Abstract
We created a Thrsp (Spot 14 or S14) null mouse (Thrsptm1cnm) to study the role of Thrsp in de novo lipid synthesis. The Thrsp null mouse exhibits marked deficiencies in de novo lipogenesis in the lactating mammary gland. We now report the Thrsp gene deletion affects body weight and glucose tolerance associated with increased insulin sensitivity. By postnatal day 150 the rate of first generation C57BL/6J backcross Thrsp null mouse weight gain slowed compared to wild type animals. This was due to changes in body fat mass. We studied mice backcrossed for 5 and 11 generations. The weight difference between the null and wild type adult mice diminished with progressive backcross generations. In conclusion the Thrsp gene is involved in the regulation of diet induced obesity and deletion of Thrsp leads to an improvement in age associated glucose tolerance.
Keywords: Spot 14, obesity, glucose tolerance, insulin sensitivity
Introduction
The regulation of body weight is currently a topic of major interest (Badman and Flier, 2005, Barsh et al., 2000, Campfield et al., 1998, Evans et al., 2004, Friedman and Halaas, 1998, Woods et al., 1998). The role of lipid synthesis in the regulation of body weight is controversial (Jiang et al., 2005, Schutz, 2004b, Schutz, 2004a) and not well explored. Nevertheless, there has been support for the “thrifty gene hypothesis” in the development of obesity (Prentice, 2001, Ravussin and Bogardus, 1990). This hypothesis is based on the concept that mammals (including humans) have evolved to maximize their ability to store excess energy when food is plentiful, and minimize energy expenditure when food is in short supply. This concept implies that one or more genes are involved in the ability to store excess calories as fat whether this fat is derived from exogenous sources or converted to fat by de novo lipogenesis.
The regulation of body fat is complex as dietary fuels are partitioned for either energy utilization or energy storage. If the intake of fuel is in excess of immediate needs, the carbohydrate calories can be stored as glycogen for immediate glucose release, or converted to fat for subsequent energy release. Conversion of carbohydrate to fat occurs in several tissues, such as liver and adipose tissue, by a process called de novo lipogenesis. It has long been known that hepatic de novo lipogenesis is highly regulated both at a transcriptional level as well as by post-transcriptional mechanisms (Munday and Hemingway, 1999, Towle and Mariash, 1986). In view of these considerations, it is important to understand the regulation of de novo lipogenesis to define those factors that regulate body weight.
We have used the Thrsp gene (Thrsp, GenBank mRNA # NM_009381) as a model for the regulation of de novo lipogenesis, since the tissue content of Thrsp mRNA varies directly with the ability of that tissue to synthesize lipids. Specifically, the level of Thrsp mRNA in white adipose tissue, brown adipose tissue, and liver is elevated when de novo fatty acid synthesis is induced by dietary and hormonal stimuli (Freake and Oppenheimer, 1987, Jump et al., 1984, Jump and Oppenheimer, 1985, Kinlaw et al., 1986, Narayan et al., 1984, Towle and Mariash, 1986). To study the role of the Thrsp protein in de novo lipogenesis, we created a Thrsp null mouse and found that the rate of hepatic lipogenesis was normal but mammary gland lipogenesis was markedly reduced (Zhu et al., 2001, Zhu et al., 2005). We showed that the reduced mammary gland lipogenesis led to a decrease in pup growth rate while suckling (Zhu et al., 2005). We hypothesized that the Thrsp null mouse would also show a defect in adult weight gain. We now show that Thrsp deficient adult animals exhibit a reduced rate of weight gain. The reduced Thrsp null mouse weight gain is resultant from decreased fat accumulation and is associated with improvement in glucose tolerance.
Materials and Methods
Animal Care and Breeding
Thrsp null and wild type mice were created as previously described (Zhu et al., 2001). Briefly, the Thrsp targeting vector was transfected into 129/SV embryonic stem (ES) cells by electroporation. Chimeric male mice produced from the ES cell injections were bred with B6D2F1/J females. Founder male mice carrying the germline mutation were backcrossed to female C57BL/6J mice to create the N1 mice. To establish a congenic line we used the following protocol: heterozygous mice from the N1 generation were backcrossed to C57BL/6J mice to create the N2 generation. Subsequent N generations were created using the same protocol, where the N generation was created by backcrossing a heterozygous N-1 mouse with a newly purchased C57BL/6J mouse. After fixing the y-chromosome, backcross breeding was continued for 11 generations. N1, N5, and N11 heterozygous progeny were interbred to create homozygous null and wild type animals for experimentation.
To create Thrsp null and wild type mice we first bred N1, N5, or N11 Thrsp heterozygous mice to generate N1, N5, or N11 Thrsp null and wild type animals. Pups were genotyped by a PCR-based methodology (see below). Null mice were obtained by breeding Thrsp null males and females with each other. Wild type mice were obtained by breeding Thrsp wild type males and females with each other. This breeding approach was used to generate the N1, N5, and N11 Thrsp null and wild type animals used in all experiments presented in this paper. Mouse genotyping was performed by multiplexed PCR as previously described (Zhu et al., 2001). Briefly, genomic DNA was prepared from mouse tails and subjected to PCR using three primers. The primers included PUP (binds only to the wild type Thrsp gene), PDN (binds to both the wild type and null Thrsp gene), and PNEO (binds only to the neomycin resistance gene located within the mutant Thrsp gene). A full description of the PCR-based screening methodology is provided in our previous publication (Zhu et al., 2001).
Mice were housed in a specific pathogen free facility with a 14 hour light and 10 hour dark cycle. Mice were given free access to food and water. Pups were weaned at post-natal day 21 (PN21). The N1 mice were fed with either Teklad 7001 (4% fat by weight, 12% fat calories) diet (Harlan, Indianapolis, Indiana) or Teklad 7004 diet (Breeder chow, 11% fat by weight, 28% fat calories). N5 and N11 mice were either fed Teklad 8640 (5% fat by weight, 14% fat calories) or Teklad 7004 diet. All mouse weights were obtained at 9 AM on the day of weighing. The 7001 and 8640 diets are low fat, and the 7004 diet is a moderate fat diet.
All animal studies were conducted in accordance with the principles and procedures outlined in the NIH guide for the Care and Use of Laboratory Animals. These procedures were approved by the local Institutional Animal Care and Use Committee.
Food intake
Food intake was determined in PN200 N1 male mice individually housed in a specially fitted cage. The mice were stood on a wire mesh platform. All feces and urine produced passed through the mesh. Feces and food particles were caught in a second, fine wire mesh beneath the platform. Urine passed through the second mesh into a collecting tube. Food was presented in a metal container housed outside the main enclosure. The container was itself enclosed within a separate container. At precisely 9AM each morning, the food remaining in the container plus any food particles found in the secondary container or caught in the second mesh below the platform was weighed on an analytical balance. The mice in the food intake study were the same animals used in the N1 weight study presented in Figure 1C. The mice were fed the moderate fat diet (Teklad 7004).
Figure 1.
