Abstract
The ubiquitously expressed 14-3-3 proteins are involved in numerous important cellular functions. The loss of 14-3-3σ is a common event in breast cancer; however, the role of other 14-3-3s in breast cancer is unclear. Recently, we found that 14-3-3ζ overexpression occurs in early stage breast diseases and contributes to transformation of human mammary epithelial cells. Here, we show that 14-3-3ζ overexpression also persisted in invasive ductal carcinoma and contributed to the further progression of breast cancer. To examine the clinical impact of 14-3-3ζ overexpression in advanced stage breast cancer, we performed immunohistochemical analysis of 14-3-3ζ expression in primary breast carcinomas. 14-3-3ζ overexpression occurred in 42% of breast tumors and was determined to be an independent prognostic factor for reduced disease-free survival. 14-3-3ζ overexpression combined with ErbB2 overexpression and positive lymph node status identified a subgroup of patients at high risk for developing distant metastasis. To investigate whether 14-3-3ζ overexpression causally promotes breast cancer progression, we overexpressed 14-3-3ζ by stable transfection or reduced 14-3-3ζ expression by siRNA in cancer cell lines. Increased 14-3-3ζ expression enhanced anchorage independent growth and inhibited stress-induced apoptosis, whereas downregulation of 14-3-3ζ reduced anchorage independent growth and sensitized cells to stress-induced apoptosis via the mitochondrial apoptotic pathway. Transient blockade of 14-3-3ζ expression by siRNA in cancer cells effectively reduced the onset and growth of tumor xenografts in vivo. Therefore, 14-3-3ζ overexpression is a novel molecular marker for disease recurrence in breast cancer patients and may serve as an effective therapeutic target in patients whose tumors overexpress 14-3-3ζ.
Keywords: 14-3-3ζ, breast cancer, apoptosis resistance, disease recurrence, prognostic marker
Introduction
The diagnosis and treatment of multiple cancer types are usually determined by a subset of clinical markers that predict disease recurrence, clinical outcome, and resistance to therapies (1). For example, overexpression of ErbB2 is a well characterized prognostic factor for adverse clinical outcome of breast cancer patients (2). The identification of additional markers, to be used alone or in combination with other established factors, would facilitate better clinical management of cancer patients. Specifically, new markers for disease recurrence, distant metastasis, and resistance to therapy may separate a subgroup of patients for more aggressive treatment earlier in the course of cancer development. Furthermore, identification of these markers may lead to the development of novel targeted therapies to provide more effective treatment options for cancer patients.
The 14-3-3 family of proteins are ubiquitously expressed and highly conserved in all eukaryotic organisms (3). In humans, seven different isoforms have been identified (ζ, β, γ, ε, η, τ and σ) (4). 14-3-3 proteins lack endogenous enzymatic activity; however, they function by forming homo or hetero dimers and binding to phosphorylated-serine/threonine motifs on their target proteins (5). Through modulation of their binding partners, 14-3-3s have been implicated to regulate a diverse number of cellular processes including apoptosis, mitogenic and stress signaling, cell cycle progression, transcription, metabolism, and cytoskeletal integrity (6, 7). Therefore, 14-3-3 proteins may impact multiple pathways involved in cancer.
One hallmark of cancer is apoptosis resistance and several members of the 14-3-3 protein family are suggested to modulate cell survival pathways (6, 8). In response to survival signals, 14-3-3 protein family members inhibit apoptosis by binding and sequestering the pro-apoptotic proteins Bad and FOXO/Forkhead away from their interaction partners and sites of action (reviewed in (9)). 14-3-3 proteins thus mediate an integral balance between cell survival and cell death.
The deregulation of 14-3-3 protein expression in normal cells may disrupt cellular homeostasis and contribute to the initiation and progression of cancer and other diseases. For example, 14-3-3σ is a known tumor suppressor and loss of 14-3-3σ is a frequent event in breast cancer (10). In contrast, elevated expression of other 14-3-3 proteins has been detected in certain cancer types but the clinical and biological significance is not clear (7). Therefore, the role of 14-3-3 proteins in human cancer needs careful investigation. Recently our laboratory discovered that 14-3-3ζ overexpression can be detected in early stage breast lesions and contributes to early stage transformation of human mammary epithelial cells (11). Here, we show that 14-3-3ζ is overexpressed in >40% of advanced stage breast cancers and was correlated with disease recurrence and poor survival in breast cancer patients. We also present data demonstrating that 14-3-3ζ overexpression confers cancer cell apoptosis resistance and may serve as a new therapeutic target.
Materials and Methods
Constructs, cell Lines, and antibodies
MCF7 and MDA-MB-435 breast cancer, and H1299 lung cancer cell lines were transfected using Lipofectamine Plus (Invitrogen, Carlsbad, CA) with plasmids expressing hemagglutinin-tagged 14-3-3ζ (pcDNA3-HA-14-3-3ζ) or empty vector as control (pcDNA3) and selected with neomycin. MCF7 and MDA-MB-435 cells were maintained in DMEM/F12 with 8% FBS. H1299 cells were maintained in RPMI with 8% FBS. The immortalized MCF10A breast cells expressing hemagglutinin-tagged 14-3-3ζ (pLNCX2-HA-14-3-3ζ) or empty vector (pLNCX2) (Clonetech, Mountain View, CA) were constructed and maintained as previously described (11). Cell lines used for analysis of 14-3-3ζ expression were obtained from our laboratory cell banks, collaborators at The University of Texas M.D. Anderson Cancer Center, and the American Tissue Culture Collection (Manassas, VA). Immunoblots were performed as previously described (11). Antibodies used were: 14-3-3ζ (C-19), 14-3-3β (C-20), and PARP (H250) (Santa Cruz, Santa Cruz, CA); β-actin (Sigma); cytochrome C (6H2.B4) (BD Biosciences, San Jose, CA); FOXO3a (9467) and procaspase 9 (C9) (Cell Signaling Technology, Danvers, MA).
