Abstract
A select set of microalgae are reported to be able to catalyse photobiological H2 production from water. Based on the model organism Chlamydomonas reinhardtii, a method was developed for the screening of naturally occurring H2-producing microalgae. By purging algal cultures with N2 in the dark and subsequent illumination, it is possible to rapidly induce photobiological H2 evolution. Using NMR spectroscopy for metabolic profiling in C. reinhardtii, acetate, formate, and ethanol were found to be key compounds contributing to metabolic variance during the assay. This procedure can be used to test algal species existing as axenic or mixed cultures for their ability to produce H2. Using this system, five algal isolates capable of H2 production were identified in various aquatic systems. A phylogenetic tree was constructed using ribosomal sequence data of green unicellular algae to determine if there were taxonomic patterns of H2 production. H2-producing algal species were seen to be dispersed amongst most clades, indicating an H2-producing capacity preceded evolution of the phylum Chlorophyta.
Keywords: Biohydrogen, Chlamydomonas reinhardtii, Chlorophyta, photobiological H2 production
Introduction
Global warming and dwindling fossil fuel supplies have led to a need to develop clean and sustainable energy supplies. Micro-algae may prove to be a source of fuels for the future owing to their ability to capture solar energy and store it as chemical energy in a range of substrates for the production of biodiesel, biomethane, and biohydrogen (Kruse et al., 2005; Chisti, 2007; Schenk et al., 2008). The green alga Chlamydomonas reinhardtii has been used extensively as a model organism and has also offered valuable information regarding the mechanisms underlying photobiological H2 production (Melis et al., 2000; Kruse et al., 2005). Although C. reinhardtii currently produces the highest continual levels of H2 from water of all eukaryotic species tested, there remains the distinct possibility that other green algae may be better suited to perform this task, as the huge biodiversity available is only beginning to be screened.
Many eukaryotic and prokaryotic micro-organisms are capable of photobiological H2 production (Boichenko and Hoffmann, 1994). The hydrogenase enzymes thought to be involved in this process in the organisms screened to date are metalloproteins that contain Ni or Fe ions at their catalytic site and catalyse the two-electron reduction of two protons to H2 (Peters et al., 1998). C. reinhardtii is able to produce H2 using two [Fe-hydrogenases, HydA1 and HydA2 (Happe and Naber, 1993; Forestier et al., 2003). Hydrogenase transcripts and their enzymes have been shown to be up-regulated by the onset of anaerobiosis (Posewitz et al., 2005) and it is under these conditions that H2 production can be observed.
Gaffron and Rubin (1942) may be credited with conducting the experiments that sparked an interest in H2 production in green algae when they found that with Scenedesmus obliquus, following a period of dark anaerobic incubation, H2 production could be observed. The H2 produced from their early experiments was only minimal and it was not until 2000 that it was reported that, by depriving C. reinhardtii of sulphur, H2 production could be observed for several days (Melis et al., 2000). Sulphur deprivation causes a reduction in photosynthetic O2 evolution and leads to the onset of anaerobiosis. It is under such anaerobic circumstances that H2 can be produced.
Photosystem II splits water using energy from light during photosynthesis to provide electrons and protons for H2 production, but at the same time also produces molecular oxygen (O2), which inhibits transcription and function of the hydrogenase enzymes (Ghirardi et al., 1997). For H2 to be continually produced, respiratory O2 uptake must match the rate that it is produced through photosynthesis to keep the culture anaerobic. Endogenous substrates such as starch and protein are consumed during sulphur deprivation and are likely to aid in maintaining an anaerobic environment through their oxidation and consumption by mitochondrial respiration. Such energy reserves may also play a more direct role in H2 production through non-photochemical reduction of the plastoquinone pool (Stuart and Gaffron, 1972). In this scenario NAD(P)H reduces the plastoquinone pool via a NAD(P)H plastoquinone reductase complex and these electrons are then transferred to PSI, where they are light energized and eventually donated to the hydrogenase via ferredoxin (Bernard et al., 2006).
In this work, an assay was developed to explore the potential of microalgae from environmental samples for an efficient H2-producing capacity. Samples collected from marine and fresh-water environments, can be tested for an ability to produce H2, prior to time-consuming isolation of axenic uniclonal species. This approach led to the discovery of new algal isolates capable of H2 production and their phylogenetic grouping suggests that potentially many more species have this ability.
Materials and methods
Acquisition of algal cultures
C. reinhardtii, WT strain CC-124, was obtained from the Chlamydomonas Centre (http://www.chlamy.org/). Tetraselmis suecica (cs-56) and Nannochloris atomus (cs-184) were obtained from the CSIRO Collection of Living Microalgae in Tasmania, Australia (CCLM). Euglena sp. was obtained from Southern Biological, Australia. Chlamydomonas coccoides was obtained from the Cawthron Institute Culture Collection of Micro-Algae, New Zealand (CICCM). All other samples were isolated from marine, brackish, and freshwater sites around Brisbane and Moreton Bay, Queensland, Australia. Global Positioning System (GPS) coordinates of sampling sites for which new isolates were brought to axenic uniclonal states and identified are shown in Table 1.
Table 1.
Global Positioning System (GPS) coordinates of sampling sites from which new algal species were isolated
| Species/GPS coordinates | South | East |
| Desmodesmus isolate 1 | 27° 36′ 10″ | 152° 53′ 58″ |
| Desmodesmus isolate 2 | 27° 32′ 32″ | 152° 55′ 11″ |
| Desmodesmus isolate 3 | 27° 29′ 40″ | 153° 24′ 09″ |
| Chlorella isolate 1 | 27° 32′ 33″ | 152° 51′ 09″ |
| Chlorella isolate 2 | 27° 04′ 26″ | 152° 35′ 56″ |
| Chlorella isolate 3 | 27° 28′ 37″ | 153° 24′ 20″ |
Isolation of axenic uniclonal cultures
Strains were made uniclonal and axenic by continual subculturing in TRIS-Acetate-Phosphate (TAP) medium (Harris, 1989) supplemented with combinations of ampicillin, chloramphenicol, doxycycline, gentamycin, streptomycin, and kanamycin. In the absence of fungal or bacterial contamination, cultures were streaked on TAP agar plates and individual colonies were cultured in liquid media (TAP) for further analysis.
