best defined SERK gene in relation to SE is the Arabidopsis SERK1 (AtSERK1) and overexpression of this SERK was shown to enhance embryogenic competence in Arabidopsis cultures (Hecht et al., 2001). SERK genes exist as gene families in many species with five SERK genes in Arabidopsis. MtSERK1 is the orthologue of AtSERK1 in the model legume, Medicago truncatula (Nolan et al., 2003).
After the earliest work on SERK1 linked it with the ability of plants to express their totipotent nature through SE (Schmidt et al., 1997; Somleva et al., 2000; Hecht et al., 2001), other studies began to indicate a broader role for this gene. Further culture studies showed that, in addition to expression during SE, SERK1 played a part in pluripotency during in vitro root formation in M. truncatula (Nolan et al., 2003) and in vitro shoot formation in sunflower (Thomas et al., 2004). In potato (Solanum tuberosum L.) SERK1 is highly expressed in microtubers which can develop into plants through organogenesis (Sharma et al., 2008). The argument for a role of SERK in pluripotency was further developed through expression data in Arabidopsis. SERK1 is expressed in the pluripotent cells of the vascular procambium, and it was speculated that SERK1 functioned in maintaining the pluripotent nature of procambial cells (Kwaaitaal and de Vries, 2007). Given that procambial cells are the origin of somatic embryos in carrot, it makes sense in a developmental context that procambial cells, which already exhibit a stem cell capacity, given the correct signals, could acquire a totipotent nature and develop into somatic embryos (Guzzo et al., 1994, 1995; Kwaaitaal and de Vries, 2007).
If SERK1 is linked to pluripotency, how is this linkage defined in the whole plant? In Arabidopsis, up-regulated SERK1 expression accompanies the initiation of lateral root growth (Kwaaitaal et al., 2005). The new organ, the lateral root, develops from the pericycle cells which become pluripotent. AtSERK1 expression is also closely linked to reproduction with expression during the development of both male and female reproductive tissues. In the ovule it is expressed during megasporogenesis, in cells of the embryo sac, and in the embryo up to the heart stage of development. Both AtSERK1 and AtSERK2 are expressed during anther development in anther primordia and then later in the tapetum and the middle layer precursors. Arabidopsis serk1/serk2 double mutant plants fail to develop a tapetal layer and are male sterile (Hecht et al., 2001; Albrecht et al., 2005; Colcombet et al., 2005; Kwaaitaal et al., 2005).
Receptor-like kinases transmit their signal by forming homodimers or heterodimers with other RLKs, in response to binding by a ligand. This ligand-induced dimerization causes phosphorylation of the intracellular kinase domains of the RLKs, which activates the next stages of the signal transduction pathway. There is potential for different levels of complexity in the signalling through variation in the binding partners of different RLKs. AtSERK1 is able to form homodimers (Shah et al., 2001) and also heterodimers with other RLKs. Heterodimerization of AtSERK1 with AtSERK2 (Albrecht et al., 2005) and AtSERK3 (Karlova et al., 2006) have been demonstrated, and also with BRASSINOSTEROID-INSENSITIVE1 (BRI1), which mediates brassinosteroid (BR) signal transduction (Karlova et al., 2006). It is established that AtSERK3 (also called BRI1-associated kinase1; BAK1) dimerizes with BRI1 in BR signal transduction (Li et al., 2002) and recently it was demonstrated that AtSERK1 can function with AtSERK3 in mediating BR signalling through BRI1 (Albrecht et al., 2008). BR can promote SE under some circumstances (Malik et al., 2008). However, signalling of AtSERK1 and AtSERK2 in anther development is not dependent on BR (Albrecht et al., 2008) and so regulation by another ligand(s) must occur in other AtSERK1 signalling pathways. Such ligands for AtSERK1 or its orthologues are yet to be identified. Current evidence shows members of the SERK family are part of both developmental and defence pathways acting in response to both steroid and peptide ligands. In particular, AtSERK3 has been shown to function in pathways that respond to the steroid ligand, BR, and the peptide ligand, flagellin (Albrecht et al., 2005; Colcombet et al., 2005; Hu et al., 2005; Chinchilla et al., 2007; He et al., 2007; Heese et al., 2007; Kemmerling et al., 2007). A similar situation occurs in tomato with the same LRR-RLK acting as a receptor protein for both BR and the peptide hormone, systemin (Montoya et al., 2002; Szekeres, 2003).
The role of MtSERK1 in cultured tissue of M. truncatula was previously studied (Nolan et al., 2003), comparing expression of SERK1 between a highly embryogenic seedline, 2HA (Rose et al., 1999), and its near isogenic, low embryogenic, progenitor line, Jemalong. These results indicated that MtSERK1 expression in culture was not only related to somatic embryogenesis, but also to organogenesis and, possibly, other forms of cellular reprogramming.
The present study was undertaken to gain insight into the roles of SERK1, not only in culture, but as it is expressed throughout the life cycle of the plant. An MtSERK1 promoter-driven GUS (prSERK1::GUS) expression construct was transformed into tissue of the embryogenic seedline, 2HA, of M. truncatula using Agrobacterium rhizogenes- and A. tumefaciens-mediated transformation. The prSERK1::GUS expression could then be visualized in the resulting transformed root clones, embryogenic cultures, regenerated plants and their progeny. This allowed a comprehensive view of MtSERK1 expression in culture and during all phases of plant development, including the formation of root nodules in response to symbiotic rhizobia bacteria. In embryogenic cultures, prSERK1::GUS expression correlates well with a change in developmental programming, as callus then somatic embryos are formed. In planta there is expression of MtSERK1 in the procambial zone of the vascular tissue and in meristem regions, in keeping with their roles in the maintenance of pluripotency. It is up-regulated during lateral root development and rhizobia-induced root nodule formation, both of which form from pluripotent stem cells. Expression was also observed during zygotic embryogenesis and in areas that may be described as ‘developmental transition zones’. These zones occur in places where there is a transition from one type of tissue to another, or between organs. The role of SERK1 appears to be linked to developmental change and the associated cellular reprogramming.
Materials and methods
The MtSERK1 promoter sequence was isolated using the Genome Walker Kit (Clontech) according to the manufacturer's manual. A sequence 1.5 kb upstream of the start codon was obtained. The sequence information has been deposited in the GenBank database under the accession number EU499307. This sequence length is similar to the length of sequence upstream from the start of the MtSERK1 orthologous gene in Arabidopsis, AtSERK1 (At1g71830). There is approximately 1460 bp of sequence between the gene upstream of the AtSERK1 at locus At1g71820 and the AtSERK1 start codon.
