Abstract
Unfolded vs. native carbon monoxide coordinated horse heart cytochrome c (h-cyt c) and a heme axial methionine mutant cyt c552 from Hydrogenobacter thermophilus (Ht-M61A) are studied by IR absorption spectroscopy and ultrafast 2D-IR vibrational echo spectroscopy of the CO stretching mode. The unfolding is induced by guanidinium hydrochloride (GuHCl). The CO IR absorption spectra for both h-cyt c and Ht-M61A shift to the red as the GuHCl concentration is increased through the concentration region over which unfolding occurs. The spectra for the unfolded state are substantially broader than the spectra for the native proteins. A plot of the CO peak position vs. GuHCl concentration produces a sigmoidal curve that overlays the concentration dependent circular dichroism (CD) data of the CO-coordinated forms of both Ht-M61A and h-cyt c within experimental error. The coincidence of the CO peak shift curve with the CD curves demonstrates that the CO vibrational frequency is sensitive to the structural changes induced by the denaturant. 2D-IR vibrational echo experiments are performed on native Ht-M61A and on the protein in low and high concentration GuHCl solutions. The 2D-IR vibrational echo is sensitive to the global protein structural dynamics on time scales from sub-picosecond to greater than one hundred picoseconds through the change in the shape of the 2D spectrum with time (spectral diffusion). At the high GuHCl concentration (5.1 M), at which Ht-M61A is essentially fully denatured as judged by CD, a very large reduction in dynamics is observed compared to the native protein within the ~100 ps time window of the experiment. The results suggest the denatured protein may be in a glassy like state involving hydrophobic collapse around the heme.
I. Introduction
Protein folding is a fundamentally important problem that has been intensely investigated for decades.1–10 Although a great deal is known about protein folding, some of the most important issues remain unresolved. The theoretical models of protein folding can be broadly divided into two categories. The first model is that protein folding proceeds through a unique predetermined sequence of metastable intermediate states of increasingly lower energy until the native state is reached.2–5 The native state is assumed to be the state of lowest free energy. The alternative folding funnel model7,8,11,12 based on energy landscape considerations, advocates a multitude of pathways and no unique sequence of intermediate states. The driving force in the funnel model is also the free energy bias towards the native state but entropy considerations are included in the description of stabilization. While experimental results have been proposed in the favor of both the models,6,10 it is fair to say that it is not clear which model is correct or whether either model is uniquely correct. Indeed, treatments of protein folding data benefit from consideration of both of these models which are not mutually exclusive.
A major difficulty in experimentally addressing the protein folding problem is the proper identification and characterization of non-native conformational states on the folding energy landscape. These conformations can be short lived, which can inhibit thorough examination. To make it possible to improve characterization of folding intermediates, considerable research has been directed toward unfolding of proteins from their native states. The utility of this approach is that a non-native state can be stabilized by the use of an appropriate amount of a denaturant such as guanidinium hydrochloride (GuHCl) or urea. These denaturants are known to destabilize proteins in different ways.13–17 The guanidinium cation may disrupt salt bridges and other Coulombic interactions within the protein.18,19 In contrast, urea is uncharged and generally denatures a protein by disrupting its secondary structures and operates at higher concentrations than GuHCl. By using a particular denaturant, it is possible to create an ensemble of non-native states of nearly identical characteristics which can then be probed experimentally.
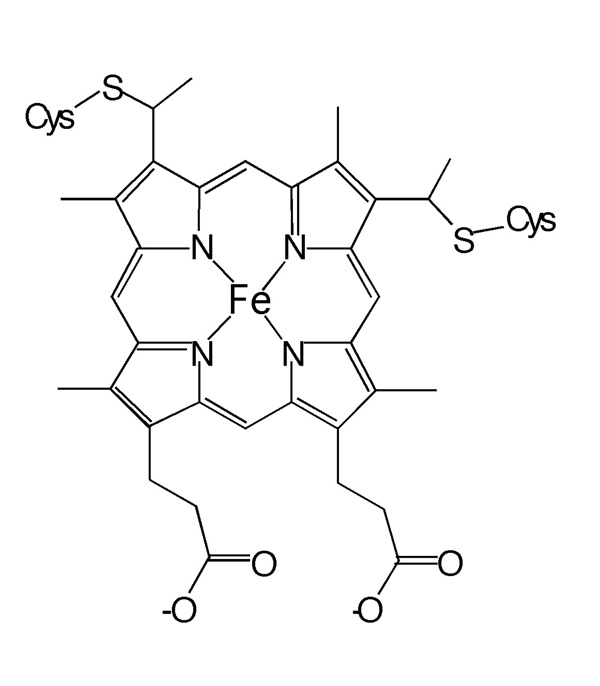
Because of their relative simplicity and high solubility, proteins in the cytochrome c (cyt c) family have served as important model systems in protein folding studies using a host of optical and magnetic resonance techniques.13,17,20–33 Class I cyts c proteins are soluble and monomeric with a single heme group covalently bound to a Cys-X-X-Cys-His motif in which the two Cys form thioether linkages to the heme and the His binds the heme iron.34 Figure 1 displays the active site with the Cys linkages shown. A range of experiments on cyts c have been directed toward distinguishing between the sequential model and the folding funnel model and to characterize the non-native states. A series of papers by Englander and coworkers (who used a hydrogen exchange method), have indicated the existence of a definite pathway of unfolding through a well-defined sequence of intermediate states.10,35 These studies seem to run counter to the folding funnel picture. Subsequent theoretical studies by Wolynes and co-workers have suggested that the hydrogen exchange experiments can be explained within the folding funnel picture.6 Comparative studies of the folding of Class I cyts c from different subfamilies suggest that proteins in this class share a common folding nucleus, which translates to a common folding mechanism.26,36
Figure 1.
The active site of Class I cyts c with a single heme group covalently bound to a Cys-X-X-Cys-His motif in which the two Cys form thioether linkages to the heme.
Class I cyts c as a group are among the most actively studied proteins in the context of chemical and thermal denaturation, in which the heme plays an important role and serves as a valuable probe.13,16,17,21–30,37–41 The thioether bonds ensure that the heme prosthetic group is closely associated with the protein in the native and unfolded configurations. The heme is axially ligated by side chains of a histidine and a methionine. The methionine-iron bond may be disrupted by binding of exogenous ligands which serve as probes of the physical properties of the heme and its environment.42 Alternatively, mutation of the axial methionine to non-ligating alanine creates a site for ligand probes to bind the heme iron.43
In this paper we report a new approach to studying the denaturation of two different class I cyts c using infrared spectroscopy. The stretching mode of CO bound to the reduced Fe of these cyts c is studied as a function of GuHCl concentration using infrared absorption spectroscopy and ultrafast two dimensional infrared (2D-IR) vibrational echo spectroscopy. Experiments are conducted on CO-coordinated ferrous cyt c552 mutant from Hydrogenobacter thermophilus (in which the heme axial ligand Met61 is replaced with Ala, Ht-M61A) and on the CO derivative of ferrous horse heart cyt c (h-cyt c). Both are Class I cyts c proteins, but h-cyt c is from the mitochondrial cyt c subfamily, and Ht cyt c552 is from the cyt c8 subfamily (sometimes called the cyts c551).44 The folding of these proteins has been proposed to involve a common folding nucleus and mechanism resulting from their similar folding topology.26,36
The linear absorption experiments display a single CO band for the native proteins that shifts to lower frequency as the GuHCl concentration is increased. For h-cyt c, a comparison of the CO absorption peak shift with GuHCl concentration is almost identical to the concentration dependence of the CD data, demonstrating that the CO vibration is sensitive to the structural changes induced by the denaturant. In addition to the absorption band shift with increasing concentration, the CO band develops a shoulder and then becomes a much broader band at high (> 5 M) GuHCl concentration. The CO band is inhomogeneously broadened, and the width reflects the range of structural configurations available to the protein. The increase in width when the protein is unfolded indicates the existence of a broader range of structures in the denatured state than in the native state.
The 2D-IR vibrational echo experiments are conducted on the CO bound to the Ht-M61A ferrous heme. 2D-IR vibrational echo spectroscopy 45–47 can probe protein conformational fluctuations under thermal equilibrium conditions on timescales ranging from subpicosecond to ~100 ps or longer.48–53 2D-IR vibrational echo spectroscopy reports on protein dynamics that occur on fast timescales. The method has recently been applied to biological problems 54,55 including the study of model enzymes,56,57 protein unfolding,50 peptide dynamics in membranes,51 and protein equilibrium fluctuations in aqueous and confined environments.48,49,52,53,58
Here we employ 2D-IR vibrational echo spectroscopy to examine the equilibrium structural fluctuations of different states of Ht-M61A by observing the change in the 2D lineshapes with time for three samples, the native protein, and the protein in low and high concentration GuHCl. Within the time window of the experiments (subpicosecond to ~100 picoseconds), the native protein displays considerable fast structural dynamics with approximately >50% of the accessible structures being sampled in <100 ps. However, the unfolded protein formed at high GuHCl concentration (5.1 M) shows a dramatic change in the structural dynamics, with only ~15% of the protein configurations being sampled in <100 ps. There is a significant decrease in the homogeneous (motionally narrowed) contribution to the 2D spectra and a substantial decrease fraction of structures sampled on the longer time scale (100 ps). The changes may indicate that the unfolded state is in a compact, hydrophobicity driven collapsed glassy state.
