Abstract
The oncogene HER2 is overexpressed in a variety of human tumors, providing a target for anti-cancer molecular therapies. Here we employed a 2’-O-methoxyethyl (MOE) splice switching oligonucleotide, SSO111, to induce skipping of exon 15 in HER2 pre-mRNA, leading to significant downregulation of full-length HER2 mRNA, and simultaneous upregulation of Δ15HER2 mRNA. SSO111 treatment of SK-BR-3 cells, which highly overexpress HER2, led to inhibition of cell proliferation and induction of apoptosis. The novel Δ15HER2 mRNA encodes a soluble, secreted form of the receptor. Treating SK-BR-3 cells with exogenous Δ15HER2 protein reduced membrane-bound HER2 and decreased HER3 transphosphorylation. Δ15HER2 protein thus has similar activity to an autoinhibitory, natural splice variant of HER2, Herstatin, and to the breast cancer drug Herceptin. Both SSO111 and Δ15HER2 may be potential candidates for the development of novel HER2-targeted cancer therapeutics.
Keywords: HER2, splice switching oligonucleotides, alternative splicing, apoptosis
Introduction
HER2/ErbB2, a member of human EGFR (epidermal growth factor receptor) family of transmembrane receptor tyrosine kinases, is an oncogene amplified and/or overexpressed in many human malignancies, especially in 25–30% breast cancers.1 HER2 overexpression correlates with enhanced tumor aggressiveness and decreased patient survival.2 Unlike the other three family members (EGFR/HER1, HER3 and HER4), HER2 has no known high affinity ligands,3 and is constitutively active as a homodimer in HER2-overexpressing cells.4 EGFR/HER1, HER3, or HER4 preferentially recruit HER2 upon ligand binding to form heterodimers and become transactivated, which leads to downstream signaling, primarily through mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathways.5 HER2-containing heterodimers, especially HER2•HER3, are the most efficient in ligand binding and signal transduction among EGFR dimers and crucial in mediating cell proliferation and survival in HER2 positive tumors.6 Given its vital role in EGFR family signaling and tumorigenesis, HER2 is an important target for cancer therapeutics. Current approaches include humanized monoclonal antibodies against HER2 extracellular domain (ECD) and tyrosine kinase inhibitors targeting the intracellular kinase domain.7 Successful examples are Herceptin/Trastuzumab,8 the first FDA-approved HER2-targeted therapeutic for HER2 positive breast cancer, and lapatinib.9 Due to development of drug resistance,10 common in cancer, and limitations of existing therapeutics,11 new strategies are still being developed.12–15
Work from this and other laboratories showed that alternative splicing not only increases proteome complexity, but also serves as a target for molecular therapies.16–19 Splice switching oligonucleotides (SSOs) can be used to manipulate alternative splicing pathways by hybridizing to pre-mRNA sequences important for splicing, thereby altering the expression of targeted genes, both in cell culture and in vivo.20 SSOs can repair aberrant splicing,21, 22 or alter normal splicing to abrogate the expression of diseasecausing genes, and even lead to production of autoinhibitors.23 Two HER2 splice variants arising from intron retention have been reported, both being soluble dominant negative inhibitors of HER2, which likely interfere with receptor dimerization.24, 25 Herstatin, generated by retention of intron 8, has been recognized as a promising anticancer drug candidate.26 Here we show that SSO-induced skipping of exon 15 in HER2 pre-mRNA results in significant reduction of full length, membrane bound HER2 protein, and concomitant apoptosis of HER2-overexpressing SK-BR-3 breast cancer cells. Importantly, the protein encoded by the novel HER2 mRNA lacking exon 15, Δ15HER2, was a HER2 inhibitor that downregulated full-length HER2 protein and reduced transphosphorylation of HER3.
Materials and Methods
Cell lines
SK-BR-3 cells were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum. MCF7 cells were cultured in modified essential medium supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate (Life Technologies, Inc.) and 0.1 mM nonessential amino acids (Sigma). Twenty-four hours prior to oligonucleotide or plasmid treatment, the cells were plated in 2 ml of medium in 6-well plates at a density of 3 × 105 cells/well, or in 1 ml of medium in 24-well plates at a density of 1 × 105 cells/well.
Oligonucleotide Treatment
2’-O-(2-Methoxyethyl)- or 2’-MOE-modified oligoribonucleotide phosphorothioate 20-mer SSO111 (GACCGTTGGACTCACGAGTG) antisense to the 5’ splice site of HER2 exon 15 (underline, targeting exon 15; non-underline, targeting intron 15) was used. A mismatched, SSO111m (GACCATTCGACACACCAGTG; mismatches underlined) and unrelated SSO (ctrl), with anti-β-globin sequence (GCTATTACCTTAACCCAG) 27 was used as a negative control. All oligonucleotides were synthesized and purified by IDT, Inc. (Coralville, IA). The cells were treated with oligonucleotides complexed with Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s directions at the concentrations indicated on the figures. Analyses were carried 24–72 hours after transfection.
Plasmid construct and purification of Δ15HER2-His protein
The Δ15HER2 sequence was reverse-transcribed and amplified from the total RNA isolated from SK-BR-3 cells treated with SSO111 for 24 hours. The forward and reverse primers used were CACCATGGAGCTGGCGGCCT and TCCAGGTCCACACAGCGGTCC, respectively. The Δ15HER2 sequence was cloned into the pcDNA™3.1, a directional TOPO expression vector (Invitrogen), which encodes six histidine residues at the carboxy terminus of the expressed protein. The Δ15HER2-His expression plasmid was transfected into MCF-7 cells with Lipofectamine™ 2000 (Invitrogen) in serum-free medium. After 48 hours, the medium was collected, concentrated, purified with HisPur™ Cobalt spin columns (Pierce), and desalted using Zeba™ Desalt spin columns to yield the soluble Δ15HER2-His protein. Purity of the protein was confirmed by SDS-PAGE and SimplyBlue™ (Invitrogen) staining, and the yield was determined by Bradford Assay.