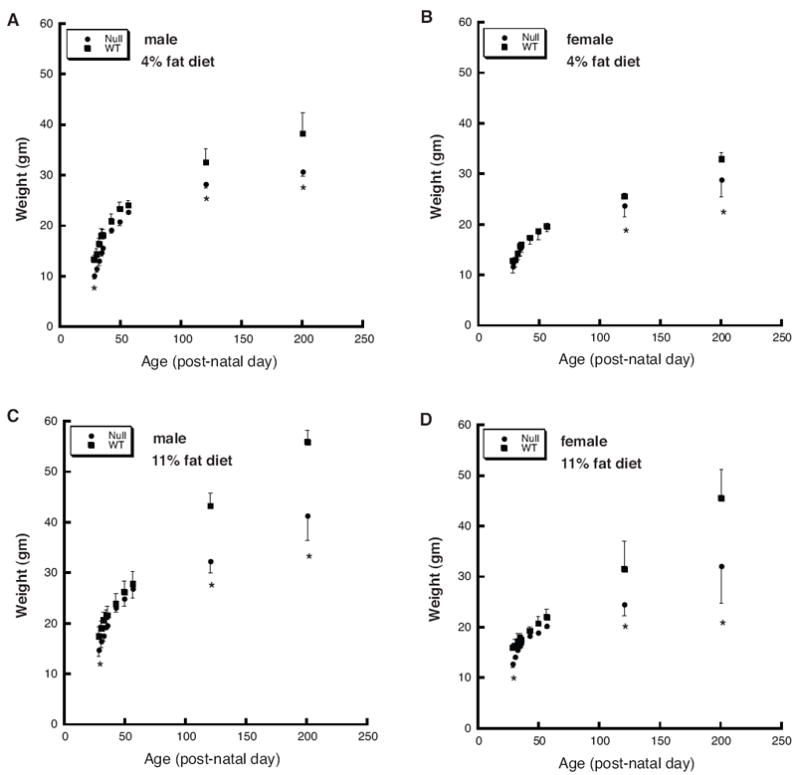
The Thrsp null mutation results in resistance to diet-induced obesity in N1 adult mice. Mouse weights were obtained from male (panels A and C) and female (panels B and D) N1 mice from birth to 200 days postnatal. Mice were maintained on a 4% (panels A and B) or 11% (panels C and D) fat diet from the time of weaning. All data represent the mean ± SD (n=3 per group). The asterisk indicates a statistically significant difference in body weights as determined by ANOVA. Weights are measured in grams (g).
Mouse densitometry
PN200 N1 generation mice maintained on the moderate fat diet were subjected to a dual-energy x-ray absorptiometry (DEXA) scan. The mice were weighed, anesthetized with inhaled isofluorine (Fort Dodge, IA), and then scanned. The DEXA scan was performed at the Mayo Clinic (Rochester, MN) in the laboratory of Dr. James Levine using a PIXImus DEXA scanner. Subcranial total body measurements were obtained from each animal. Results obtained included: bone mineral density (BMD) in mg/cm2; bone mineral content (BMC) in mg; bone area in cm2; %fat; lean tissue in grams; fat tissue in grams. Relative bone growth was determined by measuring the femur length with a ruler. The femur was visualized from the PIXImus generated body scan.
Glucose tolerance test and insulin tolerance test
PN200 N11 generation mice maintained on a 4% fat diet were used for both these studies. Food was removed at 1800 and measurements were obtained at 0900 the following day. All blood glucose levels were measured on a drop of tail blood with a Precision QID (Abbott Laboratories Inc.) glucose meter. For the glucose tolerance test (GTT) a baseline glucose reading was first obtained. Animals were then injected with 10 microliters/gm BW of 20% glucose intraperitoneally (i.p.). Glucose readings were obtained at various time points post injection. Glucose injections and all bleedings were preformed without anesthesia on hand-gentled mice.
Similar methodology was used for the insulin tolerance test (ITT). We first obtained a baseline glucose reading. Animals were then i.p. injected with 0.425 units/kg BW of recombinant human insulin (Humulin R, Eli Lilly, Indianapolis) and glucose values were obtained at various time points post injection. Insulin injections and all bleedings were preformed without anesthesia on hand-gentled mice. ITT tests were performed on PN200 animals maintained on low fat diet.
Serum triglyceride measurements
Serum triglycerides were measured using a Serum Triglyceride Determination Kit (Sigma, St. Louis, MO). Briefly, this methodology measures serum triglyceride levels by hydrolyzing triglycerides to glycerol and free fatty acids followed by measurement of the released glycerol by an enzymatic and spectrophotometric method. The measured glycerol is directly proportional to the triglyceride fatty acid content. Free endogenous glycerol is also measured and is taken into account when determining the true serum triglyceride concentrations. Blood was obtained retroorbitally from anesthetized animals and centrifuged to obtain serum. The animals used were 4 month old, male, N5 mice maintained on a moderate fat diet
Fad pad weight measurements
Five month old male N11 mice maintained on a low fat diet were sacrificed and epididymal and brown adipose fat pads dissected. The epididymal fat pad is discretely located in the male mouse abdominal cavity and is easily completely excised. The complete epididymal fat pads were excised and immediately weighed on a calibrated scale. Similarly, the complete brown fat pad was excised from the interscapular region and immediately weighed on a calibrated scale.
Energy Expenditure
Indirect calorimetry measurements were made with an open circuit, computer-controlled, system (Applied electrochemistry Inc, Pittsburgh, Pennsylvania). At 9:00 AM mice (4 Thrsp null and 5 wild-type) were placed in the chamber in a post-absorptive state and measurements of oxygen consumption, carbon dioxide production, and system temperature were taken every 2 minutes for up to 4 hours. Food and water were restricted for the entire procedure. Energy expenditure was corrected for body weight, and respiratory quotient (RQ) was measured by the ratio of carbon dioxide produced to oxygen consumed (in ml).
Statistics
All data are presented as mean±sd and represent at least 4 animals per group unless otherwise stated. Comparisons between groups were made by analysis of variance (ANOVA) and post-hoc testing used the Scheffe method. When necessary to achieve homogeneity of variances between groups, data were logarithmically transformed before performing the ANOVA.
Results
Thrsp N1 null mice exhibit resistance to diet-induced obesity
We have previously reported that Thrsp null mice demonstrate reduced weight compared to wild type mice from postnatal day 7 (PN7) until weaning (Zhu et al., 2005). We next asked whether these weight differences continued to be manifest in the adult Thrsp null animals. First generation backcross (N1) Thrsp null and wild type mice born to homozygous dams, were maintained on either a low fat diet (7001; 4% fat by weight, 12% fat calories) or a moderate fat diet (7004; 11% fat by weight, 28% fat calories) from the time of weaning. Mouse weights were obtained and plotted. We detected no significant differences in body weight in any groups tested through PN56 (Fig. 1). However, both male and female null mice, fed the 7004 diet, exhibited marked reductions in the rate of body weight gain by PN120 (Fig. 1C and D). These differences in body weight persisted and continued to through PN200. Male PN200 null mice weighed 26% less than wild type males and female PN200 null mice weighed 29.4% less than wild type females. PN200 Thrsp null mice fed the 7001 low fat diet (Fig. 1A and B) also exhibited reduced body weight compared to wild type mice (19.8% reduction for males and 12.5% reduction for females). However, the weight changes were less marked than those observed from animals fed the moderate fat diet. These data suggest that the Thrsp null N1 mice are resistant to diet induced obesity.