II. Experimental Procedures
A. Sample preparation
Preparation of Ht-M61A utilized an E. coli-based expression system.59,60 Molecular biology procedures and materials and the preparation of Ht-M61A are described in detail elsewhere.20,61,62 To prepare aqueous samples of CO-ligated Ht-M61A for IR studies, 10 mg of lyophilized protein were dissolved in 1.0 mL of pD 7.4 phosphate buffer (50 mM) in D2O. The solutions were reduced with a 5-fold excess of dithionite (Aldrich) and stirred under a CO atmosphere for 1 h. The solutions were centrifuged at 3000 relative centrifugal force for 15 minutes through a 0.45-µm acetate filter (Pall Nanosep MF) to remove particulates. CO-ligated Ht-M61A was then denatured by adding GuHCl phosphate buffer to obtain final concentrations of 3.2 M and 5.1 M GuHCl. The samples were further concentrated by repeated centrifugation (Eppendorf 5415D) over modified polyetersulfone membranes (Pall Nanosep 3K Omega) to a final protein concentration of 6–8 mM. The sample was then placed in a sample cell with CaF2 windows and a 50 µm Teflon spacer.
Horse heart cyt c (Type VI from Sigma) in 6 M GuHCl was prepared by dissolving 6 mM protein in deoxygenated GuHCl and 50 mM potassium phosphate (pD 7.4). The protein was reduced and ligated with CO as described above. Samples in the range of 0.0003–5.8 M GuHCl were prepared by combining the h-cyt c/6 M GuHCl solution with a 50 mM phosphate buffer solution. To remove light scattering sources such as dust particles, samples were filtered through a 0.45-µm acetate filter (Pall Nanosep MF), and then were concentrated by centrifugation to a final protein concentration of 6–8 mM before loading in a gas tight 50 µm path length sample cell with CaF2 windows.
B. 2D-IR Vibrational Echo Spectroscopy
The experimental setup is similar to ones that have been described previously.46,47,63 Briefly, the mid-IR pulses with center frequency adjusted to the absorption frequency of each protein sample were generated by an optical parametric amplifier pumped with a regeneratively amplified Ti:Sapphire laser. The band width and duration of the mid-IR pulses were 150 cm−1 and 110 fs, respectively. Three mid-IR pulses were sequentially time-delayed before they were crossed and focused in the sample. The vibrational echo pulse generated in the phase-matched direction was made collinear with a local oscillator pulse, dispersed through a monochromator, and detected with a 32-element HgCdTe array detector. The signal is mixed with the local oscillator to obtain full time, frequency, and phase information from vibrational echo wave packet.
The 2D-IR spectrum is obtained as a function of three variables: the emitted vibrational echo frequencies, ωm, and the variable time delays between the first and second pulses (τ) and the second and third pulses (Tw, “waiting” time). The 2D spectrum at each Tw was obtained by numerically Fourier transforming the τ scan data at each emission frequency, ωm, to give the ωτ axis. The 2D-IR data are plotted as a function of ωτ, and ωm.
C. Circular Dichroism Spectroscopy
Samples of CO-ligated Ht-M61A for circular dichroism (CD) spectroscopy were prepared as described above, except that 10 µM protein samples were prepared in 50 mM sodium phosphate buffer in H2O. GuHCl solutions were prepared from a stock solution of 8.2 M GuHCl in 50 mM sodium phosphate buffer, pH 7.0, with a final range of GuHCl concentrations from 0.0 M to 7.0 M. Refractive index measurement was used to determine the GuHCl concentration of each sample as described.64 The pH of each sample was adjusted to 7.0 prior to data collection. CD measurements were performed on an Aviv Instruments model 202 spectropolarimeter using a quartz cell with 0.100 cm path length. CD spectra of samples were recorded every 0.5 nm over a range of 224 nm to 220 nm, with an averaging time of 5 s and a bandwidth of 1.00 nm. The change in the CD signal at 222 nm was analyzed to follow the unfolding of the protein at different GuHCl concentrations. The concentration used for the CD experiments is very low compared to those used for the vibrational echo experiments. A protein concentration study of the denaturation of h-cyt c with GuHCl measured with CD over the protein concentration range 0.014 mM to 0.224 mM showed no dependence on concentration.29 The highest concentration used in that study is well below the concentration employed in the vibrational echo experiments, but it suggests that the CD results will be unchanged at even higher concentrations. NMR experiments carried out on high concentrations (1–3 mM) of the wild-type Ht-cyt c show no evidence of aggregation.65 Furthermore, as discussed below, the denaturation studies using IR absorption experiments on h-cyt c and Ht-M61A as a function of GuHCl concentration show the same denaturation curves as found with CD. These IR absorption studies were performed on samples with the same high concentration used in the vibrational echo experiments. Therefore, the high protein concentrations used in the IR experiments do not appear to influence the results.
III. Results
A. Time Independent Spectroscopy
A complete set of CO ligated h-cyt c IR absorption spectra of the CO stretching mode were taken as a function of GuHCl concentration and a small number of spectra were taken on Ht-M61A. Most of the absorption experiments were conducted on h-cyt c because it is readily available.
The heme iron of h-cyt c is axially ligated with His18 and Met80 to yield a six-coordinated form under physiological conditions. When the protein is denatured under high GuHCl or urea concentration, Met80 dissociates from the heme and heme iron (Fe2+) can bind small ligands such as CO and NO tightly in the denatured state.29 When the denaturant is diluted, the denatured protein with CO bound refolds to form CO-ligated ferrocyt c.29 Studies have shown that unfolded h-cyt c at neutral pH displays non-native ligation to the heme by His26 and His33.13,29,66–68 Partly as a result of the formation of this improperly ligated structure, the refolding of h-cyt c at neutral pH is quite complex. However, binding CO to heme in the unfolded state eliminates the possibility of forming improperly ligated structures.
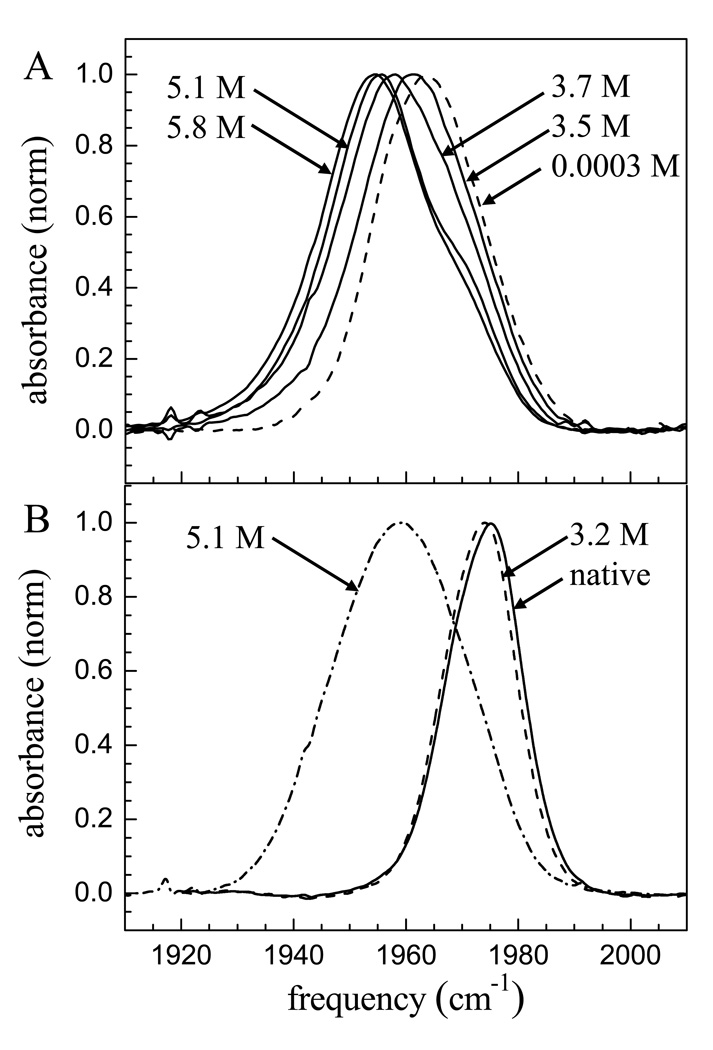
Figure 2A displays several spectra of the CO stretch of h-cyt c-CO. As the concentration of GuHCl increases the band shifts to the red (lower frequency) and broadens. In addition, there is a shoulder on the blue side of the line for the higher concentrations that decreases as the concentration increases further. Figure 2B shows spectra of the mutant protein M61A-CO, for the three GuHCl concentrations that were studied with the 2D-IR vibrational echo experiments discussed below.
Figure 2.
Vibrational absorption spectra of the stretching mode of CO bound to horse heart cyt c (A) and the mutant Ht-M61A (B) at room temperature, pD 7.4, for various concentrations of GuHCl.
An earlier study using a native Ht-M61A FT-IR spectroscopy at room temperature reported a single CO stretching band at 1974 cm−1 with 14.7 cm−1 fwhm20. In that study, the double mutant Ht-M61A/Q64N was prepared to replace Gln64 with Asn in addition to the M61A mutation. According to the NMR study20, Gln64 in Ht-M61A is expected to be oriented out of the heme pocket to be consistent with a non-hydrogen-bonding interaction with the CO ligand, while Asn64 is oriented into the active site in Ht-M61A/Q64N to donate a hydrogen bond. The CO band in a native Ht-M61A is attributed to a conformation in which the distal Gln64 is positioned out of heme pocket. Thus the CO band position in Ht-M61A is higher in frequency (~9 cm−1) than that in Ht-M61A/Q64N.
