Abstract
SPPA1 is a protease in the plastids of plants, located in non-appressed thylakoid regions. In this study, T-DNA insertion mutants of the single-copy SPPA1 gene in Arabidopsis thaliana (At1g73990) were examined. Mutation of SPPA1 had no effect on the growth and development of plants under moderate, non-stressful conditions. It also did not affect the quantum efficiency of photosynthesis as measured by dark-adapted Fv/Fm and light-adapted ΦPSII. Chloroplasts from sppA mutants were indistinguishable from the wild type. Loss of SPPA appears to affect photoprotective mechanisms during high light acclimation: mutant plants maintained a higher level of non-photochemical quenching of Photosystem II chlorophyll (NPQ) than the wild type, while wild-type plants accumulated more anthocyanin than the mutants. The quantum efficiency of Photosystem II was the same in all genotypes grown under low light, but was higher in wild type than mutants during high light acclimation. Further, the mutants retained the stress-related Early Light Inducible Protein (ELIP) longer than wild-type leaves during the early recovery period after acute high light plus cold treatment. These results suggest that SPPA1 may function during high light acclimation in the plastid, but is non-essential for growth and development under non-stress conditions.
Keywords: Anthocyanin, chloroplast, high light acclimation, NPQ, protease, SPPA
Introduction
Proteases are essential for the development and function of all living organisms. They are responsible for processing, repair, and turnover of proteins and protein complexes in all compartments of the organism. Protease activity is tightly controlled either via protease expression, protease activation, or substrate accessibility. This research addresses the function of the plastid SPPA1 protease in Arabidopsis thaliana L. (Heynh.).
Plastids contain at least 11 types of proteases (Sakamoto, 2006), which have been identified by biochemical and genetic analyses, proteomic analysis or by prediction from EST databases and expression profiling (reviewed in Adam and Clarke, 2002; Adam et al., 2006; Sakamoto, 2006). All of the plastid proteases discovered to date have homologues in a bacterium, which reflects the bacterial origin of the organelle (Adam et al., 2006). They vary in location (envelope, stroma, lumen, or thylakoid-localized) and some of their functions are known (reviewed in Adam and Clarke, 2002; Adam et al., 2006; Sakamoto, 2006). Several of the best characterized plastid proteases are Clp, FtsH, Deg, and CND41. The SPPA1 protease is always relegated to the heading of ‘other proteases’ in reviews on the subject, along with other proteases about which little is known (Ostersetzer et al., 2007).
SPPA is an ATP-independent protease IV/serine-type endopeptidase. It was first described in eubacteria where it has a signal peptide peptidase activity, but is also found in viruses, archaea, and in the chloroplasts of photoautotrophic eukaryotes (reviewed in Sokolenko, 2005). The SPPA protease family contains two conserved potentially catalytic domains. In the cyanobacterium Synechocystis sp. PCC 6803, and many of the other heterotrophic and photoautotrophic bacteria, SppA has several isoforms that vary in the number of these domains they contain (Sokolenko, 2005). In higher plants the two catalytic domains are retained in a single gene, SPPA1 (hereafter referred to as SPPA), although the structure suggests that only one of the catalytic domains is active (Lensch et al., 2001; Sokolenko, 2005). Higher-plant SPPA was first identified in A. thaliana (Lensch et al., 2001), but it is present in many other species. NCBI Unigene currently lists SPPA genes from 16 plant species and the putative catalytic sites are conserved (CM Wetzel, data not shown; Lensch et al., 2001). This indicates that SPPA is ubiquitous in higher plants and is conserved, presumably because it has a purpose. Its function in chloroplasts is still unknown. Note that SPPA1 is structurally and functionally distinct from SPase1 (At2g30440, At1g06870, At3g24590), a known plastid thylakoid signal peptide peptidase (Inoue et al., 2005).
The SPPA protease was identified as a single copy gene in A. thaliana (At1g73990). Its resultant protein is estimated at 680 amino acid residues to yield a 74.8 kDa product. After plastid import and post-translational processing the protein is approximately 66–68 kDa in size. The chloroplast protease is anchored in the thylakoid membrane, enriched in stroma lamellae, but lacks transmembrane domains seen in its bacterial counterparts (Lensch et al., 2001; Friso et al., 2004). The SPPA mRNA is light-inducible in A. thaliana, yet the protein is constitutively present suggesting post-translational control of protein accumulation (Lensch et al., 2001). SPPA protein is easily detectable through immunoblotting during pre-induction, and experiences a 4-fold increase in abundance after a 52 h high light induction period (Lensch et al., 2001). Giacomelli et al. (2006) observed a dramatic increase in SPPA abundance in the proteome of high-light-stressed plants, and loss of ClpR2 resulted in reduced Clp complex accumulation and a 10-fold increase in SPPA (Rudella et al., 2006). Its light dependence and response to the loss of Clp suggest a role in protein turnover in chloroplasts during light stress and thylakoid protein homeostasis.
When the amount of light energy absorbed by chlorophyll exceeds the capacity of the photochemical process, photoinhibition and/or irreversible photo-oxidative damage (photodamage) can occur in chloroplasts (reviewed in Oelmüller, 1989; Prasil et al., 1992; and Logan et al., 2006; Barber and Andersson, 1992; Aro et al., 1993; Mishra et al., 1996). In the latter situation, singlet chlorophyll molecules remain in an excited state long enough to cause their transition to reactive triplet chlorophyll molecules. Damage to the chloroplast occurs when triplet chlorophyll reacts with oxygen, creating singlet oxygen radicals, which react with membrane lipids and proteins. Photoinhibition is a slowly reversible decrease in photosynthetic capacity primarily caused by repairable damage to the D1 polypeptide of Photosystem II (e.g. mediated by DegP2 and FtsH proteases; Lindahl et al., 2000; Haußühl et al., 2001) and engagement of photoprotective energy quenching, while photodamage and bleaching is a more extreme, non-reversible condition of chloroplast destruction. Several systems have evolved to prevent and/or protect the chloroplast from photo-oxidative damage (reviewed in Aro and Andersson, 2001). These include oxygen radical and peroxide-scavenging systems (ascorbate, glutathione, superoxide dismutase, etc), as well as scavengers of excess light energy (carotenoids). Any process that inhibits the use of absorbed light energy relative to its rate coming in will cause ‘excitation pressure’ on the photosystem, that is, more light energy is absorbed than can be processed by photochemistry so the reaction centres become reduced or ‘closed’ (Maxwell et al., 1995; Gray et al., 1996; Huner et al., 1998). Temperature extremes, sub-optimal salt levels, heavy metals, and desiccation all contribute to inhibition of processes downstream of light absorption and can lead to photoinhibition and/or photodamage.
To determine if SPPA in chloroplasts plays a role in high light acclimation, two SPPA T-DNA insertional knockout lines were identified in A. thaliana and the sppA mutant phenotypes were characterized under normal growth and high light stress conditions compared to wild-type plants. It was determined that under non-stress conditions, lack of SPPA had no appreciable effect on plant growth and development, but the mutants maintained a different level of non-photochemical quenching of Photosystem II (NPQ) than wild-type plants. Importantly, the lack of SPPA had little effect on plant response to short-term high-light treatment, but did alter the plant's response to longer-term high-light acclimation conditions compared to wild-type plants (measured by photoinhibition, early light-inducible protein, NPQ, chlorophylls, and anthocyanins).
Materials and methods
Plant material
The A. thaliana SPPA gene sequence (At1g73990) was used to query the Salk SIGnAL T-DNA express gene mapping tool (www.signal.salk.edu) and seeds for the potential knockout lines were ordered. Putative insertion lines included 07272 from the Salk Institute (Alonso et al., 2003; ordered from the ABRC); and 142B03, 313C07, and 320F09 T-DNA mutants that were generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (MPI for Plant Breeding Research; Cologne, Germany; Rosso et al., 2003). The lines are referred to hereafter as sppA-7272, sppA-142, sppA-313, and sppA-320, respectively. All transgenic lines were in the Columbia ecotype background and Columbia-0 was used as the wild type in all experiments. Unless otherwise noted, plants were grown on basic potting soil medium with slow-release fertilizer, under 140 μmol m−2 s−1 illumination by a mix of fluorescent and incandescent bulbs at 21 °C.