Patient tissue samples
Archival paraffin embedded primary tumor samples were obtained from 121 women with invasive breast carcinoma who were diagnosed and treated from 1988 to 1991 at Cancer Hospital, Fudan University, Shanghai, China. All patients underwent radical or modified radical mastectomy with complete axillary lymph node dissection. Patients were treated with cyclophosphamide, methotrexate, and 5-fluorouracil post surgery. The women ranged in age from 25 to 81 years (mean, 50.6 years; SD, 11.2 years). The median follow-up time was 67 months. Patient data were recruited and generated under the institutional policy of Fudan University. Immunohistochemistry, 14-3-3ζ FISH, and statistical analysis are described in the Supplementary Methods.
Soft agar and apoptosis assays
Soft agar assays were preformed as previously described (12) except MCF7, MDA-MB-435 and H1299 cells (1 × 103cells/well), and MCF10A cells (5 × 103cells/well) were plated in culture media containing 0.3% Nobel agar (Sigma) overlaying a 0.5% agarose bottom layer. Cells were replenished with media weekly. MDA-MB-435 and MCF-7 cells transiently transfected with 14-3-3ζ siRNA were plated 48 hours post transfection in the same manner.
Apoptosis was analyzed using Annexin-V-FLUOS (Roche) or APO-BRDU (Phoenix Flow Systems, San Diego, CA) staining kits following the manufacturers’ protocols. Immunofluorescence staining for cytochrome C was performed as previously described (13).
RNA interference of 14-3-3ζ
Duplex siRNAs were synthesized as follows by Dharmacon Research Inc (Denver, CO): 14-3-3ζ-specific sequence- 5’-AAAGUUCUUGAUCCCCAAUGC-3’ and control sequence- 5’-CAGUCGCGUUUGCGACUGG-3’. 14-3-3ζ pooled siRNA (Dharmacon Research Inc.), containing four independent 14-3-3ζ specific siRNAs, was used to confirm key findings. Cells (1 × 106) were transfected with 100nM of siRNA duplexes using Lipofectamine Plus (Invitrogen) according to the manufacturer’s protocol.
Tumorigenicity assay
MDA-MB-435 breast cancer cells were transiently transfected with control or 14-3-3ζ siRNAs. Forty-eight hours post transfection, cells (1 × 106) were injected into the mammary fat pad of severe combined immunodeficient mice (SCID, Taconic, Oxnard, CA) as previously described (14). For immunohistochemical (IHC) analysis of tumor xenografts, mice were injected with 5 × 106 siRNA treated MDA-MB-435 cells. Tumors were collected 5 days post-injection, fixed and embedded in paraffin. IHC was performed as described in Supplementary Methods. All procedures involving mice were conducted in accordance with U.T. M.D. Anderson Cancer Center Animal Care guidelines.
Results
14-3-3ζ overexpression confers poor prognosis and is an independent marker for disease recurrence in breast cancer patients
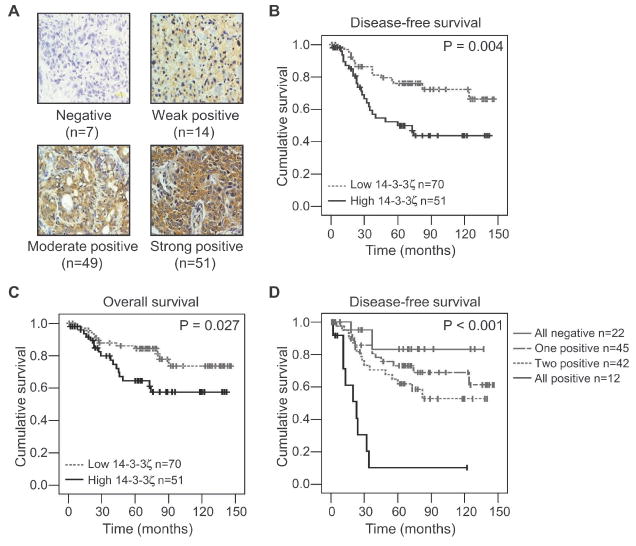
To investigate the role of 14-3-3ζ in breast cancer, we first examined 14-3-3ζ expression by immunohistochemical (IHC) analysis in primary tumor tissue sections from 121 patients with invasive breast carcinoma. These patients received adjuvant chemotherapy (cyclophosphamide, methotrexate, and 5-fluorouracil) after mastectomy with axillary node dissection and a median follow-up time of 67 months. The 14-3-3ζ antibody specificity was determined and we scored 14-3-3ζ IHC staining as negative (score 0), weak positive (score 1+), moderate positive (score 2+) and strong positive (score 3+) based on percentage of cells staining positive and staining intensity (Fig. 1A and Supplementary Fig. 1). Samples that were strong positive (score 3+) were defined as 14-3-3ζ overexpression. Compared to normal breast epithelial cells, 14-3-3ζ staining was strongly positive in 42% (n = 51/121) of patients’ breast tumor specimens (Fig. 1A, lower right panel). Positive 14-3-3ζ staining was mainly observed in the cytoplasm of the breast carcinoma cells; whereas breast epithelial cells and stromal components were negative or weakly positive for 14-3-3ζ staining under the same experimental conditions (data not shown). Strikingly, strong 14-3-3ζ staining in breast tumors was significantly associated with reduced disease-free survival (log rank, P = 0.004), which was defined as local/regional recurrence or distant metastasis (Fig. 1B). Among the 41 patients who experienced recurrence in this cohort, two patients had local/regional recurrence and 39 had distant metastasis. Strong 14-3-3ζ staining in breast tumors was also significantly associated with reduced overall survival (log rank, P = 0.027) (Fig. 1C). As expected, overexpression of ErbB2 and positive lymph node status were also associated with reduced disease-free survival in this cohort, while tumor stage 3 and tumor grade 3 were associated with reduced overall survival (Supplementary Table S1). Among several known prognostic markers of breast cancer, 14-3-3ζ overexpression was associated with ErbB2 overexpression and with stage 3 tumors (Supplementary Table S2). Next, we conducted multivariate analysis to determine if 14-3-3ζ overexpression was an independent prognostic marker for survival in the presence of other known prognostic factors for these patients (see Supplementary Table S2). Strong 14-3-3ζ expression, positive lymph node status, and ErbB2 overexpression were determined to be independent prognostic factors for disease-free survival. Tumor grade 3 was the only independent prognostic factor for overall survival in this model (Table 1). Thus, 14-3-3ζ overexpression may be a novel independent biomarker to predict breast cancer recurrence.
