Abstract
A previous article proposed an electronic structure-based polarizable potential, called the explicit polarization (X-POL) potential, to treat many-body polarization and charge delocalization effects in polypeptides. Here, we present a variational version of the X-POL potential, in which the wave function of the entire molecular system is variationally optimized to yield the minimum total electronic energy. This allows the calculation of analytic gradients, a necessity for efficient molecular dynamics simulations. In this paper, the detailed derivations of the Fock matrix and analytic force are presented and discussed. The calculations involve a double self-consistent-field procedure in which the wave function of each fragment is self-consistently optimized in the presence of other fragments, and in addition the polarization of the entire system is self-consistently optimized. The variational X-POL potential has been implemented in the Chemistry at Harvard Molecular Mechanics (CHARMM) package and tested successfully for small model compounds.
INTRODUCTION
The explicit polarization (X-POL) potential has been proposed for use as a force field for simulations of large molecules in condensed phases (note that the acronym X-POL is also synonymous for Xie’s polarization).1 The X-POL potential is based on division of a system into fragments within which the interactions are treated using molecular-orbital (MO) theory while interactions between fragments are treated using a combined quantum mechanical and molecular mechanical2, 3, 4, 5 (QM∕MM) calculation. In the QM∕MM calculation, the fragments other than the one currently under consideration are represented by MM point charges that are computed by Mulliken6 population analysis. Polarization of the wave function of each fragment is included in the context of this QM∕MM framework. Electronic structure calculations are sequentially performed for each fragment in the presence of Mulliken charges that are obtained from the wave functions of all other fragments. We iteratively cycle through all the fragments until the total energy or electron density is converged to a given tolerance.1 One of the advantages of dividing the system into fragments is that we need to construct and diagonalize Fock matrices only for small fragments, so that the computational bottleneck of diagonalizing the Fock matrix of the entire system is avoided. Since the interfragment interactions are treated using a QM∕MM method, the number of integrals to be evaluated is significantly decreased compared to a full QM calculation of the entire system. Another advantage of the X-POL potential is that it can be easily parallelized by assigning a certain number of fragments to each processor. Communication between processors will be mainly for Mulliken charges.
The X-POL potential was originally called the MO derived empirical potential (MODEL) for liquids;7 it has been applied to the Monte Carlo simulation of liquid water8 and liquid hydrogen fluoride,9 and it was recently extended to the treatment of polypeptides in which the fragments are covalently bonded.1 The wave function for each fragment is updated iteratively in the electric field from all other fragments until self-consistency is achieved for the entire system. Thus, each part of the system is treated at the same level. However, the Fock matrix used previously1, 7, 8, 9 was not derived variationally, so it is not guaranteed that the converged wave function yields the minimum electronic energy of the entire system. In fact, tests showed that the previous converged X-POL potential does not have the lowest energy for a given molecular geometry, although the difference from the true minimum is very small. The nonvariational behavior of the previous formulation makes it difficult to calculate an analytic gradient. Although it is possible to use the coupled-perturbed Hartree–Fock method10 to determine forces iteratively, this would add additional cost to the computational scheme. Approximate gradients obtained previously are not consistent with the energies, as found in the fragment MO method11 (FMO) because of the neglect of the derivative of the wave function (Pulay force).12
In this work, we derive a set of variational equations for the X-POL potential to yield an analytic gradient. The method is similar to that used by Roothaan in deriving the Hartree–Fock equations.13 We show that the Fock matrix for the variational X-POL potential has three extra terms due to the variation of Mulliken charges. We present a method for calculating analytic forces for the entire system in which the wave function of each fragment is self-consistently optimized in the presence of other fragments, and in addition the polarization of the entire system is self-consistently optimized, which is called the double self-consistent-field (DSCF) method.