To determine whether changes in fat or lean mass accounted for the observed changes in weight gain we subjected null and wild type N1 mice to an analysis of body mass by DEXA scan. The mice used in this experiment included the PN200 male N1 mice from the weight experiment presented in Fig. 1C. We determined that while the total lean mass was not different between the Thrsp null and wild type mice, the null mouse fat mass was significantly reduced by 45.4% (Fig. 2A). A visual comparison of two representative mice demonstrates the marked genotype-dependent difference in accumulated fat mass (Fig. 2 B). Measurement of long bone length revealed no significant differences in skeletal size between Thrsp null (16 mm) versus wild type (16 mm) mice. Similar data were obtained when null and wild type female mice were studied (data not shown). Thus, these data suggest that the difference in body weight observed in Thrsp null and wild type mice is the result of decreased fat accumulation in the null animals.
Figure 2.
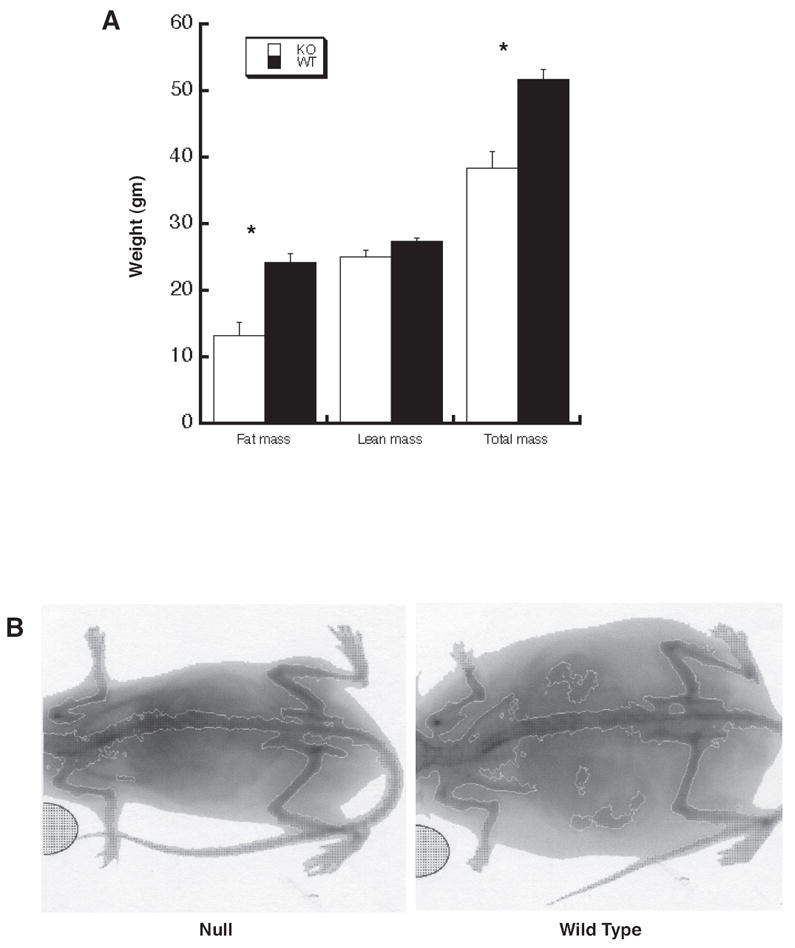
Adult Thrsp null mutant mice exhibit reduced body fat content as determined by DEXA scan. PN200 male N1 Thrsp null and wild type mice were subjected to DEXA scan to determine their percent total body fat. All data represent the mean ± SD (null n=6; wild type n=7). The asterisk indicates a statistically significant difference in as determined by t-test. B. PIXImus image of a representative N1 null (left) and wild type male (right) mouse. The light gray areas indicate body fat whereas the dark gray areas indicate boney structures.
We next investigated the possibility that the decreased fat mass in the N1 null mouse was due to diminished food intake. Thus, we measured daily food consumption of Thrsp null and wild type N1 mice for one week. Fig. 3 shows the average daily food intake for null and wild type N1 mice. Surprisingly, we observed that the null mice ingested more food than the wild type mice when adjusted for body weight. Furthermore, there was no significant difference in fecal fat content between genotypes when measured at the same time as the food intake study (data not shown). Therefore, these results suggest the Thrsp N1 null mice have a higher metabolic rate than wild type mice as the null mice ingest more food but accumulate less fat.
Figure 3.
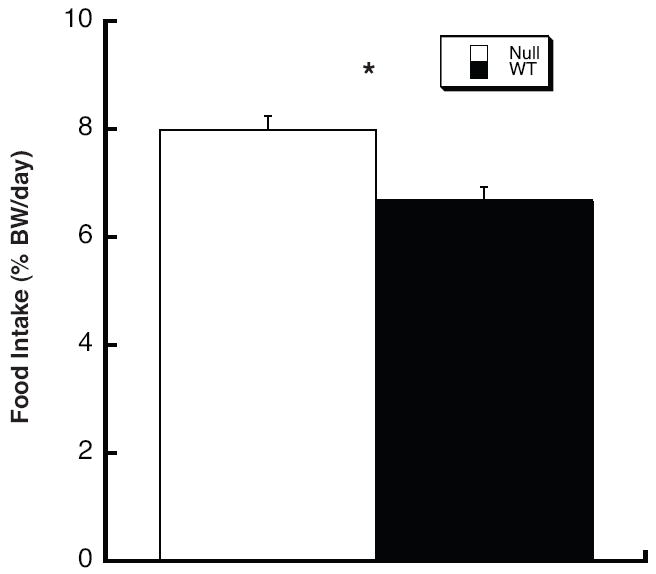
Adult Thrsp null mice exhibit greater daily food intake compared to wild type control mice. PN200 N1 male mice were used to measure daily food intake. All data represent the mean ± SEM (n = 16). The asterisk indicates a statistically significant difference by t-test (p < 0.01).
