Abstract
Potato (Solanum tuberosum) multicystatin (PMC) is a crystalline Cys protease inhibitor present in the subphellogen layer of potato tubers. It consists of eight tandem domains of similar size and sequence. Our in vitro results showed that the pH/PO4−-dependent oligomeric behavior of PMC was due to its multidomain nature and was not a characteristic of the individual domains. Using a single domain of PMC, which still maintains inhibitor activity, we identified a target protein of PMC, a putative Cys protease. In addition, our crystal structure of a representative repeating unit of PMC, PMC-2, showed structural similarity to both type I and type II cystatins. The N-terminal trunk, α-helix, and L2 region of PMC-2 were most similar to those of type I cystatins, while the conformation of L1 more closely resembled that of type II cystatins. The structure of PMC-2 was most similar to the intensely sweet protein monellin from Dioscorephyllum cumminisii (serendipity berry), despite a low level of sequence similarity. We present a model for the possible molecular organization of the eight inhibitory domains in crystalline PMC. The unique molecular properties of the oligomeric PMC crystal are discussed in relation to its potential function in regulating the activity of proteases in potato tubers.
INTRODUCTION
The soluble proteins of potato (Solanum tuberosum) are comprised primarily of protease inhibitor proteins (∼50%) and patatin (∼40%) (Pouvreau et al., 2001). The inhibitors include protease inhibitors I and II (and other Ser protease inhibitors), potato aspartate protease inhibitor, Kunitz-type protease inhibitor, potato carboxy-peptidase inhibitor, potato Cys protease inhibitor, and potato multicystatin (PMC) (Rodis and Hoff, 1984; Pouvreau et al., 2001). PMC is a Cys protease inhibitor (cystatin) that occurs in high concentrations as discrete crystals in the cortical parenchyma cells directly underneath the periderm of tubers (Hoff et al., 1972). The cuboidal or prismoidal crystals of PMC range in size from 5 to 25 μm and are present from the early stages of tuber development to the onset of sprouting from fully developed tubers. The crystalline nature of PMC facilitates its isolation to purity via sucrose density gradient centrifugation (Rodis and Hoff, 1984; Kumar et al., 1999).
In contrast with cystatins from other plant and animal sources, PMC has a high molecular mass (∼85 kD/monomer); most plant protease inhibitors vary between 8 and 25 kD (Garcia-Olmedo et al., 1987). PMC can bind and inhibit several Cys proteases (e.g., papain) simultaneously, and the term multicystatin thus refers to its multiple inhibitory domains (Walsh and Strickland, 1993). PMC consists of eight tandem cystatin domains of ∼10 kD molecular mass, which share protein sequence identities of 53 to 89% and are linked by proteolytically sensitive short linkers (Walsh and Strickland, 1993). PMC can be degraded in vitro by trypsin (Ser protease) into a 35-kD fragment and 10-kD fragments, and both types of fragment retain inhibitory activity (Rodis and Hoff, 1984; Walsh and Strickland, 1993). While the 35-kD fragment binds to two to three papain molecules, the 10-kD fragments each bind to one. Therefore, the molecular architecture of PMC was proposed to consist of eight inhibitory domains united at regions that are sensitive to proteolysis (Walsh and Strickland, 1993).
The soluble form of PMC (85 kD) is distributed throughout the tuber. When ingested by insects, the acidic pH of the midgut solubilizes the crystalline PMC, thus interfering with protein digestion (Walsh and Strickland, 1993; Orr et al., 1994). Cys proteases dominate the digestive protease profile of phytophagous coleopteran (beetle) insect larvae (Murdock et al., 1987). Inclusion of PMC in the diet of such insects at levels found in tubers significantly inhibits their growth (Orr et al., 1994). Hence, protease inhibitors in general are valued as potential insecticides (Ryan, 1990; Jongsma and Bolter, 1997; Schuler et al., 1998; Zavala et al., 2004), and transformation of plants with genes encoding Cys protease inhibitors imparts resistance against select insect pests (Irie et al., 1996).
The cystatin superfamily, which includes PMC, is a family of inhibitors that binds reversibly to the widely distributed Cys proteases, with a stoichiometry of 1:1, and with high affinity (Stubbs et al., 1990; Turk and Bode, 1991). Cystatins can be divided into several groups. The type I cystatin family (∼11 kD) consists of proteins with no disulfide bonds and includes human stefin A and B, and rat cystatin. The larger type II cystatin family (∼13 kD) consists of proteins with two conserved disulfide bonds and includes chicken egg white (CEW) cystatin and human cystatin C, D, and F. The largest family of cystatins, the type III cystatin family (∼68 to 120 kD), includes kininogen, which is made up of several glycosylated type II cystatin-like domains (reviewed in Rawlings and Barrett, 1990; Turk and Bode, 1991). More recently, several other types of cystatin-like inhibitor proteins have been characterized (Brown and Dziegielewska, 1997). Even though sequence similarity between PMC and the type I cystatin family is low, an individual domain of PMC could belong to the type I cystatin family because they both lack disulfide bonds. However, the tethered arrangement of eight domains and the natural existence in a crystalline form are unique to PMC.