THEORY
Noncovalently bound clusters
We first consider a system consisting of M molecules that are not covalently bonded to each other (Sec. 2B extends the treatment to a system in which fragments are chemically connected). The electronic interactions within each molecule are described by a QM electronic structure method, and the interactions between molecule A (A=1,2,…,M) and the other M−1 molecules are treated by using an electronic wave function for A and partial atomic charges (point charges at the nuclei) for the M−1 other molecules; the partial atomic charges for those molecules are computed by Mulliken6 population analysis of their wave functions. [This assumption for the interfragment treatment is called a QM∕MM (Refs. 2, 3, 4, 5) treatment]. The N-electron wave function of each QM fragment is constructed as a Slater determinant of the occupied molecular spin orbitals ψk as follows:
| (1) |
where denotes electron i in spin orbital k, and N is the number of explicitly treated electrons in molecule m. We assume a closed-shell wave function for each fragment. Then, each spin orbital ψk is the product of an orbital φi and a spin function (α or β), and the total electronic energy of the system is written as
| (2) |
where i and j represent orthonormal MOs, m denotes a fragment, and B represents an atom in a fragment. The terms in the square brackets are the Hartree–Fock electronic energy and the nuclear-nuclear interaction energy within fragment m. The terms in the final parentheses are QM∕MM interactions between fragment m and all other fragments; these are multiplied by 1∕2 to avoid double counting. The symbols in Eq. 2 are defined below.
In Eq. 2, Hi is the expectation value of the one-electron Hamiltonian H as follows:
| (3) |
where an asterisk denotes complex conjugation. Hi is the sum of the electronic kinetic energy and the electron-nuclei Coulomb attraction (only for the nuclei in the same molecule), and Jij and Kij are Coulomb and exchange integrals defined by
| (4) |
| (5) |
where e is the atomic unit of charge, μ and ν label electrons, and rμν is the distance between two electrons. It will be useful to define the Coulomb operator Ji and the exchange operator Ki by
| (6) |
| (7) |
The quantity Ii in Eq. 2 is the energy of interaction of orbital i with the electric potential due to the MM charges of the other fragments, that is,
| (8) |
where A∉m denotes that the sum is over nuclei that are not in the same molecule m in which the orbital φi is centered, qA is the partial atomic charge (in a.u.) on MM atom A, and rμA is the distance between atom A and electron μ. Finally, LB represents the interaction of the nucleus of QM atom B in fragment m and the partial atomic charges qA in atoms A in other fragments as follows
| (9) |
Note that the above formalism is written for theoretical methods that treat all electrons explicitly. In semiempirical MO theory, only valence electrons are explicit, and the total number of electrons and the effective nuclear charges should be adjusted accordingly.
In practice, MOs are constructed as linear combinations of atomic basis functions
| (10) |
where χp are normalized nuclear-centered basis functions, and the normalization condition is
| (11) |
Although there is actually a different set of MOs and a different set of coefficients in each molecule m and we could write and , we omit these superscripts to keep the notation manageable. We have
| (12) |
where χ is a row vector with elements χp, and ci is a column vector with elements Cpi. Let Φ denote a row vector with elements φi; then,
| (13) |
Following Roothaan’s derivation13 of the Hartree–Fock equations, we define, for any one-electron operator M,
| (14) |
| (15) |
Then, the matrix elements of the one-electron operators can be written as
| (16) |
where the obelisk denotes a Hermitian conjugate. With the above definition, the total electronic energy of Eq. 2 can be rewritten as
| (17) |
To find the linear combinations of atomic orbitals (LCAOs) that give the minimum of Eq. 17, we vary the vectors ci by infinitesimal amounts δci and find the variation of the energy. In Eq. 17, matrices H, Jj, and Kj are independent of variations of ci; however, the QM∕MM interaction matrix I in the terms of Eq. 17 referring to molecule m depends on the ci of other molecules through the Mulliken charges on the MM atoms.
The Mulliken charge on atom A in molecule m is given by
| (18) |
where ZA is the atomic number of atom A in an all-electron treatment or the effective nuclear charge equal to the number of valence electrons on neutral atom A in a valence-electron treatment, S is the overlap matrix defined as
| (19) |
and the elements of the density matrix are
| (20) |
It is useful to introduce, for each molecule m, a NBm×NBm (where NBm is the number of basis functions on molecule m) matrix with binary elements
| (21) |
where δpq and δpλ are Kronecker deltas. The Mulliken charge on atom A can be expressed as
| (22) |
where we have defined , and where λ∊m(A) means that λ is in the same molecule m as atom A. Thus, the variation of qA is given by
| (23) |
We define the QM∕MM energy as
| (24) |
One can show that variation of EQM∕MM due to variation of the Mulliken charges on atoms in fragment m is
| (25) |
where the superscript 0 denotes that the matrix element is calculated by setting the MM charges on atom A to +e.