Figure 1. 14-3-3ζ overexpression confers poor survival in breast cancer patients.
A. Representative 14-3-3ζ immunohistochemistry staining in breast cancer specimens (n=121) classified as negative (0), weak positive (1+), moderate positive (2+), and strong positive (3+) for 14-3-3ζ expression.
B and C. Disease-free (B) and Overall survival (C) rates of patients with low (0, 1+, 2+) compared to high (3+) 14-3-3ζ expressing breast cancers.
D. Disease-free survival rates of patients examining combined 14-3-3ζ expression, ErbB2 expression and lymph node (LN) status. All negative (14-3-3ζ - /ErbB2 - /LN -), One marker positive (14-3-3ζ + /ErbB2 - /LN -; 14-3-3ζ - /ErbB2 + /LN -; 14-3-3ζ - /ErbB2 - /LN +), Two markers positive (14-3-3ζ + /ErbB2 + /LN -; 14-3-3ζ - /ErbB2 + /LN +; 14-3-3ζ + /ErbB2 - /LN +), All positive (14-3-3ζ + /ErbB2 + /LN +). 14-3-3ζ and ErbB2 scores 0, 1+, and 2+ were considered negative (-) and 3+ was considered positive (+). P values for (B-D) were determined by log rank analysis.
Table 1.
Variables with independent prognostic value for clinical outcome in multivariate analysis
| DFS | Hazard Ratio (95% CI) | P value |
|---|---|---|
| 14-3-3ζ (score 3+) | 2.38 (1.27 – 4.46) | 0.007 |
| Lymph node (positive) | 2.53 (1.18 – 5.40) | 0.017 |
| ErbB2 (score 3+) |
2.10 (1.11 – 4.00) | 0.024 |
| OS |
||
| Grade 3 | 0.11 (0.02 – 0.84) | 0.033 |
DFS, disease free survival; OS, overall survival
High 14-3-3ζ expression stratified patients at high risk of metastasis
ErbB2 overexpression and regional lymph node involvement are two known prognostic markers for predicting poor clinical outcome in breast cancer patients (15). In our patient cohort, strong 14-3-3ζ expression was also identified as an independent marker for reduced disease-free survival. Therefore, we investigated whether adding 14-3-3ζ overexpression to ErbB2 overexpression and lymph node positive status would predict a subset of patients with worse clinical outcome. Patients whose tumors had 14-3-3ζ overexpression (score 3+), ErbB2 overexpression (score 3+), and positive lymph node status showed significantly decreased disease-free survival compared to the other subgroups of patients (n = 12; log rank, P < 0.001) (Fig. 1D and Supplementary Table S3). A similar trend was also demonstrated for overall survival when all three markers were positive (data not shown). We further analyzed the data using a Concordance Index to determine whether addition of 14-3-3ζ status with the known prognostic markers would improve our ability to predict patient outcome (16). In our patient cohort, the concordance index for ErbB2 overexpression and positive lymph node status was 0.63. With the addition of 14-3-3ζ, the concordance index increased to 0.67. In addition, positive status of all three markers was significantly associated with an increased risk for distant metastasis (n = 8/12; Armitage trend test, P = 0.003) (Supplementary Table S3). The data suggest that the addition of 14-3-3ζ overexpression to ErbB2 overexpression and positive lymph node status provided more power to identify patients at a high risk for breast cancer recurrence and developing distant metastasis.
Overexpression of 14-3-3ζ occurred in multiple cancer types and could result from gene amplification
We next examined whether 14-3-3ζ overexpression also occurs in other cancer types. IHC analysis of 14-3-3ζ expression on a tissue microarray containing one representative sample from 19 different tumor types with matched normal tissue demonstrated that, in addition to breast carcinomas, cancers of the lung, liver, uterus, and stomach expressed higher levels of 14-3-3ζ than their respective normal tissues (Fig. 2A and Supplementary Table S4). This finding was further supported by data from the Human Protein Atlas (www.proteinatlas.org) (17) which demonstrated 14-3-3ζ overexpression also occurred in cervical, ovarian, skin, pancreatic, prostate and urothelial tumors. In addition, 14-3-3ζ protein expression was elevated in multiple breast cancer, lung cancer, and sarcoma cell lines as compared to their non-malignant counterparts (Fig. 2B). Together, these data suggest that 14-3-3ζ overexpression may play an important role in cell transformation and tumor progression in breast and other types of cancers.
Figure 2. 14-3-3ζ is overexpressed in multiple human cancers and cancer cell lines.