Thrsp N5 and N11 wild type mice exhibit reduced susceptibility to diet-induced obesity
We next asked whether the genotype-specific weight differences observed in the N1 mice continued to be manifest in subsequent backcross generations. Fifth (N5) and eleventh (N11) generation backcross Thrsp null and wild type mice born to homozygous dams, were maintained on a moderate fat diet (7004; 11% fat by weight, 28% fat calories) from the time of weaning. Mouse weights were obtained and plotted. We detected no statistically significant differences in body weight in N5 male or female mice through PN100 (Fig. 4). However, both male and female null mice exhibited reduced rates of body weight gain beginning at PN150 (male) and PN100 (female) (Fig. 4A and B). These differences in body weight persisted and continued through PN300. Male PN300 null mice weighed 9.9% less than wild type males and female PN300 null mice weighed 16.9% less than wild type females. In contrast to these data, N11 mice fed a low (8640) fat diet exhibited no statistically significant differences in body weight and only male mice fed a moderate (7004) fat diet showed significantly lower body weight from PN120 to PN200 (Fig. 5). Finally, circulating true triglyceride levels were not different in N5 Thrsp null (0.947 ± 0.012 mg/ml) versus wild type (0.928 ± 0.015 mg/ml) mice.
Figure 4.
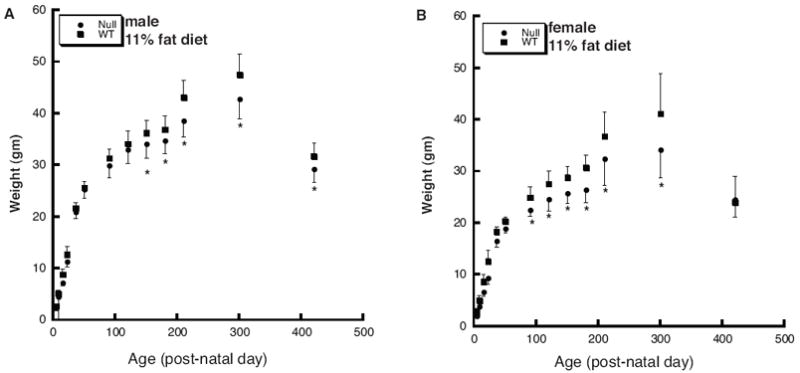
The Thrsp null mutation results in resistance to diet-induced obesity in N5 adult mice. Mouse weights were obtained from male (panel A) and female (panel B) N5 mice from birth to 420 days postnatal. Mice were maintained on an 11% fat diet from the time of weaning. All data represent the mean ± SD (wild type male n=14; wild type female n=8; null male n=13; null female n=13). The asterisk indicates a statistically significant difference in body weights as determined by ANOVA. Weights are measured in grams.
Figure 5.
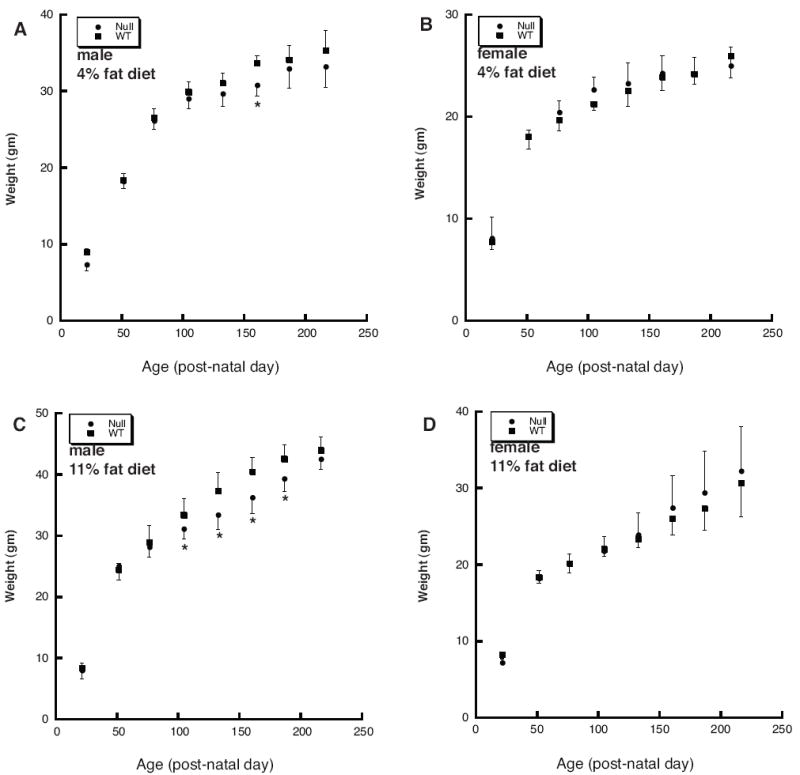
The Thrsp null mutation does not affect resistance to diet-induced obesity in N11 adult mice. Mouse weights were obtained from male (panels A and C) and female (panels B and D) N11 mice from birth to 216 days postnatal. Mice were maintained on a 4% (panels A and B) or 11% (panels C and D) fat diet from the time of weaning. All data represent the mean ± SD (n=6 for all groups fed the 4% fat diet; n=12 for all groups fed the 11% fat diet). Weights are measured in grams.
Thrsp N11 wild type mice exhibit improved glucose and insulin tolerance
Increased fat accumulation is associated with impaired glucose metabolism (McGarry, 2002). To determine whether systemic glucose clearance is affected by the Thrsp null mutation we performed both glucose tolerance and insulin tolerance tests on Thrsp null and wild type animals. The animals used for these experiments were N11 PN200 male mice that had been maintained on a low fat diet from the time of weaning. There was no statistically significant difference in weights between Thrsp null (33.3±2.7g) and wild type (35.5±2.5g) animals. We observed that the Thrsp null animals presented with reduced fasting glucose levels compared to wild type animals (Fig. 6A). Upon glucose administration, maximal blood glucose levels were significantly reduced in the null animals suggesting increased glucose clearance in the null animals compared to wild type. We similarly noted an improvement in the Thrsp null insulin tolerance test (Fig. 6B). Blood glucose levels fell to the same extent in both null and wild type animals. However, the duration of insulin-dependent repression of circulating blood glucose levels was significantly reduced in wild type versus null animals. Thus, these data suggest that N11 Thrsp null mice exhibit improved glucose utilization compared to wild type animals.
Figure 6.
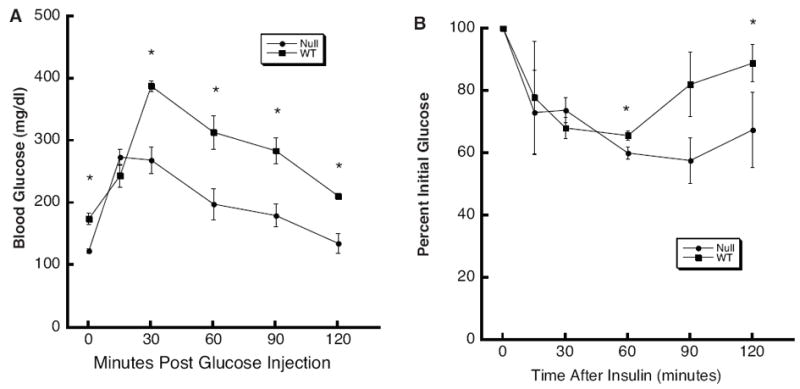
The Thrsp null mouse exhibits improved glucose and insulin tolerance. A. Glucose tolerance test. Seven month old male N11 mice maintained on a 4% fat diet were injected with 20% glucose i.p.. Blood glucose levels were measured at the indicated times post-injection. All data represent the mean ± SEM (n = 6). The asterisk indicates a statistically significant difference in blood glucose levels as determined by ANOVA (p<0.01). B. Insulin tolerance test. In a separate set of N11 mice of the same age, blood glucose levels were measured at the indicated times post insulin injection. All data represent the mean ± SEM (n = 4). The asterisk indicates a statistically significant difference in blood glucose levels as determined by ANOVA.