The variation of total electronic energy is then given by
| (26) |
Using the Hermitian property of H, I, Jj, Kj, and T(λ,p), we may rewrite Eq. 26 as
| (27) |
where superscript T denotes a transpose. We define the Fock matrix for fragment m as follows:
| (28) |
From Eq. 28, we immediately see that each fragment m is fully polarized by the rest of the system such that half of the polarization comes from the Mulliken charges of the rest of the system and the other half from charge density plus nuclei charges of the rest of the system. The variation of total electronic energy is then readily expressed as
| (29) |
We assume that the LCAOs satisfy the orthonormal condition
| (30) |
The restriction on δci resulting from the orthonormality of the MOs is obtained by varying Eq. 30, which yields
| (31) |
or
| (32) |
We multiply these restricting conditions by the Lagrangian multipliers −2εij and add them together as follows:
| (33) |
or
| (34) |
Adding Eq. 34 to the variation of total electronic energy given by Eq. 29, we obtain
| (35) |
For each fragment, the conditions for δEelec=0 for any choice of the vectors δci and , or and , are given by
| (36) |
Since εji are the elements of a Hermitian matrix,13 the two equations are equivalent:
| (37) |
Note that although the derivation above is similar to that for the Roothaan equations,13 Eq. 37 treats intermolecular interactions among fragments by a combined QM∕MM algorithm2, 3, 4, 5 in which the MM potential is derived from the corresponding molecular wave functions of other fragments.1, 7
Covalent bonding of fragments
Here, we extend the variational X-POL potential to proteins or other systems in which the QM fragments are covalently connected. The partition of a protein gives rise to one or two boundary atoms on each peptide unit connected to adjacent fragments (in general, two, but only one at the N- and C-termini). As in our previous work,1 we choose to assign two sets of hybrid orbitals (HOs) (active and auxiliary orbitals) on each boundary atom, and this boundary atom is effectively partitioned into an “active atom” and an “auxiliary atom,” each of which has half of the nuclear charge (active and auxiliary nuclei). For each fragment m, the number of electrons is equal to the number of valence electrons on non-boundary plus 2 for each boundary atom.
In the following derivation, we use the hybrid basis set, which is defined in the previous paper1 and is convenient for discussion purposes. The total electronic energy of a protein is defined as
| (38) |
in which the last term is the QM core-core repulsion energy, and Eact is the electronic energy involving only active orbitals:
| (39) |
where Nact is the number of active orbitals (equal to the number of valence orbitals on non-boundary nuclei plus 2 for each boundary atom), where we use ci to denote the coefficients that define the wave function in the hybrid basis for simplicity. To avoid double counting, all QM∕MM interactions are multiplied by 1∕2. The only exception is that orbitals on a boundary atom experience attraction from the full boundary nuclei charge when they are “active.” Consequently, they do not interact with their nuclei when they become “auxiliary” orbitals, which will be denoted a and b. The effective potential due to the auxiliary orbitals on each boundary atom is approximated by a 2×2 auxiliary charge density matrix1 so that the contribution from auxiliary orbitals can be written in the HO basis as
| (40) |
where Naux is the number of auxiliary orbitals (two or four). Note that we have used the notations a and b for auxiliary orbitals. The two-electron integrals are
| (41) |
Note that Gab does not include the interaction between auxiliary orbitals (no interaction within the MM region). Since a and b are auxiliary orbitals, Hab does not include the attraction from the corresponding boundary nuclei, which is accounted for in the fragment that a and b are treated as active orbitals.
Now, we need to find the variation of the total energy with respect to the perturbation of the MOs. To solve this problem, we first need to find the variation of the Mulliken charges of the MM atoms and the auxiliary density matrix elements. We define the transformation matrix T that transforms the MOs in the atomic basis set to the hybrid basis set
| (42) |
where superscript H denotes hybrid basis and superscript AO denotes the corresponding spherical harmonic basis. Then, the variation of Mulliken charges can be written in the hybrid basis set as
| (43) |
For each auxiliary density matrix element Pab, we define a matrix T(a,b), which connects MOs to auxiliary charge density
| (44) |
We immediately find that the T(a,b) matrix has only one nonzero element, which is . Variation of auxiliary charge density due to variation of MOs is thus
| (45) |
Consequently, the variation of electronic energy due to the interaction involving auxiliary orbitals can be written explicitly as
| (46) |
Note that the variation of active orbitals in fragment m will introduce variation of Eaux at its neighboring fragments m±1. At this point, the variation of total electronic energy can be readily written in the hybrid basis
| (47) |
where c.c. denotes complex conjugate. Following the same line of argument as in the previous derivation, we find that the Fock matrix for each fragment in the hybrid basis is
| (48) |
The optimized MOs are obtained by solving Eq. 37 in the hybrid basis.