High-resolution structures for some of the cystatins have been determined, including CEW cystatin (Bode et al., 1988; Dieckmann et al., 1993), human stefin A (Martin et al., 1995; Tate et al., 1995; Jenko et al., 2003), and human stefin B (Stubbs et al., 1990). To date, only one nuclear magnetic resonance structure for cystatin (oryzacystatin-1) from rice (Oryza sativa) is available (Nagata et al., 2000). The highly conserved amino acid residues and secondary structures in these type I and II cystatins generate an intimate contact region with the target proteases. This contact region is predominantly hydrophobic and has a tripartite nature (Stubbs et al., 1990). Cystatin binds to the target protease through a wedge-shaped edge composed of three different regions that are separated in the primary sequence. This edge is composed of the first binding loop (L1), the second binding loop (L2), and the N-terminal trunk, which together form the wedge-shaped region that is complementary to the active-site cleft of the target protease (Stubbs et al., 1990). L1 is a β-hairpin loop containing a highly conserved QXVXG motif. Hydrogen bonds form between the backbone of these conserved residues and the binding pocket residues of the target Cys proteases. However, substituting amino acids in the L1 site does not substantially affect inhibition of the target protease (Stubbs et al., 1990). L2 is also a β-hairpin loop and contains an aromatic residue, Trp or His, that interfaces with hydrophobic residues in the binding pocket of the protease. The third contact point, referred to as the N-terminal trunk, contains a conserved Gly-Gly sequence after the five or six residues that form the disordered N-terminal. Truncation of the N-terminal amino acids results in the loss of inhibition in CEW cystatin (type-II) but not in the stefin family (type-I), indicating a slight difference in mode of action among cystatins (Abe et al., 1988; Thiele et al., 1988; Stubbs et al., 1990).
The ability of PMC to inhibit Cys proteases (e.g., papain, chymopapain, and ficin) is well known (Rodis and Hoff, 1984); however, its physiological role in tuber ontogeny remains unresolved. PMC accumulation in tubers precedes patatin (45-kD storage glycoprotein) deposition during the initial stages of tuber development (Weeda et al., 2008). In storage, the PMC content of tubers declines over time, concomitant with a loss of patatin. PMC levels also decline precipitously during the early stages of plant development from potato tubers, and this likely facilitates mobilization of nitrogen reserves to support the developing plants (Weeda et al., 2009). Thus, by modulating the activities of Cys proteases, PMC may have a role in regulating tuber protein content during development, in storage, and during sprouting. Understanding the molecular structure and monomer-to-oligomer transition of PMC in relation to its function as a protease inhibitor is a prerequisite for defining how PMC is catabolized in vivo and for elucidating its potential role in regulating the protein content and nutritional value of potato. The objective of this study was to define in detail the structural characteristics of PMC, including its polymerization and the arrangement of individual domains in the multidomain crystal, relative to that of a single functional domain.
RESULTS
Limited Proteolysis of PMC
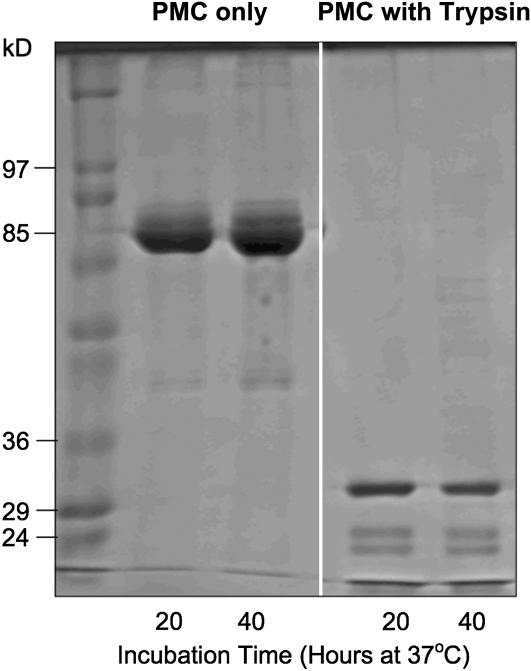
To understand the arrangement of the eight domains in native PMC, a trypsin digestion experiment was performed. As reported previously (Walsh and Strickland, 1993), tryptic digestion of PMC produced a unique profile as shown by SDS-PAGE (Figure 1). After 20 h of digestion, the 85-kD PMC was reduced to three species of distinct molecular mass, ∼35, ∼15, and ∼10 kD, which was maintained through 40 h of incubation. The corresponding bands were excised from the gel and subjected to mass spectroscopic (liquid chromatography–tandem mass spectrometry [LC-MS/MS]) and N-terminal sequence analyses. The exact molecular mass of the ∼35-kD fragment was 33.7 kD and spanned from residues Leu-382 to Arg-589 of PMC, which correspond approximately to domains 5, 6, and 7. The predominant species in both the ∼15- and ∼10-kD fragments has LGGIIVP as the N-terminal sequence, which corresponds to the N-terminal of domain 6. However the ∼15-kD band contains a small amount of peptides derived from other domains, which have similar amino acid sequences.
Figure 1.
SDS-PAGE Showing the Tryptic Digestion of PMC from Potato Tuber.
PMC (43.5 μg) was incubated with or without trypsin (50 μg) in Tris-HCl buffer (0.1 M, pH 7.4). The 85-kD native PMC was digested to 35-, 15-, and 10-kD fragments after 20 and 40 h of incubation. Lane 1: molecular mass standards; lanes 2 and 3: PMC was incubated without trypsin; lanes 4 and 5: PMC was incubated with trypsin. Each lane was loaded with 6.7 μg PMC equivalent protein.