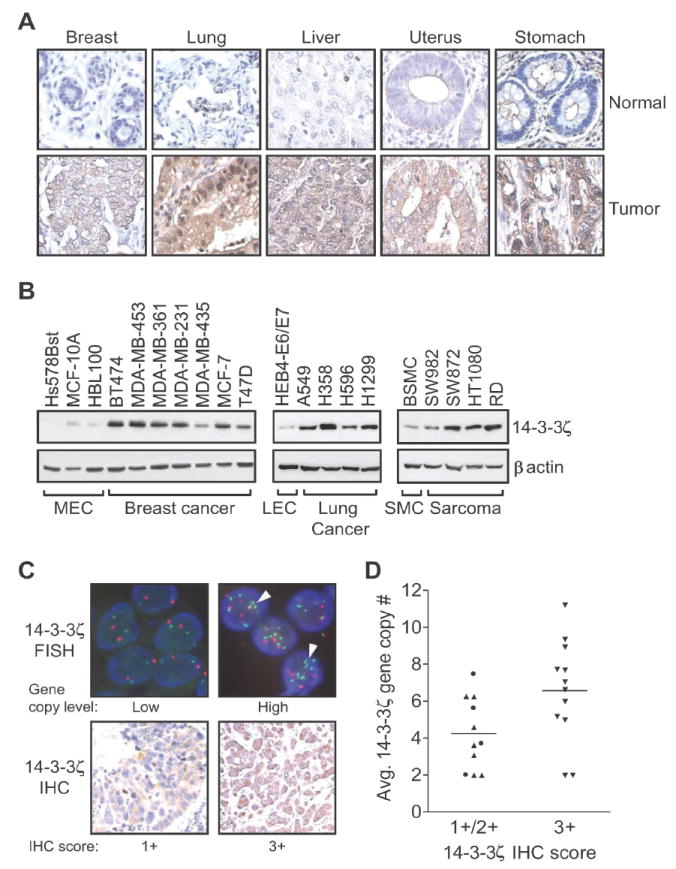
A. Immunohistochemistry staining of 14-3-3ζ expression in tumor and normal tissues on an IMGENEX Tissue Array.
B. Western blot analysis of 14-3-3ζ expression in the indicated non-tumorigenic and cancer cell lines. MEC: mammary epithelial cells, LEC: lung epithelial cells, SMC: smooth muscle cells.
C. top panels: Representative fluorescent in situ hybridization (FISH) analysis of breast tumors. The 14-3-3ζ gene and the chromosome 8 centromere are labeled with green and red fluorophores, respectively. DNA was counterstained with 4’,6-Diamidino-2-phenylindole (DAPI) (blue). Gene copy level: low (>2 to <5 copies per cell); high (≥5 copies per cell). Arrowheads indicate representative regions exemplifying gene amplification. Bottom panels: Representative immunohistochemical (IHC) staining for 14-3-3ζ expression (1+, weak; 3+, strong) relative to increased 14-3-3ζ gene copy number in breast tumor samples.
D. Correlation between average14-3-3ζ gene copy number and 14-3-3ζ IHC staining in 23 primary breast carcinomas. Tumors with 1+ (●; n=4) and 2+ (▲; n=7) IHC scores were combined and compared to 3+(▼; n=12). Bar represents mean for each group (t-test; P = 0.031).
Gene amplification is a common feature of oncogenic activation via overexpression of oncogenic proteins (18). To determine whether 14-3-3ζ overexpression in breast cancers results from gene amplification, dual-color fluorescent in situ hybridization (FISH) for the 14-3-3ζ gene (green) and the chromosome 8 centromere (red) was performed on a panel of 43 breast tumors (Fig. 2C, top). Of these 43 cases, 53.5% showed a high level (≥5) of 14-3-3ζ gene copies per cell as measured by both 14-3-3ζ gene amplification and chromosome 8 polysomy (Fig. 2C, top right; and Supplementary Table S5). We next determined whether increased 14-3-3ζ gene copy number correlated with strong (3+) 14-3-3ζ IHC staining in 23 of the 43 FISH cases. Of the 23 samples that we IHC stained for 14-3-3ζ, 14 samples had a high level (≥5) of 14-3-3ζ gene copies per cell. High level 14-3-3ζ gene copy numbers (≥5) occurred in both IHC score 2+ and 3+ samples; however, 10 of 14 (71%) high copy samples occurred in the IHC 3+ group, suggesting increased 14-3-3ζ gene copy number was associated with strong (3+) 14-3-3ζ IHC staining (Fig. 2C and D). Our data are consistent with a previous array CGH analysis showing that 8q22.3, the region in which the 14-3-3ζ gene resides, is frequently amplified in breast tumors and cell lines (19). Similar to several well-characterized human oncogenes, gene amplification and chromosome polysomy may contribute to 14-3-3ζ overexpression in breast cancers (20, 21).
14-3-3ζ overexpression conferred anchorage independent growth and apoptosis resistance
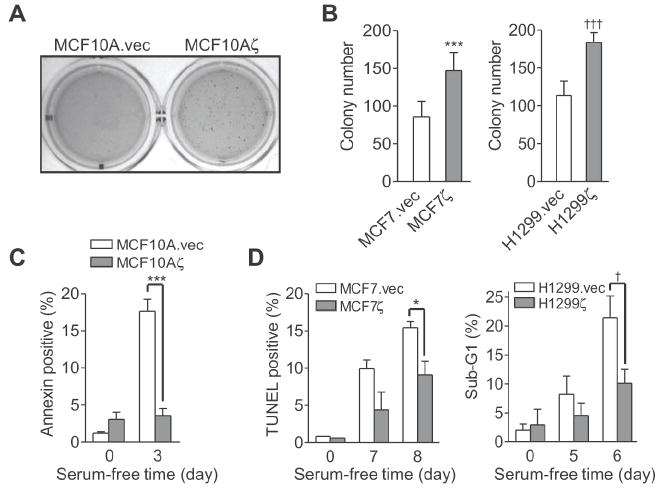
To investigate whether 14-3-3ζ overexpression promotes cell transformation, we stably expressed in MCF10A non-transformed mammary epithelial cells (MECs) a hemagglutinin (HA)-tagged 14-3-3ζ (MCF10Aζ) or an empty vector as a control (MCF10A.vec) (Supplementary Fig. S2A) (11). MCF10A cells were known to exhibit anchorage-dependent growth in vitro and do not form tumors in vivo (22). As expected, in a soft agar colony formation assay for anchorage-independent growth ability, MCF10A.vec control cells remained as single cells and did not form colonies. In contrast, MCF10Aζ cells overexpressing 14-3-3ζ formed colonies in soft agar, demonstrating that 14-3-3ζ overexpression induced anchorage-independent growth of MCF10A MECs (Fig. 3A). To further investigate the effects of 14-3-3ζ overexpression in cancer cells, we established stable transfectants using control vector or HA-tagged 14-3-3ζ expression vector in MCF7 and MDA-MB-435 breast cancer cells as well as H1299 lung cancer cells (MCF7.vec, MCF7ζ; 435vec, 435ζ; H1299.vec, H1299ζ) (Supplementary Fig. S2A). Increasing expression of 14-3-3ζ in these cell lines further enhanced the anchorage-independent growth in soft agar (Fig. 3B and Supplementary Fig. S2B). These results indicated that 14-3-3ζ overexpression contributes to transformation of MECs and further enhanced the transformation of cancer cells during anchorage independent growth.