With Ht-M61A, again there is a red shift and broadening as the concentration of GuHCl increases. While the trend in peak positions and widths is similar, they are not identical. For h-cyt c, the spectrum shifts from 1963 cm−1 (0.0003 M) to 1955 cm−1 (5.8 M) while for Ht-M61A the spectrum shifts from 1975 cm−1 (native) to 1959 cm−1 (5.1 M). In addition, the highest concentration h-cyt c peak has a width of 25 cm−1 FWHM while for Ht-M61A it is 28 cm−1 FWHM. Using CO as a reporter, Ht-M61A has a somewhat greater response to unfolding than does h-cyt c. The CO peak for Ht-M61A shifts further to the red at high GuHCl concentration and the high concentration absorption band width is larger.
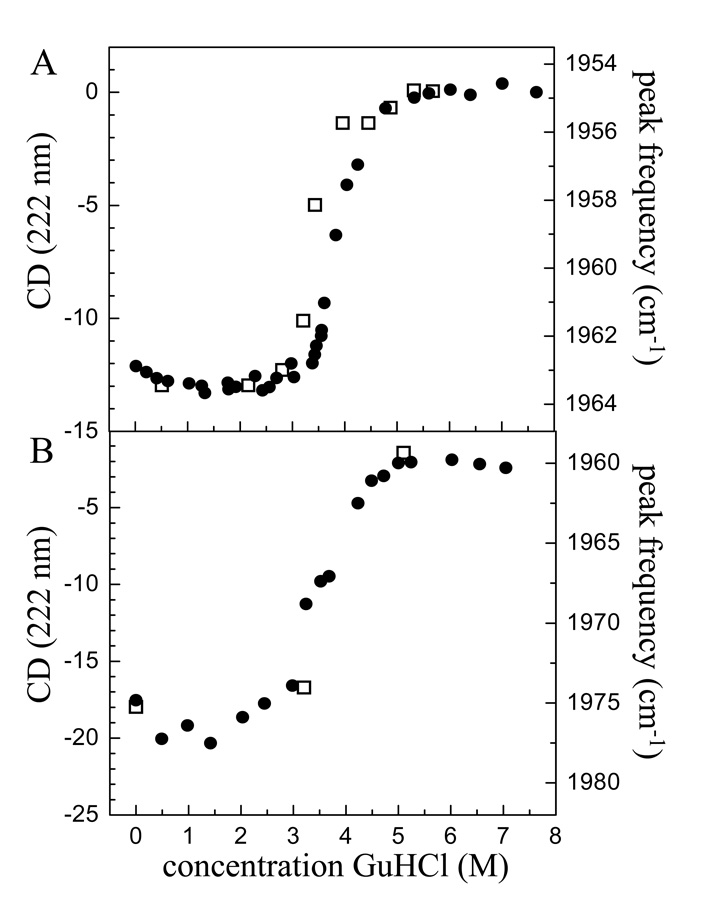
Figure 3A displays two types of data as a function of GuHCl concentration for h-cyt c. The squares are the peak positions of the CO absorption band. The circles are CD data from the literature.29 Two data sets have been scaled so that the amplitudes match at the highest and lowest GuHCl concentration. The change in the peak position with GuHCl concentration closely mimics the CD data, suggesting that the CO vibrational band frequency is sensitive to global unfolding rather than a more subtle effect of denaturant. Figure 3B shows CD data taken on Ht-M61A (circles). The signal-to-noise ratio for the CD of Ht-M61A is not as good as for the spectra of h-cyt c. The squares are the Ht-M61A-CO peak positions for the three concentrations studied below with 2D-IR experiments. While there are only three points, they appear to be consistent with the CD data. In the future this plot will be filled in. For now, we will assume that the vibrational frequency of Ht-M61A is sensitive to the extent of denaturation in the same manner as h-cyt c and the global changes in the protein structure monitored by CD. It is important to note, however, that protein unfolding is complex; Ht cyt c unfolding involves a well-populated intermediate non-native state26 and although h-cyt c unfolding can be approximated with a two-state model, close examination has shown it to be highly heterogeneous.16,32,69,70 Thus, the interpretation of how denaturation impacts the CO stretch will be revisited as models of folding and our understanding of 2D-IR as a probe of folding evolve.
Figure 3.
CD data (filled circles) and CO stretching mode absorption band shift for CO bound to (A) horse heart cyt c and (B) to the mutant Ht-M61A as a function of GuHCl concentration. The CD data for horse heart cyt c is from reference29. The close correspondence between the CD data and the vibrational band shift data shows that the CO vibrational frequency is sensitive to the same global structural changes as CD.
B. 2D-IR Vibrational Echo Spectroscopy
2D-IR vibrational echo spectroscopy directly examines structural degrees of freedom on very fast time scales, ~100 fs to ~100 ps. The vibrational echo signals are recorded by scanning τ at fixed Tw to produce a 2D spectrum. Tw is then increased, and another spectrum is taken. As Tw increases, the peak shapes in the 2D-IR spectra change. These changes are directly related to the structural evolution of the protein through the influence of the changes in structure on the CO stretch frequency. Very qualitatively, the 2D-IR experiment can be viewed as follows. The first and second pulses act to label the initial frequencies of the molecular oscillators. Between the second and third pulses, structural evolution causes the initially labeled frequencies to change (spectral diffusion). The third pulse ends the evolution period, and the vibrational echo signal reads out the final frequencies of the initially frequency-labeled molecular oscillators. As Tw increases, there is more time for structural evolution to occur, and, therefore, larger frequency changes. The structural evolution (frequency change) is reflected in the changes of peak shape of the 2D-IR spectra. The 2D-IR vibrational echo experiments have been reviewed recently.45–47
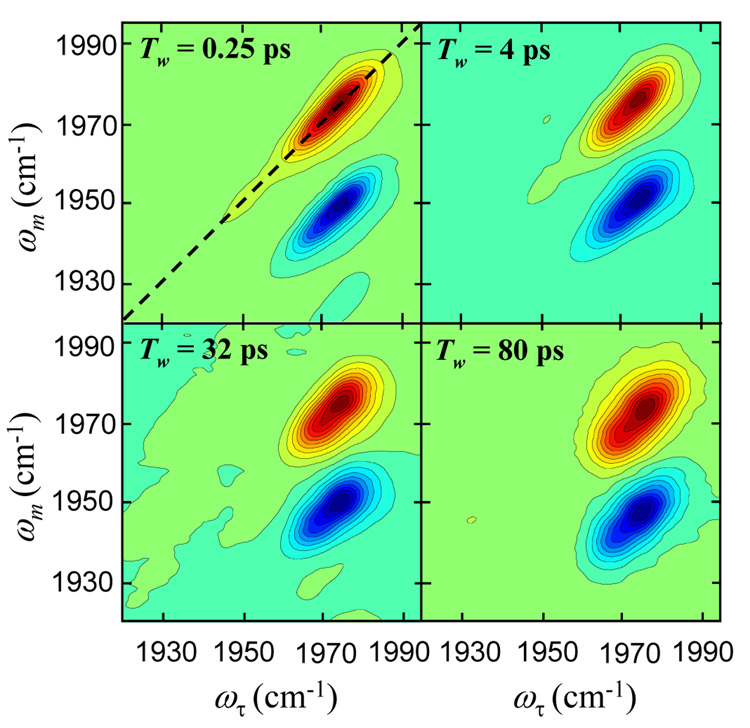
Figure 4 displays 2D-IR vibrational echo spectra for native Ht-M61A at several times, Tw. In the Tw = 0.25 ps panel, the dashed line is the diagonal. There are two bands in each of the spectra. The band on the diagonal (positive going) arises from the 0–1 vibrational transition of the CO stretching mode. The band below it, off-diagonal (negative going), arises from vibrational echo emission at the 1–2 transition frequency, which is shifted to lower frequency by the anharmonicity of the vibrational potential.71,72 Both types of bands provide the same information. Therefore, we will focus on the 0–1 transition band on the diagonal.
Figure 4.
2D-IR vibrational echo spectra of the stretching mode of CO bound to Ht-M61A at several delays, Tw. The diagonal is the dashed line shown in the upper left panel. The bands on the diagonal are positive going, and the off-diagonal bands are negative going. As Tw increases, the shape of the 2D spectrum changes, becoming less elongated. The change in shape with Tw is caused by protein structural evolution induced spectral diffusion.
At short Tw, the peaks in 2D-IR spectra reveal a strong elongation along the diagonal. For the shortest two Tws shown, there is a long tail going to low frequency. This tail arises from a low amplitude band in the absorption spectrum that is centered at ~1953 cm−1. This peak has a shorter lifetime than the main band further to the blue. It has decayed and is not visible in the Tw = 32 ps spectrum. The important feature for the main band is that as Tw increases the shape becomes less elongated along the diagonal. Elongation along the diagonal is caused by inhomogeneous broadening. At short time there are many structural configurations of the protein that have not yet been sampled by structural evolution. As time increases, structural evolution causes spectral diffusion resulting in the change in shape. The longest time for the measurements is limited to a few T1, where T1 is the vibrational lifetime. Here data could be collected out to 120 ps. The spectrum is sensitive to structural fluctuations that are a few times the longest Tw measured because some portion of slower fluctuations will occur in the experimental window if their time scale is not too slow.73 If there was no time limit on the measurement, then the 2D bands would eventually become round when spectral diffusion is complete, that is, when all possible structural configurations are sampled.