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Cells were lysed in 800 µL of TRI-reagent and total RNA was isolated as suggested by the supplier (Molecular Research Center, Inc.). For RT-PCR, the reverse transcription (RT) mixture containing 200 ng of RNA, the reverse primer only and the rTth polymerase (PerkinElmer Life Sciences), was incubated at 70°C, 15min; PCR was carried out after addition of the forward primer and 0.02mM Cy5-AP3-dCTP (GE Healthcare) at: 95°C, 3 min, 1 cycle; 22 cycles of 95°C for 30 sec, 56 °C for 30 sec, 72°C for 1 min, plus a final extension at 72°C for 7 min. The forward and reverse primers were 5’-GCTCCCCAGGGAGTATGTGAATGC-3’ and 5’-CAGAGGGCTGGCTCTCTGCTCG-3’, respectively. The PCR products (exon15-included: 307bp; exon15-skipped: 146bp) were separated on 10% non-denaturing polyacrylamide gels (Invitrogen). Bands were visualized on Typhoon™ Variable Mode Imager (GE Healthcare), and quantified with ImageQuant™ software (GE Healthcare). Yield calculations of Δ15HER2 mRNA reflect the fact that the number of Cy5-labeled cytidine nucleotides in the exon15-included band is 2.3 times higher than that in the Δ15-band. Thus, the percent of correction is higher than appears from the autoradiograms.
Western blot analysis and immunoprecipitation
Cells were washed twice with PBS and lysed in RIPA buffer (radioimmune precipitation assay buffer; 50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate) and a mixture of protease inhibitors (Sigma). Total protein (20 µg) was separated by electrophoresis on a 4–10% pre-cast Bis-Tris gel (Invitrogen) and electrotransferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked for 30 min in StartingBlock (PBS) blocking buffer (Pierce) and incubated overnight at 4°C with different primary antibodies (all at 1:4000 dilution): rabbit erbB2 polyclonal antibody (Abcam), mouse PARP monoclonal antibody (Invitrogen), rabbit erbB3 polyclonal antibody (Abcam), rabbit phospho-HER2/erbB2 (Tyr877) polyclonal antibody (Cell Signaling), rabbit phospho-HER3/erbB3 (Tyr1289) monoclonal antibody (Cell Signaling), rabbit Akt polyclonal antibody (Cell Signaling), rabbit phospho-Akt(Ser473) polyclonal antibody (Cell Signaling), or mouse β-actin monoclonal antibody (Sigma). After 1-hour incubation with horseradish peroxidase-conjugated goat-anti-rabbit (1:100,000 dilution; Abcam) or goat-anti-mouse (1:100,000 dilution; Invitrogen) secondary antibodies, blots were developed with ECL™ Plus reagents (GE Healthcare) and exposed to Kodak film. HER2, HER3, full-length PARP, cleaved PARP, Akt, and β-actin migrated at ~180, 185, 116, 85, 60, 42 kDa, respectively. β-actin was used as a loading control.
For immunoprecipitation, cells were lysed in NP-40 buffer (50 mM Tris-HCl, 100 mM NaCl, 1% nonidet-P40, 10% glycerol) supplemented with protease inhibitors. Total protein (0.5–1 mg) was incubated with the indicated antibody for 1 hour with shaking at 4°C, and with 20 µl of protein A/G plus-agarose immunoprecipitation reagent (Santa Cruz) at 4°C overnight. The immune complexes were washed three times with cold PBS, resuspended in electrophoresis sample buffer, and analyzed by Western blots.
Cell viability assay
Cells (~2×104/well) were plated in 96-well plates 24 hours prior treatment, and treated in triplicates with 100 nM of the indicated SSOs and Lipofectamine™ 2000 (Invitrogen), or purified Δ15HER2-His protein at the designated concentrations. After 72 hours, CellTiter 96® AQueous One Solution reagent (Promega) was added into each well of the 96-well plate, and incubated at 37°C for 1–4 hours. The absorbance, which is proportional to the number of viable cells, was recorded at 490nm using a 96-well plate reader. Cell viability was normalized to mock-treated cells.
Results
Skipping of HER2 exon 15 in SSO-treated SK-BR-3 cells
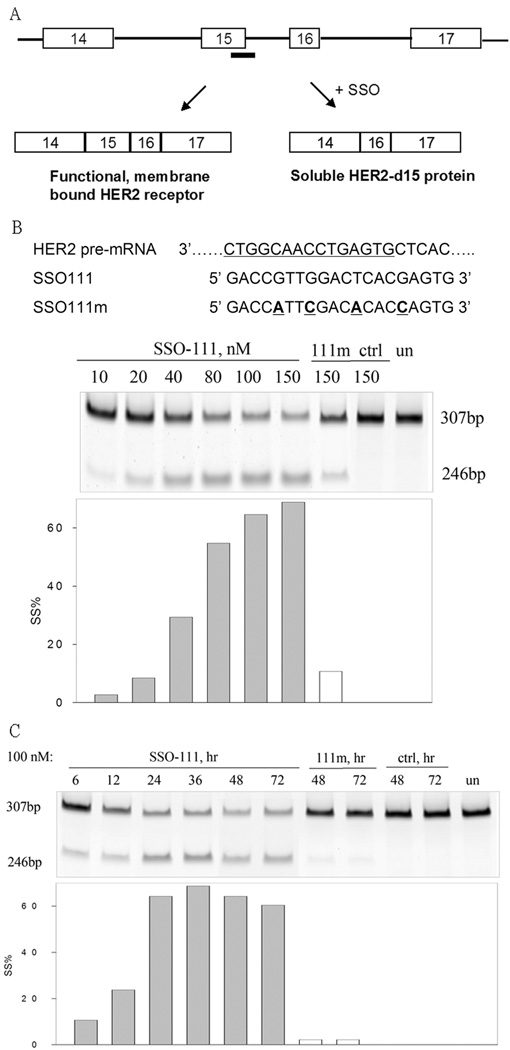
Compared to intron retention, needed for induction of the natural splice variant Herstatin, exon skipping is more amenable to SSO technology. Therefore, we analyzed the HER2 gene sequence and exon/intron organization, and reasoned that SSO-induced skipping of exon 15 would lead to downregulation of mRNA coding full length, membrane bound HER2 and simultaneous upregulation of Δ15HER2 mRNA. To test this hypothesis, HER2-overexpressing SK-BR-3 breast cancer cells were transfected with SSO111, a 20-mer 2’-O-MOE phosphorothioate oligonucleotide targeted to the 5’ splice site of exon 15 in HER2 pre-mRNA (Fig. 1A). The 2’-O-MOE modification was chosen because it prevents RNase H-mediated degradation of targeted pre-mRNA, improves SSO binding affinity, and enhances nuclease resistance and pharmacological activity of the SSO.28, 29
Figure 1.