Algal growth conditions
All algal cells were grown aerobically under constant shaking at 25 °C under fluorescent white light (400 μE m−2 s−1). Fresh water algal cells were grown under continuous light in TAP medium, pH 7.2. Marine algal strains Chlamydomonas coccoides, Tetraselmis suecica, and Nannochloris atomus were grown under a 12/12 h light/dark photoperiod in F/2 media (Guillard and Ryther, 1962). For NMR experiments, TAP media was prepared with the exception of TRIS being replaced by high concentrations of phosphate to buffer the media (NaH2PO4.H2O, 2.33 g l−1 and Na2HPO4, 3.27 g l−1).
Dark anaerobic adaptation assay to induce H2 production
To induce H2 production, algal cultures were taken at late logarithmic phase and purged of O2 in the dark, by continuous bubbling with N2 for 90 min at a rate of 500 ml min−1. Following purging of O2, cultures were transferred into sealed 650 ml bioreactors that were fitted with gas collection tubes. Gas collection tubes were pre-filled with 10 ml N2 to enable measurement of gas at a later stage. Cultures were mixed with magnetic stirrers (140 rpm) and exposed to between 400–600 μE m−2 s−1 fluorescent white light (intensity at side of bioreactor and intensity at light source).
Long-term H2 production via sulphur depletion
Long-term H2 production was initiated by the method of sulphur depletion as reported by Melis et al. (2000). In brief, algal cultures were harvested at late logarithmic phase by centrifugation (2500 g, 3 min, 25 °C) and washed three times in sulphur-depleted TAP medium. Cells were then resuspended to a density of 12.5 million cells ml−1 in sulphur-depleted TAP medium and sealed in 310 ml conical flasks fitted with gas collection tubes. Cultures were stirred with magnetic stirrers at 140 rpm and exposed to between 400–600 μE m−2 s−1 continuous fluorescent white light (measured intensity at side of bioreactor and at light source).
O2 and H2 evolution measurements
Dissolved O2 concentrations of algal cultures were measured every 5 min during N2 bubbling and during H2 production in the bioreactors using a D130 data logger system with SZ10T DO electrodes (Consort, Belgium). Gas composition was measured every 30 min by extraction with a Hamilton gas tight syringe and injection into an Agilent Micro GC3000 gas chromatograph fitted with a PlotU pre-column (3 m×0.32 mm) and MolSieve 5APlot column (10 m×0.32 mm). Argon (32.5 psi) was used as the carrier gas.
Extraction and analysis of metabolites by NMR spectroscopy
Metabolites were extracted from algal cultures by removal of 1 ml culture through the bioreactor side port with a gas tight lockable Hamilton syringe. The syringe was immediately placed in boiling water and held for 3 min to quench enzymatic activity and simultaneously to extract metabolites. The boiled culture was centrifuged (17 000 g, 3 min, 25 °C) and 500 μl of the supernatant was frozen in liquid N2 and stored at –80 °C prior to analysis.
10% D2O (v/v, final concentration) was added to 500 μl of algal extract. High-resolution proton NMR spectroscopy was performed on a 500 MHz spectrometer (Avance 500, Bruker, Karlsruhe, Germany). Standard 1D CPMG spectra with 0.5 ms T2 spin echo time and water presaturation during the relaxation delay were recorded with 512 scans. Fourier transformation was performed, and all 1H NMR spectra were manually phased and baseline corrected using TOPSPIN (Bruker Biospin, Karlsruhe, Germany), Sodium-2, 2-dimethylsilapentane-5-sulphonate (DSS) was used as external standard for chemical shift referencing.
Data analysis was carried out using the program AMIX–viewer (version 3.6.8; Bruker Biospin, Karlsruhe, Germany). The chemical shift region from 0.2–9.0 ppm of each NMR spectrum was divided into buckets (spectral integral regions) of 0.04 ppm width. Integral intensities were normalized to the total intensity of the regions examined for each spectrum. The region from 4.2–6.0 ppm was set to zero to exclude the variation introduced from the residual water signal. The resulting bucket tables were used for basic principal component analysis (PCA) without further scaling to determine the metabolites contributing to variation between NMR spectra of samples.
RNA isolation and cDNA synthesis
Total RNA was isolated from 10 ml of C. reinhardtii cultures using a Promega SV RNA isolation kit. Total RNA concentration was measured using a NanoDrop ND-1000 Spectrophotometer. First strand cDNA was synthesised using 2 μg of total RNA in a reaction volume of 20 μl using the SuperScript III RT (Invitrogen) protocol. A combination of oligo(dT) (0.2 μl of 100 μM) and random hexamers (0.05 μl of 3 μg μl−1) primers were used. Following synthesis, cDNA was diluted to a concentration of 10 μg μl−1.
Real-time quantitative RT-PCR conditions and analysis
Real-time quantitative RT-PCR (qRT-PCR) of the cDNA samples was carried out in an ABI optical 384-well plate using an ABI PRISM 7900 HT Sequence detection System (Applied Biosystems). Each reaction contained 3 μl SYBR Green 2× Master Mix, 5 ng of cDNA, and 200 nM of each gene specific primer pair to a final volume of 6 μl. PCR reagents were aliquoted into a 384 well plate using an Eppendorf epMotion 5075 liquid handler. The following standard thermal profile was used: 50 °C for 2 min; 95 °C for 10 min; 45 cycles of 95 °C for 15 s, and 60 °C for 1 min; followed by a dissociation stage of 95 °C for 2 min; 60 °C for 15 s then with a ramp rate of 2% up to 95 °C for 15 s (20 min ramp). The dissociation curves of all PCR products were compared with that of a water control to identify samples with primer dimer, and samples with primer dimers were removed from the analysis. The PCR primer efficiency (E value) of each primer pair in each individual reaction was calculated from the Rn values of each amplification plot using linear regression analysis. Primer efficiencies (E) for each gene were averaged across all samples except in cases where linear regression of amplification plots yielded an R2 value of less than 0.998, in which case the derived E-value for that sample was omitted from calculation of mean E. Amplification plots were analysed using a Rn threshold of 0.3 to give a Ct value (cycle threshold) for each gene/cDNA combination. Gene expression levels relative to 18S rRNA were calculated for each cDNA sample using the equation: relative ratio gene/18SrRNA=(E gene (–Ct gene))/(E 18SrRNA (–Ct 18SrRNA)). Primer sequences of analysed genes are shown in Table 2.