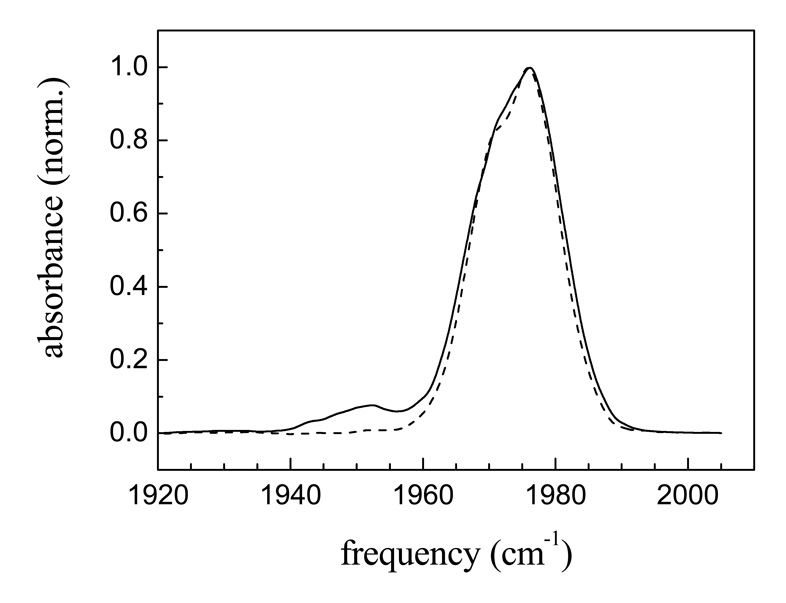
Figure 5 displays spectra that are slices along the diagonal of the 2D spectra taken at two times, Tw = 0.25 ps (solid curve) and Tw = 32 ps (dashed curve). The two spectra have been normalized. At short time (0.25 ps), the small peak at ~1953 cm−1 is apparent. At long time (32 ps), the 1953 cm−1 peak is absent because it has a shorter lifetime. For CO bound to hemes or metallo-porphyrins, it is well documented that the CO vibrational lifetime decreases essentially linearly as the frequency of the transition decreases.74–76 In both figure 2B and figure 5, the main band for the native protein shows some asymmetry. In figure 5, the 2D diagonal cut at 32 ps strongly suggests that there are two substates responsible for the absorption band. The difference between the 2D diagonal spectrum at 0.25 ps and 32 ps arises from a slight difference in lifetimes of the two substates. The spectra shown in figure 5 indicate that the native Ht-M61A protein exists in three conformational substates.
Figure 5.
Native Ht-M61A-CO spectra produced by cutting the 2D vibrational echo spectrum along the diagonal at two Tws. Solid curve, Tw = 0.5 ps; dashed curve, Tw = 32 ps. The data show that the native protein has three conformational substates (see text).
The change of peak shapes in the 2D-IR spectra with increasing Tw (see figure 4) can be employed to determine the time scales and amplitudes of various contributions to the structural evolution of the protein using methods based on diagrammatic perturbation theory.77,78 The frequency-frequency correlation function (FFCF) connects the experimental observables to the underlying dynamics. The FFCF is the joint probability distribution that the frequency has a certain initial value at t = 0 and another value at a later time t. As the structure evolves, the initial frequencies of the protein bound COs change, and the FFCF decays. Once the FFCF is known, all linear and nonlinear optical experimental observables can be calculated by time-dependent diagrammatic perturbation theory.77,78 Conversely, the FFCF can be extracted from 2D-IR spectra with additional input from linear FT-IR absorption spectra. In general, to determine the FFCF from 2D-IR and linear FT-IR spectra, full calculations of linear and nonlinear third-order response functions are performed iteratively until the calculation results converge to the experimental results.79
Here, the Center Line Slope (CLS) method is employed.80,81 The CLS method is an approach for extracting the FFCF from the Tw dependence of the 2D-IR spectra that is accurate and much simpler to implement numerically than iterative fitting methods with calculations of all response functions based on time-dependent diagrammatic perturbation theory. Furthermore, the CLS provides a more useful quantity to plot for visualizing differences in the dynamics as a function of GuHCl concentration than a series of full 2D-IR spectra.52,53,80,81 In the CLS method employed here, frequency slices through the 2D-IR spectrum parallel to the ωm axis at various ωτs are projected onto the ωm axis.80,81 These projections are a set of spectra with peak positions, , on the ωm axis. The plot of vs. ωτ forms a line called the center line. In the absence of a homogenous contribution to the 2D-IR spectrum (see below), the peak shape in the 2D-IR spectrum at Tw = 0 ps would be essentially a line along the diagonal at 45°. The slope of this center line would be 1. At very long time, the peak shape in the 2D-IR spectrum becomes symmetrical, and the center line is horizontal (slope is zero). It has been shown theoretically that the normalized FFCF is equal to the Tw dependence of the slope of the center line.80,81 The center line is called the CLS. Therefore, the CLS will vary from a maximum value of 1 at Tw = 0 ps to a minimum value of 0 at a sufficiently long time. It has also been shown theoretically that combining the analysis of the CLS with the linear FT-IR absorption spectrum, the full FFCF can be obtained, including the Tw independent homogenous component.80,81
A homogenous contribution to the peak shape in the 2D-IR spectra and to the line shape of the linear FT-IR absorption spectra can arise from three sources: very fast structural fluctuations that produce a motionally narrowed contribution to the FFCF,79,82 vibrational population relaxation, and orientational relaxation. Because the rotational diffusion time of the protein is long relative to the vibrational lifetime, the contribution from orientational relaxation is negligible. The vibrational population relaxation times (vibrational lifetimes) were measured independently using IR pump-probe experiments. The lifetime contribution to the homogeneous linewidth is small. Motional narrowing occurs when there is some portion of the structural fluctuations that are extremely fast, such that Δτ <1, where τ is the time scale of the fast fluctuations and Δ is the amplitude of the associated frequency fluctuations. Motionally narrowed fluctuations produce a Lorentzian contribution to both the 2D-IR spectrum and the linear FT-IR absorption spectrum. The Tw-independent homogeneous contribution manifests itself by broadening the 2D-IR spectrum along the ωτ axis even at Tw = 0 ps. This homogeneous broadening reduces the initial value of the CLS to a number less than 1, which permits its determination.
A multiexponential form of the FFCF, C(t), is used to model the multi-time scale dynamics of the structural evolution of the protein systems. This form has been used previously for the analysis of a number of heme proteins.20,46,48,49,52,53,58,62,83 The FFCF has the form:
| (1) |
The Δi and τi terms are the amplitudes and correlation times, respectively, of the frequency fluctuations induced by protein structural dynamics. τi reflects the time scale of a set of structural fluctuations and the Δi is the range of CO frequencies sampled due to those structural fluctuations. The experimental time window is a few T1, because lifetime decay reduces the signal to zero. The 2D-IR vibrational echo experiment is sensitive to fluctuations a few times longer than this window73 because some portion of slower fluctuations will occur in the experimental window if their time scale is not too slow. Protein structural dynamics that are sufficiently slow will appear as static inhomogeneous broadening, which is reflected in C(t) by Δs, a static term. In obtaining the FFCF from the data, the Δi and the τi are determined. However, for a motionally narrow term (Δτ< 1), only the product, , can be obtained. is called the pure dephasing time. The pure dephasing time is obtained using , where T2 is determined from the CLS with use of the linear absorption spectrum80,81, and T1 is obtained from IR pump-probe experiments. The pure dephasing linewidth is .
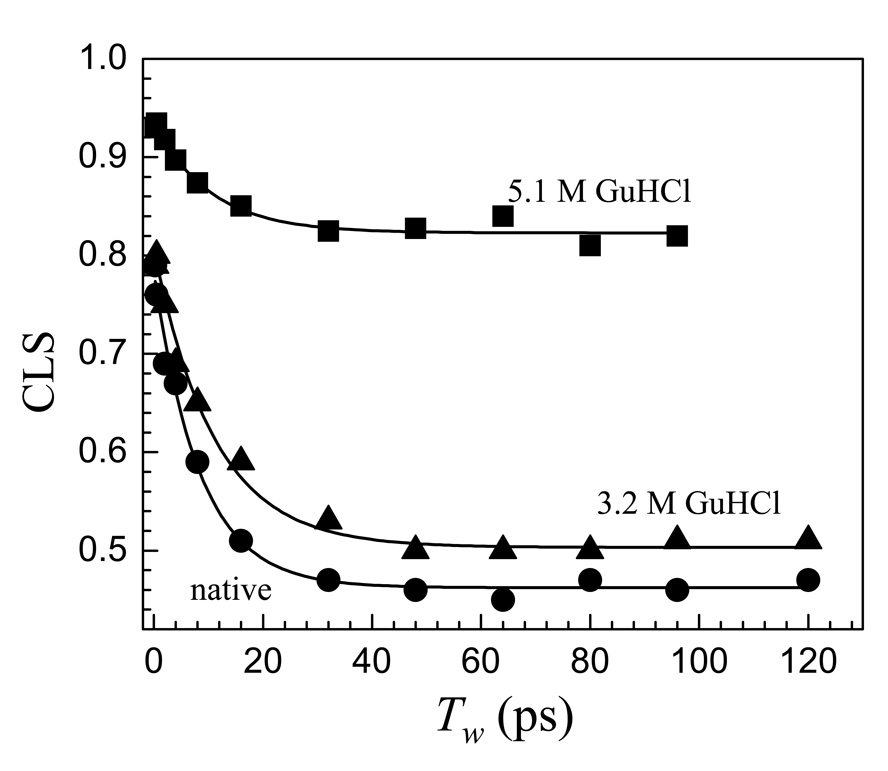
Figure 6 displays CLS data for native Ht-M61A and for the protein in 3.2 M and 5.1 M GuHCl solutions. As can be seen in figure 3, 3.2 M is at the onset of the denaturation while at 5.1 M denaturation is essentially complete. As will be discussed below, the FFCF for all three samples is well described by three terms in equation 1. As discussed above, the difference between 1 and the Tw = 0 value of the CLS is related to the homogeneous component of the dynamics. All three curves show a deviation from 1 at Tw = 0. At relatively short times, < ~40 ps, there is a decay. Following the decay, the CLS becomes horizontal. The horizontal portion of the data is described by the static term in the FFCF and indicates that there are dynamics that are too slow to be observed within the time frame of the experiment.