Skipping of exon 15 in HER2 pre-mRNA. A) SSOs were designed to target the 5’ splice site of exon 15. Skipping of exon 15 induced by SSOs generated a downstream stop codon, and results in a truncated HER2 ECD. Thick bar: SSO. Part of HER2 pre-mRNA (3’ to 5’, with intron underlined), SSO111, and SSO111m (with mismatches underlined and highlighted in bold) were shown in B). B) and C) Results from the analysis of total cellular RNA by RT-PCR are shown: B) Dose-dependence of SK-BR-3 cells treated with SSO111 at designated concentrations for 24 hours; C) Time-course of Sk-BR-3 cells treated with SSO111 for the designated time at 100 nM. The lengths of the PCR products (in base pairs) are indicated. Lower panels in B) and C): quantitative analysis of the results. Gray bars: SSO111 transfected cells; white bars: 111m-transfected cells.
RT-PCR analysis of total cellular RNA 24-hour post-treatment showed that SSO111 induced a dose-dependent skipping of exon 15, as indicated by an increase in the ratio of Δ15HER2 to full length mRNA (Fig. 1B). With 100 nM SSO111, Δ15HER2 mRNA reached ~65% of the total. The exon-skipping effect was sequence-specific, as SSO111m, with four mismatches, had a significantly reduced activity, and a globin-targeted SSO (ctrl), had no effect (Fig. 1B). Maximal skipping of exon 15 by SSO111 occurred 36 hours post-transfection and persisted for at least 72 hours (Fig. 1C).
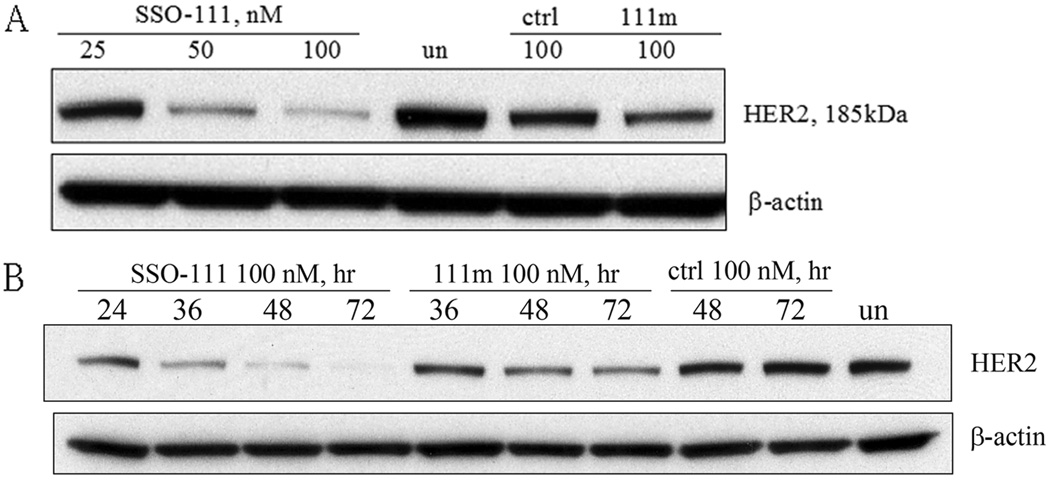
Analysis of total cellular protein by immunoblot with an N-terminal HER2 antibody showed that, consistent with the decrease of HER2 mRNA as shown in RT-PCR results, SSO111 reduced full length HER2 protein in a dose (Fig. 2A) and time (Fig. 2B) dependent manner. At 100 nM, SSO111 treatment reduced HER2 protein by 50% and 95%, at 24 and 72 hours post transfection, respectively. As expected, treatment with control SSOs had no or minimal effect on HER2 expression.
Figure 2.
Western blot analysis of HER2 protein from SK-BR-3 cells transfected with SSO111. A) dose dependence (at 48 hours). B) time course (at 100 nM). Top panels: HER2 protein; lower panels: β-actin as a loading control. See “Methods and Materials” for details.