Table 2.
Primer sequences used to amplify genes for real-time quantitative RT-PCR
| Gene | Forward 5′–3′ | Reverse 5′–3′ |
| 18S rRNA | CTTGTAAACCGCGTCGTGATG | GACGTAATCAACGCGAGCTGAT |
| HydA1 | AGGCTGACCGCGACTGGT | GCGCTCCTTGAAGATGTTGC |
| HydA2 | TGGACGAGCGCAACACG | CACGTAGTGGGTGTGCAGCA |
| HydEF | AGTACAACGTCAGCCCAGTGCT | GGGAATCTGCTTGCTTGGGT |
| PFL | CAGATGCTGCTGGAGAAGACAA | CGAACGTTTCCAAGCGTCA |
DNA isolation and sequencing
Genomic DNA was isolated from all algal species using Plant DNAzol (Invitrogen) on a pellet obtained by centrifugation (2500 g for 1 min) of 10 ml of algal culture at mid to late log phase. DNA amplification of ITS1, 5.8S, and ITS2 regions of the ribosome was performed by PCR using the following primers: Forward: 5′-GAAGTCGTAACAAGGTTTCC-3′ and Reverse: 5′-TCCTGGTTAGTTTCTTTTCC-3′. A touchdown amplification program was used where DNA was denatured at 95 °C for 4 min and amplified by 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C (dropping 0.5 °C each cycle) for 30 s and extension at 72 °C for 40 s. Amplification was continued using a further 30 cycles of denaturation at 95 °C for 30 s, annealing at 45 °C for 30 s, and extension at 72 °C for 30 s; there was a final extension period at 72 °C for 7 min prior to a 20 °C hold. The PCR product was cleaned up using the Wizard SV Gel PCR Clean–Up System (Promega). For the sequencing reactions, 30 ng of PCR product was used as template with 6.4 pmol of the above primers and made to a final volume of 10 μl with distilled water; the samples were sent to the Australian Genome Research Facility where the labelling reaction, cleanup and analysis of samples was performed on an AB3730xl sequencer.
Phylogenetic analysis
Nucleotide sequences annotated with ‘Chlorophyta, ITS, and 5.8S’ were obtained from the NCBI database. Sequences containing fewer than 200 nucleotides or in excess of 1500 nucleotides, were removed, sequences not belonging to unicellular green microalgal species were also discarded except for four colonial chlorophytes that have been shown to exhibit an H2-producing ability. In cases where sequences for multiple isolates of a species were present in the database, a single representative sequence was used. The aim was to include species with sequence data available for the ITS1, 5.8S, and ITS2 regions of the ribosome and were required manually to remove a number of species if there were insufficient sequence data in this region to allow appropriate alignment. In total, 130 sequences from the NCBI database and six sequences from locally collected algae isolated and sequenced during this study were aligned with the MUSCLE sequence alignment software (Edgar, 2004). The resulting alignment was manually inspected for quality and the end gaps trimmed.
Phylogenetic trees were constructed from the alignment using the standard Bayesian Markov-chain Monte-Carlo method implemented in MrBayes (Ronquist and Huelsenbeck, 2003). Inference was performed under the General Time Reversible substitution model, with substitution rate heterogeneity across sites modelled by a gamma distribution and some sites allowed to be invariant (GTR+Γ+inv). The parallel MrBayes implementation (Altekar et al., 2004) was used. After 8 d of total CPU time on an SGI Altix compute cluster, MrBayes had run for 1 000 000 generations and sampled 20 000 estimates of the true tree topology, branch lengths, and substitution model parameters. Convergence of Markov-chains was checked empirically by comparison of log–likelihoods across runs and the first 250 000 generations were discarded as burn-in. The resulting data provide an estimate of the true tree topology, along with an estimate of the statistical confidence of each clade in the tree. A consensus tree estimate was made with clades exhibiting Bayesian posterior probability less than 0.5 collapsed into multi–furcating nodes which represent unresolved branching order in the tree.
Results
Development of a rapid dark anaerobic adaptation assay to induce H2 production in C. reinhardtii
For biohydrogen production, hydrogenases involved in photosynthetic H2 production from water are of particular interest. In microalgae this process is, by definition, light dependent and has also been shown to be sensitive to oxygen (Greenbaum et al., 1983; Ghirardi et al., 1997). The assay presented here was therefore designed to rapidly select for these two properties. The principles of the screen are presented in Fig. 1, based on the model system Chlamydomonas reinhardtii.
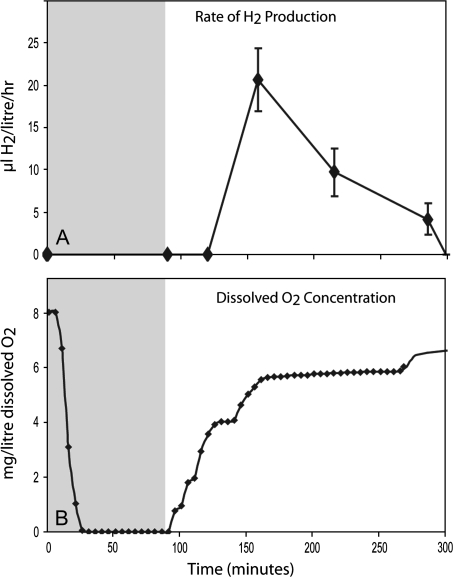
Fig. 1.
Measurements of (A) H2 production and (B) dissolved O2 concentration of the culture. The dark anaerobic adaptation period is shaded grey. C. reinhardtii, WT strain CC-124, was used for these experiments. Standard deviations are shown for four independent biological replicates in (A).
The light dependence of H2 production was confirmed by direct H2 measurement in the light and dark. Photosystem II drives the photosynthetic water splitting reaction (2H2O→4H+ +4e–+O2) which is the first step of this process. In a second step, these H+ and e– (derived either directly from water or indirectly via stored carbohydrates) are recombined to produce H2 (Fig. 1A). This only occurs at low dissolved O2 concentrations as the hydrogenase (HydA) is O2 sensitive (Ghirardi et al., 1997). The dissolved O2 concentration can be kept low by reducing PSII activity by sulphur deprivation or by sparging the solution with N2 gas. The latter process was shown to take ∼20 min, but sparging continued for a total of 90 min (Fig. 1 shaded region) to induce the expression of the hydrogenase (HydA). Subsequently the algal cultures were sealed for gas collection and illuminated. As the PSII-driven water-splitting reaction is activated by light, H+, e–, and O2 are then released. This has the result that the dissolved O2 concentration rises, but during the early stage of the light phase the levels reached do not greatly inhibit HydA mediated H2 production (Fig. 1A). When cultures were kept in constant darkness after 90 min of N2 purging no H2 could be detected (data not shown), indicating the process is light dependent.