Because the N1 generation mice showed such marked differences in fat mass between the Thrsp null and wild type animals, we speculated that while there was little significant difference in N11 total body weight, fat mass differences may still exist between the Thrsp null and wild type animals. If true then it is possible that such differences may account for the improved glucose tolerance noted in the Thrsp null animals. Figure 7 shows that the N11 generation Thrsp null animals continue to have a slight but significant reduction in fat mass as measured in two different fat depots.
Figure 7.
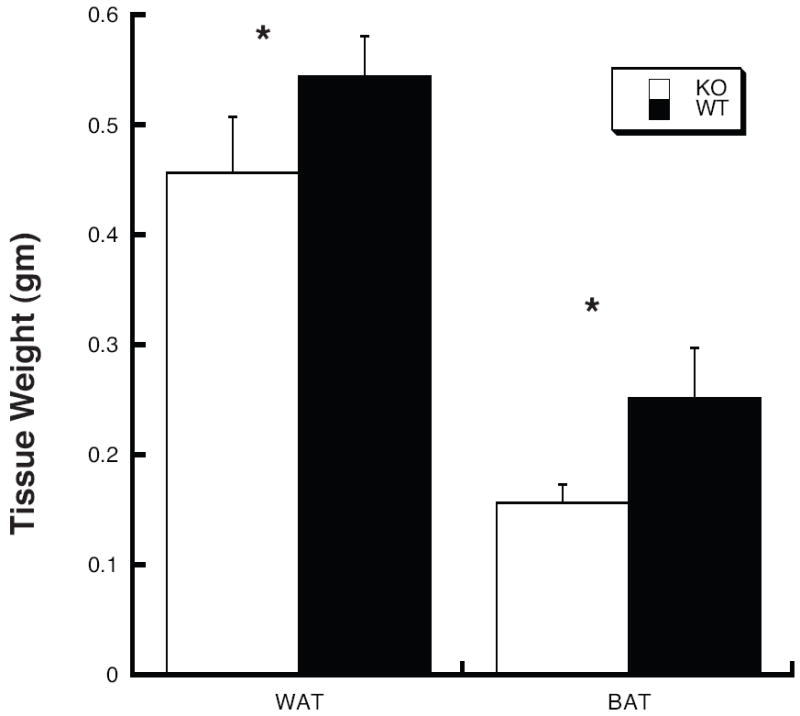
The Thrsp null mouse exhibits reduced epididymal and brown adipose fat pads. Five month old male N11 mice maintained on a 4% fat diet were sacrificed and epididymal and brown adipose fat pads dissected. Each bar represents the mean ± SD from 4 mice. The asterisk indicates a significant (p < 0.05) difference between the two genotypes by t-test. This experiment has been repeated 2 more times with similar results.
Since the N11 mice continued to show decreased fat mass and enhanced glucose tolerance and insulin sensitivity, we speculated that the mice also had an increase in energy expenditure. Therefore, we used indirect calorimetry to measure the energy expenditure and RQ of the mice. Mice were placed in the calorimetry chamber in a post-absorptive state and remained in the chamber for up to 4 hours. Chamber temperature, oxygen utilization, and carbon dioxide production were constantly monitored, but the mice were not disturbed and food and water were not present in the chamber. After placement in the chamber we noted the RQ of all mice was near 1.0. However, over the 4 hours of observation the RQ fell to just under 0.7. While we were unable to demonstrate any significant difference in oxygen consumption between the genotypes, we did observe several significant differences in metabolism. We found that the time achieve a minimum RQ was significantly less (2.4±0.2 vs 3.1±0.4 hrs, p = 0.01) in the wildtype animals compared to the null mice. Thus, in the post-absorptive state the wildtype mice achieved a “resting” state with a minimal oxygen consumption much sooner than the null mice. This was further corroborated by our finding that the null mice were also much more active in the chamber than the wild type mice. The null mice showed significantly more (4.6±0.6 vs 3.4±0.6, p < 0.01) episodes of spontaneous movement as noted by bursts in oxygen consumption.
Discussion
We have previously reported that Thrsp null suckling pups nursing on Thrsp null dams exhibit decreased neonatal weight gain compared to wild type pups nursing on wild type dams (Zhu et al., 2005). We now report that Thrsp gene deletion also results in decreased adult weight gain. However, it is likely that the mechanisms responsible for the gene-specific effects on pup weight gain are different than those in the adult. The effects of the Thrsp null mutation on neonatal weight gain are the result of decreased milk fat content in milk produced by Thrsp null dams (Zhu et al., 2005). This decrease in milk fat content is associated with a reduction in lactating mammary gland de novo lipogenesis. The effect of the Thrsp null mutation on neonatal weight gain is not intrinsic to the pup as Thrsp null pups born to heterozygous dams do not differ in weight from wild type littermates. Thus, the effects of the Thrsp gene deletion on neonatal weight gain are secondary to the effects of the Thrsp gene deletion on the dam’s lactating mammary gland.
In contrast to these neonatal weight data, we now report that deletion of the Thrsp gene results in primary effects on adult weight gain. After weaning, homozygous N1 null mouse weight rapidly catches up to wild type mouse weight (Fig. 1). This catch-up is independent of diet or sex. However, as the animals continue to age the N1 null mice begin to lag behind in weight gained. This effect is noted in both male and female Thrsp null mice and is independent on diet (Fig. 1). However, mice fed a moderate fat diet exhibited greater divergence in weight.
To assess the effects of the Thrsp null mutation on diet-induced obesity, we placed the Thrsp null mutation on a C57BL/6J background and created a congenic mouse strain. To generate this strain we backcrossed Thrsp null mice repeatedly onto the C57BL/6J background. Animals representing the first, fifth, and eleventh generation backcrossing were selected for our experimental studies. Importantly, we did not use the inbred C57BL/6J mice as our wild type control animals since these inbred animals do not properly control for the mixed genetic background possessed by the N1 and N5 mice. Rather, we generated both our wild type and null experimental animals by mating heterozygous animals and selecting resultant null and wild type littermates as the parents for our experimental animals. Therefore, the Thrsp null and wild type experimental animals were always controlled for the extent of genetic heterogeneity attributed to the initial complex genetic background of the founder mice. Use of wild type controls bred in this fashion is essential for properly evaluating the contribution of a single gene to a genetically complex phenotype such as obesity (Sigmund, 2000).