Figure 3. 14-3-3ζ overexpression promotes anchorage independent growth and resistance to apoptosis.
A. MCF10A cells stably expressing HA-14-3-3ζ (MCF10Aζ) or vector control (MCF10A.vec) were grown in soft agar for 64 days. Fresh media with 4% matrigel was added weekly.
B. Soft agar colony formation of MCF7 breast cancer cells and H1299 lung cancer cells stably expressing HA-14-3-3ζ (MCF7ζ; H1299ζ) or vector control (MCF7.vec; H1299.vec) cells (t-test; ***, P<0.001; †††, P<0.001).
C. MCF10A.vec and MCF10Aζ cells were serum starved for 3 days. Apoptosis was measured by Annexin V assay (t-test; ***, P<0.001).
D. MCF7.vec and MCF7ζ and H1299.vec and H1299ζ cells were subjected to serum starvation for the indicated times. Apoptosis was measured by TUNEL or sub-G1 analysis (t-test; *, P = 0.046; †, P = 0.011). Error bars in (B- D) represent standard deviation.
Since 14-3-3 proteins are known to modulate apoptosis and apoptosis resistance could lead to increased colony formation in soft agar, we next examined the impact of 14-3-3ζ on stress-induced apoptosis. To this end, we cultured the MCF10Aζ, MCF7ζ, 435ζ, and H1299ζ stable transfectants and their corresponding vector control cells under serum-free conditions and examined apoptosis. The transfectants overexpressing 14-3-3ζ showed decreased apoptosis under serum-starvation as compared to vector control cells (Fig. 3C and D and Supplementary Fig. S2C). These data indicated that increased expression of 14-3-3ζ conferred a survival advantage to MECs and cancer cells.
14-3-3ζ knockdown sensitizes cancer cells to stress induced apoptosis through the mitochondrial apoptotic pathway
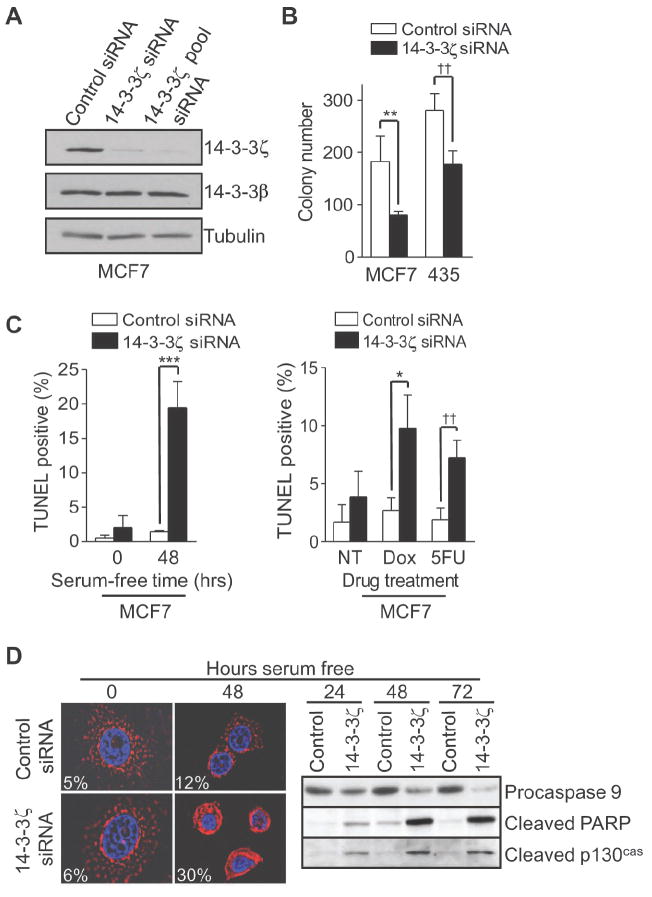
To assess the contribution of endogenously overexpressed 14-3-3ζ to transformation in cancer cells, 14-3-3ζ expression was silenced using small interfering RNA (siRNA) duplexes in MCF7 and MDA-MB-435 cancer cell lines. We used both a single siRNA oligo (14-3-3ζ siRNA) and a pool of four independent siRNA oligo sequences (14-3-3ζ pool siRNA) to target 14-3-3ζ. The 14-3-3ζ siRNAs specifically silenced 14-3-3ζ in these different cancer cell lines without affecting the expression of 14-3-3β, the isoform most homologous to 14-3-3ζ, and the inhibitory effect of 14-3-3ζ siRNA persisted for 5-7 days (Fig. 4A and Supplementary Fig. S3A). We then examined whether downregulation of 14-3-3ζ would impact on anchorage independent growth in MCF7 and MDA-MB-435 cell lines. Compared to control siRNA-treated cells, 14-3-3ζ siRNA-treated cells formed fewer colonies in soft agar (Fig. 4B and Supplementary Fig. S3B), suggesting 14-3-3ζ contributed to the transformation of these cancer cells. Additionally, 14-3-3ζ siRNA-treated cancer cells had increased apoptosis under serum starvation conditions compared to controls (Fig. 4C, left panel, and Supplementary Fig. S3C), indicating downregulation of 14-3-3ζ sensitized cancer cells to apoptosis.
Figure 4. Silencing 14-3-3ζ sensitizes cancer cells to stress induced apoptosis.
A. MCF7 cells were treated with control or 14-3-3ζ siRNA. Expression of 14-3-3ζ and 14-3-3β were determined by immunoblotting with isoform specific antibodies.
B. Soft agar colony formation assay of siRNA transfected MCF7 and MDA-MB-435 cells (t-test; **, P = 0.006; ††, P = 0.002).