After transformation studies, in order to validate the quality of the expression data, seven independent transgenic lines were used in the analyses to provide a consensus pattern of expression. The expression observed using the prSERK1::GUS reporter construct showed similar expression patterns to those previously observed using quantitative real-time PCR in 2HA embryogenic cultures (Nolan et al., 2003) and in 2HA seedling tissues (results not shown). This construct also gave expression patterns in general agreement with those observed in AtSERK1 promoter driven GUS expression studies (Hecht et al., 2001; Kwaaitaal and de Vries, 2007; see Results and Discussion for details), indicating that the length of the promoter sequence obtained was sufficient to drive expression of the reporter gene in a way that gave a reliable indication of the expression of MtSERK1 in vivo.
Cloning the MtSERK1 promoter into a binary vector
The MtSERK1 promoter was amplified from 2HA leaf genomic DNA using the Expand Long Template PCR System (Roche), the SERK forward primer 5′-CTCGAGTTCTACCCGTCCGTACACCATAAC-3′ and the SERK reverse primer, 5′-CCCGGGTTGATTAAGTAGTAAATAACCTCA-3′ to give 1560 bp of promoter sequence before the ATG start codon and 60 bp of sequence downstream from the start codon. These primers also contained added sequences for restriction digestion with the enzymes XhoI (forward primer) and XmaI (reverse primer) (underlined).
The purified PCR product was cloned into the Gateway compatible pCR®8/GW/TOPO vector (Invitrogen) and electroporated into DH10B E. coli. Colonies were grown overnight on LB agar plates containing 50 μg ml−1 spectinomycin. The orientation of the promoter sequence in the vector was determined by colony PCR using the GW1 forward primer (Invitrogen) from the vector sequence and the SERK reverse primer (above). Colonies containing the promoter sequence in the correct orientation were cultured overnight on liquid LB 100 μg ml−1 spectinomycin medium and plasmid DNA extracted using the Wizard Plus SV Miniprep DNA Purification System (Promega). DNA from the cloned region to be inserted into the binary vector was sequenced.
The binary vector chosen was pHGWFS7 (Karimi et al., 2005), that allows both GFP and GUS to be expressed under the control of the inserted promoter sequence. As pCR®8/GW/TOPO (entry) and pHGWFS7 (destination) vectors both have resistance to the same antibiotic, spectinomycin, it was necessary to prevent colonies containing the pCR®8/GW/TOPO vector from growing after the LR recombination reaction. Therefore the pCR8/GW/TOPO vector (containing the SERK promoter sequence) was first digested with PvuI and XbaI. These two enzymes digest the pCR8/GW/TOPO plasmid, with PvuI cutting within the spectinomycin resistance gene, but leave the attL1 and attL2 recombination sites and the SERK promoter sequence intact. This allows the recombination reaction to take place but prevents growth of bacteria containing the original entry vector.
Digested entry vector DNA was purified and used to set up the Clonase LR recombination reaction (Invitrogen) with the pHGWFS7 vector. The products of the LR reaction were electroporated into competent DH10B E. coli and plated onto LB medium with 100 μg ml−1 spectinomycin selection. Plasmid DNA from six colonies was extracted and checked by gel electrophoresis. Three colonies with inserts of the expected size were then checked for the presence of the SERK promoter sequence by PCR with the SERK forward and SERK reverse primers. Plasmid DNA from one colony was electroporated into Agrobacterium tumefaciens, AGL1 and A. rhizogenes, R1000, competent cells and grown on YEP medium with selection antibiotics at 27 °C. Single colonies were checked by colony PCR for the presence of the SERK promoter sequence using the SERK forward SERK reverse primers.
Agrobacterium strains
Agrobacterium strains used for transformation were A. tumefaciens, AGL-1 (Lazo et al., 1991) and A. rhizogenes, R1000 (White et al., 1985). Glycerol stocks of Agrobacteria were streaked onto YEP (1% w/v Bacto tryptone, 1% w/v Bacto yeast extract, 0.5% w/v NaCl, pH 7) agar plates containing 100 μg ml−1 ampicillin+100 μg ml−1 spectinomycin for AGL-1 or 100 μg ml−1 spectinomycin for R1000, and grown at 27 °C for 2–3 d.
Transformation with A. tumefaciens and culture of tissue
Single colonies were used to inoculate 20 ml YEP liquid medium containing selective antibiotics in 50 ml centrifuge tubes and were incubated for 24–48 h at 27 °C at 150 rpm. OD600 was measured, bacteria centrifuged at 4000 rpm for 5 min and the pellet resuspended in about 20 ml of liquid P4 medium (Thomas et al., 1990) containing 10 μM NAA and 4 μM BAP (P4 10:4 medium) to a OD600 of 0.6.
Leaf tissue from greenhouse-grown plants of the highly embryogenic 2HA seedline (Rose et al., 1999) was collected and sterilized and cut up as described in Nolan et al. (2003). Leaf explants for transformation were immersed in bacterial suspension for 5 min and then blotted dry on sterile paper towel. Non-transformed control leaf tissue was immersed in liquid P4 10:4 with no bacteria. Explants were plated onto co-cultivation medium (P4 10:4 + 10 mM glucose+100 μM acetosyringone) and grown for 2–4 d in the dark. Explants were transformed with AGL-1 containing the prSERK1::GUS binary vector or with the pHGWFS7 empty vector control.
After co-cultivation, explants were washed in sterile distilled water for 5 min followed by washing in 500 μg ml−1 timentin in sterile distilled water for 5 min, and then blotted dry on sterile paper towel. Explants were plated onto P4 10:4 medium+750 μg ml−1 augmentin+25 μg ml−1 hygromycin and incubated in the dark at 27 °C. Non-transformed control tissue was plated both on medium with antibiotics and without antibiotics as a negative and positive control. After 3 weeks of culture, tissue was transferred to P4 10:4:1 medium (P4 10:4 plus 1 μM abscisic acid) containing the same antibiotic treatments as before. Tissue was then subcultured every 4 weeks to P4 10:4:1 medium ±antibiotics as before. Somatic embryos were removed from the callus tissue at subculture, plated onto P40 medium (P4 medium without inositol) without hormones, cultured in the light (14 h day length) and transferred to fresh medium every 4 weeks. Developing plantlets were cultured in Magenta pots on filter paper bridges soaked in about 8 ml of liquid P40 medium with low (1% w/v) sucrose. When sufficiently grown, plantlets were transferred to soil and covered with plastic wrap supported on stakes to maintain humidity. The plastic wrap was removed gradually, starting after 5–7 d.