Metabolite analysis throughout the dark anaerobic adaptation assay
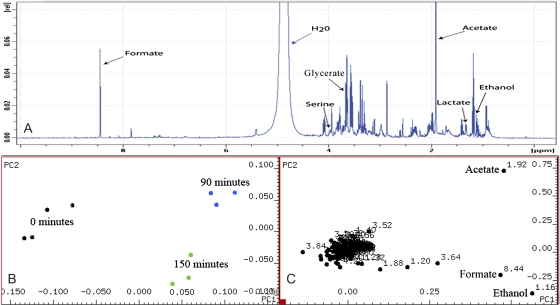
To characterize the underlying biochemical pathways in operation at different stages of the H2 production process in this assay, NMR spectroscopy was used to analyse changes in the metabolic profile of the C. reinhardtii culture at three time points. (i) At 0 min, prior to N2 purging and darkness; (ii) at 90 min, just prior to termination of N2 purging and darkness; and (iii) at 150 min, after 1 h of illumination and when H2 could first be detected in the gas collection tube attached to the bioreactor. Principal component analysis (PCA) was used to identify compounds of interest and revealed acetate, ethanol, and formate to be the main metabolites contributing to variance in the spectral data for which functional roles could be assigned. An example of a spectral profile obtained from NMR analysis as well as PCA scores and loading plots from the assay is shown in Fig. 2.
Fig. 2.
(A) NMR spectrum obtained from a sample at the end of the dark anaerobic adaptation period showing some identified metabolites. (B) Scores plot showing sample points separate into distinct groupings along principal components 1 and 2. (C) Loadings plot from assay revealing the key components leading to variance throughout the assay along principal components 1 and 2 to be ethanol, acetate, and formate. Four biological replicates were taken for analysis at the three timepoints. However, baseline distortion during spectral analysis resulted in the omission of one sample from the latter two timepoints. (This figure is available in colour at JXB online.)
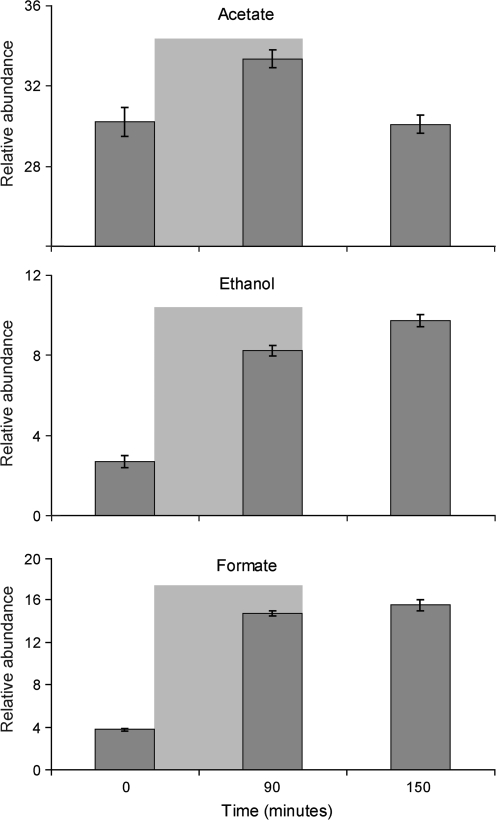
The first and second principal components revealed acetate, ethanol, and formate to be the key compounds contributing to variance throughout the assay, changes in their relative abundances are shown in Fig. 3. Under the assay conditions used, acetate levels were seen to rise by 10% by the end of the dark anaerobic adaptation period but dropped to their original level after 60 min in light-induced H2 production. Ethanol was present only in trace quantities at the beginning of the assay but rose to 200% of its original level after 90 min of dark anaerobic adaptation; it continued to rise a further 20% by the time H2 was first detected in the light. Formate too was present in small amounts at the beginning of the assay but rose to 300% of its original level at the end of the dark anaerobic adaptation phase, its levels continued to rise by approximately 5% by the time H2 was first detected at 150 min after the commencement of N2 bubbling. Principal component three revealed the compounds glycerate, lactate, alanine, and serine to be contributing to further variance in the spectral data. However, these compounds showed high standard deviations within sampling points and did not account for grouping. Their significance will not be discussed further.
Fig. 3.
Relative molar abundance in arbitrary units of acetate, ethanol, and formate throughout the dark anaerobic adaptation assay with C. reinhardtii. Standard error bars are shown from four independent biological replicates. Areas shaded grey represent the period when the culture was placed in darkness and purged with N2.
Transcriptional regulation during dark adaptation-induced H2 production
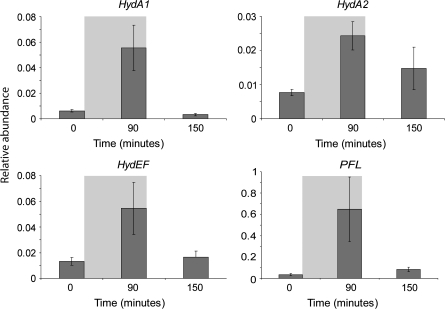
Transcript levels of HydA1, HydA2, HydEF, and a pyruvate formate lyase-encoding gene (PFL) were monitored during the dark anaerobic adaptation assay. Samples were taken for qRT-PCR analysis at three time points: (i) 0 min, prior to N2 purging and darkness; (ii) at 90 min, just prior to termination of N2 purging and darkness; and (iii) At 150 min, after 1 h of illumination and when H2 could first be detected in the gas collection tube attached to the bioreactor. Relative abundance of transcripts compared to 18S rRNA is shown in Fig. 4. Transcript levels of HydA1, HydA2, HydEF, and PFL were all induced by dark anaerobiosis then returned to near basal levels when H2 production was first observed.