C. MCF7 cells transfected with siRNA were maintained in (left panel) serum free media for 48 hours or treated with (right panel) doxorubicin (Dox; 1 μM) or 5-Fluorouracil (5FU; 1 mM) for 48 hours. Apoptotic cells were identified by TUNEL staining (t-test; ***, P = 0.001; *, P = 0.016; ††, P = 0.007).
D. MCF7 cells transfected with siRNA were maintained in serum-free medium for the indicated time points. left panel, Immunofluorescence of cytochrome C (red) was analyzed by 2D-deconvolution microscopy. Nuclei (blue) were stained with DAPI. Cytochrome C release was identified as diffuse staining within the cell. The percentage of cells with released cytochrome C is indicated. Right panel, Western blot analysis of siRNA-treated cells. Error bars in (B-D) represent standard deviation.
14-3-3ζ overexpression in breast cancers from patients receiving adjuvant chemotherapy (cyclophosphamide, methotrexate, and 5-fluorouracil) led to poor disease-free survival (Fig. 1). This raises the possibility that 14-3-3ζ overexpression in breast cancers may also contribute to resistance to standard chemotherapies via apoptosis resistance. Therefore, we investigated whether inhibiting endogenously overexpressed 14-3-3ζ in breast cancer cells would enhance their sensitivity to 5-flourouracil or doxorubicin. MCF7 cells treated with 14-3-3ζ siRNA had increased cell death compared to control siRNA treated cells in response to 5-flourouracil or doxorubicin (Fig. 4C, right panel, and Supplementary Fig. S3D), indicating 14-3-3ζ overexpression in breast cancers may contribute to chemoresistance and targeting 14-3-3ζ may sensitize cancer cells to chemotherapy.
14-3-3ζ has been shown to modulate multiple cell survival regulators. We therefore investigated the mechanism of increased cell death in 14-3-3ζ siRNA treated and serum-starved MCF7 cells. We found that 14-3-3ζ siRNA sensitized cancer cells to apoptosis under low serum by increasing cytochrome C release, subsequent reduction of procaspase 9 expression (indicative of caspase 9 activation), and caspase substrate cleavage (Fig. 4D). A similar result was seen in SKBR3 cells (Supplementary Fig. S4A). 14-3-3ζ siRNA-mediated cleavage of caspase substrates and apoptosis under low serum were inhibited by the caspase 9 specific inhibitor z-LEHD-fmk or the general caspase inhibitor z-VAD-fmk (Supplementary Fig. S4B and C). Taken together, these data suggest that 14-3-3ζ may be a key factor contributing to apoptosis and therapeutic resistance in breast cancer cells through inhibition of the mitochondrial apoptotic pathway.
14-3-3ζ contributes to tumor development in vivo by maintaining apoptosis resistance
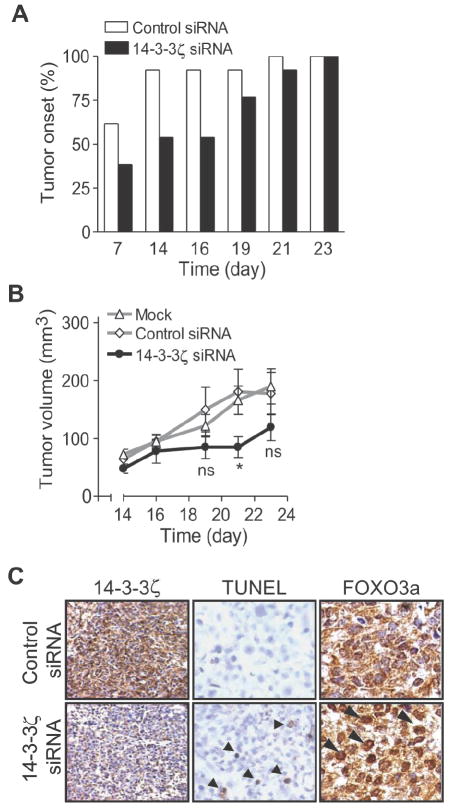
Our in vitro data indicated that 14-3-3ζ mediates anchorage independent growth and apoptosis resistance, which are important properties for tumor development. To test whether 14-3-3ζ contributes to tumorigenicity in vivo, we transfected the highly tumorigenic MDA-MB-435 cells with 14-3-3ζ siRNA or control siRNA oligonucleotides, then injected the cells into the mammary fat pads of female SCID mice (Supplementary Fig. 5A). Mice injected with 14-3-3ζ siRNA-treated cells demonstrated delayed tumor onset and reduced tumor growth as compared to those mice injected with control siRNA transfected cells (Fig. 5A and B). Thus, a brief blockade of 14-3-3ζ was sufficient to slow tumor growth of MDA-MB-435 breast cancer cells in mice, suggesting these cancer cells may be dependent on 14-3-3ζ for survival and/or growth, similar to previously reported findings on transient blockade of other oncogene expression in tumor xenografts and genetically engineered mouse models (23).
Figure 5. Knock down of 14-3-3ζ reduces tumor growth in vivo.
A and B. MDA-MB-435 cells underwent mock, control, or 14-3-3ζ siRNA transfection. Forty-eight hours post-transfection, cells were injected into the mammary fat pad of SCID mice (10 mock and 12 control or 14-3-3ζ siRNA treated mice/group). Tumor onset (A) was determined by number of mice with palpable tumors at time of measurement. Tumor volume (B) was determined on indicated days. Error bars represent SEM (ns = not significant; *, p = 0.043).
C. Immunohistochemical analysis of 14-3-3ζ and FOXO3a expression from siRNA transfected MDA-MB-435 tumor xenografts. Arrowheads in FOXO3a staining signify representative cells with increased FOXO3a localization to the nucleus compared to control. Apoptosis was measured by in situ TUNEL assay (arrowheads indicate apoptotic cells).