Transformation with A. rhizogenes
2HA seeds were soaked in concentrated sulphuric acid for 6 min, rinsed in distilled water and then placed in a wire mesh tea infuser for sterilization as described previously (Nolan et al., 2003). Seeds were plated onto filter paper soaked with sterile distilled water in 9 cm Petri dishes, sealed with Parafilm and incubated in the light for germination. Liquid cultures of R1000 containing the prSERK::GUS construct or empty vector were inoculated from agar plates and grown overnight in YEP medium+100 μg ml−1 spectinomycin. The hypocotyls of 3-d-old seedlings were pricked with a 30 gauge needle and a drop of Agrobacterium suspension injected from a 1 ml syringe, through the needle onto the surface of the hypocotyl. Seedlings were blotted with sterile filter paper, plated onto P40 agar medium in 9 cm Petri dishes, and incubated in the light (14 h photoperiod). After 4 d, seedlings were removed from the medium and washed once in sterile distilled water followed by one wash in SM4 medium (Thomas et al., 1990) containing 250 μg ml−1 timentin. They were blotted on sterile filter paper and plated onto tall (2 cm high) 9 cm Petri dishes containing SM4 agar medium+1 μM NAA+250 μg ml−1 timentin. Ten days later, transformed roots were excised and plated onto sterile filter paper soaked with 4 ml of liquid SM4 medium+250 μg ml−1 timentin+25 μg ml−1 hygromycin in 9 cm Petri dishes. Dishes were placed in the dark, sitting at a slight angle to prevent over-saturation of the filter paper with medium. Root clones were subcultured to fresh medium every 3 weeks.
Nodulation
The seed coat from seeds to be germinated was pierced with a needle to allow penetration of water and the seeds were surface-sterilized as described above. Seeds were placed in 9 cm plastic Petri dishes containing filter paper and 8 ml of sterile water and left overnight to imbibe. Seeds were the transferred to 15 cm Petri dishes containing nitrogen-free Fåhraeus medium (Fåhraeus, 1957) with 1.5% w/v agar (Grade J3; Gelita Pty Ltd.). The bottom two-thirds of the plate was sealed with Parafilm and covered with black cardboard to inhibit light penetration. Plates were incubated in a growth room at 25 °C and a 14 h photoperiod with a light intensity of 30 μmol m−2 s−1. The rhizobia bacterium strain used to inoculate the roots was Sinorhizobium meliloti, 1021. S. meliloti were grown on agar plates containing Bergensen's modified medium (BMM) (Rolfe et al., 1980) and then used to inoculate 10 ml of liquid BMM medium which was grown overnight at 28 °C in a shaker at 150 rpm. The bacterial suspension was diluted with BMM medium to an OD600 of 0.1. Five-day-old seedlings were inoculated with 20 μl of bacterial suspension 1 cm from the growing root tip and returned to the growing conditions described above for nodule formation.
GUS staining
Tissue collected for GUS staining was immersed in freshly made GUS staining solution composed of 50 mM sodium phosphate buffer (pH 7.0), 1 mM EDTA, 0.1% Triton X-100, 1 mM X-Gluc (5-Br-Cl-3-indole-β-D-glucuronic acid; Research Organics), 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide. Tissue in GUS stain was vacuum infiltrated for 2–5 min and incubated overnight, or longer (up to 2 d) at 37 °C. Tissue was cleared by immersion in either 70% v/v ethanol, 1 M NaOH or diluted Hoyers solution (100 g chloral hydrate, 7.5 g gum arabic, 5 ml glycerol, 60 ml water), with solutions changed as required until the tissue was cleared.
Histology
Tissue to be sectioned was embedded in 2% w/v DNA grade agarose which had been dissolved in water and allowed to cool to 60 °C. Agarose was left to set for 30 min before trimming to the required size for sectioning. The agarose block was attached to the sectioning platform with Superglue and sectioned using a vibratome (The Vibratome company, St Louis, MO, USA). Sections were viewed using a Zeiss Axiophot microscope.
Results
The 1.5 kb MtSERK1 promoter (prSERK1) fused to the GUS reporter gene was used to visualize prSERK1::GUS expression in A. tumefaciens-transformed cultured plant tissue, in transformed regenerated plants, in cultures and seed progeny from those plants, and in A. rhizogenes-transformed root clones. This enabled prSERK1::GUS expression to be visualized throughout all stages of culture, during somatic embryogenesis, through seedling development, in all parts of the mature plant, and during the process of rhizobia-induced nodulation.
SERK1 expression in embryogenic cultures and somatic embryos
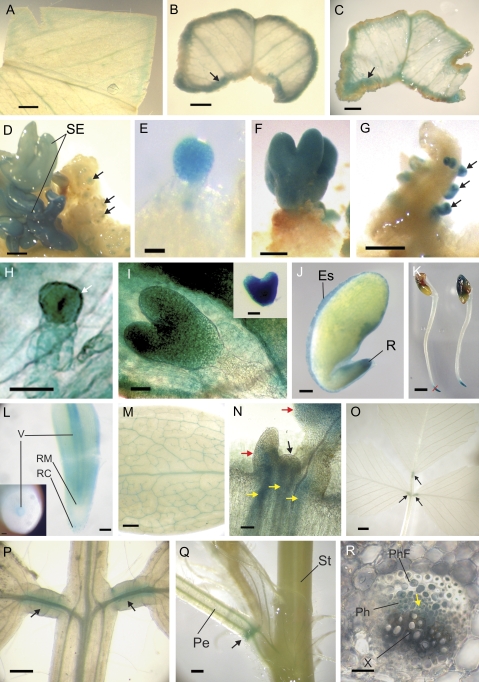
In embryogenic cultures GUS staining was evident after 2 d of culture. Expression occurred around the wounded edge of the explant and surrounding the vascular tissue within the explant (Fig. 1A). At 1 week, MtSERK1 expression increased as callus tissue formed on the edge of the explant with GUS expression forming a band encircling the explant (Fig. 1B). As the initiation of callus formation moved inwards from the edge of the explant there was concomitant prSERK1::GUS expression, but leaving the first formed callus on the edge of the explant displaying little GUS activity (Fig. 1C). Callus formation continued across the explant with associated GUS activity in newly forming callus, until the explant was covered with callusing tissue. Any somatic embryos that formed showed strong GUS activity (Fig. 1D). As callus tissue became established, the callus itself showed little GUS staining, only present as small spots in the callus (Fig. 1D).