IMPLEMENTATION
Energy
We rewrite the Fock matrix in Eq. 48 as follows:
| (49) |
with
| (50) |
Starting from an initial guess for the electron density and charges on the MM atoms, we use the DSCF algorithm14 to self-consistently variationally determine the total energy of the system by optimizing both the wave function of each individual molecular fragment in the presence of the rest of the system and the explicit polarization of the entire system. This DSCF optimization procedure is implemented here in the neglect-of-diatomic-differential-overlap15 (NDDO) approximation [which underlies such popular models as AM1 (Ref. 16) and PM3], and it involves the following steps.
-
(a)
Scale the MM charges and density matrix elements for auxiliary orbitals by 1∕2, then transform the density matrix into the AO basis [Eq. 42]. Construct F1 in the AO basis including all basis functions on the two boundary atoms and then transform it into the HO basis using the transformation matrix T. I0 can be calculated together with I. Transform I0 in the AO basis to the HO basis. Then, F2 can be calculated by adding QM core-unit charge interaction to L0 and putting the elements in the appropriate positions in the matrix. Set the auxiliary density matrix element in the HO basis to zero and transform it to the AO basis. Construct the one-electron H matrix for boundary atoms by neglecting kinetic energy and attraction of orbitals to their own nuclei and construct the two-electron G matrix using the AO basis and then transform to the hybrid basis [see Eq. 50]. Then, the F3 matrix is constructed using the elements in the transformed H and G matrices. The elements of F are then constructed by Eq. 49.
-
(b)
Diagonalize the Fock matrix F and calculate the electron density matrix. Calculate the total electronic energy associated with each fragment and check for convergence. If not converged, repeat steps (a) and (b). This represents the “inner” SCF optimization of the individual molecular wave functions.
-
(c)
When convergence is achieved, calculate the Mulliken charges for the QM fragment and build F2 and F3 for calculation on the adjacent fragments. Move to the next QM fragment and repeat the above steps.
-
(d)
After looping over all fragments, calculate the total electronic energy and test for convergence. Repeat (a), (b), and (c) until the change of the total electronic energy of the entire system satisfies a given convergence criterion. This constitutes the “system” SCF optimization of the explicit polarization of the fully interacting system.
Analytical first derivative of energy
The total energy of the system consists of the electronic energy (including, as usual, nuclear repulsion) of the QM subsystem plus the van der Waals terms (vdW) as follows
| (51) |
The first term is treated by the explicit electronic structural method discussed above, and the second term contains the vdW interactions. The latter account for the short-range exchange repulsion energy between atoms in different fragments and for long-range dispersion attractions that are omitted in the electronic energy term. The vdW energy is given by a sum of Lennard–Jones interactions as follows:
| (52) |
where standard combining rules are used to obtain the Lennard–Jones parameters from atomic parameters suchthat and σAB=(σA+σB)∕2. Computation of the energy derivatives for these semiempirical terms is straightforward.
The electronic energy of a system with atoms treated by the generalized hybrid orbital17 (GHO) approach can be written in the AO basis as
| (53) |
where density matrices P and P′ in the AO basis are calculated by multiplying the auxiliary density matrix elements by 1∕2 (to account for the 1∕2 prefactor for the auxiliary orbitals) and setting the auxiliary density matrix elements to zero (auxiliary orbitals do not interact with MM atoms because the interactions are already determined when they are treated as active orbitals), then transforming to the AO basis. The energy correction term Ecorr includes the interaction between auxiliary orbitals plus the nuclear attraction from the boundary atom where the auxiliary orbitals are located:
| (54) |
where B is a boundary atom in fragment m. corrects for the attraction from the corresponding boundary nuclei and orbital kinetic energies. In order to calculate , we first set all active density to zero and scale the auxiliary density by 1∕2, then transform the density matrix into the AO basis. Next, the two-electron matrix G is constructed using the resulting density matrix. Finally, are obtained by transforming G into the hybrid basis.