Localization, Isolation, and Crystallization of PMC
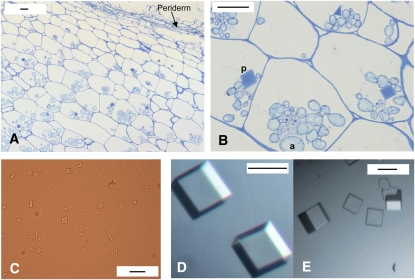
The crystalline form of PMC is readily apparent in the parenchyma tissue directly beneath the periderm of potato tubers (Figures 2A and 2B). While not all cells contain PMC crystals, their number appears limited to one per cell where present. Diffraction studies were conducted to decipher the detailed lattice interactions and arrangement of the individual domains in the natural crystalline form of PMC. However, crystals isolated directly from potato tubers (Figure 2C) failed to diffract. To improve the quality of the crystal, native crystals were solubilized and purified by an anionic exchange column (mono-Q). The crystals formed from the purified PMC (Figure 2D) were all cuboidal in shape and larger than the native crystals. The crystal lattice of PMC belonged to an orthorhombic space group, I222, with cell lengths of a = 205, b = 208, and c = 236 Å, but diffracted only up to ∼20 Å, even after 1 min of exposure to the synchrotron radiation (Advanced Light Source beamline 8.2.1). Therefore, for further characterization, one of the eight repeating units of the PMC gene was cloned. The second repeating unit (PMC-2) was selected because it contains the largest amount of consensus sequences among the eight repeating units (Figure 3). The resulting recombinant PMC-2 domain was purified, and its molecular mass was confirmed (13.5 kD) by MS analysis.
Figure 2.
Light Micrographs of Potato Multicystatin Crystals in Tubers.
(A) and (B) Periderm and underlying cortical parenchyma cells containing amyloplasts (a) and darkly stained cuboidal and pyramidal crystals of PMC (p).
(C) Native PMC crystals isolated from potato tubers on a sucrose gradient.
(D) and (E) Recrystalized PMC.
Bars = 50 μm in (A), (B), and (E) and 100 μm in (C) and (D).
Figure 3.
Amino Acid Sequence Alignment.
The recombinant PMC-2 and other repeating units of PMC were compared with oryzacystatin-I, human stefin A, human stefin B, CEW cystatin, human cystatin F, single chain monellin, and mouse latexin domain 1. The β-strands are highlighted in green, and the α-helices are highlighted in yellow.
Comparison between PMC and PMC-2
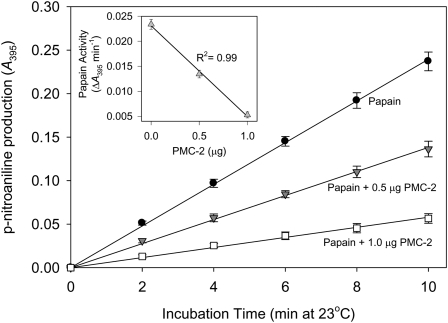
Protease inhibitory activity of recombinant PMC-2 was verified by quantifying its ability to attenuate papain-mediated degradation of N-α-benzoyl-dl-arginine-p-nitroanilide (BAPNA). PMC-2 inhibited the catalysis of BAPNA by papain in a concentration-dependent manner (Figure 4).
Figure 4.
The Ability of PMC-2 to Inhibit the Catalysis of BAPNA by Papain.
Papain protease activity was quantified by monitoring the production of p-nitroaniline from BAPNA at A395 for 10 min. Papain activity declined linearly with increasing PMC-2 concentration (inset). Error bars indicate ± se mean (n = 3).
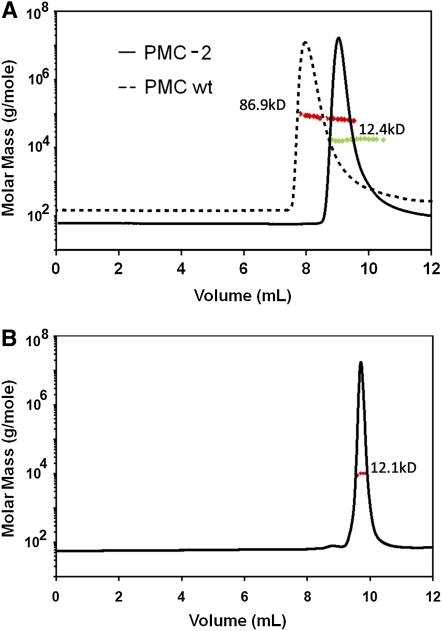
The pH-dependent oligomerization behavior of PMC and PMC-2 was then compared by monitoring elution profiles from size exclusion chromatography using multiangle static laser light scattering (690 nm) and differential refractometry, which allow determination of molecular weights across chromatographic peaks. As shown in Figure 5A, both PMC and PMC-2 exist as monomers in 20 mM acetate buffer at pH 5.5. PMC-2 also maintains its monomeric state in 20 mM phosphate buffer at pH 7.5 (Figure 5B), even though PMC aggregates or crystallizes at that pH, effectively preventing HPLC elution.
Figure 5.
Molecular Mass Determination of PMC and PMC-2.
(A) Multiangle laser light scattering elution profile of PMC (black dotted line, ∼0.2 mg/injection) and PMC-2 (black solid line, ∼0.2 mg/injection) at pH 5.5 (20 mM sodium acetate buffer). The red and green dotted lines indicate calculated molecular masses, and the average molecular masses calculated from light scattering are indicated next to the corresponding elution peaks.