Fig. 1.
prSERK-driven GUS expression during tissue culture and somatic embryogenesis, zygotic embryogenesis, and seed development, in seedling tissues and shoots. (A) Expression in embryogenic cultures can be seen after 2 d of culture at the edges of the explant and around the vascular tissue. (B) At 1 week, the edge of the explant shows stronger expression and callus formation (arrow). (C) After 3 weeks, callus tissue showing diminished expression is present on the edge of the explant. prSERK::GUS expression has moved centrally in from the edge of the explant (arrow) to the site of new callus formation. (D) By 8 weeks of culture, strongly expressing somatic embryos have formed. Callus tissue showing small spots of expression (arrows) covers the entire explant. (E) A single globular somatic embryo showing strong GUS expression. (F) Torpedo stage somatic embryo at the front with another somatic embryo behind it. (G) Recurrent somatic embryogenesis. New somatic embryos (arrows) form on the radicle of an older ‘germinated’ somatic embryo. (H) Globular stage zygotic embryo showing increased GUS staining in the protoderm (arrow). (I) Heart stage zygotic embryo showing GUS staining. Inset: strong GUS staining in heart stage zygotic embryo dissected from an ovule. (J) Developed zygotic embryo with seed coat removed. GUS staining is evident at the tip of the radicle and in the cellular endosperm layer. (K) Whole uncleared 2-d-old seedlings. GUS expression remains at the tip of the elongating radicle after germination. (L) 75 μm thick longitudinal vibratome section through the root apex of a 2-d-old seedling, which corresponds to the region below the red line drawn across the root pictured in (K). The inset shows a cross-section of root from a 2-d-old seedling taken further up the root in the region that does not show GUS staining in K. In the root tip, expression is visible behind the area of the root apical meristem (RM) including the cortex and the epidermal cells, with stronger expression in the vascular tissue (V). Expression is also seen in the peripheral cells of the root cap (RC). Inset: further up the root expression is confined to the vascular tissue. (M) Cotyledon from a 4-week-old seedling showing GUS staining in vascular tissue. (N) Longitudinal section through shoot apex of a 1-week-old seedling shows expression around vascular tissue below the SAM (yellow arrows) and at the proximal end of the leaf primordia and early leaves (red arrows). GUS staining is not apparent in the top cell layers of the SAM (black arrow). (O) The trifoliate leaf shows expression at the secondary pulvini (arrows). (P) A closer view of secondary pulvini showing expression surrounding the vascular tissue (arrows). (Q) Primary pulvinus showing GUS staining (arrow) at the node joining the petiole with the stem. (R) Cross-section of a vascular bundle in the petiole. A very low level of GUS stain is present in the procambial zone (arrow). Scale bars: (K) bar=2 mm; (B, C, D, O) bar=1 mm; (A, F, G, J, M, Q) bar=0.5 mm; (P) bar=0.25 mm; (L) bars=100 μm; (E, H, I, N) bar=50 μm; (R) bar=25 μm. Es, endosperm; Pe, petiole; Ph, phloem; PhF, phloem fibres; R, radicle; RC, root cap; RM, root apical meristem; SE, somatic embryo; St, stem; V, vascular tissue; X, xylem.
All somatic embryos exhibited strong prSERK1::GUS expression from the early globular stage through to the cotyledonary stage of development (Fig. 1E, F). Control cultures from plants transformed with the empty (promoterless) binary vector did not show any GUS expression either in callus tissue or in somatic embryos. As the embryo developed from the embryo phase to the germination phase, prSERK1::GUS expression was lost.
M. truncatula somatic embryos often show a high level of recurrent somatic embryogenesis (RSE). When this occurs, somatic embryos at different stages of development, or even small plants can cease ‘normal’ development and new somatic embryos will begin to form on the older tissue. During RSE, prSERK1::GUS expression was strongly up-regulated again in the newly developing embryos. This switch in expression was particularly evident when a developing plant, showing strongly diminished levels of expression, switched to RSE (Fig. 1G), providing further evidence of the association of SERK1 expression with the switch to embryogenic pathways.
MtSERK1 expression during zygotic embryogenesis
As in SE, MtSERK1 was expressed during zygotic embryo development. However, it did not show identical expression patterns. Expression was seen at the globular stage of embryo development with stronger expression in the protoderm (Fig. 1H). The strongest expression occurred at the heart stage (Fig. 1I) and then decreased as the embryo developed. In the late maturation phase of development, embryo expression always occurred in the radicle with varied expression in the cotyledons (Fig. 1J). The radicle expression was in the provascular strand and the epidermis (results not shown). The layer of cellular endosperm surrounding the embryo also showed expression (Fig. 1J).
Expression during seedling development
GUS expression was examined in seedlings germinated from transformed seed. Immediately after germination the radicle elongated rapidly, and this showed strong GUS staining at the tip (Fig. 1K). Sectioning of the root tip revealed that the root tip expression was primarily in the region behind the root apical meristem and the peripheral cells of the root cap. The tissue 1–2 mm behind the root apical meristem showed GUS expression in the cortical and epidermal cells, with stronger expression in the vascular tissue (Fig. 1L). Further up the seedling root (Fig. 1L inset), GUS staining was only observed in the vascular tissue. The cotyledons showed strong GUS staining throughout the vascular tissue (Fig. 1M). GUS was also strong in the vascular tissue immediately below the shoot apical meristem (SAM) and extended into the lower part of the leaf primordia and early leaves around the SAM. Within the SAM itself, virtually no expression was observed in the top few layers of cells but was present in the underlying cells of the rib zone and peripheral zone (Fig. 1N).
Expression during vegetative development and nodulation
Shoot:
In the mature plant, what was striking about the general pattern of prSERK1::GUS expression was that it often occurred in a transition zone between one type of tissue and another or one organ and another. The most apparent expression in the shoot was at the pulvinus, both at the primary pulvinus, where the petiole of the trifoliate leaf joins the stem and at the secondary pulvinus where each foliole of the trifoliate leaf joins the petiole (Fig. 1O, P, Q). Sections taken through the petiole showed a low level of GUS staining in the procambial zone (Fig. 1R).