The gradient of the total electronic energy is composed of the derivative of each term in Eq. 53. For convenience of discussion, we use E1, E2, and E3 to denote QM, QM∕MM, and correction energies, which are defined by
| (55) |
Consequently,
| (56) |
where XA is the Cartesian coordinate of atom A.
The derivative of E1 can be explicitly written as follows:
| (57) |
The first term, which is the contribution from Hartree–Fock energy, is calculated in the usual fashion in the AO basis. The second term, which comes from the basis transformation of the density matrix,16 can be expressed as
| (58) |
The derivative of E2 can be calculated in the same way by replacing P with P′ and 2H+G with 2I.
Similarly, the derivative of correction energy E3 can be divided into a Hartree–Fock contribution and the contribution from the derivative of the transformation matrix. The Hartree–Fock contribution can be evaluated by calculating the force between two boundary atoms using standard methods except that the density matrix in the AO basis is obtained by the transformation of density matrix in the HO basis with active density matrix elements set to zero and auxiliary density matrix elements multiplied by 1∕2. The contribution from the derivative of transformation matrix can be evaluated in the same way as for E1.
RESULTS AND DISCUSSION
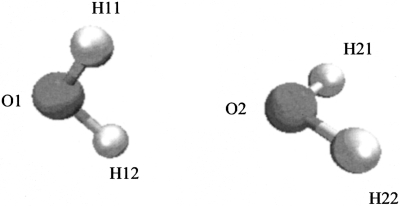
We have implemented the variational X-POL potential in the CHARMM (Ref. 18) package (version c33al). In the current implementation of variational X-POL, Austin Model 1 (AM1) (Ref. 16) is chosen as the QM method and Mulliken charges are used to approximate the electrostatic potential for QM∕MM interactions. Standard Lennard–Jones parameters in the CHARMM force field18 are used for amino acids and the values reported in Ref. 8 are adopted for water.8 The validity and implementation of the variational X-POL potential were tested for a water dimer (see Fig. 1). In a subsequent paper, we present results from molecular dynamics (MD) simulation of a solvated protein employing the X-POL potential. Table 1 lists the Mulliken charges calculated by AMI by using the self-consistent but not variational X-POL potential,1, 7 and by using the variational X-POL potential using the same, fixed input Cartesian coordinates. In X-POL calculations, each water molecule is defined as a QM fragment. All three calculations give similar Mulliken charges with the largest deviation being 0.01.
Figure 1.
Atom numbers assigned to the water dimer with each of the monomers treated as a QM fragment.
Table 1.
Mulliken population charges for the water dimer from X-POL. AM1 denotes a full AM1 calculation for the water dimer, SX-POL denotes the self-consistent but not variational X-POL potential. X-POL denotes the variational result obtained using the equations of the present article. Cartesian coordinates (in Å) used for the water dimer are O1(−6.302 51,2.127 23,0.823 74), H11(−6.891 82,2.556 39,1.444 04), H12(−5.736 54,1.587 07,1.375 23), O2(−5.610 42,3.122 60,−1.661 72), H21(−6.136 34,2.539 42,−2.209 02), and H22(−5.804 20,2.830 43,−0.771 04).
| Atom | AM1 | SX-POL | X-POL |
|---|---|---|---|
| O1 | −0.4008 | −0.4056 | −0.4095 |
| H11 | 0.2056 | 0.2028 | 0.2047 |
| H12 | 0.2056 | 0.2028 | 0.2047 |
| O2 | −0.4104 | −0.4001 | −0.4109 |
| H21 | 0.1840 | 0.1873 | 0.1852 |
| H22 | 0.2160 | 0.2129 | 0.2257 |
Table 2 shows excellent agreement of analytic and numerical forces for the water dimer. Since the variational and nonvariational X-POL potentials give very similar Mulliken charges, it is not surprising that forces calculated by finite differences are very similar for both potentials.
Table 2.