We observed that as the genetic background of both the Thrsp null and wild type mice becomes more identical to the C57BL/6J background, the Thrsp-dependent differences in body weight diminish in magnitude. Thus, while the effects of the Thrsp null mutation on weight gain were marked in our first generation backcross progeny, we noted that subsequent generations of backcrossing resulted in a blunting of the observed weight phenotype. Weights of approximately 200-day old males fed a moderate fat diet were reduced by 19.8% in N1, 10.5% in N5, and 7.5% in N11 null mice (Figs. 1, 4 and 5). Even more striking blunting of the weight phenotype was observed in female mice.
Interestingly, the blunting of weight differences associated with backcrossing was almost entirely due to reductions in wild type weight gain. Thrsp null mouse weights did not change as a result of backcross generation. Weights of 200-day old Thrsp null males fed a moderate fat diet were 41.4 ± 4.9 g in N1, 38.7 ± 3.2 g in N5, and 42.7 ± 1.7 g in N11 animals (Figs. 1, 4 and 5).
The founder heterozygote Thrsp null mice were genetically 50% C57BL/6J, 25% 129/J, and 25% B6D2F1/J (Zhu et al., 2001). As we continued to backcross our mouse strains, the 129 and B6D2F1 genetic contributions diminished. B6D2F1/J mice demonstrate a significantly greater postnatal weight than C57BL/6J mice (Bogue et al., 2006). Thus, it is not surprising that the weights of wild type backcross progeny decreased with each successive backcross generation as the contribution of the B6D2F1 strain to the genetic background also decreased with each successive backcross. That the Thrsp null mouse weight remained constant within each generation suggests that in the context of the heterogeneous 129/J-B6D2F1/J-C57BL/6J genetic background, the Thrsp gene contributes to weight gain induced by a moderate fat diet. It should be noted that our N11 mice did not become as obese on the moderate fat diet as C57BL/6J mice do when placed on a very high fat diet (Lin et al., 2000). It is possible that we may have seen a larger difference between null and wild-type mice in the N11 generation if placed on a standard high fat (45%) diet.
DEXA scan analysis revealed that differences in N1 mouse weights were entirely attributed to differences in fat mass (Fig. 2). These data suggest that deletion of the Thrsp gene results in a decreased ability to accumulate fat. De novo lipid synthesis in white adipose tissue is not reduced in Thrsp null animals (data not shown). Furthermore, de novo lipogenesis is inhibited in rodents fed a high fat diet (Wilson et al., 1990). Thrsp null mice fed a moderate fat (11%) diet exhibit greater gene specific divergences in weight than null mice fed a low fat diet (Fig. 1). As a moderate fat diet will inhibit de novo lipogenesis to a greater extent than a low fat diet, it is unlikely that the observed weight differences are due to reduced lipogenesis in the null animals. Thus, we propose that the decreased fat mass observed in the Thrsp null animals is either the result of increased metabolism or decreased storage of ingested fat. The Thrsp null N1 mice exhibiting reduced weight gain actually eat more food than wild type animals (Fig. 3). In addition, null mice exhibited decreased weight gain when fed either a low or moderate fat diet. Lastly, we found by indirect calorimetry the Thrsp null mice continued to show more activity and had elevated oxygen utilization longer than the wild-type mice in the post-absorptive state. Thus, these data support the hypothesis that metabolism is increased in the Thrsp null animals. However, additional experiments will be necessary to test this hypothesis.
The role of Thrsp in regulating adult body weight is further supported by studies in humans and livestock. Fasting results in reduction in mRNA levels of many genes involved in de novo lipid synthesis including Thrsp (Jump and Oppenheimer, 1985). Corresponding reductions in de novo lipogenesis are also observed. We have previously reported that down-regulation of adipose tissue Thrsp mRNA levels by fasting is blunted in obese humans (Kirschner and Mariash, 1999). These data led us to propose that expression of the Thrsp gene may contribute to human fat accumulation in humans. Intriguingly, recent data reveal a genetic association between Thrsp gene mutations and abdominal fat accumulation in the chicken (Wang et al., 2004). Additionally, Thrsp gene expression is highly correlated with the intramuscular fat content of beef cattle (Wang et al., 2008). Together with the findings reported here, these data support a role for the Thrsp gene in regulating fat accumulation in adipose tissue.
The mouse Thrsp gene resides on chromosome 7. Several studies have assessed chromosome 7 for the presence of obesity determining genetic loci (Cheverud et al., 2004, Diament and Warden, 2004, Warden et al., 1993, Warden et al., 1995, Yi et al., 2004). Intriguingly, one of these studies linked the region of mouse chromosome 7 encoding the Thrsp gene to body fat mass accumulation (Diament and Warden, 2004). Subcongenic cross experiments suggested that the identified body fat mass quantitative trait locus (QTL) contains at least two obesity genes (Diament and Warden, 2004). We speculate that at least one interacting locus is located near the Thrsp gene on chromosome 7.
The concept of genetic epistasis where multiple genes contribute to the observed phenotype is generally accepted in the genetic study of obesity (Warden et al., 2004). Indeed, mouse studies have demonstrated epistatic interactions between mutant genes and other alleles that are dependent on the genetic background of the mouse strain used for the experiment (Coleman and Hummel, 1973, Harris et al., 2001, Hummel et al., 1972). In the studies presented here we observed reduced diet-induced weight gain in Thrsp null mice when compared to wild type animals (Figs. 1, 4, and 5). The magnitude of these differences however, was markedly reduced with increasing backcross generations. Importantly, the reduction was entirely due to decreased weight gain of the wild type mice. The weights of the Thrsp null animals changed little with increasing backcross generations. One possibility is that these observed effects of Thrsp on body weight are due to genetic epistasis. In epistasis, the epistatic gene masks the action of a second hypostatic gene(s). Our data suggests Thrsp may act as a hypostatic gene in regulating body weight. We hypothesize that on the mixed genetic background (containing 129/J, B6D2F1/J, and C57BL/6J alleles) of our early generation (N1 and N5) backcross progeny an epistatic allele is missing. Thus, in the Thrsp N1 and N5 wild type mice, the hypostatic Thrsp gene is unmasked and induces the development of diet-induced obesity. The N1 and N5 Thrsp null mice are resistant to diet-induced obesity due to the absence of Thrsp. Our Thrsp null (Thrsptm1cnm) mice were subsequently backcrossed to the C57BL/6J strain. We observed a progressive loss of the weight gain phenotype as the backcross generations increased. By backcross generation 11 (N11 mice), the Thrsptm1cnm mouse genetic background is almost entirely derived from the C57BL/6J strain and the wild type weight gain phenotype is concomitantly severely reduced in magnitude. We hypothesize that a Thrsp masking epistatic allele resides on the C57BL/6J genetic background. While this is one possibility, we recognize other possibilities also exist to explain our findings.