To determine the basis of the tumor inhibition following 14-3-3ζ downregulation with siRNA, tumor xenografts were collected from 14-3-3ζ siRNA-treated MDA-MB-435 cells and control siRNA transfected cells. Since our previous data established that blocking 14-3-3ζ sensitized cancer cells to apoptosis in vitro, we examined apoptosis in these tumors by TUNEL staining. Indeed, tumor xenografts from 14-3-3ζ siRNA-treated cells showed increased apoptosis compared to xenografts from control siRNA transfected cells (Fig. 5C, middle panel).
We next examined possible molecular mechanisms for the increased apoptosis in vivo. One of the previously reported in vitro mechanisms for 14-3-3ζ-mediated apoptosis resistance involves 14-3-3ζ binding to phosphorylated FOXO3a transcription factor and subsequent sequestration of FOXO3a in the cytoplasm to prevent it from trans-activating pro-apoptosis genes (24). We therefore investigated whether 14-3-3ζ was associated with FOXO3a in breast cancer cells and whether 14-3-3ζ knockdown increased FOXO3a transcriptional activity. Immunoprecipitation of 14-3-3ζ from breast cancer cell lysates demonstrated association with FOXO3a (Supplementary Fig. 5B). Further, 14-3-3ζ knockdown led to increased FOXO3a transcriptional activity in serum free conditions compared to controls (Supplementary Fig. 5C). We next examined FOXO3a sub-cellular localization by IHC in tumor xenografts treated with control or 14-3-3ζ siRNA. Compared to control siRNA treated xenografts, 14-3-3ζ siRNA-treated tumor xenografts showed increased FOXO3a nuclear localization correlating with their increased apoptosis (Fig. 5C, arrowheads in lower right panel). Together, our data indicated that 14-3-3ζ overexpression inhibited the pro-apoptosis function of FOXO3a in vivo and contributes to the tumorigenic potential of MDA-MB-435 cancer cells.
Discussion
In this study, we demonstrated that 14-3-3ζ overexpression significantly associated with disease recurrence and poor survival in breast cancer patients. Overexpression of 14-3-3ζ induced anchorage independent growth and conferred a survival advantage under stress conditions, whereas knockdown of 14-3-3ζ reduced tumor growth and sensitized cells to chemotherapeutic treatment, suggesting that 14-3-3ζ promotes malignancy through regulation of cancer cell survival. Our findings linked a clinically relevant prognostic marker (14-3-3ζ) with biological outcome (apoptosis resistance) and allow for identification of patients who may be resistant to standard chemotherapies and require more aggressive treatment strategies.
The highly significant association between 14-3-3ζ overexpression and decreased disease-free survival of breast cancer patients suggest the clinical potential of 14-3-3ζ as a marker of cancer recurrence. One of the major challenges in breast cancer treatment is to identify patients at high risk who would benefit from otherwise potentially unnecessary and toxic systemic therapy. Our data demonstrated that the addition of 14-3-3ζ expression status to other well-known breast cancer prognostic factors stratified a subgroup of patients at a high risk for disease recurrence with distant metastasis who need more intensive or alternative adjuvant therapy. Consistent with our findings, 14-3-3ζ overexpression was also found to be associated with decreased survival in lung cancer patients (25). Notably, overexpression of 14-3-3ζ in our patient population (>40%) was a more frequent event than alterations of currently established molecular markers of poor patient outcome in breast cancer patients (e.g., <30% for ErbB2/HER2 and 15% for urokinase plasminogen activator) (26). We further determined that increased expression of 14-3-3ζ was associated with ErbB2 overexpression and stage 3 breast disease. These associations indicate that overexpression of 14-3-3ζ may cooperate with known oncogenes leading to more aggressive breast cancers. Our studies also demonstrated that gene amplification and chromosome 8 polysomy could account for up to 80% of 14-3-3ζ protein overexpression in breast tumors. Similarly, amplification of 14-3-3 genes have been discovered in urothelial carcinomas (27), suggesting amplification may be a common mechanism leading to 14-3-3ζ overexpression in multiple cancers. Mechanisms independent of increased gene copy number, such as modulation of 14-3-3ζ gene transcription, protein translation, or RNA and protein stability may also contribute to increased 14-3-3ζ expression. Although a larger cohort study will be needed to validate these findings before being applied in a clinical setting, our novel findings indicated 14-3-3ζ as an important molecular marker for disease recurrence in breast cancer patients.
Our data demonstrated that 14-3-3ζ overexpression provided an increased survival advantage in response to anchorage independent growth and low serum conditions in breast epithelial and breast cancer cells. In contrast, inhibiting 14-3-3ζ expression sensitized breast cancer cells to chemotherapeutics in vitro and transient blockade of 14-3-3ζ reduced the growth of breast tumor xenografts in vivo. These data suggest that 14-3-3ζ may serve as a therapeutic target for chemo-sensitization and effective tumor inhibition. Interestingly, reducing 14-3-3ζ expression in lung cancer cells sensitized the cells to ionizing radiation, cisplatin, and anoikis (25, 28, 29), whereas, high expression of 14-3-3ζ in breast cancer cells predicted poor response to tamoxifen treatment (30). We have recently found that 14-3-3ζ down-regulates the tumor suppressor gene p53 via activation of Akt and Mdm2 mediated p53 protein degradation, conferring anoikis resistance in mammary epithelial cells (11). Additionally, 14-3-3 is known to modulate multiple survival signals including Akt downstream signals. Therefore, targeting 14-3-3ζ or targeting downstream pathways regulated by 14-3-3ζ may sensitize cells to apoptosis and serve as effective anti-cancer strategies in patients whose tumors overexpress 14-3-3ζ.