Each line was grown and individual segregant plants were screened for homozygosity of the T-DNA insert by PCR. Primers and annealing temperatures (*) used for PCR screening of T-DNA insert lines were as follows. Touchdown PCR cycles: 94 °C for 3 min, 3 reps of (94 °C for 1 min, 65 °C for 0.5 min, 72 °C for 1 min), 35 reps of (94 °C for 1 min, annealing temperature *°C for 0.5 min, 72 °C for 1 min), 72 °C for 10 min. Primer sets and annealing temperatures were: sppA-7272, Left primer 5′-CAAGACGCTTCCCTTACGAAC-3′, Right primer 5′-CGGAAGAAGAGTCAAATTGGG-3′, T-DNA LB primer 5′-TGGTTCACGTAGTGGGCCATCG-3′, Annealing temperature 55 °C; sppA-142, Left primer 5′-TTGTCTTTCTCGACTCCAAGC-3′, Right primer 5′-TATCTTGGATGTGCATGCAAC-3′, T-DNA LB primer 5′-CCCATTTGGACGTGAATGTAGACAC-3′, Annealing temperature 50 °C; sppA-313, Left primer 5′-AAGAAGTTGCACAGGGCAGAG-3′, Right primer 5′-TCTTGACGGATCAATGTACGC-3′, T-DNA LB primer 5′-CCCATTTGGACGTGAATGTAGACAC-3′, Annealing temperature 50 °C; sppA-320, Left primer 5′-CAAGACGCTTCCCTTACGAAC-3′, Right primer 5′-CGGAAGAAGAGTCAAATTGGG-3′, T-DNA LB primer 5′-CCCATTTGGACGTGAATGTAGACAC-3′, Annealing temperature 50 °C. DNA extraction and PCR were carried out with the REDExtract-N-Amp Plant PCR Kit (Sigma). All four lines were successfully amplified and homozygous plants were identified for each. PCR products were sequenced to verify the insert location. A single plant from each line was chosen for seed collection and further experiments. Subsequent analyses of SPPA mRNA and protein (described below) revealed that sppA-7272 and sppA-313 were significantly leaky, so data pertaining to these two lines are not included in this paper (data not shown).
Analysis of SPPA mRNA expression
Because SPPA mRNA is induced by high light (HL) treatment (Lensch et al., 2001), wild type and putative knockout plants were shifted to high light (600 μmol photons m−2 s−1) for 10 h and 24 h to induce gene expression.
Characterization of SPPA mRNA expression was carried out by non-quantitative RT-PCR. Total RNA was extracted using the Sigma Spectrum Plant Total RNA kit with on-column DNase treatment, then quality was assessed by gel electrophoresis. RNA concentration was determined using a Nanodrop spectrometer and 0.5 μg of total RNA was reverse-transcribed using Invitrogen Superscript III RNase H- reverse Transcriptase with oligo dT primers. cDNA was amplified by PCR (TaKaRa Ex-Taq Polymerase) using three different sets of primers (Fig. 1; see Supplementary Table S1 at JXB online). Conditions were optimized so that the wild-type cDNA amplification was maximized (i.e. beyond the linear range) to ensure that even low template levels in the putative mutants would amplify enough to be detected (not shown). All primer sets flanked introns so that any contaminating DNA would be detected by its product size (none detected except in genomic DNA positive controls, not shown).
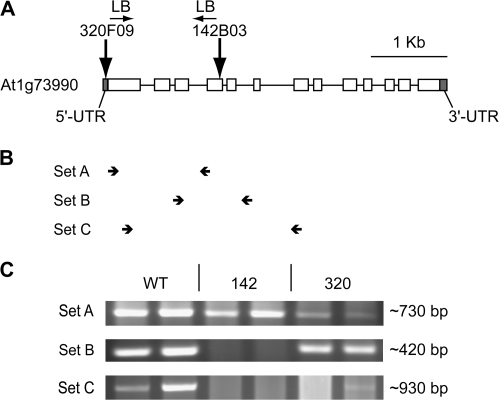
Fig. 1.
Location of T-DNA inserts in At1g73990 in homozygous plant lines. (A) Site of T-DNA insertion in lines sppA-320 (320F09) and sppA-142 (142B03). (B) Location of RT-PCR primers sets (not to scale). (C) Ethidium bromide-stained agarose gels of RT-PCR products corresponding to the primer sets shown in (B). RT-PCR products from two independent RNA extractions are shown for each genotype. Product size is indicated.
RLM 5′-RACE (Ambion FirstChoice RLM-RACE kit) was carried out according to manufacturer's instructions. This method clones only full-length and capped transcription products. RNA from two different plants each of WT and sppA-320 were used, and two WT plus five sppA-320 RACE clones were sequenced. The SppA gene-specific forward primer was 5′-TCCGCAGCGGCTTTATACAGAAGA-3′, the outer reverse primer was 5′-TGGTCAAGACGCTTCCCTTACGAA-3′, and inner reverse primer was 5′-TCCAGCAGCTGTTCCTGTTTCTCT-3′.
Production of anti-SPPA antiserum
An anti-peptide antiserum was produced in rabbits using the peptide (C)LGVEKDKKLPTVDYKKYS (Pocono Rabbit Farm and Laboratory, Inc., Pennsylvania). Preimmune and post-immune sera were analysed by Western blots of thylakoids isolated from wild-type and putative mutant plants to verify specificity of the antiserum (Fig. 2A).
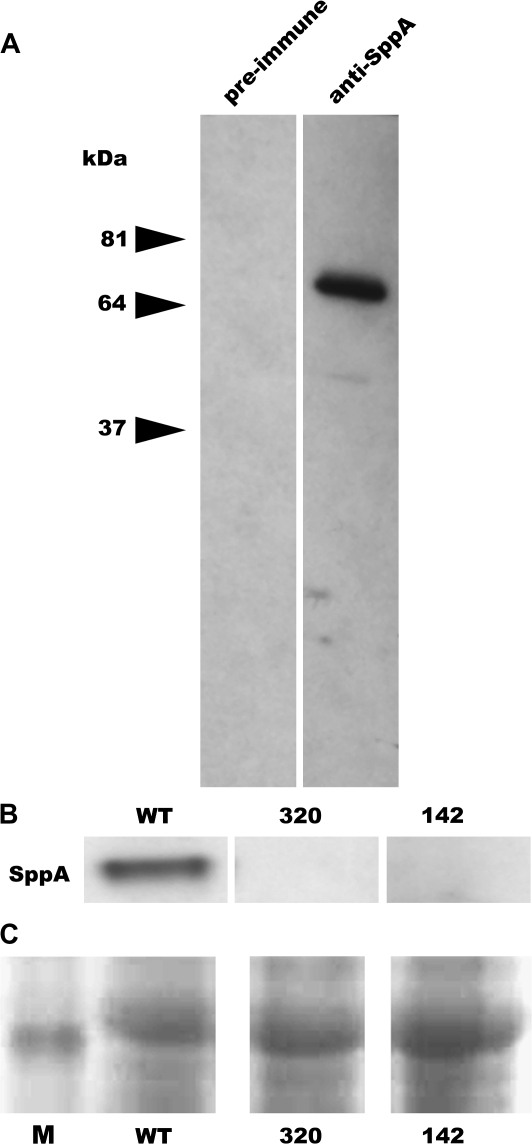
Fig. 2.
Western blot hybridization with anti-SPPA antiserum. (A) Thylakoids isolated from WT leaves were electrophoretically separated and blotted. One blot was probed with the preimmune serum and the other with anti-peptide antiserum to demonstrate specificity of the anti-SPPA antiserum. (B) Western blot analyses of thylakoid proteins isolated from WT, sppA-320, and sppA-142 leaves probed with anti-SPPA antiserum showing that SPPA protein is not detectable in the T-DNA insertion lines. Samples were loaded on an equal chlorophyll basis. (C) Portion of Ponceau S-stained blot showing the relative amount of protein loading and transfer from lanes in (B) (M=band of apparent molecular mass of ∼50 kDa from BenchMark Protein Ladder).
Protein preparation and Western blot analysis
Thylakoids were isolated from high-light treated leaves (48 h, 850 μmol m−2 s−1) according to Casazza et al. (2001), with the following modifications: BSA was omitted because it runs at approximately the same position as SPPA protein on SDS-gels and interfered with Western blot analysis, and the Sigma Plant Protease Inhibitor Cocktail was included in all buffers. Chlorophyll was measured in 80% acetone (Arnon, 1949) and 2 μg of chlorophyll was run per lane on SDS-PAGE.