Fig. 4.
Transcript abundance (×10−4) of HydA1, HydA2, HydEF, and PFL compared to 18S rRNA during dark adaptation induced H2 production. Standard errors from four biological replicates are shown. Grey shaded area represents the time in which the culture was purged with N2 in the dark.
Ability of local unicellular green microalgal species to produce H2
With the establishment of the above-described assay in place, the aim was to discover new algal species capable of H2 evolution. Unicellular green microalgae were collected from multiple sites in Brisbane, Australia and ordered through collection centres. The ability of these cultures to produce H2 in the light following dark anaerobic adaptation with N2 purging for 90 min was tested. All cultures were monitored for up to 10 h following sealing in bioreactors and exposure to light. Table 3 lists the algal name, source, and whether they were found to produce H2 under the test conditions used.
Table 3.
Species tested for H2 production via N2 bubbling assay
| Algal strain | Source | H2 detected |
| Unidentified isolates 1–9 | Fresh water river | No |
| Unidentified isolates 10–14 | Brackish river | No |
| Chlorella isolate 1 | Brackish river | No |
| Chlorella isolate 2 | Fresh water dam | No |
| Euglena sp. | Southern Biological | No |
| Tetraselmis suecica cs-56 | CCLM (marine species) | No |
| Nannochloris atomus cs-184 | CCLM (marine species) | No |
| Chlamydomonas coccoides | CICCM (marine species) | No |
| Desmodesmus isolate 1 | Fresh water river | Yes |
| Desmodesmus isolate 2 | Brackish river | Yes |
| Desmodesmus isolate 3 | Fresh water river | Yes |
| Chlorella isolate 3 | Fresh water river | Yes |
| Chlorella vulgaris | Fresh water river | Yes |
| C. reinhardtii CC-124 | Chlamydomonas Center | Yes |
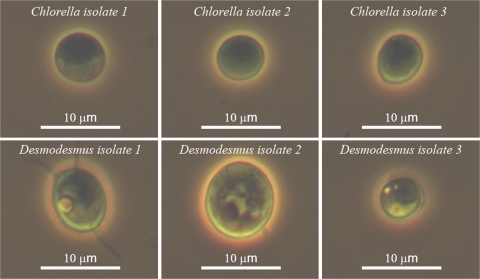
From sampling at 60 marine, brackish, and freshwater locations, 21 green unicellular microalgal cultures were successfully cultivated to densities of over 1×106 cells ml−1 in the laboratory. From these 21 cultures, five species were found that were able to produce H2 under our test conditions, four of which represent new species based on morphological characteristics and by sequence comparison of ribosomal ITS1, 5.8S, and ITS2 regions to data available at the National Center for Biotechnology Information (NCBI). Light microscopic images of new species isolated during this study are shown in Fig. 5. Their growth characteristics under constant light (400 μE s−1 m−2) at 25 °C on an orbital shaker (100 rpm) in 250 ml flasks (100 ml culture) are shown in Table 4.
Fig. 5.
Microscopic images of six putative new algal species isolated during this study. (This figure is available in colour at JXB online.)
Table 4.
Growth characteristics of new algal species isolated during this study
| Species | Size(μm) | Doubling time | Maximum cell density (cells ml−1 culture) |
| Chlorella isolate 1 | 2–8 | 6 | 155×106 |
| Chlorella isolate 2 | 2–7 | 8 | 150×106 |
| Chlorella isolate 3 | 2–9 | 9 | 115×106 |
| Desmodesmus isolate 1 | 5–14 | 17 | 20×106 |
| Desmodesmus isolate 2 | 5–12 | 16 | 20×106 |
| Desmodesmus isolate 3 | 2–7 | 16 | 60×106 |
Species of Desmodesmus are closely related to Scenedesmus and all isolates made during this study were seen to be capable of H2 production. Chlorella appeared to be the dominant fresh water genus around the Brisbane area and a number of times the same species (Chlorella isolate 3) was cultured from separate locations. Interestingly, although ribosomal sequence data matched perfectly between Chlorella isolate 3 isolates and they were indistinct under the light microscope, we could not detect H2 production from certain isolates while it was possible from others following N2 bubbling or sulphur depletion.
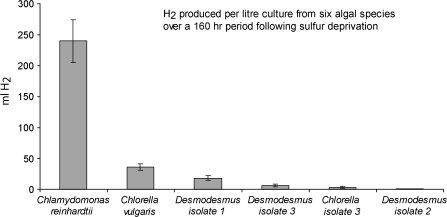
H2 production following sulphur deprivation
Due to the fact that only trace amounts of H2 were being produced during our N2 bubbling assay, the sulphur deprivation method as first described by Melis et al. (2000) was used quantitatively to determine the levels of H2 achievable with our new isolates (Fig. 6). H2 produced from a 1.0 l culture over a 160 hour period following sulphur deprivation was observed to be 241 ml with C. reinhardtii, 36 ml with Chlorella vulgaris, 18 ml with Desmodesmus isolate 1, 7 ml with Desmodesmus isolate 3, 3 ml with Chlorella isolate 3, and 0.025 ml with Desmodesmus isolate 2. Two additional species isolated to uniclonal states during this study, Chlorella isolate 2 and Chlorella isolate 1, were not found to be capable of producing H2 under either of the methods tested. It should be noted that these measurements were taken in a medium well suited for photobiological H2 production in C. reinhardtii and that further improvements in H2 yield may be achieved for each isolate upon species-dependent media optimization.
Fig. 6.
Comparison of H2 produced from six different algal species over a 160 hr period following sulphur deprivation. Standard deviations are shown for three biological replicates.