(B) Multiangle laser light scattering elution profile of PMC-2 at pH 7.5 (20 mM sodium phosphate buffer). Elution profile is shown as molecular mass versus elution volume. The black solid line represents changes in refractive index on an arbitrary scale that is proportional to protein concentration. The red dotted line indicates calculated molecular masses, and the average molecular mass calculated from light scattering is indicated next to the elution peak.
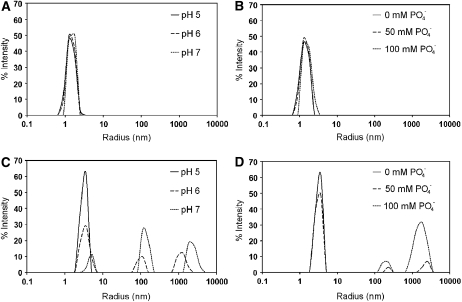
In addition, both wild-type PMC and recombinant PMC-2 were subjected to dynamic light scattering studies to determine their tendencies to form oligomers upon gradual increase of phosphate ions and pH. The results showed that PMC-2 is consistently monomeric from pH 5.0 to 7 (Figure 6A) and from 0 to 100 mM phosphate (Figure 6B). However, wild-type PMC shows a rapid transition to a higher molecular mass species upon incremental increase of pH (Figure 6C). PMC also transitions to a higher molecular mass upon incremental increase of the phosphate concentration (Figure 6D). The new molecular mass profiles induced by increased phosphate concentration or pH were stable even after several hours.
Figure 6.
Dynamic Light Scattering Data.
(A) Molecular radius profile of PMC-2 at various pH values (pH 5, 100 mM Na acetate buffer; pH 6, 100 mM Na cacodylate buffer; pH 7, 100 mM HEPES buffer).
(B) Molecular radius profile of PMC-2 at various concentrations of phosphate ions.
(C) Molecular radius profile of PMC at various pH values (same pH values and buffers as in [A]).
(D) Molecular radius profile of PMC at various concentrations of phosphate ions.
The y axis represents the percentage of the scattered light, and the x axis represents the radius of the PMC or PMC-2 molecules.
Crystal Structure of PMC-2
The PMC-2 domain was crystallized in the monoclinic space group, C2, diffracting to 2.5 Å resolution (Table 1). Our attempt to solve the structure with the nuclear magnetic resonance coordinate of oryzacystatin (1EQK) was not successful. Therefore, SeMet-substituted PMC-2 preparation was crystallized in the same space group, and its structure was determined by the selenomethionyl multiwavelength anomalous diffraction (MAD) method (Hendrickson, 1991). Most of the backbone and side chain residues were fitted using the Se-MAD map, and the final R-factor for PMC-2 was 21.2% (Rfree = 24.6) for 34,671 (10 to 2.7 Å) unique reflections (Table 1). The asymmetric unit of the crystal contains 14 independent PMC-2 molecules, which is somewhat more molecules than typical crystals. The global structures of these 14 PMC-2 molecules in the asymmetric unit are all essentially superimposable, as demonstrated by the small average root mean square deviations (rmsd) of 0.14 Å.
Table 1.
Crystallographic Data for PMC-2
| Data | Native C2 | SeMet C2 | Native P212121 |
|---|---|---|---|
| Wavelength (Å) | 1.0332 | 0.97930 (W1) | 1.54 |
| 0.97943 (W2) | |||
| 0.95370 (W3) | |||
| Resolution (Å) | 20–2.7 | 50–2.8 | 20–2.5 |
| Space group | C2 | C2 | P212121 |
| Cell dimensions (Å) | a = 210.85 | a = 211.56 | a = 25.46 |
| b = 85.71 | b = 85.92 | b = 54.53 | |
| c = 96.86 | c = 96.97 | c = 79.15 | |
| β = 100.60 | β = 100.59 | ||
| Asymmetric unit | 14 molecules | 14 molecules | 1 molecule |
| Total observations | 103,238 | 63,398 (W1) | 11,023 |
| 61,781 (W2) | |||
| 62,337 (W3) | |||
| Unique reflections | 40,548 | 33,670 (W1) | 3,847 |
| 29,699 (W2) | |||
| 30,875 (W3) | |||
| Completeness (%) | 88.4 (69.6) | 80.6 (48.2) | 94.8 (90.4) |
| 74.1 (35.9) | |||
| 77.1 (43.6) | |||
| Rsymab | 5.5 (11.4) | 5.3 (11.6) | 3.0 (7.4) |
| 5.9 (13.6) | |||
| 5.7 (10.6) | |||
| Refinement | |||
| Resolution (Å) | 10–2.7 | 10–2.5 | |
| Number of reflections (>2σ) | 34,671 (75.6%) | 3,837 (94.5%) | |
| Rcrystc | 21.2 | 19.3 | |
| Rfreed | 24.6 | 23.2 | |
| rmsd bonds (Å) | 0.014 | 0.016 | |
| rmsd angles (°) | 3.185 | 3.82 | |
| Number of atoms | |||
| Protein | 10,038 | 718 | |
| Water | 0 | 64 |
Numbers in parentheses refer to the highest-resolution shell.
Rsym = ΣIh – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalent reflections.