Root:
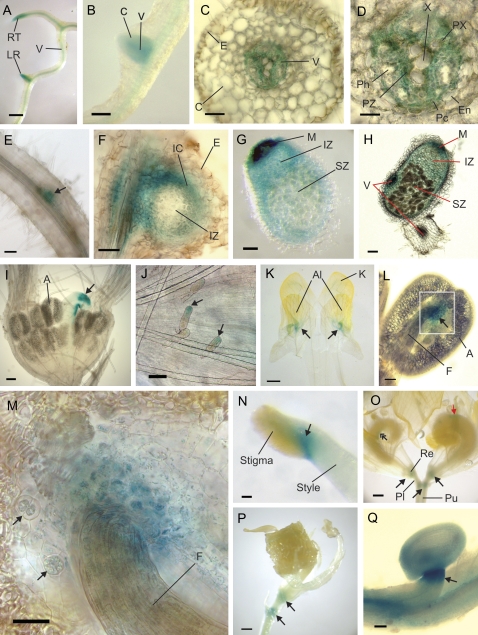
Root expression of prSERK1::GUS was studied using tissue from A. tumefaciens-transformed whole plants and from A. rhizogenes-transformed root clones. The A. rhizogenes-transformed root system is often used as a model for plant root gene expression (Colditz et al., 2007; Boualem et al., 2008). In root tissue of the mature plant, as in the seedling, expression was evident throughout the vascular tissue with stronger expression at the root tip and at the site of lateral root formation (Fig. 2A). In the emerging lateral root it was observed strongly in the vascular tissue and to a lesser degree in the cortex (Fig. 2B). The cortical expression diminished as the lateral root developed. In cross-section (Fig. 2C, D) GUS staining in the root vascular tissue was evident in the pericycle and the cells of the procambial zone. A. rhizogenes-transformed root clones did not show strong GUS staining. When GUS staining was easily visible, its expression pattern mirrored that in the root of the whole plant with expression in the vascular tissue and at the site of lateral root formation (Fig. 2E).
Fig. 2.
prSERK-driven GUS expression during root, nodule, and flower development. (A) In the root there is generalized expression in the vascular tissue with up-regulated expression at the root tip and the site of lateral root formation. (B) Emerging lateral roots show expression in both the vascular tissue and the cortex. (C) Cross-section of a mature root shows that the cortical expression of the emerging lateral root has been lost. Expression is limited to the vascular tissue. (D) A closer view of the root vascular tissue shows expression in the pericycle and the procambial tissue. (E) A hairy root from A. rhizogenes transformation shows lower expression than in that observed in A. tumefaciens-transformed regenerated plants and their progeny. Expression is observed at the site of lateral root formation (arrow). (F) Section through a root nodule, 3 weeks after inoculation with rhizobia showing expression around the vascular tissue and the inner cortex but not in the epidermal cells or infection zone. (G, H) In the mature nodule, expression can be seen throughout the nodule, except in the epidermal cells, but is strongest in the meristem and vascular tissue. (I) The floral meristem shows no expression in the male tissues, with the only expression coming from the developing pistil (arrow) and the glandular trichomes (arrows) on the external part of the meristem (J). (K) Flattened petals from a bud with an incision separating the two halves of the keel petal. Expression occurs in the region where the alae petals (on top) join the keel (arrows). (L) In the stamen there is expression at the junction point where the anther joins the filament (arrow). (M) A closer view of the boxed region in (L), showing expression at the anther/filament junction. Two cells can be seen to be in division (arrows). (N) Expression at the junction of the stigma with the style (arrow) in the female reproductive organs. (O) Expression in the flower occurs at major junction sites of the peduncle with the pedicel and the pedicel with the receptacle (arrows). There is also expression at the adaxial suture line of the ovary (double arrow) and sometimes on other parts of the ovary wall (red arrow). (P) Expression at the major junction sites of the flower persists as the seedpod develops (arrows). (Q) The older flower shows expression in the ovary wall and ovules, with up-regulated expression at the junction site between the ovule and ovary wall (arrow). Scale bars: (A, K, O, P) bar=0.5 mm; (B) bar=0.2 mm; (E, G, H, I) bar=100 μm; (C, F, J, L, N, Q) bar=50 μm. (D, M) bar=25 μm. A, anther; Al, alae petal; C, cortex; E, epidermis; En, endodermis; F, filament; IC, inner cortex; IZ, infection zone; K, keel petal; LR, lateral root; M, meristem; Pc, pericycle; Ph, phloem; Pl, pedicel; Pu, peduncle; PX, protoxylem; PZ, procambial zone; Re, receptacle; RT, root tip; SZ, symbiotic zone; V, vascular tissue, X, xylem.
Nodulation:
After inoculation with rhizobial bacteria, the roots of legumes can develop nitrogen-fixing root nodules. The inoculation of prSERK1::GUS transformed seedlings with S. melitoti indicated that MtSERK1 expresses in nodule development. Similar to the pattern of expression observed during lateral root formation, prSERK1::GUS expression was first up-regulated around the vascular tissue at the site of nodule formation. Expression spread around the edge of the developing nodule in the area of the cortical cells and vascular bundles. Expression was not observed in the infection zone or in the epidermal cells (Fig. 2F). As the nodule developed, expression spread to the cells of the infection zone. There was strong expression in the nodule meristem and vascular tissue (Fig. 2G, H).
Expression during floral development
The floral meristem showed a small spot of GUS staining on the wall of the developing pistil (Fig. 2I), with some limited expression in the glandular trichomes (Fig. 2J). At this stage, there was no expression in the ovules. Bud petals showed expression of prSERK1::GUS around the area where the alae petals are attached to the keel (Fig. 2K). This expression only appeared at the bud stage and was lost during flower development. GUS staining of the stamen was limited to the junction site of the anther with the filament (Fig. 2L). Recent cell divisions occur at this site (Fig. 2M). Expression in the ovary was primarily in the ovary wall at the adaxial suture site where the developing ovules join the ovary wall, but was sometimes observed on other parts of the ovary wall. Later in flower development, expression was observed at the transition zone between the stigma and style (Fig. 2N). As observed in the leaf, GUS staining in the inflorescence was present at the major organ junctions; the site of the peduncle with the pedicel, and the pedicel with the receptacle (Fig. 2O) and this expression pattern persisted during seed pod development (Fig. 2P). As the flower aged, GUS staining was observed in the ovule wall and particularly at the junction of the ovule with the ovary wall (Fig. 2Q).
Discussion
Previous work indicated MtSERK1 expression was part of the somatic embryogenesis pathway, but also played a broader role in the development of M. truncatula cultured tissue (Nolan et al., 2003). This work has further examined the role of SERK1 in embryogenic cultures and investigates prSERK1::GUS expression during all stages of legume development, including the formation of nitrogen-fixing root nodules.