Crude membrane proteins were isolated by sequential extraction from leaves ground in liquid nitrogen. The first extraction, of soluble proteins, was made in ‘S’ buffer containing 100 mM TRIS-HCl pH 7.5, 100 mM NaCl, 5 mM EDTA pH 8.0, 5 mM DTT, 10 mM β-mercaptoethanol, and 1 μl per ml Plant Proteinase Cocktail (Sigma). The homogenate was pelleted at 16 000 g for 10 min, and the supernatant containing soluble proteins was collected. For the second extraction, the pellet was resuspended in ‘M’ buffer containing the same components as the ‘S’ buffer plus 1% SDS. The homogenate was pelleted at 16 000 g for 10 min, and the supernatant containing the membrane proteins was collected. For assays based on an equal fresh weight basis, leaves were weighed prior to grinding and 5 μl of buffer was added per 1 mg of tissue, and 20 μl of extract plus 20 μl of 2× Laemmli buffer was used for gel electrophoresis.
SDS-PAGE (14% resolving gels; Laemmli, 1970) was used to separate proteins prior to electrotransfer to nitrocellulose for Western blotting. Apparent molecular weights were estimated using the BenchMark Prestained Protein Ladder (Invitrogen). Loading levels and protein transfer were verified by Ponceau S staining of blots prior to immunodetection. Proteins were immunodetected using the Pierce SuperSignal West Femto chemiluminescence system, with goat-anti rabbit-HRP secondary antibodies. SPPA antiserum is described above. Anti-ELIP (early light inducible protein) antisera were purchased from AgriSera (Sweden).
Plant microscopy
Chloroplast morphology of A. thaliana wild-type and mutant plants was examined by transmission electron microscopy. Tissues were fixed at room temperature under vacuum with 2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) for 90 min at decreasing pressures. After releasing the vacuum the samples were kept overnight at 4 °C. Samples were rinsed three times with the same buffer (15 min each) and post-fixed with 1% OsO4 in buffer for 2 h at room temperature as described by Constan et al. (2004). Fixed tissue was then dehydrated progressively in an ethanol series and embedded in a low viscosity epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and examined in a JEOL 100CX transmission electron microscope (JEOL USA, Peabody, MA).
High light acclimation of mature plants
Plants were grown under long day conditions (20 h light 120 μmol m −2 s−1, 22 °C; 4 h dark, 19 °C; mix of fluorescent and incandescent lights). When 3 weeks old (mature but pre-bolting), plants were moved to a different growth chamber with identical settings except that the light intensity was at 850 μmol m−2 s−1. For 7 d, dark-adapted (22 °C, 60 min) Fv/Fm values (see below) were measured and photographs were taken daily. At the end of the acclimation period whole rosettes were harvested, split into two samples, weighed, and used either for chlorophyll or anthocyanin measurements. Alternatively, intact plants were used for chlorophyll fluorescence measurements (see below).
Pigment measurements
Chlorophyll a and b levels were measured after the extraction of plant material in dimethylformamide (DMF) in the dark, with spectrophotometric calculations using the equations of Porra et al. (1989). Anthocyanins were measured at A530 according to Guisti and Wrolstad (2000) using two dilutions of each sample. Multiple measurements of each treatment were averaged for reported values and each experiment was repeated three times.
NPQ, Fv/Fm, and ΦPSII measurements of mature plants
The methods of Golan et al. (2006) were followed except that an FMS1 instrument was used (Hansatech, UK). Four independently grown sets of plants were measured, with 2–4 plants per genotype per set sampled after overnight dark-adaptation. NPQ was calculated as . Dark-adapted (22 °C), maximal quantum yield of Photosystem II (Fv/Fm) was calculated as (Fm–Fo)/Fm. The realized quantum yield of Photosystem II versus irradiance (ΦPSII; also known as ) was calculated as . General chlorophyll fluorescence nomenclature and calculations were based on Maxwell and Johnson (2000).
Acute high light plus cold treatments of detached leaves
Plants grown under short days (8 h light/22 °C, 16 h dark/19 °C) conditions were used, and the time of initiation of the experiment was 2 h into the light cycle. Mature leaves (6–8 per sample) were detached from the plants and floated on 4–7 °C water in full sunlight (HL; 1600–1900 μmol photons m−2 s−1) for 4 h and then returned to room-temperature (21–22 °C) at approximately 10 μmol photons m−2 s−1 (LL) for the indicated amount of time. Dark-adapted (22 °C, 30 min) Fv/Fm measurements were taken pretreatment, after the HL+cold treatment (after dark-adaptation time), after 24 h LL recovery, and after 72 h LL recovery. In some initial experiments Fv/Fm was also measured after 2 h and 4 h recovery but the values were no different from the zero recovery samples and were not included in subsequent analyses (not shown). Control experiments were essentially the same as above except the treatments were either LL+cold or LL at 22 °C prior to the LL recovery period. Membrane proteins were isolated from the treated leaves and Western blot analyses of SPPA and ELIP were carried out as described above.
Statistical analysis
Time-course data from pulse-modulated chlorophyll fluorescence measurements were analysed using a linear mixed effects (LME) model to account for the associations between repeated observations taken under different growth conditions (Fitzmaurice et al., 2004; Finucane et al., 2007). Light treatment and genotype were incorporated as fixed effects, and a random intercept for each specimen was estimated to account for the clustering. The ‘xtmixed'procedure within Stata version 10.0 was used to fit these models. To aid interpretation of interactions a series of pairwise t tests was conducted using Microsoft Excel. Pigment analyses were analysed by 2-way ANOVA and pairwise t tests using Microsoft Excel.
Results
Identification of null sppA T-DNA knockout plants
The locations of the T-DNA inserts are shown in Fig. 1A. Line sppA-320 has the T-DNA left border junction in the 5′-UTR of SPPA. Line sppA-142 has the T-DNA left border junction in exon 4 of the SPPA gene. There is a 30 bp DNA insertion between the sppA-142 T-DNA and the plant sequence that is unidentifiable; BLAST searches did not pick up any nucleotide sequences similar to the insertion. Analysis of the segregation of resistance to sulphadiazine, the selectable maker used in the GABI-Kat T-DNA insertion lines, indicated that there was just one T-DNA insertion each in the genomes of sppA-320 and sppA-142 (confirmation report, GABI-Kat).
Because SPPA mRNA is induced by high light (HL) treatment (Lensch et al., 2001), putative knockout plants were shifted to HL for 24 h to induce SPPA gene expression. Characterization of SPPA mRNA expression was carried out by RT-PCR for WT and lines sppA-142 and sppA-320. Eight separate plants were used for RNA extractions and RT-PCR assay (four from ambient growth light level and four from after HL treatment). The location of primer sets is indicated in Fig. 1B. As shown in Fig. 1C, WT plants had detectable RT-PCR products with all primer sets and plants. Primer set A, downstream of the sppA-320 T-DNA insert site, but upstream of the insert site for sppA-142, produced products for sppA-320 and sppA-142 indicating expression at least through to this location on the gene. Nested primer sets B and C, which flanked the sppA-142 T-DNA insert site, produced products for most sppA-320 samples but not for any sppA-142 samples (in Fig. 1C, lane 5, the white is a smear with no distinct bands while lane 6 has a faint band). The primer set C RT-PCR products in sppA-320 and WT were sequenced and found to be identical to the WT sequence (not shown), suggesting that small amounts of the mRNA from this region are synthesized in some mutant plants.
RLM-5′-RACE of cDNA from WT (two sequenced clones) and sppA-320 (five sequenced clones) revealed that a 277 bp section of the T-DNA insert was expressed at the 5′ end of the sppA-320 mRNA (not shown). Several out-of-frame ATG codons are present in the additional sequence. Despite the small amount of abnormal SPPA mRNA apparently produced in some sppA-320 plants, protein for the gene product was never detected in any Western blots run for various experiments with the genotype (Figs 2B, 8; data not shown). This suggests that translation of normal SPPA product is disrupted in the chimeric sppA-320 mutant mRNA. The RLM-5′RACE also revealed that WT SPPA mRNA is approximately 36 bp longer than has been reported to date in public databases (e.g. Genbank NM_106058).
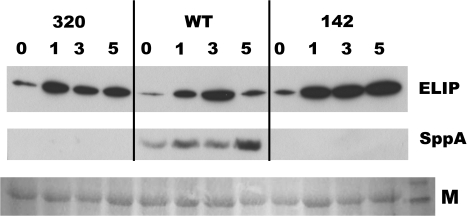
Fig. 8.