Rcryst = Σ|Fobs – Fcalc|/ΣFobs, where summation is over the data used for refinement.
Rfree was calculated as for Rcryst using 5% of the data that was excluded from refinement.
To confirm the heterogeneity of the primary sequence among the repeating units, we unambiguously identified the individual amino acids in electron density maps. In all 14 molecules in the asymmetric unit, the corresponding electron density for the first three amino acids from the N terminus and the last four amino acids from the C terminus were not visible, probably due to their disordered nature. The residues preceding the second residue of the conserved GG pair were flexible, as was previously observed for the structures of other cystatins (Bode et al., 1988). Additionally, instead of two Asp residues in positions 61 and 62, as previously reported (Waldron et al. 1993), the electron density map indicated that only one Asp was present. Therefore, it is likely that there is only one Asp, and the corrected local sequence is in good agreement with the other repeats (Figure 3).
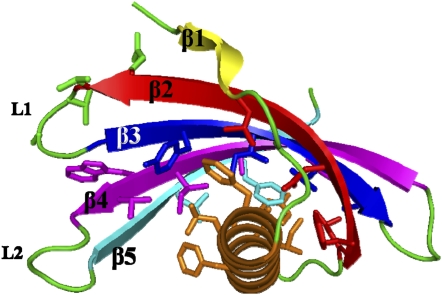
The global shape for PMC-2 is illustrated in Figure 7. The five-stranded antiparallel β-sheet (β1-β5) wraps around a central five-turn α-helix, with the β-strands in an almost perpendicular orientation to the helical axis. The α-helix consists of residues Asn-13 to Asn-32, and the five β-strands (β1 to β5) consist of Val-6 to Val-8, Glu-36 to Val-48, Ile-51 to Asp-61, Lys-65 to Lys-76, and Phe-81 to Val-90, respectively.
Figure 7.
Ribbon Diagram Representing the Crystal Structure of PMC-2.
The individual residues are depicted with the same colors as the secondary structural elements to which they belong (see Figure 3).
The tightly packed core of PMC-2 is mainly composed of nonpolar, nonhydrogen-bonding hydrophobic side chains contributed by the α-helix and β-strands. Seven of the 20 amino acids in the core are identical among PMC repeats, and the differences among the remaining 10 residues are conservative. Specifically, Phe-16, Leu-19, Ala-20, Phe-22, Ala-23, Ile-24, and Tyr-27 are from the α-helix; Leu-35, Phe-37, Asn-40, and Val-43 are from the β2-strand; Tyr-53, Ile-55, Leu-57, and Ala-59 are from the β3-stand; Tyr-69, Ala-71, Ile-73, and Val-75 are from the β4-strand; and Val-84 and Phe-87 are from the β5-strand. A unique polar residue, Asn-40, is conserved among repeating units, except in the first unit (PMC-1), where residue 40 is Val. Noticeably, both of the conserved residues, Tyr-53 and Tyr-69, with their polar hydroxyl groups, are located on the edge of the hydrophobic core and are partially exposed to the solvent. The significance of Tyr-53 will be discussed later.
As observed in other cystatin structures (Murzin, 1993; Nagata et al., 2000), the inner two β-strands, β3 and β4, are coiled smoothly, forming regular antiparallel hydrogen bonds, but some parts of the outer two β-strands, β2 and β5, have irregular backbone hydrogen bonds. Three bulges were identified, which are formed by the hydrogen bonds from the backbone amide nitrogen atoms of both Leu-41 and Asn-42 to the carbonyl oxygen of Thr-56, from both Val-38 and Glu-39 to Ala-58, and from both Val-85 and Glu-86 to Lys-72. Together, these bulges produce a tight coiling of the β-sheet, allowing it to wrap around the helix.
Structural Differences among Asymmetric Units
Comparisons of the asymmetric units reveal several structural differences. The average rmsd values (Cα atoms only) are ∼0.14 Å among pairwise comparisons; thus, there are only small global changes for each molecule. In general, the primary sequences of the secondary structural elements, except for β1, are highly conserved. The most significant differences in primary sequence among the repeating units in PMC occur in β1 and its N-terminal overhang, and the N- and C-side loops of the α-helix, which are also reflected in the structural differences among the 14 molecules in the asymmetric unit. The largest differences among the molecules in the asymmetric unit occur in the loop that is located in the N-terminal side of the α-helix. In addition, the loop connecting β3 and β4 shows some degree of variation. These regions are the most heterogeneous in terms of primary sequence among various other cystatins. The type-I cystatins have a long insertion in the same loop area connecting β3 and β4 (Figure 3).
We were also able to crystallize PMC-2 in the orthorhombic space group of P212121, containing only one molecule in an asymmetric unit. These crystals were extremely sensitive to any kind of manipulation and diffracted to 2.5 Å resolution. The corresponding refined structure was superimposed to the 14 molecules of the C2 crystal with an average rmsd of 0.99 Å.
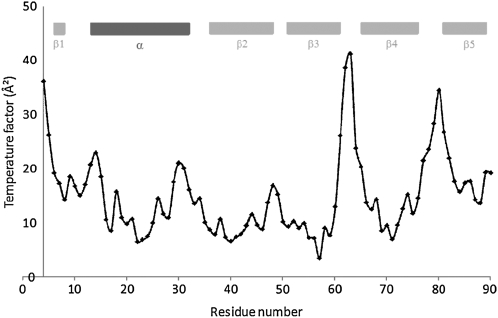
Graphs of backbone atom temperature (B) factors of the P212121 crystal structure are shown in Figure 8 and generally correlate with the structural differences among molecules in the asymmetric unit of the C2 crystal structure. The largest B factors for PMC-2 structure correspond to the N-terminal trunk, L1, and L2, that is, all tripartite inhibitory components, as discussed later. The corresponding density for the first three residues, Met-Pro-Gly, is not visible, and the following two residues, namely the GG trunk, have a high temperature factor.