Growth inhibition and apoptosis of SK-BR-3 cells caused by SSO111
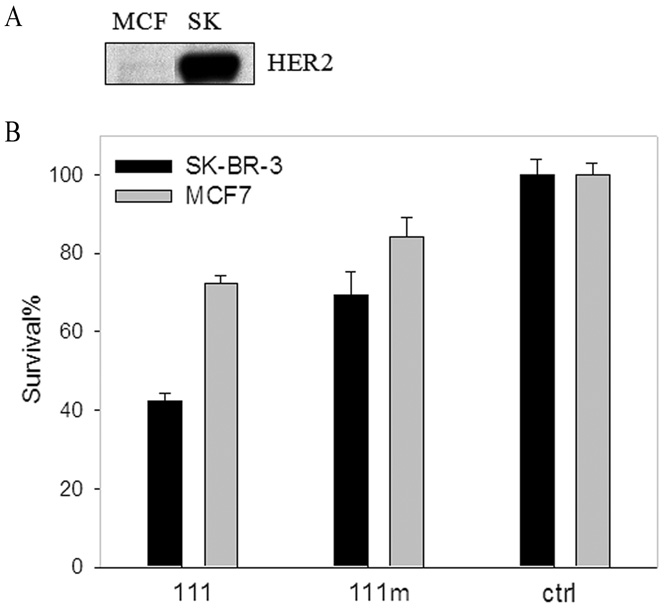
Since survival of SK-BR-3 cells is dependent on HER2 overexpression, we hypothesized that SSO111 treatment may lead to cell death much more readily than MCF-7 cells, which express low levels of HER2 (Fig. 3A).30, 31 Indeed, 72-hour incubation with 100 nM SSO111 inhibited survival of SK-BR-3 cells by ~60%, compared to only 25% in MCF7 cells (Fig. 3B), relative to control SSO. Mismatch SSO111m had a slight effect, which was again more pronounced in SK-BR-3 than in MCF7 cells (Fig. 3B). It is worth noting that SSO111 did induce exon 15 skipping in MCF7 cells (data not shown). Therefore, the lack of SSO111 effects in MCF7 cells was not due to poor uptake of the SSO into those cells and/or its poor splice switching efficiency, and confirmed that SSO111-induced cell growth inhibition is sequence and HER2 level dependent.
Figure 3.
Growth inhibition of SK-BR-3 cells by SSO111 treatment. A) Western blot demonstrating relative HER2 expression in MCF7 and SK-BR-3 cell lines (20 µg protein/lane). B) MTS assay of SK-BR-3 (black bar) and MCF7 (gray bar) cells after 72 hours incubation with 100 nM SSO111. Shown are the mean ± standard deviation of triplicates.
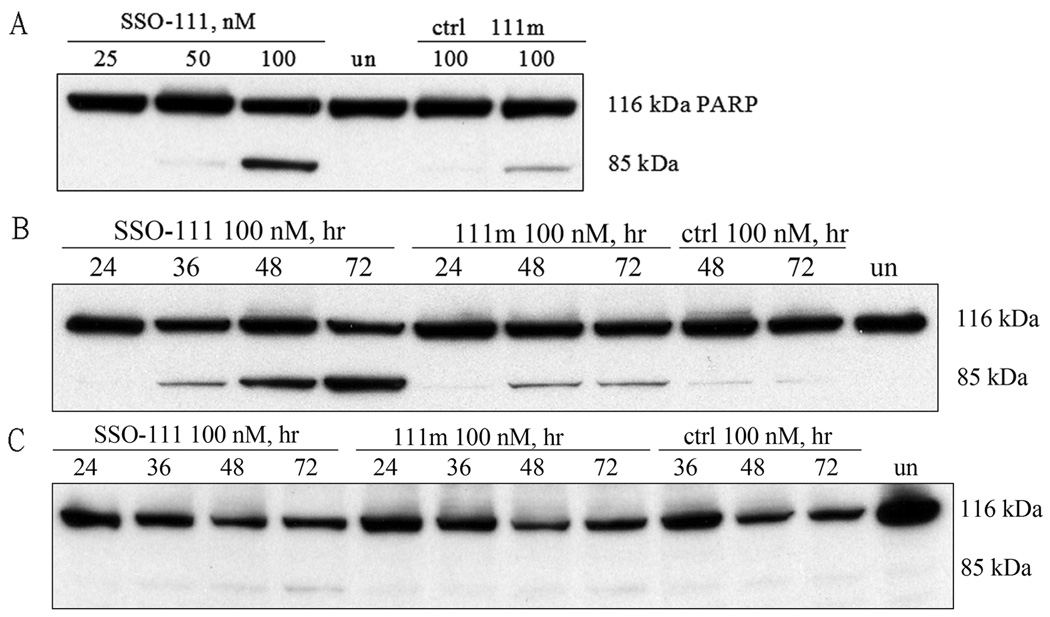
We found also that the cytotoxic effect of SSO111 on SK-BR-3 cells was due to its ability to induce apoptosis. SSO111 treatment showed sequence specific, dose (Fig. 4A) and time (Fig. 4B) dependent induction of PARP cleavage, an early marker for apoptosis. In contrast, SSO111 induced only minimal PARP cleavage in MCF7 cells (Fig. 4C), again demonstrating its HER2-dependence in SK-BR-3 cells.
Figure 4.
PARP cleavage as a marker for apoptosis after SSO111 treatment. In SK-BR-3 cells: A) dose dependence (at 48 hours); B) time course (at 100 nM). In MCF7 cells: C) time course (at 100 nM). Full-length PARP: 116 kDa; cleavage product: 85 kDa.