Analytic and numerical forces (kcal mol−1 Å−1) for the water dimer calculated by X-POL. Anal. denotes analytic gradient, Numer. denotes forces calculated by finite differences from displacements of atom positions, and Diff. denotes the deviation of analytic gradient from the numerical one. SX-POL and X-POL are defined in Table 1. Cartesian coordinates for the water dimer are also given in Table 1.
| SX-POL | X-POL | ||||
|---|---|---|---|---|---|
| Dim. | Atom | Numer. | Anal. | Numer. | Diff. |
| X | O1 | 1.939 851 | 1.909 172 | 1.909 175 | −0.000 003 |
| Y | O1 | 2.290 872 | 2.253 172 | 2.253 176 | −0.000 004 |
| Z | O1 | 0.029 756 | 0.056 810 | 0.056 811 | −0.000 001 |
| X | H11 | 2.678 915 | 2.671 901 | 2.671 899 | 0.000 002 |
| Y | H11 | −1.948 362 | −1.939 838 | −1.939 839 | 0.000 001 |
| Z | H11 | −3.559 570 | −3.536 848 | −3.536 849 | 0.000 001 |
| X | H12 | −2.623 117 | −2.611 412 | −2.611 413 | 0.000 001 |
| Y | H12 | −2.501 712 | −2.494 665 | −2.494 663 | 0.000 002 |
| Z | H12 | −3.246 610 | −3.224 742 | −3.224 742 | 0.000 000 |
| X | O2 | −5.501 806 | −5.510 548 | −5.510 546 | −0.000 002 |
| Y | O2 | −7.480 413 | −7.492 851 | −7.492 850 | −0.000 001 |
| Z | O2 | −13.727 177 | −13.757 475 | −13.757 482 | −0.000 007 |
| X | H21 | 1.798 564 | 1.798 540 | 1.798 539 | 0.000 001 |
| Y | H21 | 2.002 657 | 2.002 396 | 2.002 397 | 0.000 000 |
| Z | H21 | 1.760 802 | 1.764 064 | 1.764 064 | 0.000 000 |
| X | H22 | 1.707 371 | 1.742 347 | 1.742 346 | 0.000 001 |
| Y | H22 | 2.633 766 | 2.682 457 | 2.682 455 | 0.000 001 |
| Z | H22 | −8.711 621 | −8.816 759 | −8.816 765 | 0.000 006 |
It is instructive to compare the present method to some previous approaches. For example, one can compare the present method to the NDDO fragment SCF method of Ferenczy et al.19 in which a system is divided into a subsystem (S) and an environment (E) with a block diagonal Fock matrix. The environmental orbitals are obtained from a zeroth-order calculation similar to our first SCF iteration. In the fragment SCF method, the density matrix of the environment and the electronic integrals of both the subsystems and the environment are used to construct the final Fock matrix of the subsystem. This is analogous to the second iteration of the subsystem in X-POL, but in the fragment SCF method, the self-consistent subsystem is not rotated through the entire system to make the whole system self-consistent.19 In the fragment SCF method, one would not get an analytic gradient if one switches S and E because the method is not variational. A second fundamental difference between these methods is that X-POL uses combined QM∕MM methodology where the MM representation is obtained from a self-consistent QM calculation. The MM representation of the environment, which has also been employed in the local SCF approximation20 and the generalized hybrid orbital method17, 21, 22 (which have some similarities to the present method in the treatment of a QM system with its environment), makes the method efficient for large molecules. On the other hand, all electronic integrals are explicit computed in the fragment SCF calculation.19 In a different fragment orbital approach in Ref. 11, the fragmental electronic structures were iteratively converged, but the system energy was not obtained variationally. The use of QM∕MM methodology also distinguishes the present method from the divide-and-conquer procedure of Lee et al., which constructs a fully antisymmetric wave function for the entire system along with explicit evaluation of all one- and two-electron integrals.23