Expression in embryogenic culture
Setting up cultured tissue allows the study of MtSERK1 expression in a system where previously differentiated cells are induced to dedifferentiate and enter into cell division, callus and embryo formation. In this system, explants taken from leaves, with very low expression, show an up-regulation of MtSERK1 expression within the first 2 d of culture (Nolan et al., 2003). Part of this expression comes from the vascular tissue and surrounding area, where the basal level of expression in the procambial region is probably up-regulated and the area of expression spreads to surrounding cells. The rest comes from new sites of expression at the wounded edge of the explant. At 1 week after the initiation of culture, the initial callus tissue at the edge of the explant shows strong MtSERK1 expression. However, rather than remaining high in the newly formed callus tissue, MtSERK1 expression is down-regulated as the callus becomes established and moves inwards to cells forming new callus. Over subsequent weeks MtSERK1 expression and callus formation spread across the entire explant. This wave pattern of expression in cultures strongly suggests that MtSERK1 is part of a signalling pathway that mediates developmental changes in cells in response to culture conditions. These developmental changes involve the initiation of cell division and cellular reprogramming required for callus formation. Once developmental change is established, the requirement for MtSERK1 signalling is diminished and MtSERK1 expression is down-regulated, even though callus proliferation continues. The observation that GUS expression tends to be present in isolated spots in later callus tissue may be an indication of cellular reprogramming to an embryogenic pathway. The strong GUS staining observed in somatic embryos adds further evidence to the established role of SERK1 in SE induction and development (Schmidt et al., 1997; Somleva et al., 2000; Hecht et al., 2001; Nolan et al., 2003; Thomas et al., 2004).
Previous work showed that during this phase of callus formation and development, MtSERK1 expression is also up-regulated in cultures of Jemalong, the non-embryogenic progenitor of the embryogenic 2HA line used in this study (Nolan et al., 2003). At the time, it was proposed that Jemalong cultures may initiate SE, but this pathway is blocked at a stage before embryos are visible. The current study indicates a second explanation for these results. In Jemalong, as in 2HA, SERK1 may mediate cellular reprogramming towards callus formation, but then Jemalong cultures are largely unable to initiate SE, which requires a fresh round of SERK1 expression when SE is triggered.
Once somatic embryos have developed and undergo germination to form small plants, MtSERK1 expression retracts to a very low basal level, suggesting that the differentiation pathways in the young plant have been established. However, when a small regenerated plant undergoes RSE, some of its cells cease the normal differentiation process, and once again, dedifferentiate and reprogramme into the embryogenesis pathway. The development of somatic embryos from these cells is via direct somatic embryogenesis, and once again MtSERK1 expression is induced in the new embryo. This new induction of expression is a further indication of a link between MtSERK1 expression and cellular reprogramming to an embryogenic pathway.
Zygotic embryogenesis
As the embryo progresses from globular to heart stage, the blueprint is set for shoot and root apical meristems and the different tissue types lying between these meristems. MtSERK1 is expressed during these early stages of embryogenesis, with expression peaking at the heart stage when these cellular patterns are first established. This is similar to the pattern of AtSERK1 expression during Arabidopsis zygotic embryogenesis, where expression was also observed in the suspensor, the outer cell layer of the globular stage embryo, and in the heart stage of development, but not later (Hecht et al., 2001; Kwaaitaal et al., 2005). DcSERK of carrot also shows expression in the early embryo but this ceases after the globular stage (Schmidt et al., 1997). M. truncatula and Arabidopsis also both show SERK1 expression in the endosperm (Hecht et al., 2001). As well as supplying nutrients to the developing seed, the endosperm is also an integrator of seed growth and development (Berger et al., 2006). Two other LRR-RLKs expressed in the endosperm of Arabidopsis seeds, HAIKU2 (IKU2) and EXTRA SPOROGENOUS CELLS (EXS) function in controlling seed size (Canales et al., 2002; Luo et al., 2005). Like these LRR-RLKs, SERK1 may function in the control of seed development.
In the developed embryo, MtSERK1 expression is evident in the provascular strands and epidermis of the radicle. After germination, strong expression continues in the tip of the elongating radicle. There SERK1 expression is observed in the root cap and in the young cells just behind the root apical meristem. Behind the root apical meristem, expression occurs in the epidermal and cortical cells and more strongly in the vascular tissue. Further up the root where the cells are older, expression is lost from the epidermis and cortex and is confined to the vascular tissue.
Thus the fundamental expression pattern of MtSERK1 in the root tissue is initiated in the seed before germination, and continues in the root tip and in the vascular tissue of the root throughout plant development. Expression in the root apex allows for a continued role of MtSERK1 in developmental change as the plant grows, as the pluripotent stem cells divide and differentiate into the various root tissues.
Expression during plant development
MtSERK1 shows expression in all organs of the plant, but this expression is under distinct temporal and positional control. The type of basal expression throughout the plant vascular tissue is always associated with cells of the procambial zone. In roots, this expression is higher than in shoots and is also present in the cells of the root pericycle. Therefore MtSERK1 is expressed in cells that have the potential to become meristematic under the correct signals. AtSERK1 is similarly expressed in the vascular tissue of Arabidopsis, particularly in the procambium and pericycle (Hecht et al., 2001; Kwaaitaal and de Vries, 2007).
Seedlings show an overall higher and more widespread expression of MtSERK1 than do adult plants. For example, the cotyledons have a high level of vascular expression that is not present in trifoliate leaves. The same is true for the vascular tissue of the stem, whose MtSERK1 expression decreases substantially as the plant develops. As the plant develops from a juvenile to an adult state, MtSERK1 expression in the vegetative shoot is almost undetectable through GUS staining except in the pulvini. These small regions enable movement of the leaf in response to signals such as light or stress.
In the stamen, GUS staining occurred at the junction of the anther with the filament. Arabidopsis SERKs 1 and 2 are both expressed in the tapetal cell layer of the anther and are necessary for microspore formation (Albrecht et al., 2005; Colcombet et al., 2005). MtSERK1 was not expressed in the tapetum or in any of the cells involved in microspore development and so probably does not have a similar role in male fertility. The transition site between the stigma and the style in the female reproductive tissues also shows MtSERK1 expression, as does the area joining the ovule with the ovary wall. The border area between two different organs or tissue types could be conceived as an area of developmental change as there is a transition from one tissue to another.
Other regions of MtSERK1 expression in the plant more clearly indicate a role for this gene in developmental change. It is expressed in shoot, root, and floral meristem regions. Developmental change in the forms of lateral root initiation and rhizobia-induced nodule development are both associated with up-regulated SERK1 expression. SERK expression has been shown to be induced during lateral root initiation in Arabidopsis (Kwaaitaal et al., 2005) and rice (Ito et al., 2005). A role for SERK in nodulation has not previously been reported.