The mice used in our studies were suckled on homozygous null or wild type dams. As we previously reported, null dams produce milk deficient in lipid and the null neonates are reduced in weight for the first few weeks after birth (Zhu et al., 2005). Energy-rich milk is composed of protein, lactose and lipid at the most basic level. The suckling mammalian neonate acquires a large amount of the energy needed for growth and development from the lipid constituents of milk (Edmond et al., 1985). Studies, including our own, have shown that as a neonate, energy deprived rat pups exhibit marked reductions in weight gain (Zhu et al., 2005, Del Prado et al., 1997, Faust et al., 1980, Nagasawa et al., 1989). Interestingly, as adults these animals may become resistant to diet induced obesity (Faust et al., 1980). Thus, the effects of reduced energy in Thrsp null dam milk and the consequent reduced neonatal weight gain of the suckling pups, may contribute to the weight phenotype observed in the adult Thrsp null mice. The effects of a reduced lipid neonatal diet on adult weight gain are even less studied in humans. Not surprisingly, recommendations for optimal neonatal fat intake are controversial. However, it is well accepted that neonatal nutrition profoundly affects long-term human physiology (Petry et al., 2001). It is thus hypothesized that nutritional status during early neonatal mammalian development may permanently set specific metabolic characteristics that are manifest into adulthood.
Our observation of an increase in spontaneous movements in the Thrsp null mice is similar to that seen in other knockout mice that are also more glucose tolerant and resistant to obesity (Pfluger et al., 2008, Ayala et al., 2008, Ivanova et al., 2008). We were initially expecting to find an increase in resting energy expenditure and were surprised by the fact that oxygen consumption was not significantly different among the genotypes. It is not clear why the null mice exhibited more activity as a mechanism of expending energy. Further studies to examine the types of activity and the extent of time spent in increased activity would be interesting.
The marked increase in glucose tolerance and insulin sensitivity observed in our adult N11 animals (Fig. 6) can not be explained simply on a difference in weight. We chose to study glucose tolerance and insulin sensitivity in animals maintained on a low fat diet to minimize any influence of differences in weight. Indeed, the weights of the wild type and null animals were the same. Therefore, the observed increase in insulin sensitivity and improved glucose tolerance of the null animals (Fig. 6) is a reflection of the loss of the Thrsp gene. Furthermore, the reduction in fat pad size likely contributes to this phenotype. What is not clear, however, is whether this effect is a direct effect of the absence of Thrsp in these animals, or a reflection of the reduced lipid consumed as neonates. If this glucose tolerance phenotype is a result of the reduced lipid content of the milk consumed as a neonate, it could have profound public health implications related to infant feeding and the development of type 2 diabetes independent of adult weight gain (Levin, 2006, Plagemann, 2006). Clearly, further experiments are required to sort out these issues.
Table 1.
Summary of Experimental Data
| Experiments Conducted | Backcross Generation | Animal Characteristics | Figures |
|---|---|---|---|
| Weight | N1 | Male and Female Thrsp null and wild type mice Weight in grams from birth to PN200 Fed a 4% fat diet from weaning |
1A, 1B |
| N1 | Male and Female Thrsp null and wild type mice Weight in grams from birth to PN200 Fed an 11% fat diet from weaning |
1C, 1D | |
| N5 | Male and Female Thrsp null and wild type mice Weight in grams from birth to PN420 Fed an 11% fat diet from weaning |
4 | |
| N11 | Male and Female Thrsp null and wild type mice Weight in grams from birth to PN216 Fed a 4% fat diet from weaning |
5A, 5B | |
| N11 | Male and Female Thrsp null and wild type mice Weight in grams from birth to PN216 Fed an 11% fat diet from weaning |
5C, 5D | |
| DEXA scan | N1 | Male Thrsp null and wild type mice Age PN200 Fed an 11% fat diet from weaning (same animals as presented in Figure 1C) |
2 |
| Food Intake | N1 | Male Thrsp null and wild type mice Age PN200 Fed an 11% fat diet from weaning (same animals as presented in Figure 1C) |
3 |
| GTT | N11 | Male Thrsp null and wild type mice Age PN200 Fed a 4% fat diet from weaning |
6A |
| ITT | N11 | Male Thrsp null and wild type mice Age PN200 Fed a 4% fat diet from weaning |
6B |
| Fad Pad | N11 | Male Thrsp null and wild type mice Age PN150 Fed a 4% fat diet from weaning |
7 |
Acknowledgments
The authors wish to thank Mark Margosian for his expert technical assistance and Dr James Levine for his expertise and help with the PIXImus measurements.
This work was supported, in part, by NIH P30-DK50456 and T32-DK007203.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayala JE, Bracy DP, Hansotia T, Flock G, Seino Y, Wasserman DH, Drucker DJ. Insulin action in the double incretin receptor knockout mouse. Diabetes. 2008;57:288–97. doi: 10.2337/db07-0704. [DOI] [PubMed] [Google Scholar]
- Badman MK, Flier JS. The gut and energy balance: visceral allies in the obesity wars. Science. 2005;307:1909–14. doi: 10.1126/science.1109951. [DOI] [PubMed] [Google Scholar]
- Barsh GS, Farooqi IS, O’rahilly S. Genetics of body-weight regulation. Nature. 2000;404:644–51. doi: 10.1038/35007519. [DOI] [PubMed] [Google Scholar]
- Bogue M, Grubb S, Maddatu T. Mouse Phenome Database. The Jackson Laboratory; 2006. http://www.jax.org/phenome. [Google Scholar]
- Campfield LA, Smith FJ, Burn P. Strategies and potential molecular targets for obesity treatment. Science. 1998;280:1383–7. doi: 10.1126/science.280.5368.1383. [DOI] [PubMed] [Google Scholar]
- Coleman DL, Hummel KP. The influence of genetic background on the expression of the obese (Ob) gene in the mouse. Diabetologia. 1973;9:287–93. doi: 10.1007/BF01221856. [DOI] [PubMed] [Google Scholar]
- Diament AL, Warden CH. Multiple linked mouse chromosome 7 loci influence body fat mass. Int J Obes Relat Metab Disord. 2004;28:199–210. doi: 10.1038/sj.ijo.0802516. [DOI] [PubMed] [Google Scholar]
- Edmond J, Auestad N, Robbins RA, Bergstrom JD. Ketone body metabolism in the neonate: development and the effect of diet. Fed Proc. 1985;44:2359–64. [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–61. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Faust IM, Johnson PR, Hirsch J. Long-term effects of early nutritional experience on the development of obesity in the rat. J Nutr. 1980;110:2027–34. doi: 10.1093/jn/110.10.2027. [DOI] [PubMed] [Google Scholar]