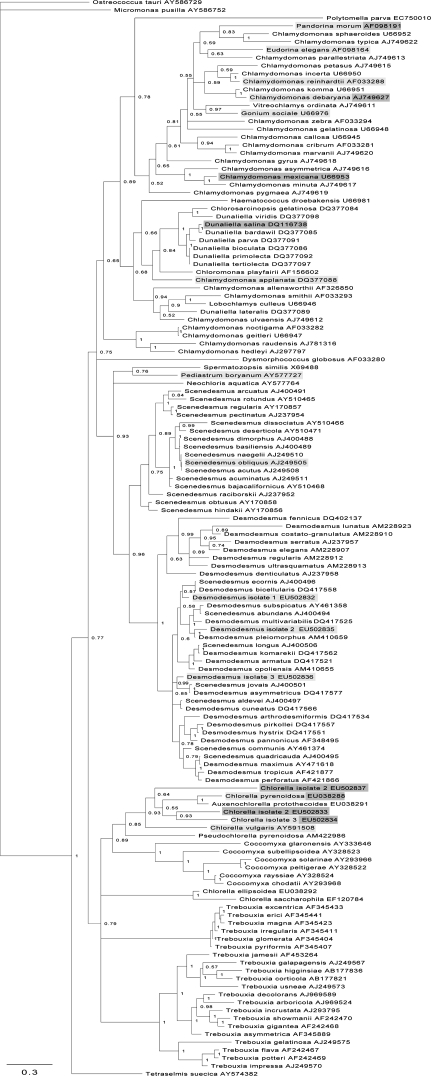
Phylogenetic analysis of H2-producing algae
To target future research in a similar direction, the aim was to determine if there were taxonomic patterns of H2 production amongst green microalgae. To this end, a phylogenetic tree was constructed using all available sequence data spanning ribosomal regions ITS1, 5.8S, and ITS2 for any unicellular Chlorophyte (Fig. 7). An additional four colonial species (Gonium sociale, Pediastrum boryanum, Eudorina elegans, and Pandorina morum) were added, due to their known ability to produce H2. Species with an ability to produce H2 are lightly shaded, species observed not to be capable of producing H2 are darkly shaded, and species that contain strains that are able and strains that are unable to produce H2 are shown in both shades. These species were observed during this study, during studies by Brand and Wright (1989) or in work reviewed by Boichenko and Hoffmann (1994), which also lists additional green algal species capable of H2 production, but are not on the phylogenetic tree shown here due to a current lack of sequence information.
Fig. 7.
Phylogenetic tree of green microalgae constructed from sequence data spanning ribosomal regions ITS1, 5.8S, and ITS2. Numbers represent clade credibility values and branch length represents substitutions per site. Species lightly shaded have been shown to be capable of producing H2 and those darkly shaded have been tested and not observed to produce H2. Species in both shades appear to have strains that can produce H2 and strains that cannot.
The phylogenetic data show that the ability to produce H2 lies predominantly within clades that house the genera Chlamydomonas, Chlorella, Scenedesmus, and Desmodesmus. To our knowledge there have been no reports of H2 production observed in clades that house species of the genera Dunaliella, or Trebouxia. Algal cells belonging to the genera Trebouxia and Coccomyxa can live freely but are predominantly associated with fungi as lichen photobionts; studies focus largely on their roles as symbionts with little work done on their H2 production capacity. However, Coccomyxa lacustris has been found to be capable of H2 production (Boichenko and Hoffmann, 1994). To our knowledge, from the genus Dunaliella, only Dunaliella salina has been tested and was not found to produce H2 (Healey, 1970).
H2-producing algae appear to be dispersed throughout the phylum Chlorophyta rather than being limited to within a specific clade. It appears that there are species within a genus that are capable of producing H2 and some that are not. In addition, it appears that some strains of the same species have this ability while others do not. These variations may therefore be related to specific anaerobic/low light habitats in which the organisms require the photobiological H2 production capacity to survive.
Discussion
Use of microalgae for photobiological H2 production
Using microalgae for the production of biohydrogen could begin to address the current problems associated with the use of crops for fuel production, as microalgal bioreactors can be sited on non-arable land and so do not compete with food production (Kruse et al., 2005; Schenk et al., 2008). Out of 21 separate green algal cultures tested, five putative new species were discovered that could produce H2 under our test conditions. This represents 24% of the tested population. Difficulties were experienced growing marine samples in the laboratory and, consequently, the samples tested for H2 production represented predominantly fresh water and brackish species. However, given the relative abundance of H2 producers, there is considerable potential to identify naturally occurring algal strains that have significantly higher H2 production capacities than C. reinhardtii and could assist in the generation of this clean and renewable fuel source from water.
It is important to realize that the screening assay for H2 production was optimized for C. reinhardtii and that other ways to induce algae to produce H2 may be more suitable for other species. For example, some species, found not to produce H2 under the conditions used here, may still be capable of H2 production. Optimizing media conditions may also enhance a species-specific H2 production capacity further. Our detection system requires that over 0.3 μl H2 be present in 10 ml headspace for a signal to be observed. If this level is not reached or photosynthesis and respiration occur at rates that do not allow the hydrogenase system to be active then potentially positive species will not be detected. Furthermore, dark anaerobic adaptation time may vary between species, with some requiring far longer than the 90 min used here. For example, Brand and Wright (1989) reported adaptation times of up to 20 h being required for a number of species they tested. It would therefore be preferable to employ longer adaptation times in future to capture a wide range of biological diversity with regard to photobiological H2 production.
Analysis of taxa capable of H2 production and their phylogenetic relationship indicated that some species within a single genus and some strains of a particular species can produce H2 and others cannot (Fig. 7). There have been a high number of photobiological H2 producers detected in the genera Chlamydomonas, Chlorella, Scenedesmus, Desmodesmus, and Ankistrodesmus; future work may profit by obtaining species of these genera from global culture collections and testing them for their ability to produce H2. The Chlamydomonas Centre (http://www.chlamy.org/) currently houses 803 strains of Chlamydomonas, and it would be interesting to test these strains for altered levels of H2 production. However, it should be noted that culture collections often favour slow-growing organisms and so there is clear merit in collecting from natural habitats where dominant species have been actively selected for.
Whether the ability to produce H2 comes from prokaryotic ancestors using H2 as an energy source millions of years ago in a more reducing atmosphere, or if it has been an adaptive strategy for aquatic organisms to survive brief periods of anaerobicity remains to be conclusively determined. H2-producing algae are found in dispersed clades, suggesting that the trait originated from a common ancestor and is not a recent acquisition within the phylum Chlorophyta. The ability to produce H2 appears to be lost within certain genera and species, so some lineages may have experienced reduced selection for maintenance of H2 production. Environments offering a competitive advantage to H2-producing algae would likely encounter periods of anaerobiosis accompanied by sunlight; these conditions may be imposed by desiccation, algal blooms or a drastic increase in other heterotrophic micro-organisms or within specific habitat zones within a vertical water column. The ability of marine algae to produce H2 has also been demonstrated (Greenbaum et al., 1983; Miura et al., 1986; Schnackenberg et al., 1995; Guan et al., 2004), showing that green algae from a wide variety of habitats have this function.