Representative Western blot analysis of ELIP and SPPA protein in sppA-320, WT, and sppA-142 leaves. Detached leaves were floated on water in an ice-bath in full sunlight for 4 h, then were allowed to recover in LL at 21–22 °C for the indicated number of hours (0, 1, 3, 5). Membrane proteins were isolated from leaves at the indicated time points for the Western blots (N=3 leaves per time point per genotype). Anti-ELIP1 antiserum was used to probe ELIP levels (top panel) and anti-SPPA antiserum was used to measure SPPA levels on the same blot (middle panel). The gel was loaded on an equal fresh weight basis. A portion of the Ponceau S-stained blot was imaged (M; bottom panel) to show protein loading and transfer (rightmost lane is ∼50 kDa band of the BenchMark Protein Ladder).
SPPA protein is constitutively present in mature plants (Lensch et al., 2001). A peptide antigen corresponding to a unique domain in SPPA was used to generate polyclonal antibodies. The antiserum detected a band at approximately 66 kDa in thylakoid extracts from wild-type leaves that is absent in the Western blot probed with preimmune serum (Fig. 2A). A Western blot of thylakoids from wild-type and putative null mutants is shown in Fig. 2B. Lines sppA-320 and sppA-142 lack detectable protein while the WT thylakoids have predicted-size protein.
The combined results of the characterization of the putative mutant lines support the conclusion that sppA-320 and sppA-142 are null for the expression of normal full-length SPPA protein. Because SPPA is a single-copy gene in A. thaliana, these plant lines were used to assess the physiological effects of loss of SPPA in the mutants compared to the wild type.
Analysis of sppA T-DNA mutants under non-stress conditions
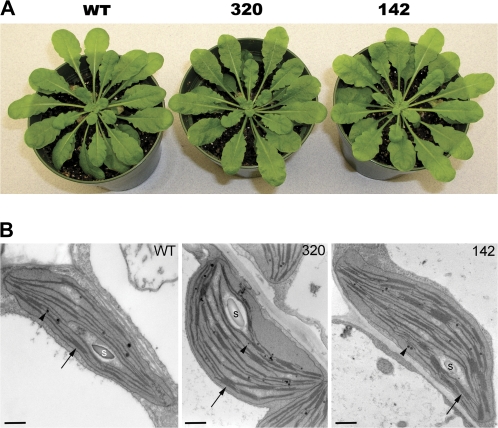
Mutant and wild-type plants were grown under moderate conditions (continuous light at 140 μmol photons m−2 s−1, 22 °C) and under these conditions there was no apparent difference in phenotype between the mutants and wild type (Fig. 3A). Plants grown under short day (8/16h light/dark, 22/19 °C) conditions also did not display any genotype-specific differences. They took the same time to flower and had wild-type levels of leaf production and rosette size (Table 1). To examine whether chloroplast differentiation is affected by the loss of SPPA, greened cotyledons of all genotypes were examined by transmission electron microscopy. As shown in Fig. 3B, chloroplasts from both sppA-142 and sppA-320 mutant plants appear to form structurally normal chloroplasts as compared to wild-type chloroplasts, containing thylakoid membranes with typical stacked grana. Also present are starch granules and plastoglobuli. This demonstrates that SPPA is not required for plant growth and development under average, non-stressful conditions.
Fig. 3.
Comparison of wild type and SPPA null mutant plants. (A) Mature, prebolting plants of each genotype used in this study. All plants were the same age and from the same generation seed batches. (B) Transmission electron micrographs showing plastid ultra structure from WT, sppA-320, and sppA-142 mutant green cotyledons. Starch granules (S), grana (arrow), plastoglobuli (arrow head). Scale bar=500 nm.
Table 1.
Average rosette sizes and time to bolting for all three genotypes
| WT | sppA-320 | sppA-142 | |
| Days to boltinga | 31.69±0.34 | 32.43±0.33 | 31.69±0.34 |
| Number of rosette leaves ±SEa | 8.03±0.23 | 7.90±0.21 | 8.23±0.23 |
| Rosette diameter (cm) ±SEa | 8.03±0.32 | 7.87±0.22 | 7.33±0.27 |
| Leaf density (g cm−2) ±SEb | 0.0138±0.0007 | 0.0129±0.0007 | 0.0118±0.0007 |
There were no statistically significant differences between the genotypes (P >0.05). SE=standard error of the mean.
N=32 per genotype.
N=7–8 per genotype.
Analysis of sppA mutant responses to high light acclimation
Wild-type and SPPA mutant plants were grown under moderately low light (LL; 120 μmol m−2 s−1) then shifted to higher light (HL; 850 μmol m−2 s−1) for 7 d. Standard measures of HL acclimation in plants are a loss in total chlorophyll and a shift to a higher chlorophyll a/b ratio, which indicates a loss of Chl b from reduced antennae size (Yang et al., 1998; Jackowski et al., 2003). After 7 d in HL all three genotypes are visually indistinguishable (Fig. 4A). Under HL they had statistically similar losses in total chlorophyll [Fig. 4B; 2-way ANOVA showed no genotype×light interaction, F(2,30)=0.49, P=0.61; genotype was not a significant source of variation, F(2,30)=0.96, P=0.40; but light level was a significant source of variation F(1,30)=8.75, P=0.006]. They also had similar increases in chlorophyll a/b ratios under HL [Fig. 4C; 2-way ANOVA showed no genotype×light interaction, F(2,30)=0.58, P=0.57; genotype was a significant source of variation, F(2,30)=4.23, P=0.03; but light level accounted for the greatest source of variation F(1,30)=96.67, P<0.0001]. Further analysis by pairwise t tests revealed that the between-genotype variation was due to sppA-142 having a higher Chl a/b ratio under LL than the others (df10, P=0.01–0.02) but all HL-treated genotypes had the same Chl a/b ratios (df10, P=0.42–0.75). The data indicate that loss of SPPA did not affect the plant's ability to adjust antennae size in response to HL acclimation.
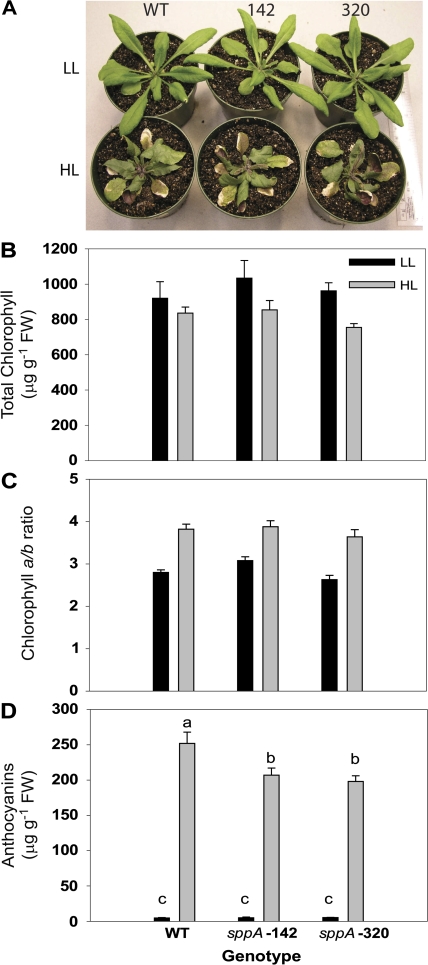
Fig. 4.
The effect of HL acclimation on leaf pigments. (A) Photograph of plants after 7 d of either low light (LL) or high light (HL) treatment. Several leaves on each HL-treated plant are turned abaxial side up to show anthocyanin accumulation. (B) Total chlorophyll levels in LL- and HL-treated plants. (C) The chlorophyll a/b ratios in HL-treated plants compared to LL. (D) Anthocyanin accumulation in plants after HL treatment compared to LL. N=6–14 per genotype, and the error bars are standard errors of the means.
Anthocyanins were barely detectable in plants grown under LL, and there were no differences between genotypes (Fig. 4D; pairwise t tests, df32, all P > 0.05). In the HL shifted plants, massive amounts of anthocyanins accumulated in all genotypes [Fig. 4A, D; 2-way ANOVA indicated a genotype×light interaction F(2,30)=6.57, P=0.004; light level accounted for the greatest amount of variation F(1,30)=555.69, P < 0.0001; and between genotype variation accounted for a significant amount of variation F(2,30)=6.38, P=0.004]. Pairwise t tests revealed that WT plants accumulated 25% more anthocyanins than the mutant genotypes (Fig. 4D; df32, P=0.006–0.02).