Another possible comparison is to polarized MM, which is reviewed24, 25 elsewhere. Empirical treatments of molecular polarization are not unique, giving rise to various representations and functional forms of polarization energy. Notable examples include the atomic interacting point dipole method,26, 27, 28 the shell model,29 and the chemical potential equalization approach employing fluctuating charges.30, 31 The present explicit polarization method treats molecular polarization, which is a property of the electronic structure, by a wave-function approach.1 In MM, it is very difficult, if not impossible, to include charge transfer energy terms, whereas some charge transfer components are already incorporated in the present X-POL potential through interactions via the boundary orbitals.1 Charge transfer, when its explicit consideration becomes necessary or desirable,32, 33, 34, 35 can be conveniently modeled by the X-POL potential by including the charge transfer partners in the same fragment. The X-POL method also has the advantage over polarized MM that the QM treatment employed in X-POL can be applied to bond breaking and chemical reactions for which polarized MM is inapplicable.34, 35
CONCLUSION
We have presented a variational methodology to be employed in a new generation force field, called X-POL, to be ultimately used for complex problems such as protein simulations. The methodology involves using electronic structure theory and the combined QM∕MM method in conjunction with a variational principle. The system is partitioned into fragments, and interactions within each fragment are treated using electronic structure theory, while interactions between fragments are treated using a combined QM∕MM method. The semiemprical AM1 model is chosen as the QM method in our current implementation, although the theory and future work are not restricted to semiempirical models and can be applied to ab initio molecular orbital and density function theory. Mulliken charges are used in the present implementation to generate the electrostatic potential for calculating QM∕MM interactions, although dipoles and multipolar terms could be included straightforwardly for systems in which anisotropic polarization beyond that which can be adequately modeled by distributed monopoles is a concern. Because the wave function of the system is variationally optimized, the gradient of the electronic energy can be calculated analytically, which provides a stable and efficient method for calculating the forces needed for MD simulations. Tests for a water dimer show that the methodology presented here gives correct analytic forces in the current implementation.
Although the X-POL method is presented and named here in light of the previously stated goal1 of developing a next-generation force field, using direct electronic structure methods to go beyond MM, it will sometimes be convenient to distinguish the underlying QM method from the X-POL force field. In that case, the underlying method may be classified as a combined QM∕QM method to distinguish it from combined QM∕MM methods, and we have used a double self-consistent-field (DSCF) method to optimize the wave function.
Key elements in the present implementation of the X-POL and DSCF algorithms are the GHO boundary and the interaction of a given fragment with its environment by a QM∕MM formalism. Although the GHO method was originally developed at the NDDO level,17, 21 it was later extended to ab initio Hartree–Fock theory,36 to a tight-binding density functional theory including overlap,37 and to density functional theory.22 The DSCF optimization technique presented here at the NDDO level, can also be extended to higher levels of theory and used in various contexts.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health (GM46736), the Office of Naval Research under Grant No. N00012-05-01-0538, and the National Science Foundation under Grant No. CHE07-04974.
References
- Xie W. and Gao J., J. Chem. Theory Comput. 10.1021/ct700167b 3, 1890 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Rev. Comput. Chem. 7, 119 (1995). [Google Scholar]
- Warshel A. and Karplus M., J. Am. Chem. Soc. 10.1021/ja00771a014 94, 5612 (1972). [DOI] [Google Scholar]