Our data suggest overexpression of 14-3-3ζ may have an oncogenic role in breast cancer. In contrast, 14-3-3σ can act as a tumor suppressor and its expression is lost in many breast cancers (31). 14-3-3σ also inhibits Akt activity after DNA damage (32) while our data have shown that 14-3-3ζ overexpression increases Akt activation (11). Thus, different 14-3-3 isoforms may have specific, non-overlapping and even opposing biological functions. Although silencing 14-3-3ζ, without altering 14-3-3β levels, decreased transformation and tumorigenesis in breast cancer cells, our data do not exclude the possibility that overexpression of other canonical 14-3-3 isoforms may also contribute to tumorigenicity. It is known that 14-3-3 proteins can form homodimers or heterodimers with other isoforms (33, 34). In the context of cancer, overexpression of 14-3-3ζ may interfere with the balance of various dimmer compositions within the cell. This would lead to substrate bias for 14-3-3ζ dimers, which may explain why increasing 14-3-3ζ, even in the presence of other 14-3-3 proteins, leads to enhanced transformation. It may also explain why decreasing only 14-3-3ζ with siRNA, even in the presence of other 14-3-3 proteins, dramatically decreases tumorigenic properties in vitro and in vivo. Our findings call for the development of 14-3-3ζ-targeting therapies for a more effective, personalized cancer care of patients whose tumors overexpress 14-3-3ζ and are at high risk of having disease recurrence with metastasis.
Supplementary Material
Acknowledgments
Grant Support: National Institutes of Health (P30-CA 16672 to The University of Texas M.D. Anderson Cancer Center; RO1-CA109570, RO1-CA119127, PO1-CA099031 project 4, P50-CA116199 project 4 (D.Y); Department of Defense Center of Excellence (BC050006) subproject to D.Y.
We thank Dr. A. Sahin for supplying tumor samples for FISH/IHC analysis. We also thank X. Du, K. Matias, J. Hannay, V. Hawthorne, T. Chen, and S. Zhang for reagents, suggestions, and manuscript reading, and W. Schober in the UTMDACC Flow Cytometry Core for technical assistance.
References
- 1.Chatterjee SK, Zetter BR. Cancer biomarkers: knowing the present and predicting the future. Future Oncol. 2005;1:37–50. doi: 10.1517/14796694.1.1.37. [DOI] [PubMed] [Google Scholar]
- 2.Press MF, Pike MC, Chazin VR, et al. Her-2/neu expression in node-negative breast cancer: direct tissue quantitation by computerized image analysis and association of overexpression with increased risk of recurrent disease. Cancer Res. 1993;53:4960–70. [PubMed] [Google Scholar]
- 3.Aitken A. 14-3-3 proteins on the MAP. Trends Biochem Sci. 1995;20:90–7. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- 4.Aitken A. 14-3-3 proteins: A historic overview. Semin Cancer Biol. 2006;16:162–72. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–97. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 6.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signaling and apoptosis. Bioessays. 2001;23:936–46. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 7.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–13. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Porter GW, Khuri FR, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 sigma gene is associated with 5’ CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353–7. [PubMed] [Google Scholar]
- 11.Danes CG, Wyszomierski SL, Lu J, Neal CL, Yang W, Yu D. 14-3-3 zeta down-regulates p53 in mammary epithelial cells and confers luminal filling. Cancer Res. 2008;68:1760–7. doi: 10.1158/0008-5472.CAN-07-3177. [DOI] [PubMed] [Google Scholar]
- 12.Tan M, Yao J, Yu D. Overexpression of the c-erbB-2 gene enhanced intrinsic metastatic potential in human breast cancer cells without increasing their transformation abilities. Cancer Res. 1997;57:1199–205. [PubMed] [Google Scholar]
- 13.Tan M, Jing T, Lan KH, et al. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9:993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 14.Tan M, Lan K-H, Yao J, et al. Selective Inhibition of ErbB2-Overexpressing Breast Cancer In vivo by a Novel TAT-Based ErbB2-Targeting Signal Transducers and Activators of Transcription 3-Blocking Peptide. Cancer Res. 2006;66:3764–72. doi: 10.1158/0008-5472.CAN-05-2747. [DOI] [PubMed] [Google Scholar]
- 15.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. The Oncologist. 2004;9:606–16. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 16.Kattan MW. Evaluating a new marker’s predictive contribution. Clin Cancer Res. 2004;10:822–4. doi: 10.1158/1078-0432.ccr-03-0061. [DOI] [PubMed] [Google Scholar]
- 17.Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–32. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science. 1991;254:1153–60. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 19.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 21.Escot C, Theillet C, Lidereau R, et al. Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci U S A. 1986;83:4834–8. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller FR, Soule HD, Tait L, et al. Xenograft model of progressive human proliferative breast disease. J Natl Cancer Inst. 1993;85:1725–32. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- 23.Felsher DW. Reversibility of oncogene-induced cancer. Curr Opin Genet Dev. 2004;14:37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 25.Fan T, Li R, Todd NW, et al. Up-regulation of 14-3-3 zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 26.Rogers CE, Loveday RL, Drew PJ, Greenman J. Molecular prognostic indicators in breast cancer. Eur J Surg Oncol. 2002;28:467–78. doi: 10.1053/ejso.2002.1258. [DOI] [PubMed] [Google Scholar]
- 27.Heidenblad M, Lindgren D, Jonson T, et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi W, Martinez JD. Reduction of 14-3-3 proteins correlates with increased sensitivity to killing of human lung cancer cells by ionizing radiation. Radiat Res. 2003;160:217–23. doi: 10.1667/rr3038. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Zhao J, Du Y, et al. Down-regulation of 14-3-3 zeta suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci U S A. 2008;105:162–7. doi: 10.1073/pnas.0710905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frasor J, Chang EC, Komm B, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–40. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson AT, Evron E, Umbricht CB, et al. High frequency of hypermethylation at the 14-3-3sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000;97:6049–54. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Wen YY, Zhao R, et al. DNA damage-induced protein 14-3-3 sigma inhibits protein kinase B/Akt activation and suppresses Akt-activated cancer. Cancer Res. 2006;66:3096–105. doi: 10.1158/0008-5472.CAN-05-3620. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem Biophys Res Commun. 2003;300:679–85. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- 34.Verdoodt B, Benzinger A, Popowicz GM, Holak TA, Hermeking H. Characterization of 14-3-3 sigma dimerization determinants: requirement of homodimerization for inhibition of cell proliferation. Cell Cycle. 2006;5:2920–6. doi: 10.4161/cc.5.24.3571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.