To determine if the loss of SPPA affected protection from photoinhibition, dark-adapted maximal quantum yield of Photosystem II photochemistry (Fv/Fm) was measured daily during the acclimation process. Figure 5A displays the observed means of photoinhibition susceptibility by genotype and time, and indicates that there was no significant difference between genotypes [LME model genotype×time interaction, Chi2(df10)=7.68, P=0.66]. All genotypes displayed a drop in Fv/Fm values after 1 d of treatment, then recovered during the course of HL acclimation [LME model for time, Chi2(df5) = 56.59, P < 0.0001], while there was not a statistically significant difference in photoinhibition susceptibility between genotypes, [Chi2(df2) = 3.10, P=0.21]. The drop in Fv/Fm during the first day is modest compared to reports for plants or leaf discs subject to more extreme shifts (e.g. higher light, or HL plus low temperatures; Havaux and Niyogi, 1999; Havaux et al., 2005; Dall'Osta et al., 2007), but is similar to that observed by Giacomelli et al. (2006). The drop in total chlorophyll and rise in Chl a/b ratio (Fig. 4B, C) plus accumulation of anthocyanins (Fig. 4A, D) in HL-treated plants demonstrates that the light shift conditions in this study were sufficient to cause bona fide acclimation.
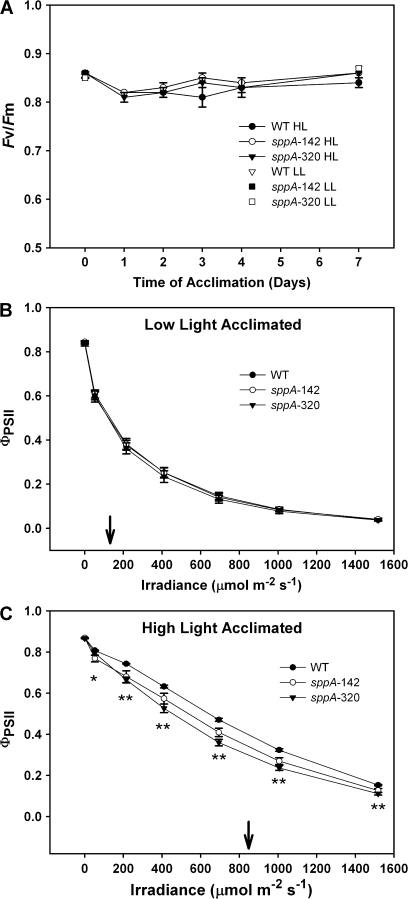
Fig. 5.
Pulse-modulated chlorophyll fluorescence measurements of quantum yield of Photosystem II. (A) Dark-adapted maximal quantum yield of Photosystem II (Fv/Fm) versus time of acclimation for HL plants. Included are the Fv/Fm values for day 0 and day 7 for LL plants. (B) The quantum yield of Photosystem II [ΦPSII; ] versus irradiance of LL acclimated plants. (C) The quantum yield of Photosystem II [ΦPSII; ] versus irradiance of HL acclimated plants. Pairwise t test results are indicated by: * (WT >sppA-142 but not sppA-320), or ** (WT > sppA-142 and sppA-320); see Results for details. In (B) and (C) the arrow indicates the acclimation light level. N=7–14 per genotype, and the error bars are standard errors of the means.
Realized Photosystem II photochemical efficiency [light-adapted quantum yield of Photosystem II; ΦPSII; ] was not affected by loss of SPPA in LL-treated plants [Fig. 5B; LME model genotype×irradiance interaction, Chi2(df12) = 4.86, P=0.96; LME model for genotype, Chi2(df2) = 0.20, P=0.90]. In contrast, in HL acclimated plants, loss of SPPA resulted in a slight but statistically significantly lower ΦPSII than WT in response to increasing irradiance [Fig. 5C; LME model genotype×irradiance interaction, Chi2(df12) = 87.65, P <0.0001] pairwise t tests were used to interpret the genotype×irradiance interaction: there was no significant difference among genotypes in dark-adapted plants (df12–13, P=0.44–0.97), WT is significantly greater than sppA-142 at irradiance of 53 μmol photons m−2 s−1 (df13, P=0.03; * in Fig. 5C), and WT was significantly greater than both mutant genotypes at all measured higher irradiances (df13, P <0.001–0.04; ** in Fig. 5C). At all measured irradiances there was no significant difference between sppA-142 and sppA-320 (df12, P=0.08–0.63). Both WT and mutant genotypes showed the expected upward shift in ΦPSII at higher irradiance levels after acclimation to HL.
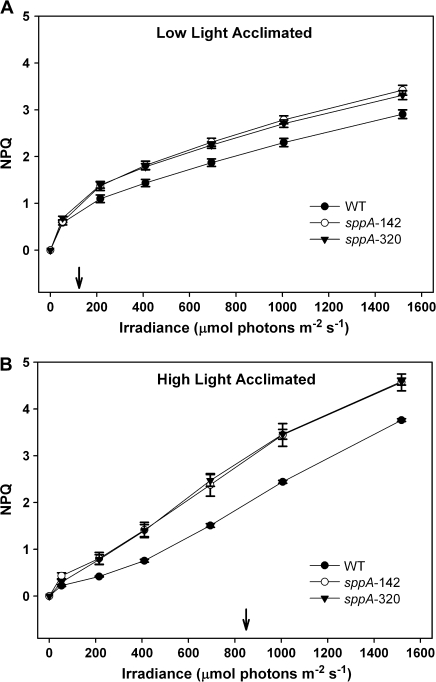
For further assessment of HL acclimation, non-photochemical quenching (NPQ) levels were measured. Figure 6A shows that the NPQ levels in LL-acclimated mutant lines were statistically higher than in the WT at all assay irradiances from 50 to 1500 μmol m−2 s−1 [LME model genotype×irradiance interaction, Chi2(df12)=75.07, P <0.0001; pairwise t tests, df13, all P < 0.05 to 0.01]. Figure 6B shows that the NPQ levels in HL-acclimated insertional mutants were also statistically higher than in the WT at all irradiances from 200 to 1500 μmol m−2 s−1 [LME model for genotype×irradiance interaction, Chi2(df12)=106.65, P <0.0001; pairwise t tests, df14, all P < 0.05–0.001]. Both WT and SPPA mutants respond to HL acclimation via NPQ to a similar extent (i.e. compare the shift in curves between Fig. 6A, B). This indicates that loss of SPPA results in engagement of a higher baseline level of NPQ regardless of acclimation light level, and that all genotypes are capable of altering NPQ in response to HL acclimation.
Fig. 6.
Pulse-modulated chlorophyll fluorescence measurements of non-photochemical quenching (NPQ) of WT and mutant plants. NPQ in response to irradiance of mature pre-bolting plants acclimated to (A), low light or (B), high light for 7 d. The arrow indicates the acclimation light level. For LL plants (A), NPQ was statistically higher in the mutants than the WT at all irradiances from 50–1500 μmol m−2 s−1. For HL acclimated plants (B), NPQ was statistically higher in the mutants than the WT at all irradiances from 200–1500 μmol m−2 s−1. N=7–14 per genotype, and the error bars are standard errors of the means.
Analysis of sppA mutants after acute high light stress
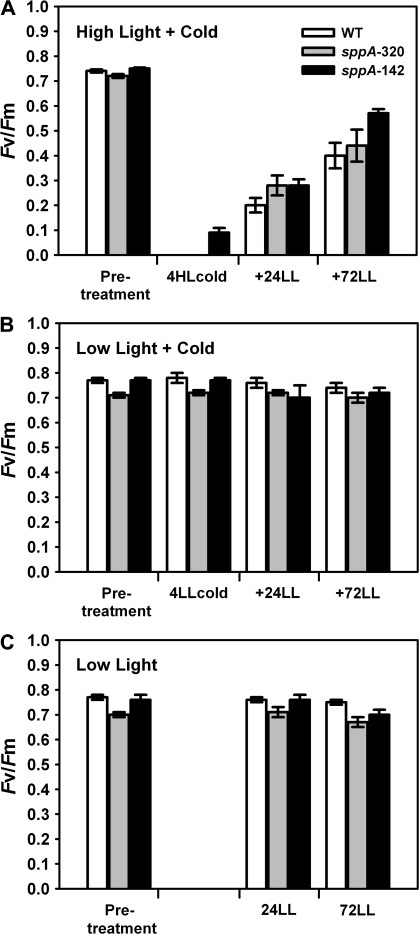
The effect of lack of SPPA was assessed in leaves that were treated with acute HL plus cold and then allowed to recover in low light (LL). Mature detached leaves were floated in 4–7 °C water in full sunlight (1600–1900 μmol photons m−2 s−1) for 4 h and then returned to room-temperature (21–22 °C) at approximately 10 μmol photons m−2 s−1 for the indicated amount of time.