Figure 6.
The center line slope (CLS) data for native Ht-M61A-CO in aqueous solution and in 3.2 M and 5.1 M GuHCl solutions. The dynamics in the ~100 ps experimental time window of the denatured protein (5.1 M GuHCl) are substantially changed from those of the native protein.
First consider the CLS data for the native state qualitatively. During the 120 ps of observation, the CLS has decayed to 0.46. This means that over half of the spectroscopic line has been sampled by spectral diffusion. Part of this sampling is described by the homogeneous component. The homogeneous component is caused by exceedingly fast fluctuations on an atomic distance scale that may be thought of as thermally populated low frequency vibrations.58,62,83 The dynamics on the tens of picosecond time scale involve much larger distance scales. A reasonable question is what is the distance scale of motions that occur on these time scales? Incoherent quasielastic neutron scattering experiments on native bovine α-lactalbumin observed collective motions on the tens of picosecond time scale.84 The correlation length for such fluctuations was reported to be 18 Å.84 Therefore, the fluctuations that cause the decay of the CLS should represent motions of amino acids and collections of amino acids. The fact that the CLS becomes flat and that 46% of the inhomogeneous absorption line is not sampled within the experimental time window means that approximately half of the accessible protein structural configurations interconvert on times scales significantly slower than 100 ps.
At the onset of denaturation (see figure 6, 3.2 M GuHCl), the CLS data changes but not by a great deal. The absorption spectrum (figure 2B) is also almost the same as that of the native protein. Quantitative analysis of the data (see below) clearly delineates the small differences. However, the fully denatured protein (see figure 6, 5.1 M GuHCl) shows dramatic changes from the native protein. The absorption spectrum is significantly different (see figure 2B). In the CLS data, the homogeneous component is substantially reduced and the decay plateaus with <20% of the spectroscopic line sampled. Therefore, on time scales of 100 ps, there is a substantial change, the nature of which is discussed below.
The CLS decay results are quantified in Table 1. The table gives the parameters obtained from the CLS determination of the FFCF and the vibrational lifetime, T1, for each sample. The numbers in parentheses give the fraction of the particular contribution of the FFCF to the entire absorption linewidth. There are three terms in the FFCF (see equation 1). The first data column has the values of the pure dephasing linewidths, . This component of the FFCF is determined by the difference between 1 and the Tw = 0 value in figure 6. The next two columns are the decay constant and the amplitude factor that give rise to the decays that are seen in all three curves in figure 6. The column labeled Δs is the amplitude of the static term, which sets the value of the long time plateau in the data. Comparing the results for the native protein (0.0 M GuHCl) to the protein at the onset of denaturation (3.2 M GuHCl) as can be seen from the CD and IR spectral shift data in figure 3, the FFCF parameters are very similar except for τ1. The decay time τ1 is longer for the slightly denatured protein, and Δs is a little larger. These differences can be seen in the data in figure 6.
Table 1.
FFCF parameters and vibrational lifetimes, T1 of CO band bound to Ht-M61A. Percentages are fractional contributions to the absorption linewidths.
| GuHCl Conc. | Γ (cm−1) | τ1 (ps) | Δ1 (cm−1) | Δs (cm−1) | T1 (ps) |
|---|---|---|---|---|---|
| 0.0 M | 2.7 (22%) | 8 | 4.2 (32%) | 3.9 (46%) | 29 |
| 3.2 M | 2.5 (21 %) | 11 | 4.0 (29%) | 3.5 (50%) | 27 |
| 5.1 M | 1.5 (7%) | 10 | 4.7 (11%) | 10.3 (82%) | 23 |
As can be seen in figure 6, the large change in the dynamics occurs when the protein is fully denatured in 5.1 M GuHCl. Looking at the parameters in Table 1, the pure dephasing linewidth, Γ, becomes much narrower ( becomes longer) by almost a factor of two. The decay time constant, τ1, is slower than in the native protein, although it does not change significantly from the value in 3.2 M GuHCl. In addition to the large change in the homogeneous component, the other large change is in the amplitude of the static term, Δs. This term goes from < 4 cm−1 to > 10 cm−1. For the denatured protein, the static term is responsible for 82% of the absorption linewidth compared to 46% for the native protein. This is seen in figure 6 as the much higher value for the plateau for the denatured protein. The results demonstrate that denaturation changes the dynamics within the experimental window. The increase in the inhomogeneous linewidth shows that there are a wider range of structure in the denatured state, and within the time window of the experiment, a smaller fraction of these structures are sampled compared to the native state. Denaturation causes a large fraction of the dynamics to occur on time scales long compared to several hundred picoseconds because measurements within the time window of ~100 ps are sensitive to structural fluctuations that occur on time scales out to several times the experimental window.73
IV. Discussion
Could the change in the Ht-M61A dynamics be caused by the increase in viscosity of 5.1 M GuHCl compared to water? A detailed study of the viscosity dependence of CO vibrational dephasing for a number of heme proteins including Ht-M61A has been reported.85 For many orders of magnitude increase in viscosity over aqueous protein solutions, it was determined that the influence of viscosity on the observed spectral diffusion is very mild. For Ht-M61A, the dependence on viscosity goes as η0.13, where η is the solution viscosity.85 Furthermore, the homogeneous component is essentially unchanged by viscosity. In going from water to 5.1 M GuHCl, the viscosity only increases by 40%.86 Given the very weak dependence of the vibrational dephasing on viscosity η0.13 and the lack of influence of the viscosity on the homogeneous component, the change in viscosity is not responsible for the observed change in protein dynamics at 5.1 M GuHCl.
Insight into the influence of denaturation can be obtained by examining the results of placing Ht-M61A and other heme proteins in a glassy trehalose solvent.62 All of the heme proteins showed the same behavior. The homogenous component of the FFCF (the extremely fast motions) remained essentially unchanged. However, the slower times scale dynamics within the experimental window, picoseconds to tens of picoseconds, were eliminated in the trehalose glass. Molecular dynamics simulations of the myoglobin-CO mutant H64V-CO (the distal histidine replaced with a valine) were conducted to understand the influence of a trehalose glassy solvent on protein dynamics and the vibrational echo observables.62 In the simulations, an aqueous protein solution was equilibrated and then the solvent was immobilized. The dynamics of the H64V-CO were investigated and the FFCFs were determined for the immobilized solvent and a normal aqueous solution. The simulations reproduced the nature of the experimental results semi-quantitatively. Like the experiments, the simulations showed that an ultrafast homogenous component remained with the immobile solvent, but the slower time scale dynamics within the experimental and simulation windows were shut down. In addition, analytical theory has been used to understand the viscosity dependence of protein dynamics using a breathing sphere model.87,88 The understanding that emerged from the simulations and theory showed that elimination of dynamics on the tens of picosecond time scale is caused by locking up the surface topology of the protein. The internal motions of the protein structure depend on the ability of the surface topology to change.
The experiments in trehalose show how slowing or eliminating protein motions manifest themselves in the experimental observables. However, there are major differences between the trehalose experimental results and the results for denatured Ht-M61A shown in figure 6. The key features of the trehalose experiments are that the homogeneous component is unchanged while the observable fluctuations on slower time scale are eliminated.62 In contrast the denatured Ht-M61A data show a substantial change in the homogeneous component and the decay over the first few tens of picoseconds is not eliminated. The comparison between the linear spectra is significant. When Ht-M61A is put into glassy trehalose, the linewidth (fwhm) of the linear IR spectrum does not change, and the peak position is only shifted by 3 cm−1 to higher frequency in trehalose compared to aqueous solution.62 This demonstrates that the basic structure of the protein and the variations in structure about the average are almost unchanged when the surface topology is locked up. The ~10 ps time scale dynamics are pushed out to a long time beyond the experimental time window, but the structure is unchanged. This behavior is very different from the results for Ht-M61A in 5.1 M GuHCl. The Ht-M61A band shifts substantially to lower frequency and broadens considerably in going from the native to denatured protein. The shift in the position and the change in width demonstrate that major structural changes have taken place in contrast to Ht-M61A in trehalose. Therefore, the changes in the FFCF observed for denatured Ht-M61A are caused by a change in structure, not by locking up the original native structure.