Soluble Δ15HER2-His protein downregulates HER2 protein and inhibits tyrosine phosphorylation of HER3
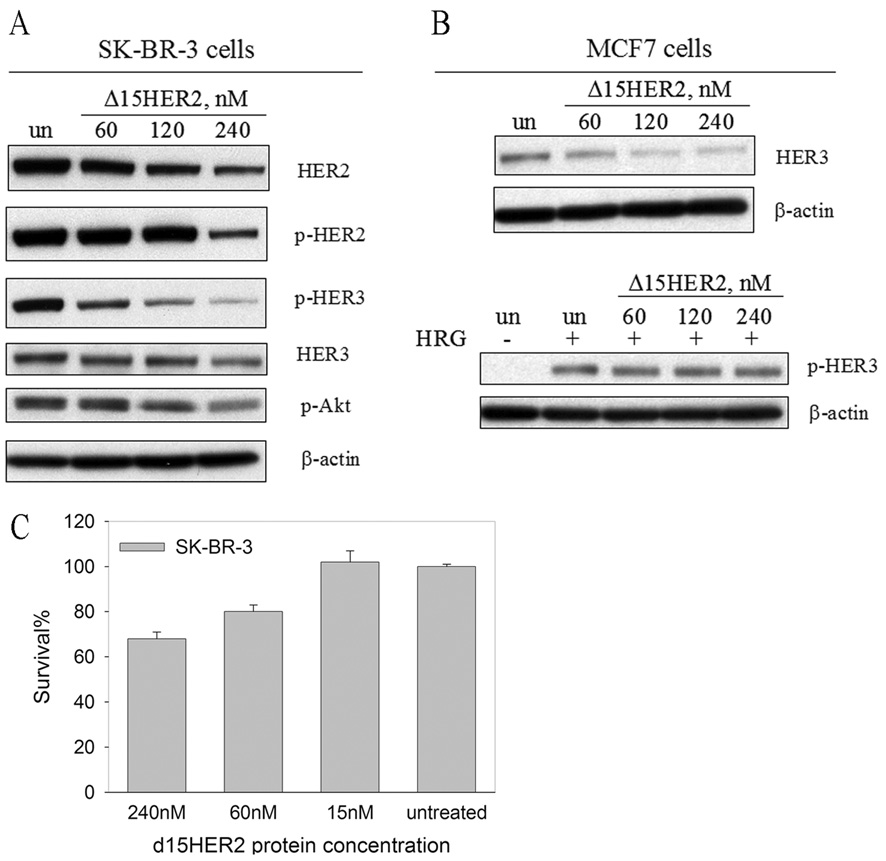
SSO111 led to generation of the novel Δ15HER2 mRNA. Δ15HER2 should encode a soluble secreted protein, since skipping of exon 15 in HER2 pre-mRNA generates a stop codon in exon 16 and truncates the protein before the transmembrane exon 17. Retaining most of the HER2 ECD sequence (579 out of ~649 amino acids), Δ15HER2 protein may be able to associate with HER2 and HER3, as does the full HER2 ECD,32 form unproductive dimers, and interfere with HER2-mediated signal transduction. To test this hypothesis, we generated Δ15HER2 cDNA from the total RNA of SK-BR-3 cells after SSO111 treatment, cloned, expressed and purified a C-terminal 6×His tag version in MCF-7 cells. This allowed for protein expression without the induction of HER2 dependent apoptosis. The purified Δ15HER2 protein was added, at 60, 120 or 240 nM, to the culture media of SK-BR-3 cells; after 48 hours incubation, the cells were analyzed by Western blot for HER2, HER3, and their phosphorylation status. The data (Fig. 5A) show that increasing concentrations of Δ15HER2-His protein decreased total HER2 protein in the cells by up to 80%; phosphorylated HER2 was slightly affected at 60 and 120 nM, and was decreased by ~80% with 240 nM Δ15HER2-His. In agreement with the established importance of HER2 in HER3 phosphorylation in SK-BR-3 cells, we found that phosphorylated HER3 decreased in a dose-dependent manner after treatment with Δ15HER2-His protein in parallel with HER2 protein. In contrast, total HER3 was essentially unaffected by Δ15HER2-His (See below and the Discussion for interpretation of these results). To examine whether downstream PI3K signaling pathway was affected, we analyzed the phosphorylation level of Akt after heregulin stimulation. Preincubation of SK-BR-3 cells with Δ15HER2-His protein resulted in a dose-dependent reduction of phosphorylated Akt (Fig 5A), suggesting the PI3K pathway activation was inhibited.
Figure 5.
Effects of Δ15HER2-His protein on HER2/HER3 receptors. Western blot analysis of (A) SK-BR-3 and (B) MCF7 cell lysates after treatment with purified Δ15HER2-His protein at the designated concentrations. For analysis of phosphorylated Akt (p-Akt) in A) and phosphorylated HER3 (p-HER3) protein in B), cells were stimulated with heregulin (15 min, 20 ng/ml). C) Growth inhibition of SK-BR-3 cells by Δ15HER2-His protein treatment after 72 hours incubation analyzed by MTS assay. Shown are the mean ± standard deviation of triplicates.
To investigate the effect of Δ15HER2-His protein on HER3 without the influence of HER2, exogenous Δ15HER2-His protein was added to MCF7 cell culture. In MCF7 cells, basal phosphorylated HER3 levels are low, so heregulin was added to activate HER3 tyrosine phosphorylation, via dimerization with low levels of HER4. The Δ15HER2-His protein had only a minimal effect on HER3 phosphorylation (Fig. 5B), suggesting its inability to inactivate HER4-mediated transactivation. However, in stark contrast to SK-BR-3 cells, total HER3 protein was decreased by up to 70% after Δ15HER2-His protein treatment, suggesting that Δ15HER2-His can bind monomeric HER3 (see Fig. 5B and Discussion).