- Warshel A. and Levitt M., J. Mol. Biol. 10.1016/0022-2836(76)90311-9 103, 227 (1976). [DOI] [PubMed] [Google Scholar]
- Field M. J., Bash P. A., and Karplus M., J. Comput. Chem. 10.1002/jcc.540110605 11, 700 (1990). [DOI] [Google Scholar]
- Mulliken R. S., J. Chem. Phys. 10.1063/1.1740588 23, 1833 (1955). [DOI] [Google Scholar]
- Gao J., J. Phys. Chem. B 10.1021/jp962833a 101, 657 (1997). [DOI] [Google Scholar]
- Gao J., J. Chem. Phys. 10.1063/1.476802 109, 2346 (1998). [DOI] [Google Scholar]
- Wierzchowski S. J., Kofke D. A., and Gao J., J. Chem. Phys. 10.1063/1.1607919 119, 7365 (2003). [DOI] [Google Scholar]
- Pople J. A., Krishnan R., Schlegel H. B., and Binkley J. S., Int. J. Quantum Chem., Quantum Chem. Symp. 13, 225 (1979). [Google Scholar]
- Kitaura K., Sugiki S., Nakano T., Komeiji Y., and Uebayasi M., Chem. Phys. Lett. 10.1016/S0009-2614(01)00099-9 336, 163 (2001). [DOI] [Google Scholar]
- Pulay P., Mol. Phys. 10.1080/00268976900100941 17, 197 (1969). [DOI] [Google Scholar]
- Roothaan C. C. J., Rev. Mod. Phys. 10.1103/RevModPhys.23.69 23, 69 (1951). [DOI] [Google Scholar]
- Gao J., J. Comput. Chem. 18, 1062 (1997). [Google Scholar]
- Pople J. A., Santry D. P., and Segal G. A., J. Chem. Phys. 10.1063/1.1701475 43, S129 (1965). [DOI] [Google Scholar]
- Dewar M. J. S., Zoebisch E. G., Healy E. F., and Stewart J. J. P., J. Am. Chem. Soc. 10.1021/ja00299a024 107, 3902 (1985). [DOI] [Google Scholar]
- Amara P., Field M. J., Alhambra C., and Gao J., Theor. Chem. Acc. 10.1007/s002140000153 104, 336 (2000). [DOI] [Google Scholar]
- Brooks B. R., Bruccoleni R. E., Olafson B. D., States D. J., Swaminathan S., and Karplus M., J. Comput. Chem. 10.1002/jcc.540040211 4, 187 (1983). [DOI] [Google Scholar]
- Ferenczy G. G., Rivail J.-L., Surján P. R., and Náray-Szabo G., J. Comput. Chem. 10.1002/jcc.540130706 13, 830 (1992). [DOI] [Google Scholar]
- Thery V., Rinaldi D., Rivail J.-L., Maigret B., and Ferenczy G. G., J. Comput. Chem. 10.1002/jcc.540150303 15, 269 (1994). [DOI] [Google Scholar]
- Gao J., Amara P., Alhambra C., and Field M. J., J. Phys. Chem. A 10.1021/jp9809890 102, 4714 (1998). [DOI] [Google Scholar]
- Pu J., Gao J., and Truhlar D. G., ChemPhysChem 10.1002/cphc.200400602 6, 1853 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-S., Lewis J. P., and Yang W., Comput. Mater. Sci. 10.1016/S0927-0256(98)00029-9 12, 259 (1998). [DOI] [Google Scholar]
- Rick S. W. and Stuart S. J., Rev. Comput. Chem. 10.1002/0471433519.ch3 18, 89 (2002). [DOI] [Google Scholar]
- Ponder J. W. and Case D. A., Adv. Protein Chem. 10.1016/S0065-3233(03)66002-X 66, 27 (2003). [DOI] [PubMed] [Google Scholar]
- Vesely F. J., J. Comput. Phys. 10.1016/0021-9991(77)90028-6 24, 361 (1977). [DOI] [Google Scholar]
- Thole B. T., Chem. Phys. 10.1016/0301-0104(81)85176-2 59, 341 (1981). [DOI] [Google Scholar]
- Gao J., Habibollahzadeh D., and Shao D., J. Phys. Chem. 10.1021/j100044a039 99, 16460 (1995). [DOI] [Google Scholar]
- Sprik M. and Klein M. L., J. Chem. Phys. 10.1063/1.455722 89, 7556 (1988). [DOI] [Google Scholar]
- Rappé A. K. and W. A.GoddardIII, J. Phys. Chem. 10.1021/j100161a070 95, 3358 (1991). [DOI] [Google Scholar]
- York D. M. and Yang W., J. Chem. Phys. 10.1063/1.470886 104, 159 (1996). [DOI] [Google Scholar]
- Van der Vaart A. and K. M.Merz, Jr., J. Am. Chem. Soc. 121, 9180 (1999). [Google Scholar]
- Mo Y. and Gao J., J. Phys. Chem. B 110, 2916 (2006). [Google Scholar]
- Gao J., Ma S., Major D. T., Nam K., Pu J., and Truhlar D. G., Chem. Rev. (Washington, D.C.) 10.1021/cr050293k 106, 3188 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenich A. V., Olson R. M., Chamberlin A., Cramer C. J., and Truhlar D. G., J. Chem. Theory Comput. 3, 2055 (2007). [DOI] [PubMed] [Google Scholar]
- Pu J., Gao J., and Truhlar D. G., J. Phys. Chem. A 10.1021/jp036755k 108, 632 (2004). [DOI] [Google Scholar]
- Pu J., Gao J., and Truhlar D. G., J. Phys. Chem. A 10.1021/jp049529z 108, 5454 (2004). [DOI] [Google Scholar]