Regulation of photobiological H2 production
Green algae appear to produce H2 under anaerobic circumstances through a co-ordinated interplay of photosynthesis, respiration, and fermentation (Hemschemeier et al., 2008). During the initial dark anaerobic adaptation period in our assay, the culture is bubbled with N2 and purged of all other gases, importantly O2 and CO2. When O2 levels approached zero, transcripts for C. reinhardtii HydA1, HydA2, and HydEF were increased and when exposed to light, H2 may be produced for a number of hours.
Anaerobiosis may not be the only switch to induce cells to utilize the H2 production machinery. The hydrogenase enzymes receive electrons from ferredoxin, which accepts electrons from Photosystem I (Florin et al., 2001). Generally, under aerobic conditions reduced ferredoxin will be employed to reduce NADP+ to NADPH via ferredoxin-NADP+ reductase (FNR) to supply reducing equivalents to the Calvin cycle. If the Calvin cycle is not functional due to a lack of CO2 then the competition for H+ and e– is diminished. Under these conditions the hydrogenase could function as the terminal electron acceptor in the photosynthetic electron transport chain. In our assay, dissolved CO2 in the culture was removed through the process of N2 bubbling during the initial 90 min of the assay. Consequently, the expressed hydrogenase enzymes may offer a route for the release of H+ and e– from the cell in the form of H2 gas, to maintain the H+ gradient across the thylakoid membrane required for ATP synthesis. The depletion of CO2, in addition to anerobiosis, may therefore be a driving factor for H2 production in this assay.
The system does not continually produce hydrogen as is the case after sulphur depletion in the approach reported by Melis et al. (2000) due to eventual photosynthetic O2 production and possibly also respiratory CO2 generation competing with the hydrogenase enzymes and their efficacy. It has been noted previously that high CO2 can adversely affect H2 production while low levels of CO2 can enhance its production (Cinco et al., 1993). Purging a culture of CO2 probably results in down-regulation of many Calvin cycle intermediates, the importance of down–regulating the Calvin cycle for H2 production has been shown in a mutant studied by Hemschemeier et al. (2008). The C. reinhardtii mutant, CC2803, which is deficient in Rubisco, could produce H2 in the absence and presence of sulphur due to cells reaching an anoxic state very quickly when placed in a sealed vessel. It is also likely that this mutant uses the hydrogenase system as its electron valve during linear photosynthetic electron transport, as CO2 cannot be reductively assimilated without Rubisco and competition for electrons would be diminished.
Cross-talk of fermentation and photobiological H2 production
It is difficult accurately to determine the ratio of H+ and e– derived directly from the water-splitting reaction of Photosystem II and those derived from the endogenously stored products of photosynthesis. However, as H2 production was not observed in the dark, the H2 production process appears to be light dependent, with the electron source ultimately coming from the oxidation of water. However, the importance of fermentation toward H2 production and cell survival should be considered. Evidence for the importance of endogenous starch reserves being important for high levels of H2 production can be seen with the C. reinhardtii mutant stm6, developed by Schonfeld et al. (2004). This mutant accumulates starch to a higher degree than the wild-type and can produce H2 at a higher rate for a longer time under sulphur deprivation (Kruse et al., 2005). Another mutant isolated by Posewitz et al. (2004) was detected due to its low levels of H2 production. When analysed, it was found to have a non-functional copy of an isoamylase gene and, consequently, much lower starch reserves. These findings highlight the importance of starch in particular for H2 production and suggest the role of fermentation to be key also to H2 production. During our assay there is evidence that fermentation takes place, with the products ethanol and formate being up-regulated during dark anaerobic adaptation and subsequent light-induced H2 production. Other studies have found additional products such as malate (Mus et al., 2007), glycerol, acetate, and CO2 (Gfeller and Gibbs, 1984) to be additional fermentation products, and small amounts of lactate, serine, and alanine have also been measured in this study.
The importance of fermentation for cell survival during anaerobiosis may be due to a number of reasons: (i) the oxidation of NADH, which is important for further glycolysis to generate ATP; (ii) the production of ATP through the conversion of pyruvate to acetate; and (iii) the fermentative production of CO2 as the Calvin cycle cannot operate in the absence of CO2; CO2 can be generated through fermentation of pyruvate to acetate, acetaldehyde, or ethanol. Fermentative pathways in C. reinhardtii give rise to a number of end products. Key products that can be detected in the light during anaerobic incubation include acetate, ethanol, formate, and H2. The flexibility of having multiple fermentative pathways allows C. reinhardtii to fine-tune its response to its changing environment. Use of pathways involving the enzymes pyruvate decarboxylase (PDC) or pyruvate ferredoxin oxidoreductase (PFO) can relieve reductive stress on the photosynthetic apparatus by production of CO2, which can serve as an electron sink in the Calvin cycle. The production of formate may be used in times of higher energy demand or to relieve reductive stress from the build-up of NADH. Formate and the PFL transcript were strongly up-regulated at the end of the dark anaerobic adaptation period, showing that the production of formate is a key response to anaerobicity. Once algal cells are exposed to light the products of fermentation appears to change slightly and favour production of H2 along with the production of ethanol and formate, over the time period analysed. H2 production may therefore aid algal survival during anaerobicity by relieving reductive stress on the photosynthetic apparatus and decreasing the build–up of toxic fermentative end-products. An obvious strategy to increase H2 production with algal cells would be to down–regulate competing fermentative pathways. However, production of the different fermentative products may be finely balanced to maximize cell fitness under anaerobic circumstances and adjusting these may have deleterious effects that would compromise overall H2 production. Ethanol and formate may serve as energy storage compounds that can be assimilated later and utilized as a carbon source during aerobic times and a cell may be programmed to accumulate these products prior to releasing H2. It is difficult to determine without experimental evidence whether knocking out fermentative pathways other than the route to H2 would increase overall H2 production or would result in an energy crisis and lead to quicker cell death.