- Freake HC, Oppenheimer JH. Stimulation of S14 mRNA and lipogenesis in brown fat by hypothyroidism, cold exposure, and cafeteria feeding: evidence supporting a general role for S14 in lipogenesis and lipogenesis in the maintenance of thermogenesis. Proc Natl Acad Sci U S A. 1987;84:3070–4. doi: 10.1073/pnas.84.9.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–70. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- Harris RB, Mitchell TD, Yan X, Simpson JS, Redmann SM., Jr Metabolic responses to leptin in obese db/db mice are strain dependent. Am J Physiol Regul Integr Comp Physiol. 2001;281:R115–32. doi: 10.1152/ajpregu.2001.281.1.R115. [DOI] [PubMed] [Google Scholar]
- Hummel KP, Coleman DL, Lane PW. The influence of genetic background on expression of mutations at the diabetes locus in the mouse. I. C57BL-KsJ and C57BL-6J strains. Biochem Genet. 1972;7:1–13. doi: 10.1007/BF00487005. [DOI] [PubMed] [Google Scholar]
- Ivanova EA, Bechtold DA, Dupre SM, Brennand J, Barrett P, Luckman SM, Loudon AS. Altered metabolism in the melatonin-related receptor (GPR50) knockout mouse. Am J Physiol Endocrinol Metab. 2008;294:E176–82. doi: 10.1152/ajpendo.00199.2007. [DOI] [PubMed] [Google Scholar]
- Jiang G, Li Z, Liu F, Ellsworth K, Dallas-Yang Q, Wu M, Ronan J, Esau C, Murphy C, Szalkowski D, Bergeron R, Doebber T, Zhang BB. Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J Clin Invest. 2005;115:1030–8. doi: 10.1172/JCI23962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump DB, Narayan P, Towle H, Oppenheimer JH. Rapid effects of triiodothyronine on hepatic gene expression. Hybridization analysis of tissue-specific triiodothyronine regulation of mRNAS14. J Biol Chem. 1984;259:2789–97. [PubMed] [Google Scholar]
- Jump DB, Oppenheimer JH. High basal expression and 3, 5,3’-triiodothyronine regulation of messenger ribonucleic acid S14 in lipogenic tissues. Endocrinology. 1985;117:2259–66. doi: 10.1210/endo-117-6-2259. [DOI] [PubMed] [Google Scholar]
- Kinlaw WB, Schwartz HL, Towle HC, Oppenheimer JH. Opposing effects of glucagon and triiodothyronine on the hepatic levels of messenger ribonucleic acid S14 and the dependence of such effects on circadian factors. J Clin Invest. 1986;78:1091–6. doi: 10.1172/JCI112665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner LS, Mariash CN. Adipose S14 mRNA is abnormally regulated in obese subjects. Thyroid. 1999;9:143–8. doi: 10.1089/thy.1999.9.143. [DOI] [PubMed] [Google Scholar]
- Levin BE. Metabolic imprinting: critical impact of the perinatal environment on the regulation of energy homeostasis. Philos Trans R Soc Lond B Biol Sci. 2006;361:1107–21. doi: 10.1098/rstb.2006.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Thomas TC, Storlien LH, Huang XF. Development of high fat diet-induced obesity and leptin resistance in C57Bl/6J mice. Int J Obes Relat Metab Disord. 2000;24:639–46. doi: 10.1038/sj.ijo.0801209. [DOI] [PubMed] [Google Scholar]
- Mcgarry JD. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- Narayan P, Liaw CW, Towle HC. Rapid induction of a specific nuclear mRNA precursor by thyroid hormone. Proc Natl Acad Sci U S A. 1984;81:4687–91. doi: 10.1073/pnas.81.15.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry CJ, Ozanne SE, Hales CN. Programming of intermediary metabolism. Mol Cell Endocrinol. 2001;185:81–91. doi: 10.1016/s0303-7207(01)00627-x. [DOI] [PubMed] [Google Scholar]
- Pfluger PT, Kirchner H, Gunnel S, Schrott B, Perez-Tilve D, Fu S, Benoit SC, Horvath T, Joost HG, Wortley KE, Sleeman MW, Tschop MH. Simultaneous deletion of ghrelin and its receptor increases motor activity and energy expenditure. Am J Physiol Gastrointest Liver Physiol. 2008;294:G610–8. doi: 10.1152/ajpgi.00321.2007. [DOI] [PubMed] [Google Scholar]
- Plagemann A. Perinatal nutrition and hormone-dependent programming of food intake. Horm Res. 2006;65(Suppl 3):83–9. doi: 10.1159/000091511. [DOI] [PubMed] [Google Scholar]
- Prentice AM. Obesity and its potential mechanistic basis. Br Med Bull. 2001;60:51–67. doi: 10.1093/bmb/60.1.51. [DOI] [PubMed] [Google Scholar]
- Ravussin E, Bogardus C. Energy expenditure in the obese: is there a thrifty gene? Infusionstherapie. 1990;17:108–12. doi: 10.1159/000222456. [DOI] [PubMed] [Google Scholar]
- Schutz Y. Concept of fat balance in human obesity revisited with particular reference to de novo lipogenesis. Int J Obes Relat Metab Disord. 2004a;28(Suppl 4):S3–S11. doi: 10.1038/sj.ijo.0802852. [DOI] [PubMed] [Google Scholar]
- Schutz Y. Dietary fat, lipogenesis and energy balance. Physiol Behav. 2004b;83:557–64. doi: 10.1016/j.physbeh.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Sigmund CD. Viewpoint: are studies in genetically altered mice out of control? Arterioscler Thromb Vasc Biol. 2000;20:1425–9. doi: 10.1161/01.atv.20.6.1425. [DOI] [PubMed] [Google Scholar]
- Towle HC, Mariash CN. Regulation of hepatic gene expression by lipogenic diet and thyroid hormone. Fed Proc. 1986;45:2406–11. [PubMed] [Google Scholar]
- Wang X, Carre W, Zhou H, Lamont SJ, Cogburn LA. Duplicated Spot 14 genes in the chicken: characterization and identification of polymorphisms associated with abdominal fat traits. Gene. 2004;332:79–88. doi: 10.1016/j.gene.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reverter A, Tan SH, De Jager N, Wang R, Mcwilliam SM, Cafe LM, Greenwood PL, Lehnert SA. Gene expression patterns during intramuscular fat development in cattle. J Anim Sci. 2008 doi: 10.2527/jas.2008-1082. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Warden CH, Yi N, Fisler J. Epistasis among genes is a universal phenomenon in obesity: evidence from rodent models. Nutrition. 2004;20:74–7. doi: 10.1016/j.nut.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Blake WL, Salati LM, Clarke SD. Potency of polyunsaturated and saturated fats as short-term inhibitors of hepatic lipogenesis in rats. J Nutr. 1990;120:544–52. doi: 10.1093/jn/120.6.544. [DOI] [PubMed] [Google Scholar]
- Woods SC, Seeley RJ, Porte D, Jr, Schwartz MW. Signals that regulate food intake and energy homeostasis. Science. 1998;280:1378–83. doi: 10.1126/science.280.5368.1378. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, Mariash CN. The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology. 2005;146:3343–50. doi: 10.1210/en.2005-0204. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Mariash A, Margosian MR, Gopinath S, Fareed MT, Anderson GW, Mariash CN. Spot 14 gene deletion increases hepatic de novo lipogenesis. Endocrinology. 2001;142:4363–70. doi: 10.1210/endo.142.10.8431. [DOI] [PubMed] [Google Scholar]