Figure 8.
Backbone Temperature Factor Plots of PMC-2 Crystal Structure.
Besides the N terminus, residues 61 to 63 and 78 to 80 show temperature factors of their corresponding Cα atoms above 25 Å2. The corresponding secondary structural elements (see Figures 3 and 7) are indicated with bars.
PMC-2 Pull-Down Assay
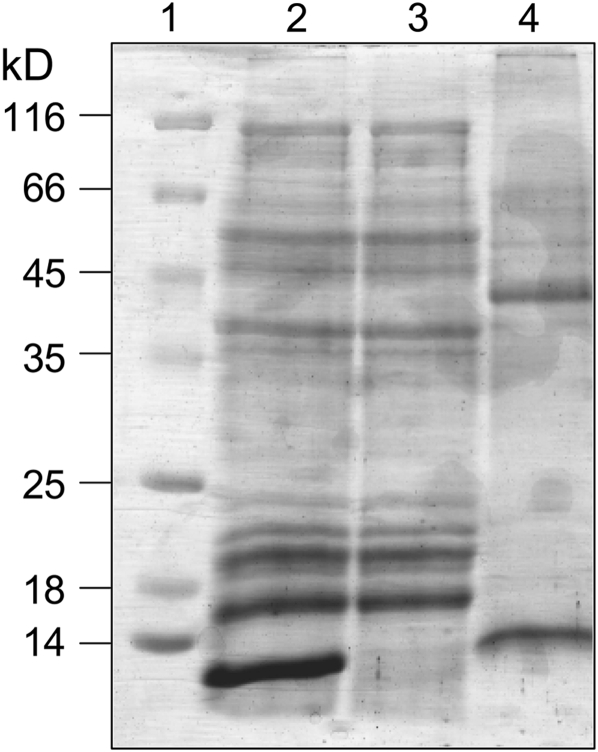
Identification of the potato Cys proteases inhibited by PMC has been difficult, as tuber extracts are extremely susceptible to proteolysis. To identify the interacting enzymes or proteases, purified His-tagged PMC-2 was added to fresh potato extract, and the mixture was then applied to Ni2+-NTA resin, followed by extensive washing to remove noninteracting proteins. Proteins binding to PMC-2 were eluted with 8 M urea, leaving the PMC-2 adsorbed to the resin. Analysis of the eluted proteins by SDS-PAGE revealed one predominant protein band of ∼45 kD, a PMC-2 band of ∼10 kD, and a number of less intense bands (Figure 9). LC-MS/MS analysis detected three peptides, IDSYEDVPVNNEK, AVAHQPVSIAIEAGGR, and NVASSSGLCGLATEPSYPVK, which were used to identify the predominant protein as a 466–amino acid putative Cys protease from S. tuberosum (Avrova et al., 1999).
Figure 9.
SDS-PAGE Analysis of the PMC-2 Pull-Down Experiment.
Lane 1: molecular weight markers. Lane 2: tuber extract with added H6-PMC-2. Lane 3: sample, as in lane 2, after passage through a Ni2+-NTA column. Lane 4: fraction eluted from the column with 8 M urea.
DISCUSSION
The primary structure of PMC is composed of eight superimposable repeating units, with slightly different primary sequences (Figure 3). The size of the individual domains of the native PMC molecule, ∼10 kD, is similar to that of type-I and type-II cystatins. It has been reported that proteolytic fragmentation of PMC does not destroy its ability to inhibit papain, but the fragments no longer crystallize at basic pH and in the presence of phosphate (Walsh and Strickland, 1993). In addition to the 10-kD fragment, there exist ∼35- and ∼15-kD fragments that are resistant to further digestion (Figure 1). It has been proposed that the ∼35-kD fragment could be due to sequence differences in the interdomain regions or to differences in presentation or accessibility of specific regions to trypsin (Walsh and Strickland, 1993). Our MS analysis of the ∼35-kD fragment, followed by careful sequence inspection, shows that it contains domains 5, 6, and 7. Its resistance to digestion is likely due to its compact structure, since there are several potential cutting sites inside this fragment (Figure 3). In addition, the N-terminal amino acid sequence shows that the major portion of both the ∼15- and ∼10-kD fragments contain domain 6.
The crystalline form of PMC dissolves in sodium acetate buffer at pH 5.0 (Rodis and Hoff, 1984). Our static and dynamic light scattering data confirm the monomeric nature of PMC at pH 5.5 (Figure 6). This low pH solubility of PMC has been suggested as a mechanism of defense, by which monomerization of PMC occurs in the mildly acidic midgut of insects (Walsh and Strickland, 1993). Cys proteases dominate the midgut of coleopteran larvae (Murdock et al., 1987). When ingested by such insects, PMC is solubilized in the acidic midgut to cause an effective inhibition of Cys proteases. Thus, PMC interferes with digestion of protein and effectively retards larval growth. For example, PMC is known to inhibit the growth of the western cornworm (Orr et al., 1994).