In conclusion, it has been shown that, by using a simple assay, it is possible to test new algal isolates for an ability to produce H2, whereby a positive result will warrant further work to isolate the species to an axenic state and test via more quantitative methods. Five green microalgal species have been isolated that have the ability to produce H2. A protocol applicable to NMR spectroscopy has also been developed that simultaneously extracts metabolites and quenches enzymatic activity from a culture of C. reinhardtii and it has also been shown that the fermentation products ethanol and formate accumulate during dark anaerobic adaptation and continue to do so to a minor extent during subsequent illumination when H2 can be detected. Future research efforts should aim to discover algal species that can produce economically viable levels of H2 and understand the mechanisms by which they perform this task.
Acknowledgments
We would like to thank Alistair Grinham and Erin Ahern for their kind help in sampling and culturing of new strains throughout this work. This work was supported by the Australian Research Council.
References
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov Chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Bernard L, Desplat C, Mus F, Cuine S, Cournac L, Peltier G. Agrobacterium tumefaciens type II NADH dehydrogenase characterization and interactions with bacterial and thylakoid membranes. FEBS Journal. 2006;273:3625–3637. doi: 10.1111/j.1742-4658.2006.05370.x. [DOI] [PubMed] [Google Scholar]
- Boichenko VA, Hoffmann P. Photosynthetic hydrogen production in Prokaryotes and Eukaryotes: occurrence, mechanism, and functions. Photosynthetica. 1994;30:527–552. [Google Scholar]
- Brand JJ, Wright JN. Hydrogen production by eukaryotic algae. Biotechnology and Bioengineering. 1989;33:1482–1488. doi: 10.1002/bit.260331116. [DOI] [PubMed] [Google Scholar]
- Cinco RM, Macinnis JM, Greenbaum E. The role of carbon dioxide in light-activated hydrogen production by Chlamydomonas reinhardtii. Photosynthesis Research. 1993;38:27–33. doi: 10.1007/BF00015058. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnology Advances. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Tsologlou A, Happe T. A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. Journal of Biological Chemistry. 2001;276:6125–6132. doi: 10.1074/jbc.M008470200. [DOI] [PubMed] [Google Scholar]
- Forestier M, King P, Zhang L, Posewitz M, Schwarzer S, Happe T, Ghirardi ML, Seibert M. Expression of two [Fe]–hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. European Journal of Biochemistry. 2003;270:2750–2758. doi: 10.1046/j.1432-1033.2003.03656. [DOI] [PubMed] [Google Scholar]
- Gaffron H, Rubin J. Fermentative and photochemical production of hydrogen in algae. Journal of General Physiology. 1942;26:219–240. doi: 10.1085/jgp.26.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller RP, Gibbs M. Fermentative metabolism of Chlamydomonas reinhardtii. I. Analysis of fermentative products. Plant Physiology. 1984;75:212–218. doi: 10.1104/pp.75.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi MI, Togasaki RK, Seibert M. Oxygen sensitivity of algal H2 production. Applied Biochemistry and Biotechnology. 1997;63:141–151. doi: 10.1007/BF02920420. [DOI] [PubMed] [Google Scholar]
- Greenbaum E, Guillard RRL, Sunda WG. Hydrogen and oxygen photoproduction by marine algae. Photochemistry and Photobiology. 1983;37:649–655. [Google Scholar]
- Guan Y, Zhang W, Deng M, Jin M, Yu X. Significant enhancement of photobiological H2 evolution by carbonylcyanide m-chlorophenylhydrazone in the marine green alga Platymonas subcordiformis. Biotechnology Letters. 2004;26:1031–1035. doi: 10.1023/B:BILE.0000032961.71564.00. [DOI] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Canadian Journal of Microbiology. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Happe T, Naber JD. Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. European Journal of Biochemistry. 1993;214:475–481. doi: 10.1111/j.1432-1033.1993.tb17944.x. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. A comprehensive guide to biology and laboratory use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Healey FP. Hydrogen evolution by several algae. Planta. 1970;91:220–226. doi: 10.1007/BF00385481. [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Fouchard S, Cournac L, Peltier G, Happe T. Hydrogen production by Chlamydomonas reinhardtii: an elaborate interplay of electron sources and sinks. Planta. 2008;227:397–407. doi: 10.1007/s00425-007-0626-8. [DOI] [PubMed] [Google Scholar]
- Kruse O, Rupprecht J, Bader K, Thomas-Hall S, Schenk PM, Finazzi G, Hankamer B. Improved photobiological H2 production in engineered green algal cells. Journal of Biological Chemistry. 2005;280:34170–34177. doi: 10.1074/jbc.M503840200. [DOI] [PubMed] [Google Scholar]
- Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiology. 2000;122:127–136. doi: 10.1104/pp.122.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y, Ohta S, Mano M, Miyamoto K. Isolation and characterization of a unicellular marine green alga exhibiting high activity in dark hydrogen production. Agricultural and Biological Chemistry. 1986;50:2837–2844. [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii. Journal of Biological Chemistry. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- Posewitz MC, Smolinski SL, Kanakagiri S, Melis A, Seibert M, Ghirardi ML. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. The Plant Cell. 2004;16:2151–2163. doi: 10.1105/tpc.104.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz MC, King PW, Smolinski SL, Smith RD, Ginley AR, Ghirardi ML, Seibert M. Identification of genes required for hydrogenase activity in C. reinhardtii. Biochemical Society Transactions. 2005;33:102–104. doi: 10.1042/BST0330102. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Thomas-Hall SR, Stephens E, Marx U, Mussgnug JH, Posten C, Kruse O, Hankamer B. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenergy Research. 2008;1:20–43. [Google Scholar]
- Schnackenberg J, Ikemoto H, Miyachi S. Photosynthesis and hydrogen evolution under stress conditions in a CO2-tolerant marine green alga, Chlorococcum littorale. Journal of Photochemistry and Photobiology. 1996;34:59–62. [Google Scholar]
- Schonfeld C, Wobbe L, Borgstadt R, Kienast A, Nixon PJ, Kruse O. The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. Journal of Biological Chemistry. 2004;279:50366–50374. doi: 10.1074/jbc.M408477200. [DOI] [PubMed] [Google Scholar]
- Stuart TS, Gaffron H. The mechanism of hydrogen photoproduction by several algae. II. The contribution of photosystem II. Planta. 1972;106:101–112. doi: 10.1007/BF00383990. [DOI] [PubMed] [Google Scholar]