Dark-adapted Fv/Fm measurements were taken pretreatment, after the HL treatment, and over a time-course during recovery for each leaf as a measure of photoinhibition. Figure 7A demonstrates that all genotypes had a similar loss of Fv/Fm and recovery, and that line sppA-142 was slightly less affected by the treatment. Maximal quantum efficiency was not affected by control treatments (Fig. 7B, C).
Fig. 7.
Pulse-modulated chlorophyll fluorescence measurements of dark-adapted Fv/Fm of detached leaves from WT (white bars), sppA-320 (grey bars) and sppA-142 (black bars) showing quantum yield after treatments. (A) Detached leaves were floated on water in an ice-bath in full sunlight for 4 h (+4HLcold) followed by treatment with LL at 22 °C for 72 h with dark-adapted Fv/Fm measurements taken at the indicated time points (pretreatments=0). (B) Leaves were detached and floated on ice water under ambient LL levels, otherwise as for (A). (C) Leaves were detached and floated on 22 °C water under ambient LL levels for 4 h, otherwise as for (A). N=6 for each treatment and genotype; error bars are standard errors of the means.
In order to understand if the A. thaliana SPPA mutants had altered photoprotection, the level of stress-related ELIP1 protein was measured in the treated leaves. There are two copies of the ELIP gene in A. thaliana (At3g22840 and At4g14690). From the literature (Adamska et al., 1993, 1996), it is expected that, once leaves are removed from HL, ELIP will start to be degraded by a partially characterized but unidentified protease (Adamska et al., 1996). Figure 8 shows that for both mutant lines, ELIP1 levels remained higher relative to wild-type plants during early recovery from HL treatment. After 24 h of LL recovery, ELIP was not detectable in any of the genotypes (data not shown). Pretreatment leaves of all genotypes lacked detectable ELIP1, and analysis of several blots revealed that ELIP2 followed the same basic pattern of abundance as ELIP1 although the overall abundance was much lower (data not shown). The same blots were immunodetected with anti-SPPA antiserum, verifying that SPPA was present in all wild-type samples but absent from the mutants (Fig. 8). Pretreatment wild-type leaves had detectable SPPA (data not shown; but see Fig. 2). Repetition of the experiment three times gave similar results, with the exception that, in one repetition after particularly high irradiance, the wild-type sample showed less extensive degradation of ELIP after 5 h and all genotypes retained detectable ELIP after 24 h recovery in LL (data not shown); this result is in agreement with results from Adamska et al. (1993) which demonstrated that the rate of ELIP degradation in LL correlates with the HL irradiance level.
Discussion
Loss of wild-type SPPA protease in A. thaliana via T-DNA insertional mutagenesis in two separate plant lines had no noticeable effect on plant health under non-stress conditions, but altered their responses to high light acclimation. These results suggest that the thylakoid-associated protease is involved with plastid stress responses.
This study shows that, when plants are well watered, fertilized, and grown under moderate conditions they do not require the activity of chloroplast protease SPPA for normal growth and development. Dark- and light-adapted photosynthetic quantum efficiency was unaffected by loss of SPPA under these conditions (Fig. 5A, B). This suggests that SPPA has no essential function under these conditions, or that there is another protease with functional redundancy that compensates for loss of SPPA. Despite the fact that SPPA may be part of a 270–500 kDa complex (Lensch et al., 2001; Majeran et al., 2008), its loss does not have any obvious deleterious effects. In contrast to our results with SPPA, loss of plastid proteases Clp and FtsH result in abnormal phenotypes and affect the general growth and development of leaves (Lindahl et al., 2000; Shikanai et al., 2001; Kuroda and Maliga, 2003; Rudella et al., 2006) indicating a much more central role in plastid function. Lack of an effect under moderate growth conditions led us to examine the effect of SPPA loss on plants under photoinhibitory stress during high light acclimation.
Acclimation of a plant to high light levels moderates potential high-light damage. It takes several days and causes changes in pigments (chlorophylls, carotenoids, and anthocyanins), leaf morphology, and thylakoid proteins (Bailey et al., 2001; Tanaka and Tanaka, 2005; Ballottari et al., 2007; Dall'Osto et al., 2007). In this study, loss of SPPA protease did not have an extreme effect on plant acclimation. However, it did alter the level of anthocyanin accumulation, which was lower in the mutants than in the wild-type plants after acclimation to high light. Stress-induced anthocyanin accumulation in vacuoles is a widespread phenomenon and there is experimental evidence for its role in photoprotection (Havaux and Kloppstech, 2001; Hughes et al., 2007; Merzlyak et al., 2008; reviewed in Close and Beadle, 2003); however, Esteban et al. (2008) found no correlation between foliar anthocyanin and photoprotection. Expression of genes for the anthocyanin biosynthetic pathway is activated by high light stress and is regulated in part by H2O2 (Vanderauwera et al., 2005). Results from this study suggest either that the wild-type plants experienced greater stress than the SPPA mutants leading to more anthocyanin accumulation, or that the mutants are unable to mount as great a response as the wild type when acclimating to high light.
Non-photochemical quenching of chlorophyll fluorescence results from excitation pressure on Photosystem II (reviewed in Müller et al., 2001). It involves a response to the trans-thylakoid pH gradient, the xanthophyll cycle and other carotenoids, the PsbS protein, and other undefined aspects of photoinhibition. Therefore it was of great interest to observe that loss of SPPA resulted in higher-than-wild type levels of NPQ in response to increases in irradiance, under both ambient light and after acclimation to high light. Realized (light-adapted) quantum efficiency of Photosystem II was also affected by loss of SPPA in high-light grown plants, with mutants unable to maintain as high a Photosystem II quantum efficiency as the wild type when challenged with high measuring irradiance. It is not clear what component of NPQ or Photosystem II quantum efficiency is affected by SPPA, and is the subject of current study.
One might argue that the higher level of anthocyanin in WT plants partially blocked the induction of NPQ under high light by altering the light absorption properties of the leaves. This explanation loses support from the fact that the mutants had higher NPQ than WT even in pretreatment or low light growth conditions, when anthocyanin levels were low and equal between genotypes. Thus screening of incident light by anthocyanins is not a robust explanation for the difference in NPQ at high light levels. It may, however, contribute to the differences in realized Photosystem II quantum efficiency with mutant leaves losing efficiency due to less screening by anthocyanins.
In this study, when detached leaves from wild-type plants were subjected to high light in combination with cold temperatures they had an expected dramatic decrease in quantum yield as excitation pressure built up on Photosystem II (Fig. 7; Gray et al., 1996; Ivanov et al., 2006). A mere 4 h of this treatment caused irreversible damage as demonstrated by severe loss of pigmentation and structural integrity of the leaves after their return to recovery conditions (not shown). However, the remaining functional tissue recovered quantum efficiency at the same rate, regardless of the presence or absence of SPPA. This result suggests that SPPA does not have a direct positive role in photoprotection under acute stress, i.e. its presence is not required for it, because if it was required then the mutants would be expected to be more susceptible to photoinhibitory stress than wild-type plants. The fact that all genotypes are equally susceptible to the stress suggests that SPPA function may be related more indirectly to one of the photoprotective mechanisms.
A related result was observed in cyanobacteria Synechocystis sp. PCC 6803 ΔsppA null mutants (Pojidaeva et al., 2004; Sokolenko, 2005). Cells that lacked SppA did not undergo normal bleaching and loss of phycobilisomes when shifted from low to higher light, and retained more phycobilisome linker proteins. In fact, although the authors did not measure photosynthesis, they showed that mutant cells had more robust growth than the wild-type cells after the shift to high light conditions. The authors concluded that SppA was responsible for degradation of the linker proteins during normal high light acclimation and antennae size adjustment. Unfortunately, they did not provide evidence for direct interaction between SppA and the proposed substrate, so specificity of the protease activity cannot be concluded.