Our light scattering data also confirm that monomeric PMC tends to polymerize as phosphate ion and pH increase (Figures 5 and 6). The ability to structurally modify PMC through changes in cellular pH and phosphate levels is likely to be biologically significant, providing a mechanism by which the activities of Cys proteases can be modulated to facilitate key physiological processes. Through maintaining neutral pH and/or a relatively high concentration of phosphate ion, tubers can store a substantial amount of inactive crystalline PMC, which can then be easily activated not only for defense but to facilitate protease-dependent aspects of tuber development, such as protein deposition, protein mobilization, and programmed cell death. For example, changes in inorganic phosphate (Pi) concentration in the cytosol depend on flux of bound phosphorus from various reservoirs (e.g., starch-bound, phytic acid, sugar phosphates, proteins, nucleic acids, and phospholipids), which is tightly regulated and thus dependent on the stage of tuber development (Samotus and Schwimmer, 1962a, 1962b).
Early in development, tubers actively synthesize and accumulate starch and protein. Potato starch is phosphorylated, and this, along with synthesis of other P-containing carbohydrates, lipids, proteins, and nucleic acids during this metabolically active growth phase, results in relatively low levels of Pi in the cytosol of young, developing tuber cells (Samotus and Schwimmer, 1962b). A relatively low level of cytosolic Pi favors the monomeric (active) form of PMC, which in turn would inhibit Cys protease activity and thus facilitate protein synthesis and deposition in the developing tubers. Indeed, PMC accumulation precedes the deposition of patatin, a major storage glycoprotein (Weeda et al., 2008). Tissue probed with anti-PMC has demonstrated the uniform distribution of monomeric PMC throughout the tuber during early development (Kumar et al., 1999). Moreover, in vitro studies have shown that proteolysis of patatin by tuber-derived Cys proteases is inhibited by PMC (Kumar et al., 1999). When tubers reach full size, synthesis of starch and other P-containing metabolites slows or ceases altogether as a sink for Pi, potentially effecting an increase in cytosolic Pi (Samotus and Schwimmer, 1962b) and favoring the crystalline inactive storage form of PMC, which is easily isolated from fully mature tubers at harvest (Rodis and Hoff, 1984).
Structural modifications of PMC may also impact the ability to wound-heal. Wounding of tubers results in a decline in pH at the wound site due to vacuolar disruption. The resultant PMC monomer potentially inhibits endogenous tuber proteases at the wound surface, which would otherwise degrade key enzymes required for wound healing. In support of this, tubers progressively lose the ability to develop a functional wound periderm as they age in storage (Kumar and Knowles, 2003, 2007) and simultaneously lose PMC, which increases Cys protease activity (Kumar et al., 1999). During sprouting, starch mobilization from seed tubers likely increases cytosolic phosphate, thus favoring the inactive crystalline form of PMC. This would facilitate protein catabolism by Cys proteases (Kitamura and Maruyama, 1985; Michaud et al., 2006), thereby providing a source of nitrogen to support early plant development. Hence, changes in pH and Pi concentrations during development may contribute to structural changes in PMC, which in turn modulate protease activity to regulate protein accumulation during tuber development or protein mobilization from seed tubers during sprouting. The ability of monomeric PMC to remain effective as a Cys protease inhibitor would also depend on the presence of PMC-targeting proteases, which are likely expressed at different stages of tuber ontogeny (e.g., developing tubers, dormant, and nondormant).
To study the characteristics of the individual domains of PMC, such as the oligomerization and inhibition activity, the second repeating unit (PMC-2) was cloned as a representative domain of PMC. PMC-2 has the greatest percentage of conserved residues among the eight repeating units and shows the highest level of sequence identity to oryzacystatin-1 (47%). PMC-2 maintains its papain inhibition activity and forms a tight complex with one specific potato Cys protease (accession number CAB53515) but does not exhibit the pH- and PO4−-dependent oligomerization behavior of PMC (Figures 5 and 6).
Comparison of PMC with Other Cystatins and the Sweet-Tasting Protein Monellin
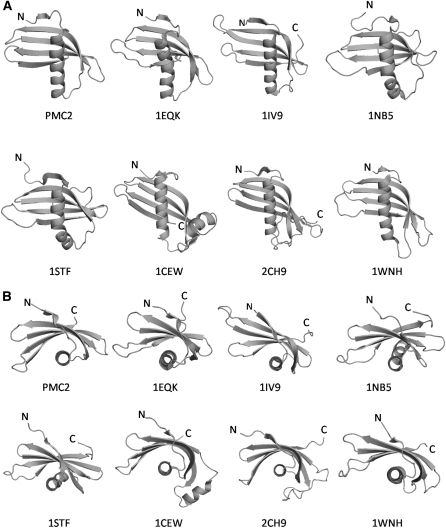
A detailed comparison with available structures in the Protein Data Bank database was performed using a Dali search (Holm and Sander, 1993). As expected, the PMC-2 structure is similar to the other known cystatin structures. However, to our surprise, the most similar structure was a single-chain monellin (1MOL, 1IV9), an intensely sweet protein from the African serendipity berry (Dioscorephyllum cumminisii) (Z score = 12.2). The other similar structures were oryzacystatin-1 (1EQK; Z score = 12.1), a CEW cystatin (1YVB; Z score = 11.1), human cystatin F (2CH9; Z score = 11.0), human stefin A (1NB5; Z score = 11.0), human stefin B (1STF; Z score = 10.6), and the first domain of the mouse latexin (1WNH; Z score = 9.6). The structural similarity of latexin, a carboxypeptidase inhibitor, confirms the previous notion that PMC could have evolved from an ancestral cystatin-like protein as a consequence of a gene duplication event (Aagaard et al., 2005). However, latexin has two tightly packed cystatin-like domains and the smallest amount of structural and sequential similarity to PMC among all cystatins examined, and it will thus not be included in the following detailed discussion.