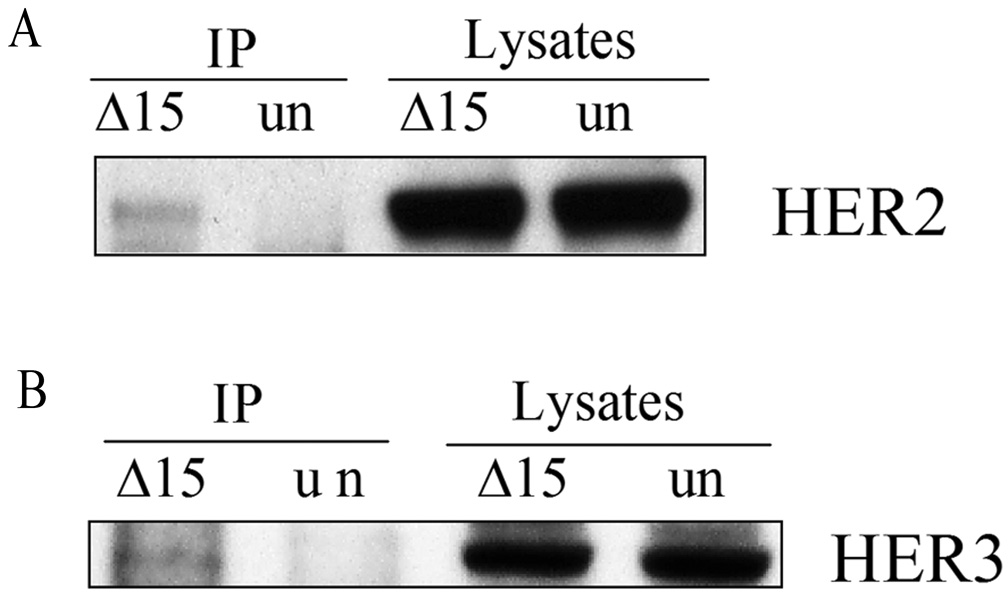
Importantly, Δ15HER2-His protein treatment affected viability of SK-BR-3 cells in a dose-dependent manner (Fig. 5C). The effect on MCF-7 cells was negligible at 240 nM (85% viable), suggesting that Δ15HER2-His protein specifically kills HER2-dependent cells. Furthermore we confirmed that Δ15HER2-His exerted its function through direct interaction with membrane bound HER2. This was shown by transfection of SK-BR-3 cells with the Δ15HER2-His expression plasmid and immunoprecipitation of the total lysates with an anti-His antibody 48 hours post transfection. Full-length HER2 protein was detected in the immunoprecipitates immunoblotted with a HER2 antibody (Fig. 6A), demonstrating that Δ15HER2-His and HER2 proteins interact. In MCF7 cells, expressed Δ15HER2-His also co-immunoprecipitated HER3 (Fig. 6B), confirming a direct interaction of the protein with its target in these cells.
Figure 6.
Interaction between Δ15HER2-His protein and HER2/HER3. A) SK-BR-3 and B) MCF7 cell lysates were collected 48 hours after transfection with Δ15HER2-His plasmid, the protein was immunoprecipitated with anti-His tag antibody and blotted with anti-HER2 antibody and anti-HER3 antibody, respectively.
Discussion
Traditional anti-HER2 methods have achieved successes, but with limitations. For example, antibody-based therapeutics are limited to targeting HER2 molecules displayed on cell surface, while tyrosine kinase inhibitors may target kinases other than HER2.7 As an alternative strategy, use of splice switching oligonucleotides (SSOs) can regulate gene expression by interfering with pre-mRNA splicing. In this work, we used an SSO to induce skipping of exon 15 in HER2 pre-mRNA. SSO111 reduced the level of full-length HER2 mRNA, which resulted in dose-dependent suppression of HER2 protein level. SSO111 induced significant growth inhibition and apoptosis in HER2-overexpressing SK-BR-3 cells. The lack of apoptosis in MCF7 cells suggests that specificity of the SSO effects depends on HER2 overexpression. The biological effects observed here are consistent with other reports in which HER2 was inhibited by various approaches such as monoclonal antibodies,33 antisense oligonucleotides,15 or RNA interference (RNAi).13 The latter two also regulate gene expression at mRNA level. Traditional antisense oligonucleotides require high concentrations (e.g. 1 µM15). Synthetic siRNAs, mediator for RNAi, have to limit numbers and/or forms of chemical modifications, which are required for improving in vivo performance, to stay compatible with the RNAi machinery.34
In SSO111-treated SK-BR-3 cells, the upregulated Δ15HER2 mRNA provides a positive and specific readout for the activity of SSO111, which is a remarkable attribute of SSO approach. Δ15HER2 protein was not detected either in the lysates or extracellular media with Western blot. It is not due to the instability of Δ15HER2 mRNA, since inhibition of NMD (nonse-mediated decay)35 through RNA interference36 didn’t change the level of Δ15HER2 mRNA (data not shown). It is possible that cells that highly expressed the protein from SSO treatment underwent apoptosis and were therefore eliminated from analysis.
With Δ15HER2 cloned from SSO111 treated cells, our studies revealed properties reminiscent of HER2 inhibitors, e.g. Herceptin and Herstatin; both selectively toxic to HER2-dependent cells. Binding of Herceptin and Herstatin to the ECD of cell surface HER2 leads to endocytosis and degradation of HER2 receptor.37, 38 Similarly, Δ15HER2-His reduced total HER2 protein level in SK-BR-3 cells after 48-hour incubation. The fact that phosphorylated HER2 was only affected at high concentrations of Δ15HER2-His suggests preferential binding of Δ15HER2-His to monomeric HER2 molecules on the cell surface. Only at large excess was Δ15HER2-His able to disrupt HER2 homodimer formation and decrease the constitutive activation. Note that SK-BR-3 cells express ~2 million HER2 molecules per cell.39
Interestingly, Δ15HER2 protein treatment has different effects on HER3 protein in MCF7 and SK-BR-3 cells, which may be due to their relative HER2 expression levels. In SK-BR-3 cells, HER3 transphosphorylation occurs mainly through HER2 heterodimer formation. Thus, the reduction in HER3 phosphorylation mirrors the reduction of monomeric HER2 by Δ15HER2-His, as observed with Herceptin.40 In MCF7 cells, HER3 expression is about two-fold that in SK-BR-3 cells, while HER2 is ~100-fold lower.39 Therefore, more monomeric HER3 is available for Δ15HER2-His binding, which explains the more significant reduction of HER3 protein in MCF7 cells. The minor effect of Δ15HER2-His on HER3 phosphorylation in MCF7 cells suggests that Δ15HER2-His may not inhibit HER3 transactivation by EGFR receptors other than HER2.