One class of proteins that has been proposed to play a role in photoprotection is ‘early light inducible protein’ (ELIP). RNA hybridization, microarray, and cDNA analyses of plants subjected to diverse stimuli show that ELIP mRNA increases in abundance in response to more than just high light stress. These include treatment with salt, heat, abscisic acid, cold, desiccation, aluminium, high CO2, and senescence (reviewed in Adamska, 2001; Binyamin et al., 2001; Harari-Steinberg et al., 2001; Bhalerao et al., 2003). ELIPs are part of a LHC superfamily that includes the light harvesting proteins, PsbS, HLIP, and Seps (reviewed in Montané and Kloppstech, 2000; Adamska, 2001).
A critical role for ELIPs in photoprotection was suggested in work done by Hutin et al. (2003). They used the chaos mutant of A. thaliana, which lacks the cpSRP43 pathway of protein processing leading to a lack of ELIP import and accumulation. These mutants were more susceptible to photoinhibition than wild-type plants, and over-expression of ELIP in the chaos background rescued the mutants and afforded greater photoprotection. Heddad et al. (2006) showed that ELIP1 location and the timing of accumulation correlate with the degree of Photosystem II photodamage, supporting a function in protection and/or the repair process. However, reports of work with ELIP1 and ELIP2 single and double T-DNA knockout A. thaliana plants (Casazza et al., 2005; Rossini et al., 2006) clearly demonstrated that ELIP is not essential for photoprotection, as the mutants were just as capable of responding to high light stress as the wild-type plants. The double mutants did show a slight pigmentation phenotype, with lower levels of chlorophyll and slightly altered xanthophyll ratios. These apparently incongruous sets of results may be interpreted as in agreement if one assumes that the chaos mutant is impaired in more than just ELIP processing, thus a lack of ELIP along with other undefined components led to increased susceptibility to photoinhibition that was compensated for by ELIP over-expression. The more-specific ELIP double knockout plants may have experienced compensation by other photoprotective mechanisms that were impaired or overwhelmed in the chaos mutant, i.e. over-expression of a photoprotective component may lead to increased photoprotection, but eliminating one photoprotective component may just lead to compensation by other mechanisms. The picture of ELIP function becomes more complicated by the fact that ELIP2 overexpression in wild-type A. thaliana leads to a decrease in chlorophyll and photosystem accumulation, and concomitant down-regulation of chlorophyll biosynthesis (Tzvetkova-Chevolleau et al., 2007). Clearly, further investigation of the exact function of ELIPs is required.
The SPPA mutants in this study did not show the wild-type pattern of ELIP1 degradation after acute high light stress (Adamska et al., 1993, 1996). When wild-type leaves are returned from HL to LL, ELIP1 abundance drops after 3–5 h recovery time. By contrast, SPPA mutant leaves retain high levels of ELIP1. Note that none of the genotypes had detectable ELIP1 or ELIP2 in pretreatment leaves, thus SPPA mutants are not constitutively expressing ELIP, only retaining it longer than the wild type after a stress event. This suggests a role for ELIP in recovery from HL stress instead of in the process itself, because all three genotypes had comparable amounts of ELIP during the stress period yet had very different early post-stress recovery phenotypes. From this study it cannot be determined if there is a direct protease–substrate interaction between SPPA and ELIP. If SPPA can proteolyse ELIP, it must do so in concert with another protease because, after 24 h of post-HL recovery, ELIP levels are undetectable in mutant as well as wild-type leaves. Lack of SPPA could generate a general stress level that maintains ELIP; for example, preacclimation to moderate stress can increase a plant's protection from photoinhibition (Anderson et al., 1995; Maxwell et al., 1995). Further assays are being carried out to address these questions, as well as to examine the phenotypes of intact plants under longer-term acclimation to moderate high light. Interestingly, Havaux and Kloppstech (2001) found the opposite pattern in a mutant deficient in the xanthophyll cycle (npq1). It had inhibited NPQ and accumulated more anthocyanins than wild-type plants. During long-term acclimation, ELIP was induced, but disappeared in the npq1mutant and remained stable in wild-type plants.
In summary, T-DNA insertional mutagenesis of the single-copy SPPA gene in A. thaliana had no measurable effect on growth and development under stable, optimal conditions. Lack of SPPA did not affect the ability of detached leaves to recover from acute high light plus cold stress, but did cause a delay in the degradation of ELIP relative to wild-type leaves after the stress abated. Loss of SPPA appears to cause an altered balance between two photoprotective mechanisms during high light acclimation: mutant plants maintained a higher level of NPQ, while wild-type plants accumulated more anthocyanin. From the data in this study it can be concluded that SPPA has a role in photoacclimation and the exact nature of that role is under investigation.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences and PCR conditions for RT-PCR assays.
Supplementary Material
Acknowledgments
The authors would like to thank Danny Schnell (University of Massachusetts- Amherst) and Sam Beale (Brown University) for helpful discussions; Nicholas Horton (Smith College) for statistical consulting; and Betty Jean Zann and Anna Naito for technical assistance. Funding was provided by DOE (DE-FG02-02ER15333) and Smith College CFCD and Blakeslee Funds (CMW); Nancy Kershaw Tomlinson Memorial and Mary Eddy Schlesinger Funds (LDH); and Smith College STRIDE funds (LDH, RC).
References
- Adam Z, Clarke AK. Cutting edge of chloroplast proteolysis. Trends in Plant Science. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- Adam Z, Rudella A, van Wijk KJ. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology. 2006;9:234–240. doi: 10.1016/j.pbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Adamska I. The Elip family of stress proteins in the thylakoid membranes of Pro- and Eukaryota. In: Aro E-M, Andersson B, editors. Regulation of photosynthesis. Dordrecht: Kluwer Academic Publishers; 2001. pp. 487–505. [Google Scholar]
- Adamska I, Kloppstech K, Ohad I. Early light-inducible protein in pea is stable during light stress but is degraded during recovery at low light intensity. Journal of Biological Chemistry. 1993;268:5438–5444. [PubMed] [Google Scholar]
- Adamska I, Lindahl M, Roobal-Bóza M, Andersson B. Degradation of the light-stress protein is mediated by an ATP-independent, serine-type protease under low-light conditions. European Journal of Biochemistry. 1996;236:591–599. doi: 10.1111/j.1432-1033.1996.00591.x. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Anderson MD, Prasad TK, Stewart CR. Changes in isozyme profiles of catalase, peroxidase, and glutathione reductase during acclimation to chilling in mesocotyls of maize seedlings. Plant Physiology. 1995;109:1247–1257. doi: 10.1104/pp.109.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E-M, Andersson B, editors. Regulation of photosynthesis. Dordrecht: Kluwer Academic Publishers; 2001. [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochimica et Biophysica Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Bailey S, Walters RG, Jansson S. Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta. 2001;213:794–801. doi: 10.1007/s004250100556. [DOI] [PubMed] [Google Scholar]
- Ballottari M, Dall'Osto L, Morosinotto T, Bassi R. Contrasting behaviour of higher plant photosystem I and II antenna systems during acclimation. Journal of Biological Chemistry. 2007;282:8947–8958. doi: 10.1074/jbc.M606417200. [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends in Biochemical Sciences. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, et al. Gene expression in autumn leaves. Plant Physiology. 2003;131:430–442. doi: 10.1104/pp.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binyamin L, Falah M, Portnoy V, Soudry E, Gepstein S. The early light-induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta. 2001;212:591–597. doi: 10.1007/s004250000423. [DOI] [PubMed] [Google Scholar]
- Casazza AP, Rossini S, Rosso MG, Soave C. Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Molecular Biology. 2005;58:41–51. doi: 10.1007/s11103-005-4090-1. [DOI] [PubMed] [Google Scholar]
- Casazza AP, Tarantino D, Soave C. Preparation and functional characterization of thylakoids from Arabidopsis thaliana. Photosynthesis Research. 2001;68:175–180. doi: 10.1023/A:1011818021875. [DOI] [PubMed] [Google Scholar]
- Close DC, Beadle CL. The ecophysiology of foliar anthocyanin. The Botanical Review. 2003;69:149–161. [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K. A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiology. 2004;136:3605–3615. doi: 10.1104/pp.104.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Osta L, Cazzaniga S, North H, Marion-Poll A, Bassi R. The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. The Plant Cell. 2007;19:1048–1064. doi: 10.1105/tpc.106.049114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Fernández-Marín B, Becerril JM, García-Plazaola JI. Photoprotective implications of leaf variegation in E. dens-canis L. and P. officinalis L. Journal of Plant Physiology. 2008;165:1255–1263. doi: 10.1016/j.jplph.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Finucane MM, Samet JH, Horton NJ. Translational methods in biostatistics: linear mixed effect regression models of alcohol consumption and HIV disease progression over time. Epidemiologic Perspectives and Innovations. 2007;4 doi: 10.1186/1742-5573-4-8. doi:10.1186/1742-5573-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice GM, Laird NM, Ware JH. Applied longitudinal analysis. New York: John Wiley and Sons; 2004. [Google Scholar]