Looking at Table 1, the denatured protein’s FFCF has a much smaller homogeneous linewidth (Γ), somewhat slower intermediate time scale dynamics (τ1), and a much larger static component (Δs) compared to the native protein. One possible view of the denatured state is that it is comprised of a large number of structures that are each only slightly different from the native protein. Such a range of structures could account for the large increase in the linewidth of the denatured protein. However, this picture is inconsistent with the data. First, in addition to the increase in linewidth, the absorption spectrum of the fully denatured protein is shifted substantially to the red, thus the average structure has changed. Furthermore there is a large change in Γ, discussed in detail below, which demonstrates a substantial change from the native structure. The change in the intermediate time scale dynamics, ~10 ps, occurs at the onset of denaturation (see Table 1), indicating a change in structure. However, the large change in Γ, the change in the absorption peak position, and the increase in absorption linewidth are very different for the fully denatured protein than the mildly denatured and native proteins, demonstrating additional substantial changes in structure for the fully denatured protein.
Γ is reduced dramatically, by almost a factor of 2, in going from the native to the denatured protein. Γ is the motionally narrow component of the FFCF. For a motionally narrowed contribution, the homogeneous linewidth is Γ = Δ2τ/π, so it is not possible to determine from the experiments whether Δ has changed or τ has changed, or both. The 2D-IR vibrational echo experiments only determine the product, Δ2τ.
Although it is not possible to determine Δ and τ from experiment, it is useful to examine a possible scenario using the results of molecular dynamics simulations. MD simulations have determined the FFCF for myoglobin-CO (MbCO).89 In the MD simulations, Δ and τ for the motionally narrowed component were determined. From MD simulations of MbCO we know that the homogeneous component is caused by the very fast localized fluctuations of very small groups of atoms. These motions can be viewed as very low frequency vibrations, such as torsions of side chains. The MbCO MD simulation gave the motionally narrowed τ = 0.14 ps. Note that kBT corresponds to 0.16 ps, where kB is Boltzmann’s constant and T is the absolute temperature. So these very fast thermal fluctuations of small local modes have τ ≈ h/kBT. It is reasonable to assume that τ is approximately the same for Mb and cyt c. Both Mb and cyt c are small globular heme proteins, and their very local motions might be expected to be similar.
Because the motionally narrowed component is due to very local motions of small groups of atoms, for heuristic purposes we will assume that τ does not change when the protein dentures. This is an assumption that will permit the discussion of a plausible explanation for the reduction in Γ when the protein unfolds. With the assumption that τ = 0.14 ps, we can obtain Δ for the native and denatured states. The values are 42 cm−1 and 31 cm−1, respectively. This decrease in Δ could be consistent with a hydrophobically collapsed state (see below) for the denatured protein in which the packing is more dense. The denser packing of the collapsed state could limit the amplitude of the local motions thereby reducing Δ. Of course this argument is not rigorous, but it is indicative of a possible explanation for the large decrease in Γ with denaturation.
A hydrophobically collapsed denatured state is consistent with the funnel model of protein folding.7,8,11,12 As discussed in the Introduction, the opposite view of protein folding from the funnel model is folding or unfolding through a well defined set of intermediates.2–5,10 In the sequential unfolding model, cyt c unfolds with as many as five intermediates through a sequence of unravelings of particular structural motifs.10,35 The denatured protein is unbundled, with all regions exposed to solvent. Could this model be consistent with the IR experiments reported above? It is reasonable to assume that if the protein denatures in this manner, then there would be a wide variety of conformations that could result in the observed increase in the line width. The change in structure could result in an overall shift in the average frequency, although a red shift is generally associated with an increase electric field at the CO or increased back bonding from the iron-heme to the CO π* anti-bonding molecular orbital.58,76,83,90,91 In MbCO, when the distal His moves away from the CO, the result is a blue shift.
The change in the 2D-IR results may not be consistent with a protein that is denatured through a sequence of unraveling intermediates. The denatured system should be more extended and floppy than the native protein. It is unlikely that such a system would have slower dynamics on the ten picosecond time scale than the native protein. In a recent experiment, it was shown that cleaving a single disulfide bond in a heme protein led to a somewhat faster decay of the FFCF.53 If large dissociative structural changes occur, then it would seem unlikely that the dynamics would slow. Furthermore, such a denatured state could have a wide range of configurations, but they might be expected to be sampled readily, which is inconsistent with the very large static component, Δs, of the FFCF found for the denatured state. Finally, consider the heuristic argument given above for the homogeneous component, Γ, and again assume that τ is unchanged. If this is the case, a less compact and less dense structure would yield a larger Δ, and therefore a larger Γ, in contrast to the experimentally observed decrease in Γ for the denatured protein.
The arguments presented above, while qualitative, indicate that the state of the denatured protein is more consistent with a compact collapsed state suggested by funnel models than an unraveled expanded state suggested by the hierarchical unfolding models. The Ht-M61A results show that the denatured protein dynamics within the experimental window sample only a small portion of the total structural space. The implication is that motions over distances from a few Ångstroms to less than or equal to ~20 Å84 are slower compared to the native protein. However, the dynamics are not slowed to the extent that occurs when the protein’s surface topology is completely locked up by a glassy solvent. The results shown in figure 6 do not seem to be consistent with a picture in which the denatured protein has lost its native structure by unraveling, significantly expanding in size,92 and becoming floppy, i.e., taking on a “random coil” form. The initial experiments presented in figure 6 may indicate that the unfolded state of Ht-M61A is a compact, hydrophobicity driven collapsed state.
In the context of the funnel model, theoretical studies7,8 have suggested the existence of a collapsed and glassy unfolded state in close proximity of the native state, particularly with respect to the size of the protein. In the case of proteins in the cyt c family, theory6 and experiments9 indicate that the initial stage of folding is the formation of a hydrophobicity induced collapsed state with most of the native contacts not formed. These contacts form subsequently along the folding pathway. For unfolding the predicted scenario is reversed. It has been suggested that for cyt c the hydrophobic heme pocket plays an important role in stabilization of the hydrophobic collapsed state.6
The theoretical study shows that because the heme-induced hydrophobic collapse occurs before a large fraction of native contacts are formed, the collapsed state can accommodate many non-native, possibly hydrophobic contacts.6 The hydrophobic collapsed state is predicted to be an important intermediate state. A similar conclusion was reached earlier by Takahashi and co-workers who used small angle x-ray scattering and resonance Raman to study folding of cyt c.9,66 It was found that along the folding pathway, the size of the protein decreases first without much change in the helical content. Subsequent to the collapse, the secondary structure is acquired.
The large static component of the FFCF for time scales longer than a few hundred ps for Ht-M61A were observed for strongly denaturing conditions. Traditionally, denatured proteins were considered to exist in an extended conformation. A number of studies, however, have revealed residual structure in denatured proteins (for review, see reference 70). Gray, Winkler and coworkers have characterized distributions of dansyl fluorophore-heme distances in Saccharomyces cerevisiae iso-1-cyt c, a Class I cyt c, through a range of denaturing conditions and during folding. Their data reveal that denatured cyt c at equilibrium exists in a range of compact and extended conformations. In addition, both compact and extended conformations are observed in kinetic folding studies. These data indicate that even under strongly denaturing conditions, a significant portion of the ensemble of cyt c structures exists in a collapsed state.32,69,70 It has been conjectured that the collapsed state can be glassy with slow collective dynamics.8 The large static component observed by the 2D-IR vibrational echoes when the denatured state is formed demonstrates that there is only a small amount of structural dynamics in the experimental time window (intermediate distance scale). The reduction in dynamics may be a signature of the hydrophobicity induced collapsed state.
V. Concluding Remarks
Here we have examined the dynamics of denatured cyt c mutant Ht-M61A in comparison to the protein in its native form using ultrafast 2D-IR vibrational echo spectroscopy and linear IR absorption experiments. The similarity in trend between CO peak shift curve and the CD curve in cyt c demonstrates that the vibrational stretching frequency of CO bound to cyt c is sensitive to the same types of changes in structure as a CD experiment.
The vibrational absorption lines are inhomogeneously broadened. The inhomogeneous broadening means that the ensemble of proteins has a heterogeneous distribution of structures. The 2D-IR vibrational echo experiment measures spectral diffusion, which occurs because of the interconversion of one protein structure to another. In terms of an energy landscape picture, under a particular set of equilibrium conditions, the protein occupies a broad rough minimum on the energy landscape. (There may be more than a single minimum that gives rise to structural substates93,94 as discussed for the native protein in connection with figure 5.) The roughness is caused by a collection of local minima between barriers of a range of heights. Some of the barriers are low enough that transitions are constantly taking place between the minima that correspond to variations in the protein structure within the experimental time window. Spectral diffusion is caused by transitions among the local minima. The time dependence of the spectral diffusion yields the time evolution of the protein structure.
In the time window of the 2D-IR vibrational echo experiments (~100 fs to ~100 ps), native Ht-M61A displays significant structural dynamics. Somewhat greater than 50% of the accessible structures that give rise to the inhomogeneous CO absorption line are sampled. When the protein is denatured in 5.1 M GuHCl, the inhomogeneous line is substantially broader than that for the native protein, which indicates a broader range of structures. The dynamics of the denatured protein in the time experimental time window sample only a small fraction, less than 20%, of the increased range of structures. Therefore, a large fraction of the structural dynamics is occurring on time scales much greater than 100 ps.