In summary, SSO111 downregulated HER2 mRNA through exon 15 skipping, which resulted in the inhibition of proliferation and induction of apoptosis in HER2-overexpressing tumor cells. Furthermore, SSO111 served as a platform for the identification of a novel HER2 ECD variant; the protein encoded by the exon 15-skipped HER2 mRNA, Δ15HER2, was able to downregulate HER2 and HER3, interfere with HER2-mediated transactivation of HER3, and specifically kill HER2-dependent cells. Both SSO111 and the Δ15HER2 protein should be further explored in vivo as potential candidates for the development of novel HER2-targeted cancer therapeutics.
Acknowledgements
The authors are grateful to Dr. H. Shelton Earp and Dr. Zefeng Wang for helpful discussion. This work was supported by National Institutes of Health grant [PO1-GM059299 to R.K.] and through a sponsored research agreement with Ercole Biotech, Inc.
Abbreviations
- EGFR
epidermal growth factor receptor
- HRG
heregulin
- MAPK
mitogen-activated protein kinase
- MOE
2’-O-methoxyethyl
- PARP
poly(ADP-ribose) polymerase
- PI3K
phosphoinositide 3-kinase
- SSO
splice switching oligonucleotide
Footnotes
Financial disclosure: RK declares a potential conflict of interest as an employee of AVI BioPharma, Inc.
- - A novel strategy was utilized to regulate HER2 protein expression, i.e. using splice switching oligonucleotides to alter splicing of HER2 pre-mRNA.
- - A novel HER2 truncated protein was generated through the mRNA variant identified with splice switching oligonucleotides, which was found to inhibit HER2 and HER2-mediated HER3 transactivation.
References
- 1.Hynes N, Stern D. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochem. Biophys. Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61:1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- 3.Tzahar E, Yarden Y. The ErbB-2/HER2 oncogenic receptor of adenocarcinomas: from orphanhood to multiple stromal ligands. Biochim. Biophys. Acta. 1998;1377:M25–M37. doi: 10.1016/s0304-419x(97)00032-2. [DOI] [PubMed] [Google Scholar]
- 4.Lonardo F, Di Marco E, King C, Pierce J, Segatto O, Aaronson S, Di Fiore P. The normal erbB-2 product is an atypical receptor-like tyrosine kinase with constitutive activity in the absence of ligand. The New Biologist. 1990;2:992–1003. [PubMed] [Google Scholar]
- 5.Rubin I, Yarden Y. The basic biology of HER2. Annals of Oncology. 2001;12 Suppl. 1:S3–S8. doi: 10.1093/annonc/12.suppl_1.s3. [DOI] [PubMed] [Google Scholar]
- 6.Graus-Porta D, Beerli R, Daly J, Hynes N. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. The EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moasser M. Targeting the function of the HER2 oncogene in human cancer therapeutics. Oncogene. 2006;26:6577–6592. doi: 10.1038/sj.onc.1210478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colomer R, Shamon L, Tsai M, Lupu R. Herceptin: from the bench to the clinic. Cancer Investigation. 2001;19:49–56. doi: 10.1081/cnv-100000074. [DOI] [PubMed] [Google Scholar]
- 9.Geyer C, Forster J, Lindquist D, Chan S, Romieu C, Pienkowski T, Jagiello-Gruszfeld A, Crown J, chan A, Kaufman B, Skarlos D, Campone M, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 10.Kute T, Lack C, Willingham M, Bishwokama B, Williams H, Barrett K, Mitchell T, Vaughn JP. Development of Herceptin resistance in breast cancer cells. Cytometry Patt A. 2004;57A:86–93. doi: 10.1002/cyto.a.10095. [DOI] [PubMed] [Google Scholar]
- 11.Brodowicz T, Wiltschke C, budinsky A, Krainer M, Steger G, Zielinski C. Soluble HER-2/neu neutralizes biologic effects of anti-HER2/neu antibody on breast cancer cells in vitro. Int. J. Cancer. 1997;73:875–879. doi: 10.1002/(sici)1097-0215(19971210)73:6<875::aid-ijc19>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Bartsch R, Wenzel C, Zielinski C, Steger G. HER2-positive breast cancer: hope beyond Trastuzumab. Biodrugs. 2007;21:69–77. doi: 10.2165/00063030-200721020-00001. [DOI] [PubMed] [Google Scholar]
- 13.Yang G, Cai K, Thompson-Lanza J, Bast RJ, Liu J. Inhibition of breast and ovarian tumor growth through multiple signaling pathways by using retrovirus-mediated small interfering RNA against Her-2/neu gene expression. J Biol Chem. 2004;279:4339–4345. doi: 10.1074/jbc.M311153200. [DOI] [PubMed] [Google Scholar]
- 14.Choudhury A, Charo J, Parapuram S, Hunt R, Hunt D, Seliger B, Kiessling R. Small interfering RNA (siRNA) inhibits the expression of the Her2/Neu gene, upregulates HLA class I and induces apoptosis of Her2/Neu positive tumor cell lines. Int. J. Cancer. 2004;108:71–77. doi: 10.1002/ijc.11497. [DOI] [PubMed] [Google Scholar]
- 15.Roh H, Pippin J, Green D, Boswell C, Hirose C, Mokadam N, Drebin J. HER2/neu antisense targeting of human breast carcinoma. Oncogene. 2000;19:6138–6143. doi: 10.1038/sj.onc.1204001. [DOI] [PubMed] [Google Scholar]
- 16.Wood M, Yin H, McClorey G. Modulating the Expression of Disease Genes with RNA-Based Therapy. PLoS Genetics. 2007;3:e109. doi: 10.1371/journal.pgen.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Blanco MA. Making antisense of splicing. Current Opinion in Molecular Therapy. 2005;7:476–482. [PubMed] [Google Scholar]