The high expression of SERK1 during nodule initiation and development indicates that SERK does play a part in this process. Nodule formation entails highly regulated plant-bacteria and intra-plant signalling. Signalling within the plant occurs over both short distances within the root zone, and long distances via regulation from the shoot. Other LRR-RLKs such as MtDMI2 and MtSUNN are part of the nodulation signalling regulation, with MtDMI2 positioned in the root and MtSUNN being part of the long-distance signal pathway from the shoot controlling autoregulation of nodulation (reviewed in (Kinkema et al., 2006). How SERK1 fits in to the nodulation process, at this stage is unclear. It is however, highly expressed in nodules, in the cortex and vascular tissue at first and then becomes concentrated in the vascular tissue and the meristem.
Lateral roots are initiated from the pericycle which expresses MtSERK1. Therefore, the newly dividing cells of the early lateral root maintain the expression of a gene already expressed in their founder cells. M. truncatula plants form indeterminate nodules, which are initiated from the inner cortical cells and maintain a meristem throughout their life cycle. As the cortical cells of the root do not express SERK1, SERK1 expression must be switched on in the dividing cortical cells during nodule development.
Role of SERK1 during development
From the data obtained here the pattern of SERK1 expression in M. truncatula during plant development is in keeping with a role for SERK1 in pluripotency and cellular reprogramming to new developmental directions. Conceptually, this is consistent with the historical role of SERK1 in totipotency where cellular reprogramming involves embryo induction (Schmidt et al., 1997; Somleva et al., 2000; Hecht et al., 2001; Nolan et al., 2003; Thomas et al., 2004). What is somewhat surprising is the involvement of SERK expression in what appears to be every phase of plant organ development.
There has been a greatly increased understanding of the SERK family in recent years (He et al., 2007; Heese et al., 2007; Kemmerling et al., 2007; Sasaki et al., 2007; Albrecht et al., 2008; Hink et al., 2008; Sharma et al., 2008; Singla et al., 2008; Song et al., 2008). What has become apparent is that, although there is some overlap of function between the different SERKs, there is also specificity. So far, evidence indicates that SERK genes may function in pairs in a particular pathway, and they cannot be substituted by other SERK genes (Albrecht et al., 2008). If SERK genes work in pairs and different SERKs mix in different combinations, there is scope to greatly increase the flexibility of the signalling processes they may mediate during the plant's life cycle.
AtSERK1 functions (in partnership with AtSERK3) in BR-mediated signalling, but there is also evidence that BR is not the only ligand for this RLK (Albrecht et al., 2008). Whether or not BR signalling is responsible for the role of AtSERK1 in SE is unknown at this time. Evidence that BR can enhance SE in some cases (Malik et al., 2008) may indicate a connection. It is known from in vitro expression studies that expression of MtSERK1 can be induced by auxin and augmented by cytokinin (Nolan et al., 2003) during the process of cell division and differentiation leading to SE. Evidence of BR signalling through auxin signal transduction proteins (Nakamura et al., 2006; Vert et al., 2008) provides a point of crossover between the auxin and BR hormone signalling pathways.
There is also evidence based on immunoprecipitation experiments that AtSERK1 can form complexes with the MADS box transcription factor, AGAMOUS-LIKE15 (AGL15) (Karlova et al., 2006). Like AtSERK1, overexpression of AGL15 enhances SE in cultured tissues (Harding et al., 2003; Thakare et al., 2008). AGL15 reduces gibberellic acid (GA) levels by inducing a GA2 oxidase (Harding et al., 2003; Wang et al., 2004), which inactivates GA. GA is more commonly associated with enhanced seedling growth and reduced somatic embryogenesis (Henderson et al., 2004) and not differentiation induction. Analysis of the sequence of the MtSERK1 promoter reveals a binding recognition site for AGL15, and GA2 oxidase is up-regulated in SE induction in M. truncatula (Mantiri et al., 2008).
The concepts and information obtained from the in vitro studies is generally consistent with what was observed in relation to SERK expression in the vascular associated procambium, lateral root, and nodule induction where differentiation is influenced by auxin and cytokinin (Beveridge et al., 2007). The developmental knowledge is extensive in all these cases and it would appear that SERK1 expression is linked to developmental change. However, each system requires closer examination of how SERK1 is involved. This also applies to the primary meristem regions of the root and shoot and the nodule meristem. The organ junctions where SERK1 expression was observed are much less studied. However, it can be seen in the case of the anther and filament junction (Fig. 2M) that cells that are GUS expressing are engaged in mitotic activity. If this is characteristic of other junctions there is again a link to differentiation from recently divided cells.
The major recurring theme in the SERK1 expression studies is that SERK1 expression is characteristic of cells embarking on a new developmental programme. As a receptor kinase such a role is quite feasible but how SERK1 is involved in pathways integrated with hormonal and cellular differentiation requires detailed molecular and cytological investigation in specific developmental systems.
Acknowledgments
This work was supported by the Australian Research Council grant (no. CEO348212) to the University of Newcastle Node of the Centre of Excellence for Integrative Legume Research.