- Friso G, Ytterberg AJ, Giacomelli L, Peltier JB, Rudella A, Sun Q, van Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts; new proteins, functions and a plastid proteome database. The Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli L, Rudella A, van Wijk KJ. High light response of the thylakoid proteome in Arabidopsis wild type and the ascorbate-deficient mutant vtc2-2. A comparative proteomics study. Plant Physiology. 2006;141:685–701. doi: 10.1104/pp.106.080150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti MM, Wrolstad RE. Characterization and measurement of anthocyanins by UV-visible spectroscopy. In: Wrolstad RE, Acree TE, Decker EA, Penner MH, Reid DS, Schwartz SJ, Shoemaker CF, Smith D, Sporns P, editors. Current protocols in food analytical chemistry. New York: John Wiley and Sons; 2000. Unit F1.2. [Google Scholar]
- Golan T, Müller-Moule P, Niyogi KK. Photoprotection mutants of Arabidopsis thaliana acclimate to high light by increasing photosynthesis and specific antioxidants. Plant, Cell, and Environment. 2006;29:879–887. doi: 10.1111/j.1365-3040.2005.01467.x. [DOI] [PubMed] [Google Scholar]
- Gray GR, Savitch LV, Ivanov AG, Huner NPA. Photosystem II excitation pressure and development of resistance to photoinhibition. II. Adjustment of photosynthetic capacity in winter wheat and winter rye. Plant Physiology. 1996;110:61–71. doi: 10.1104/pp.110.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari-Steinberg O, Ohad I, Chamovitz DA. Dissection of the light signal transduction pathways regulating the two Early Light-Induced Protein genes in Arabidopsis. Plant Physiology. 2001;127:986–997. [PMC free article] [PubMed] [Google Scholar]
- Haußühl K, Andersson B, Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO Journal. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Kloppstech K. The protective functions of carotenoids and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta. 2001;213:953–966. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- Havaux M, Niyogi KK. The violaxanthin cycle protects plants from photooxidative stress by more than one mechanism. Proceedings of the National Academy of Sciences, USA. 1999;96:8762–8767. doi: 10.1073/pnas.96.15.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porifirova S, Rey P, Dörmann P. Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. The Plant Cell. 2005;17:3451–3469. doi: 10.1105/tpc.105.037036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Norén H, Reiser V, Dunaeva M, Andersson B, Adamska I. Differential expression and localization of Early Light Inducible Proteins in Arabidopsis. Plant Physiology. 2006;142:75–87. doi: 10.1104/pp.106.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NM, Morley CB, Smith WK. Coordination of anthocyanin decline and photosynthetic maturation in juvenile leaves of three deciduous tree species. New Phytologist. 2007;175:675–685. doi: 10.1111/j.1469-8137.2007.02133.x. [DOI] [PubMed] [Google Scholar]
- Huner NPA, Öquist G, Sarhan F. Energy balance and acclimation to light and cold. Trends in Plant Science. 1998;3:224–230. [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proceedings of the National Academy of Sciences, USA. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Baldwin AJ, Shipman RL, Matsui K, Theg SM, Ohme-Takagi M. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. Journal of Biological Chemistry. 2005;171:425–430. doi: 10.1083/jcb.200506171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AG, Sane PV, Krol M, Gray GR, Balseris A, Savitch LV, Öquist G, Huner NPA. Acclimation to temperature and irradiance modulates PSII charge recombination. FEBS Letters. 2006;580:2797–2802. doi: 10.1016/j.febslet.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Jackowski G, Olkiewicz P, Zelisko A. The acclimative response of the main light-harvesting chlorophyll a/b protein complex of photosystem II (LHCII) to elevated irradiances at the level of trimeric subunits. Journal of Photochemistry and Photobiology B. 2003;70:163–170. doi: 10.1016/s1011-1344(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. The plastid clpP1 protease gene is essential for plant development. Nature. 2003;425:86–89. doi: 10.1038/nature01909. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lensch M, Hermann RG, Sokolenko A. Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. Journal of Biological Chemistry. 2001;276:33645–33651. doi: 10.1074/jbc.M100506200. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. The Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan BA, Kornyeyev D, Hardison J, Holaday AS. The role of antioxidant enzymes in photoprotection. Photosynthesis Research. 2006;88:119–132. doi: 10.1007/s11120-006-9043-2. [DOI] [PubMed] [Google Scholar]
- Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ. Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Molecular and Cellular Proteomics. 2008;7:1609–1638. doi: 10.1074/mcp.M800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Falk S, Huner NPA. Photosystem II excitation pressure and development of resistance to photoinhibition. I. Light-harvesting complex II abundance and zeaxanthin content in Chlorella vulgaris. Plant Physiology. 1995;107:687–694. doi: 10.1104/pp.107.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence: a practical guide. Journal of Experimental Botany. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- Merzlyak MN, Melø TB, Naqvi KR. Effect of anthocyanins, carotenoids, and flavonols on chlorophyll fluorescence excitation spectra in apple fruit: signature analysis, assessment, modeling, and relevance to photoprotection. Journal of Experimental Botany. 2008;59:349–359. doi: 10.1093/jxb/erm316. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Mishra NP, Kambourakis S, Orfanopoulos M, Ghanotakis DF. Generation and trapping of singlet oxygen during strong illumination of a photosystem II core complex. Plant Science. 1996;115:151–155. [Google Scholar]
- Montané M-H, Kloppstech K. The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene. 2000;258:1–8. doi: 10.1016/s0378-1119(00)00413-3. [DOI] [PubMed] [Google Scholar]
- Müller P, Li X-P, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiology. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R. Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochemistry and Photobiology. 1989;49:229–239. [Google Scholar]
- Ostersetzer O, Kato Y, Adam Z, Sakamoto W. Multiple intracellular locations of Lon protease in Arabidopsis: rvidence for the localization of AtLon4 to chloroplasts. Plant and Cell Physiology. 2007;48:881–885. doi: 10.1093/pcp/pcm052. [DOI] [PubMed] [Google Scholar]
- Pojidaeva E, Zinchenko V, Shestakov SV, Sokolenko A. Involvement of the SppA1 peptidase in acclimation to saturating light intensities in Synechocystis sp. strain PCC 6803. Journal of Bacterioogy. 2004;186:3991–3999. doi: 10.1128/JB.186.12.3991-3999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 1989;975:384–394. [Google Scholar]
- Prasil O, Adir N, Ohad I. Dynamics of photosystem II: mechanism of photoinhibition and recovery process. Topics in Photosynthesis. 1992;11:295–348. [Google Scholar]
- Rossini S, Casazza AP, Engelmann ECM, Havaux M, Jennings RC, Soave C. Suppression of both ELIP1 and ELIP2 in Arabidopsis thaliana does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiology. 2006;141:1264–1273. doi: 10.1104/pp.106.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ. Down-regulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. The Plant Cell. 2006;18:1704–1721. doi: 10.1105/tpc.106.042861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. Protein degradation machineries in plastids. Annual Review of Plant Biology. 2006;57:599–621. doi: 10.1146/annurev.arplant.57.032905.105401. [DOI] [PubMed] [Google Scholar]
- Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant and Cell Physiology. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- Sokolenko A. SppA peptidases: family diversity from heterotrophic bacteria to photoautotrophic eukaryotes. Physiologia Plantarum. 2005;123:391–398. [Google Scholar]
- Tanaka R, Tanaka A. Effects of chlorophyllide a oxygenase overexpression on light acclimation in Arabidopsis thaliana. Photosynthesis Research. 2005;85:327–340. doi: 10.1007/s11120-005-6807-z. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Franck F, Alawady AE, Dall'Osto L, Carrière F, Bassi R, Grimm B, Nussaume L, Havaux M. The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. The Plant Journal. 2007;50:795–809. doi: 10.1111/j.1365-313X.2007.03090.x. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-H, Webster J, Adam Z, Lindahl M, Andersson B. Induction of acclimative proteolysis of the light-harvesting chlorophyll a/b protein of photosystem II in response to elevated light intensities. Plant Physiology. 1998;118:827–834. doi: 10.1104/pp.118.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.