- 18.van Ommen G, van Deutekom J, Aartsma-Rus A. The therapeutic potential of antisense-mediated exon skipping. Current Opinion in Molecular Therapy. 2008;10:140–149. [PubMed] [Google Scholar]
- 19.Aartsma-Rus A, van Ommen G. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sazani P, Graziewicz M, Kole R. Splice switching oligonucleotides as potential therapeutics. In: Crooke ST, editor. Antisense Drug Technology. 2nd Edition. Boca Raton: CRC Press LLC; 2008. pp. 89–114. [Google Scholar]
- 21.Jearawiriyapaisarn N, Moulton H, Buckley B, Roberts J, Sazani P, Fucharoen S, Iversen P, Kole R. Sustained Dystrophin Expression Induced by Peptide-conjugated Morpholino Oligomers in the Muscles of mdx Mice Molecular Therapy, vol. 2008 doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercatante D, Bortner C, Cidlowski J, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. J. Biol. Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 23.Graziewicz M, Tarrant T, Buckley B, Roberts J, Fulton L, Hansen H, Ørum H, Kole R, Sazani P. An Endogenous TNF-α ntagonist Induced by Splice-switching Oligonucleotides Reduces Inflammation in Hepatitis and Arthritis Mouse Models Molecular Therapy, vol. 2008 doi: 10.1038/mt.2008.85. [DOI] [PubMed] [Google Scholar]
- 24.Doherty J, Bond C, Jardim A, Adelman J, Clinton G. The Her-2/neu receptor tyrosine kinase gene encodes a secreted autoihibitor. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10869–10874. doi: 10.1073/pnas.96.19.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aigner A, Juhl H, Malerczyk C, Tkybusch A, Benz C, Czubayko F. Expression of a truncated 100 kDa HER2 splice variant acts as an endogenous inhibitor of tumour cell proliferation. Oncogene. 2001;20:2101–2111. doi: 10.1038/sj.onc.1204305. [DOI] [PubMed] [Google Scholar]
- 26.Stix G. Blockbuster Dreams. Scientific American. 2006;294:60–63. doi: 10.1038/scientificamerican0506-60. [DOI] [PubMed] [Google Scholar]
- 27.Sierakowska H, Sambade M, Agrawal S, Kole R. Repair of thalassemic human β-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altmann K, et al. Second generation of antisense oligonucleotides: from nuclease resistance to biological efficacy in animals. Chimia. 1996;50:168–176. [Google Scholar]
- 29.Levin A, Yu R, Geary R. Basic principles of the pharmacokinetics of antisense oligonucleotide drugs. In: Crooke ST, editor. Antisense Drug Technology. Boca Raton: CRC Press LLC; 2008. pp. 183–215. [Google Scholar]
- 30.Kallioniemi O, Kallioniemi A, Kurisu W, Thor A, Chen L, Smith H, Waldman F, Pinkel D, Gray J. erbB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis G, Figari I, Fendly B, Wong W, Carter P, Gorman C, Shepard H. Differential responses of human tumor cell lines to anti-p185HER2 monoclonal antibodies. Cancer Immunology Immunotherapy. 1993;37:255–263. doi: 10.1007/BF01518520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzahar E, Pinkas-Kramarski R, Moyer J, Klapper L, Alroy I, Levkowitz G, Shelly M, Henis S, Eisenstein M, Ratzkin B, Sela M, Andrews G, et al. Bivalence of EGF-like ligands drives the ErbB signaling network. The EMBO J. 1997;16:4938–4950. doi: 10.1093/emboj/16.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudziak R, Lewis G, Winget M, Fendly B, Shepard H, Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitize human breast tumor cells to tumor necrosis factor. Molecular and Cellular Biology. 1989;9:1165–1172. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C, Li M, Chen C, Yao Q. Small interfering RNA therapy in cancer: mechanism, potential targets, and clinical applications. Expert Opin Ther Targets. 2008;12:637–645. doi: 10.1517/14728222.12.5.637. [DOI] [PubMed] [Google Scholar]
- 35.Maquat LE. Nonsense-mediated mRNA decay in mammals. Journal of Cell Science. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, Furic L, DesGroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3' UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 37.Cuello M, Ettenberg S, Clark A, Keane M, Posner R, Nau M, Dennis P, Lipkowitz S. Down-regulation of the erbB-2 receptor by Trastuzumab (Herceptin) enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2. Cancer Research. 2001;61:4892–4900. [PubMed] [Google Scholar]
- 38.Hu P, Zhou T, Qian L, Wang J, Shi M, Yu M, Yang Y, Zhang X, Shen B, Guo N. Sequestering ErbB2 in endoplasmic reticulum by its autoinhibitor from translocation to cell surface: an autoinhibition mechanism of ErbB2 expression. Biochem Biophys Res Commun. 2006;342:19–27. doi: 10.1016/j.bbrc.2006.01.115. [DOI] [PubMed] [Google Scholar]
- 39.Aguilar Z, Akita R, Finn R, Ramos B, Pegram M, Kabbinavar F, Pietras R, Pisacane P, Sliwkowski M, Slamon D. Biologic effects of heregulin/neu differentiation factor on normal and malignant human breast and ovarian epithelial cells. Oncogene. 1999;18:6050–6062. doi: 10.1038/sj.onc.1202993. [DOI] [PubMed] [Google Scholar]
- 40.Yakes F, Chinratanalab W, Ritter C, King W, Seelig S, Arteaga C. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Research. 2002;62:4132–4141. [PubMed] [Google Scholar]