Glossary
Abbreviations
- SE
somatic embryogenesis
- NAA
1-naphthalene acetic acid
- BAP
6-benzylaminopurine
- LRR-RLK
leucine-rich repeat receptor-like kinase
- GUS
β-Glucuronidase
- SERK
somatic embryogenesis receptor-like kinase
- BR
brassinosteroid
- GFP
green fluorescent protein
- RSE
recurrent somatic embryogenesis
- SAM
shoot apical meristem
References
- Albertini E, Marconi G, Reale L, Barcaccia G, Porceddu A, Ferranti F, Falcinelli M. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiology. 2005;138:2185–2199. doi: 10.1104/pp.105.062059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. The Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiology. 2008;148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Grini PE, Schnittger A. Endosperm: an integrator of seed growth and development. Current Opinion in Plant Biology. 2006;9:664–670. doi: 10.1016/j.pbi.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Mathesius U, Rose RJ, Gresshoff PM. Common regulatory themes in meristem development and whole-plant homeostasis. Current Opinion in Plant Biology. 2007;10:44–51. doi: 10.1016/j.pbi.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. MicroRNA166 controls root and nodule development in Medicago truncatula. The Plant Journal. 2008;54:876–887. doi: 10.1111/j.1365-313X.2008.03448.x. [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Current Biology. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. The Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz F, Niehaus K, Krajinski F. Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta. 2007;226:57–71. doi: 10.1007/s00425-006-0466-y. [DOI] [PubMed] [Google Scholar]
- de Oliveira Santos M, Romano E, Yotoko KSC, Tinoco MLP, Dias BBA, Aragao FJL. Characterisation of the cacao somatic embryogenesis receptor-like kinase (SERK) gene expressed during somatic embryogenesis. Plant Science. 2005;168:723–729. [Google Scholar]
- Fåhraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. Journal of General Microbiology. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- Guzzo F, Baldan B, Levi M, Sparvoli E, Loschiavo F, Terzi M, Mariani P. Early cellular events during induction of carrot explants with 2,4-D. Protoplasma. 1995;185:28–36. [Google Scholar]
- Guzzo F, Baldan B, Mariani P, Lo Schiavo F, Terzi M. Studies on the origin of totipotent cells in explants of Daucus carota L. Journal of Experimental Botany. 1994;45:1427–1432. [Google Scholar]
- Harding EW, Tang WN, Nichols KW, Fernandez DE, Perry SE. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiology. 2003;133:653–663. doi: 10.1104/pp.103.023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Current Biology. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiology. 2001;127:803–816. [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences, USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiology. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Shah K, Russinova E, de Vries SC, Visser A. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophysical Journal. 2008;94:1052–1062. doi: 10.1529/biophysj.107.112003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L, Yang Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defence response against fungal infection. Planta. 2005;222:107–117. doi: 10.1007/s00425-005-1534-4. [DOI] [PubMed] [Google Scholar]
- Ito Y, Takaya K, Kurata N. Expression of SERK family receptor-like protein kinase genes in rice. Biochimica et Biophysica Acta. 2005;1730:253–258. doi: 10.1016/j.bbaexp.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends in Plant Science. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. The Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Current Biology. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Scott PT, Gresshoff PM. Legume nodulation: successful symbiosis through short- and long-distance signalling. Functional Plant Biology. 2006;33:707–721. doi: 10.1071/FP06056. [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M, de Vries SC, Russinova E. Arabidopsis thaliana Somatic Embryogenesis Receptor Kinase 1 protein is present in sporophytic and gametophytic cells and undergoes endocytosis. Protoplasma. 2005;226:55–65. doi: 10.1007/s00709-005-0111-9. [DOI] [PubMed] [Google Scholar]
- Kwaaitaal MACJ, de Vries SC. The SERK1 gene is expressed in procambium and immature vascular cells. Journal of Experimental Botany. 2007;58:2887–2896. doi: 10.1093/jxb/erm103. [DOI] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio-Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik MR, Wang F, Dirpaul JM, Zhou N, Hammerlindl J, Keller W, Abrams SR, Ferrie AMR, Krochko JE. Isolation of an embryogenic line from non-embryogenic Brassica napus cv. Westar through microspore embryogenesis. Journal of Experimental Botany. 2008;59:2857–2873. doi: 10.1093/jxb/ern149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang X-D, VandenBosch KA, Rose RJ. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiology. 2008;146:1622–1636. doi: 10.1104/pp.107.110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ. Cloning the tomato Curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. The Plant Cell. 2002;14:3163–3176. doi: 10.1105/tpc.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. The Plant Journal. 2006;45:193–205. doi: 10.1111/j.1365-313X.2005.02582.x. [DOI] [PubMed] [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiology. 2003;133:218–230. doi: 10.1104/pp.103.020917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe BG, Gresshoff PM, Shine J. Rapid screening for symbiotic mutants of Rhizobium and white clover. Plant Science Letters. 1980;19:277–284. [Google Scholar]
- Rose RJ, Nolan KE, Bicego L. The development of the highly regenerable seed line Jemalong 2HA for transformation of Medicago truncatula: implications for regenerability via somatic embryogenesis. Journal of Plant Physiology. 1999;155:788–791. [Google Scholar]
- Santa-Catarina C, Hanai LR, Dornelas MC, Viana AM, Floh EIS. SERK gene homolog expression, polyamines and amino acids associated with somatic embryogenic competence of Ocotea catharinensis Mez. (Lauraceae) Plant Cell, Tissue and Organ Culture. 2004;79:53–61. [Google Scholar]
- Sasaki G, Katoh K, Hirose N, Suga H, K-i Kuma, Miyata T, Su Z-H. Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergiiand Nitella axillaris: extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene. 2007;401:135–144. doi: 10.1016/j.gene.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Shah H, Gadella TWJ, van Erp H, Hecht V, de Vries SC. Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. Journal of Molecular Biology. 2001;309:641–655. doi: 10.1006/jmbi.2001.4706. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Millam S, Hein I, Bryan GJ. Cloning and molecular characterisation of a potato SERK gene transcriptionally induced during initiation of somatic embryogenesis. Planta. 2008;228:319–330. doi: 10.1007/s00425-008-0739-8. [DOI] [PubMed] [Google Scholar]
- Shimada T, Hirabayashi T, Endo T, Fujii H, Kita M, Omura M. Isolation and characterization of the somatic embryogenesis receptor-like kinase gene homologue (CitSERK1) from Citrus unshiu Marc. Scientia Horticulturae. 2005;103:233–238. [Google Scholar]
- Singla B, Khurana JP, Khurana P. Characterization of three somatic embryogenesis receptor kinase genes from wheat, Triticum aestivum. Plant Cell Reports. 2008;27:833–843. doi: 10.1007/s00299-008-0505-1. [DOI] [PubMed] [Google Scholar]
- Somleva MN, Schmidt EDL, de Vries SC. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Reports. 2000;19:718–726. doi: 10.1007/s002999900169. [DOI] [PubMed] [Google Scholar]
- Song DH, Li GJ, Song FM, Zheng Z. Molecular characterization and expression analysis of OsBISERK1, a gene encoding a leucine-rich repeat receptor-like kinase, during disease resistance responses in rice. Molecular Biology Reports. 2008;35:275–283. doi: 10.1007/s11033-007-9080-8. [DOI] [PubMed] [Google Scholar]
- Szekeres M. Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends in Plant Science. 2003;8:102–104. doi: 10.1016/S1360-1385(03)00010-4. [DOI] [PubMed] [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiology. 2008;146:1663–1672. doi: 10.1104/pp.108.115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Meyer D, Himber C, Steinmetz A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiology and Biochemistry. 2004;42:35–42. doi: 10.1016/j.plaphy.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Thomas MR, Johnson LB, White FF. Selection of interspecific somatic hybrids of Medicago by using Agrobacterium-transformed tissues. Plant Science. 1990;69:189–198. [Google Scholar]
- Tucker MR, Araujo A-CG, Paech NA, Hecht V, Schmidt EDL, Rossell J-B, de Vries SC, Koltunow AMG. Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. The Plant Cell. 2003;15:1524–1537. doi: 10.1105/tpc.011742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proceedings of the National Academy of Sciences, USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. The Plant Cell. 2004;16:1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White FF, Taylor BH, Huffman GA, Gordon MP, Nester EW. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. Journal of Bacteriology. 1985;164:33–44. doi: 10.1128/jb.164.1.33-44.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]