In terms of the energy landscape picture, the results on the denatured protein in 5.1 M GuHCl indicate that the free energy landscape has become rather rugged or rough due to the appearance of deep minima, accompanied by barriers, in the region of the free energy landscape that contains the denatured state. Glassy behavior is a consequence of this ruggedness. Note that if ε is the variance that characterizes the ruggedness of the landscape, then configuration diffusion in the landscape varies as exp(-(ε/kBT)2).95 Thus ruggedness can slow down configurational fluctuations drastically, giving rise to the much slower spectral diffusion in the denatured protein. The observed vibrational dynamics could be consistent with a hydrophobicity driven collapsed glassy state with such a rugged free energy landscape. Because glassy dynamics are predicted by landscape theory as due to the appearance of ruggedness, it may be possible to estimate the ruggedness with inherent structure analysis96 and to explore the possible role of salt concentration in augmenting the ruggedness.
Clearly it will be important to fill in the 2D-IR vibrational echo data for concentrations of GuHCl between 3.2 M and 5.1 M and concentrations >5.1 M. The only limitations on obtaining additional data is that the experiments are complex and the mutant protein must be prepared in substantial quantity. While Ht-M61A under denaturing conditions may be in a hydrophobic collapsed state, there may be well defined non-native structures between those of the native state and the fully denatured state.97 This is already suggested by the data shown in figure 6 for the mildly denatured protein. These could be manifested as steps in the changes in dynamics as a function of GuHCl concentration. In addition, it will be important and interesting to perform the 2D-IR vibrational echo experiments on Ht-M61A using urea as a denaturant. Urea is believed to have a different mechanism for denaturation,13–17,98 and it is possible that the denatured state will be distinct and display different dynamics from the protein denatured with GuHCl.
The results presented in this paper are an important target for simulations. The time scales of the experiments, ~100 ps, are readily amenable to simulation. The relationship between simulations and the calculation of the 2D-IR vibrational echo spectra has been developed and applied to MbCO.58,62,83,99,100 Comparison of the simulation results for the native and denatured protein would provide a bench mark for simulations that are employed to understand protein folding and folding intermediates. If the simulations can reproduce the observed behavior, then they should be able to provide a molecular level view of the structures responsible for the change in the dynamics observed for the denatured state.
Acknowledgements
We thank Professor Abani K. Bhuyan, School of Chemistry, University of Hyderabad, India for providing the CD data shown in figure 3A. This work was supported by the National Institutes of Health (2 R01 GM-061137-05), the Air Force Office of Scientific Research (F49620-01-1-0018), and the National Science Foundation (DMR 0652232). S.K. was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, basic Research Promotion Fund) (KRF-2006-C00038). K.L.B. acknowledges support from the National Institutes of Health (R01- GM63170).
References
- 1.Daggett V, Fersht A. Nature Reviews Molecular Cell Biology. 2003;4:497. doi: 10.1038/nrm1126. [DOI] [PubMed] [Google Scholar]
- 2.Daggett V, Fersht AR. Trends Biochem. Sci. 2003;28:18. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 3.Laurents DVaBRL. Biophys J. 1998;75:428. doi: 10.1016/S0006-3495(98)77530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dill KA, Shortle D. Ann. Rev. Biochem. 1991;60:795. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 5.Wallace LA, Matthews CR. BioPhys. Chem. 2002;101–102:113. doi: 10.1016/s0301-4622(02)00155-2. [DOI] [PubMed] [Google Scholar]
- 6.Weinkam P, Zong CH, Wolynes PG. Proc. Nat. Acad. Sci. U.S.A. 2005;102:12401. doi: 10.1073/pnas.0505274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasai M, Wolynes PG. Phys. Rev. Lett. 1990;65:2740. doi: 10.1103/PhysRevLett.65.2740. [DOI] [PubMed] [Google Scholar]
- 8.Chahine J, Nymeyer H, Leite VBP, Socci ND, Onuchic JN. Phys. Rev. Lett. 2002;88:168101. doi: 10.1103/PhysRevLett.88.168101. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc. Nat. Acad. Sci. U.S.A. 2002;99:1329. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang L, Bedard S, Krishna MMG, Lin Y, Englander SW. Proc. Nat. Acad. Sci. U.S.A. 2002;99:12173. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onuchic JN, LutheySchulten Z, Wolynes PG. Ann. Rev. Phys. Chem. 1997;48:545. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 12.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins-Structure Function and Genetics. 1995;21:167. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 13.Elove GA, Bhuyan AK, Roder H. Biochemistry. 1994;33:6925. doi: 10.1021/bi00188a023. [DOI] [PubMed] [Google Scholar]
- 14.Fedurco M, Augustynski J, Indiani C, Smulevich G, Antalik M, Bano M, Sedlak E, Glascock MC, Dawson JH. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2004;1703:31. doi: 10.1016/j.bbapap.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R, Yadav S, Ahmad F. Biochemistry. 1996;35:11925. doi: 10.1021/bi961079g. [DOI] [PubMed] [Google Scholar]
- 16.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. J. Mol. Bio. 2006;357:1009. doi: 10.1016/j.jmb.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 17.Russell BS, Melenkivitz R, Bren KL. Proc. Natl. Acad. Sci. USA. 2000;97:8312. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JS, Scholtz JM. Biochemistry. 1996;35:7292. doi: 10.1021/bi960341i. [DOI] [PubMed] [Google Scholar]
- 19.Monera OD, Cyril MK, Hodges RS. Protein Sci. 1994;3:1984. doi: 10.1002/pro.5560031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massari AM, McClain BL, Finkelstein IJ, Lee AP, Reynolds HL, Bren KL, Fayer MD. J. Phys. Chem. B. 2006;110:18803. doi: 10.1021/jp054959q. [DOI] [PubMed] [Google Scholar]
- 21.Bren KL, Kellogg JA, Kaur R, Wen X. Inorg. Chem. 2004;43:7934. doi: 10.1021/ic048925t. [DOI] [PubMed] [Google Scholar]
- 22.Jones CM, Henry ER, Hu Y, Chan CK, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Proc. Natl. Acad. Sci. USA. 1993;90:11860. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinibaldi F, Howes BD, Piro MC, Caroppi P, Mei G, Ascoli F, Smulevich G, Santucci R. J. Biol. Inorg. Chem. 2006;11:52. doi: 10.1007/s00775-005-0057-6. [DOI] [PubMed] [Google Scholar]
- 24.Kumar R, Prabhu NP, Bhuyan AK. Biochemistry. 2005;44:9359. doi: 10.1021/bi050384b. [DOI] [PubMed] [Google Scholar]
- 25.Droghetti E, Smulevich G. J. Biol. Inorg. Chem. 2005;10:696. doi: 10.1007/s00775-005-0027-z. [DOI] [PubMed] [Google Scholar]
- 26.Travaglini-Allocatelli C, Gianni S, Brunori M. Trends in Biochemical Sciences. 2004;29:535. doi: 10.1016/j.tibs.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Rischel C, Jorgensen LE, Foldes-Papp Z. J. Phys. Cond. Matter. 2003;15:1725. [Google Scholar]
- 28.Hagen SJ, Latypov RF, Dolgikh DA, Roder H. Biochemistry. 2002;41:1372. doi: 10.1021/bi011371a. [DOI] [PubMed] [Google Scholar]
- 29.Bhuyan AK, Udgaonkar JB. J. Mol. Bio. 2001;312:1135. doi: 10.1006/jmbi.2001.4993. [DOI] [PubMed] [Google Scholar]
- 30.Panda M, Benavides-Garcia MG, Pierce MM, Nall BT. Protein Science. 2000;9:536. doi: 10.1110/ps.9.3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimenez R, Romesberg FE. J. Phys. Chem. B. 2002;106:9172. [Google Scholar]
- 32.Lyubovitsky JG, Gray HB, Winkler JR. J. Am. Chem. Soc. 2002;124:5481. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 33.Winkler JR. Curr. Opin. Chem. Biol. 2004;8:169. doi: 10.1016/j.cbpa.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Scott RA, Mauk AG. Cytochrome C: A Multidisciplinary Approach. Sausalito, CA: University Science Books; 1996. [Google Scholar]
- 35.Krishna MMG, Lin Y, Rumbley JN, Englander SW. J. Mol. Bio. 2003;331:29. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 36.Ptitsyn OB. J. Mol. Biol. 1998;278:655. doi: 10.1006/jmbi.1997.1620. [DOI] [PubMed] [Google Scholar]
- 37.Varhac R, Antalik M, Bano M. J. Biol. Inorg. Chem. 2004;9:12. doi: 10.1007/s00775-003-0492-1. [DOI] [PubMed] [Google Scholar]
- 38.Russell BS, Bren KL. J. Biol. Inorg. Chem. 2002;7:909. doi: 10.1007/s00775-002-0381-z. [DOI] [PubMed] [Google Scholar]
- 39.Roccatano D, Daidone I, Ceruso MA, Bossa C, Di Nola A. Biophys J. 2003;84:1876. doi: 10.1016/S0006-3495(03)74995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Droghetti E, Oellerich S, Hildebrandt P, Smulevich G. Biophys J. 2006;91:3022. doi: 10.1529/biophysj.105.079749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobson CM, Sali A, Karplus M. Angewandte Chemie-International Edition. 1998;37:868. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JLR, Chapman SK. Dalton Trans. 2005:13. doi: 10.1039/b413046d. [DOI] [PubMed] [Google Scholar]
- 43.Bren KL, Gray HB. J. Am. Chem. Soc. 1993;115:10382. [Google Scholar]
- 44.Ambler RP. Biochim. Biophys. Acta. 1991;1058:42. doi: 10.1016/s0005-2728(05)80266-x. [DOI] [PubMed] [Google Scholar]
- 45.Zheng J, Kwak K, Fayer MD. Acc. of Chem. Res. 2007;40:75. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- 46.Finkelstein IJ, Zheng J, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys. Chem. Chem. Phys. 2007;9:1533. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 47.Park S, Kwak K, Fayer MD. Laser Phys. Lett. 2007;4:704. [Google Scholar]
- 48.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc. Natl. Acad. Sci. USA. 2007;104:2637. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massari AM, Finkelstein IJ, Fayer MD. J. Am. Chem. Soc. 2006;128:3990. doi: 10.1021/ja058745y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung HS, Khalil M, Smith AW, Ganim Z, Tokmakoff A. Proc. Natl. Acad. Sci. USA. 2005;102:612. doi: 10.1073/pnas.0408646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee P, Kass I, Arkin IT, Zanni MT. Proc. Natl. Acad. Sci. USA. 2006;103:3528. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Proc. Nat. Acad. Sci. U.S.A. 2007;104:16116. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa H, Kim S, Kwak K, Wakasugi K, Fayer MD. Proc. Nat. Acad. Sci. U.S.A. 2007;104:19309. doi: 10.1073/pnas.0709760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rella CW, Rector KD, Kwok AS, Hill JR, Schwettman HA, Dlott DD, Fayer MD. J. Phys. Chem. 1996;100:15620. doi: 10.1103/PhysRevLett.77.1648. [DOI] [PubMed] [Google Scholar]
- 55.Fayer MD. Ann. Rev. Phys. Chem. 2001;52:315. doi: 10.1146/annurev.physchem.52.1.315. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Chen J, Hochstrasser RM. J. Phys. Chem. B. 2006;110:7545. doi: 10.1021/jp057564f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bredenbeck J, Helbing J, Kumita JR, Woolley GA, Hamm P. Proc. Natl. Acad. Sci. USA. 2005;102:2379. doi: 10.1073/pnas.0406948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merchant KA, Noid WG, Akiyama R, Finkelstein I, Goun A, McClain BL, Loring RF, Fayer MD. J. Am. Chem. Soc. 2003;125:13804. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karan EF, Russell BS, Bren KL. J. Biol. Inorg. Chem. 2002;7:260. doi: 10.1007/s007750100292. [DOI] [PubMed] [Google Scholar]
- 60.Fee JA, Chen Y, Todaro TR, Bren KL, Patel KM, Hill MG, Gomez-Moran E, Loehr TM, Ai J, Thöny-Meyer L, Williams PA, Stura E, Sridhar V, McRee DE. Protein Sci. 2000;9:2074. doi: 10.1110/ps.9.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen X, Bren KL. Biochemistry. 2005;44:5225. doi: 10.1021/bi047556+. [DOI] [PubMed] [Google Scholar]
- 62.Massari AM, Finkelstein IJ, McClain BL, Goj A, Wen X, Bren KL, Loring RF, Fayer MD. J. Am. Chem. Soc. 2005;127:14279. doi: 10.1021/ja053627w. [DOI] [PubMed] [Google Scholar]
- 63.Asbury JB, Steinel T, Fayer MD. J. Luminescence. 2004;107:271. doi: 10.1016/j.jlumin.2003.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pace CN, Scholtz JM. Measuring the Conformational Stability of a Protein. In: Creighton TE, editor. Protein Structure: A Practical Approach. Oxford: 1997. p. 299. [Google Scholar]
- 65.Karan EF, Russell BS, Bren KL. J. Biol. Inorg. Chem. 2002;7:260. doi: 10.1007/s007750100292. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi S, Yeh SR, Das TK, Chi-Kin C, Gottfried DS, Rousseau DL. Nature Struc. Bio. 1997;4:44. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 67.Elove GA, Chaffote AF, Roder H, Goldberg ME. Biochemistry. 1992;31:6876. doi: 10.1021/bi00145a003. [DOI] [PubMed] [Google Scholar]
- 68.Sosnick TR, Mayne L, Hiller R, Englander SW. Nature Struct. Biol. 1994;1:149. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 69.Pletneva EV, Gray HB, Winkler JR. J. Mol. Biol. 2005;345:855. doi: 10.1016/j.jmb.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 70.Pletneva EV, Gray HB, Winkler JR. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18397. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rector KD, Kwok AS, Ferrante C, Tokmakoff A, Rella CW, Fayer MD. J. Chem. Phys. 1997;106:10027. [Google Scholar]
- 72.Golonzka O, Khalil M, Demirdoven N, Tokmakoff A. Phys. Rev. Lett. 2001;86:2154. doi: 10.1103/PhysRevLett.86.2154. [DOI] [PubMed] [Google Scholar]
- 73.Bai YS, Fayer MD. Phys. Rev. B. 1989;39:11066. doi: 10.1103/physrevb.39.11066. [DOI] [PubMed] [Google Scholar]
- 74.Dlott DD, Fayer MD, Hill JR, Rella CW, Suslick KS, Ziegler CJ. J. Am. Chem. Soc. 1996;118:7853. [Google Scholar]
- 75.Hill JR, Dlott DD, Rella CW, Peterson KA, Decatur SM, Boxer SG, Fayer MD. J . Phys. Chem. 1996;100:12100. [Google Scholar]
- 76.Hill JR, Rosenblatt MM, Ziegler CJ, Suslick KS, Dlott DD, Rella CW, Fayer MD. J. Phys. Chem. 1996;100:18023. [Google Scholar]
- 77.Mukamel S. Principles of Nonlinear Optical Spectroscopy. New York: Oxford University Press; 1995. [Google Scholar]
- 78.Mukamel S. Ann. Rev. Phys. Chem. 2000;51:691. doi: 10.1146/annurev.physchem.51.1.691. [DOI] [PubMed] [Google Scholar]
- 79.Asbury JB, Steinel T, Kwak K, Corcelli SA, Lawrence CP, Skinner JL, Fayer MD. J. Chem. Phys. 2004;121:12431. doi: 10.1063/1.1818107. [DOI] [PubMed] [Google Scholar]
- 80.Kwak K, Park S, Finkelstein IJ, Fayer MD. J. Chem. Phys. 2007;127:124503. doi: 10.1063/1.2772269. (17 pages) [DOI] [PubMed] [Google Scholar]
- 81.Kwak K, Rosenthal DE, Fayer MD. J. Chem. Phys. 2008 doi: 10.1063/1.2927906. accepted. [DOI] [PubMed] [Google Scholar]
- 82.Woutersen S, Pfister R, Hamm P, Mu Y, Kosov DS, Stock G. J. Chem. Phys. 2002;117:6833. [Google Scholar]
- 83.Merchant KA, Noid WG, Thompson DE, Akiyama R, Loring RF, Fayer MD. J. Phys. Chem. B. 2003;107:4. [Google Scholar]
- 84.Bu Z, Neumann DA, Lee SH, Brown CM, Engelman DM, Han CC. J. Mol. Bio. 2000;301:525. doi: 10.1006/jmbi.2000.3978. [DOI] [PubMed] [Google Scholar]
- 85.Finkelstein IJ, Massari AM, Fayer MD. Biophys. J. 2007;92:3652. doi: 10.1529/biophysj.106.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawahara K, Tanford C. J. Biological Chem. 1966;241:3228. [PubMed] [Google Scholar]
- 87.Rector KD, Jiang J, Berg M, Fayer MD. J. Phys. Chem. B. 2001;105:1081. [Google Scholar]
- 88.Rector KD, Engholm JR, Rella CW, Hill JR, Dlott DD, Fayer MD. J. Phys. Chem. A. 1999;103:2381. [Google Scholar]
- 89.Merchant KA, Thompson DE, Xu Q-H, Williams RB, Loring RF, Fayer MD. Biophys. J. 2002;82:3277. doi: 10.1016/S0006-3495(02)75669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Augspurger JD, Dykstra CE, Oldfield E. J. Am. Chem. Soc. 1991;113:2447. [Google Scholar]
- 91.Hill JR, Dlott DD, Fayer MD, Rella CW, Rosenblatt MM, Suslick KS, Ziegler CJ. J. Phys. Chem. 1996;100:218. [Google Scholar]
- 92.Tsong TY. J. Biol. Chem. 1974;249:1988. [PubMed] [Google Scholar]
- 93.Frauenfelder H, Parak F, Young RD. Ann. Rev. Biophys. Biophys. Chem. 1988;17:471. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 94.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 95.Zwanzig R. Proc. Nat. Acad. Sci. U.S.A. 1988;85:2029. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Debenedetti PG, Stillinger FH. Nature. 2001;410:259. doi: 10.1038/35065704. [DOI] [PubMed] [Google Scholar]
- 97.Russell BS, Melenkivitz R, Bren KL. Proc. Nat. Acad. Sci. U.S.A. 2000;97:8312. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gianni S, Brunori M, Travaglini-Allocatelli C. Protein Sci. 2001;10:1685. doi: 10.1110/ps.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams RB, Loring RF, Fayer MD. J. Phys. Chem. B. 2001;105:4068. [Google Scholar]
- 100.Finkelstein IJ, Goj A, McClain BL, Massari AM, Merchant KA, Loring RF, Fayer MD. J. Phys. Chem. B. 2005;109:16959. doi: 10.1021/jp0517201. [DOI] [PubMed] [Google Scholar]