The folding topology of the compared cystatin structures with high Z scores was identical, and their individual secondary structural elements started and ended at similar positions (Figures 3 and 10). Not only do they share a common fold, but they also have a high degree of sequence identity. The structure-based alignment (Figure 3) shows that PMC-2 has 42 residues identical to oryzacystatin-1, 21 and 20 identical to CEW cystatin and stefin A, respectively, 16 and 15 identical to monellin and stefin B, respectively, and eight identical to those in latexin. The α-helix of all of these proteins is perpendicular to the direction of the β-strands, in contrast with the common parallel arrangement present in other proteins (Janin and Chothia, 1980; Murzin, 1993).
Figure 10.
Ribbon Diagrams Representing the Structures of Various Cystatins.
Oryzacystatin-I (1EQK), single-chain monellin (1IV9), human stefin A (1NB5), human stefin B (1STF), CEW cystatin (1CEW), human cystatin F (2CH9), and mouse latexin domain 1 (1WNH). PMC-2 has a straight α-helix, as do monellin (1IV9), CEW (1CEW), and human cystatin F (2CH9). However, the orientation angle between the α-helix and β-sheet in PMC-2 is perpendicular ([A], side view; [B], top view).
In spite of the similarities in topology, there were two cases that showed significant differences between PMC-2 and these other proteins (Figure 10). The most significant global differences were found between PMC-2 and CEW cystatin. The differences between these two proteins can be best characterized as rotational, with the helical axis fixed. The differences become larger at the end of the β-strands, reaching a maximum difference of 6.7 Å between the corresponding backbone atoms. The structures of PMC-2 and oryzacystatin-I also show a global difference. One end of the β-sheet in PMC-2, composed of the N-side of β2, the C-side of β3, and the N-side of β4, bends more toward the C-end of the α-helix compared with the same area in oryzacytatin-I. Consequently, the overall β-sheet of oryzacystain-I is slightly unwrapped relative to PMC-2.
There were also many differences between PMC-2 and the other cystatin proteins, which will be discussed below in a manner from the N to the C terminus direction. The electron density of PMC-2 starts from Gly-4, which is absolutely conserved in all cystatin molecules. The disordered character of the preceding three residues is similar to that observed in the structures of CEW cystatin and oryzacystatin-I. However, in stefin A and B, the corresponding N-terminal residues are ordered (Stubbs et al., 1990).
The β1 of PMC-2 is the shortest strand and is composed of three backbone hydrogen bonds from Val-6–Asn-7–Val-8 to Val-43 and Glu-45, followed by a big bulge made by Pro-9–Asn-10–Pro-11. The existence of the bulge after the β1-strand is common among the high Z score cystatin proteins, but the size of the bulge observed in PMC-2 is bigger than in the others, followed in size by latexin, which has two Pro residues in the corresponding sequence. The α-helix of PMC-2 was straight and all the hydrogen bonds exist in a regular α-helix pattern in contrast with the kinked conformation observed in the middle of the helices of oryzacystatin-I, stefin A, and stefin B. Therefore, the α-helix of PMC-2 was more similar to CEW cystatin and monellin than to the other cystatin proteins.
The sequence differences in the loop between the α-helix and β2 among the compared cystatins (Figure 3) are reflected by structural differences (Figure 10). However, the backbone conformation of PMC-2 around the loop connecting the α-helix and β2 was similar to that in stefin A and B, and quite different from that in CEW cystatin, which has two more residues in the corresponding loop.
The first β-turn loop, L1 of PMC, which connects β2 and β3 and contains the QXVXG motif, showed an almost superimposable conformation to oryzacystatin-I (1EQK) and also to stefin A and B in complex forms with protease (1NB5 and 1STF); however, it differed from CEW cystatin. The residues in the QXVXG motif showed only slightly higher temperature factors, and the φ-ψ dihedral angles of Val-48 are in a slightly strained conformation. Previously, the equivalent Val residue (Val-55) in the stefin B:papain complex (1STF) was shown to be in a similarly strained conformation and different from free CEW cystatin (Stubbs et al., 1990). Thus, it has been suggested that the strained conformation is due to complex formation and that stefin B will have a more relaxed conformation as a free inhibitor (Stubbs et al., 1990). Therefore, the observed strained conformation of the Val residue in the QXVXG motif in our free PMC-2 structure suggests its inherent nature as opposed to the previous proposal referring to complex formation.
There is also significant sequence heterogeneity among molecules in the loop region connecting β3 and β4 (Figures 3 and 10). The loops in type II cystatin are longer than those in type I cystatin. The length for the corresponding loop in PMC-2 is between those of type-I and type-II cystatins. Similar to stefin A and B, PMC-2 does not have the long insertion between β3 and β4 observed in the CEW cystatin and human cystatin F that forms a flexible helix. In PMC-2, that loop is also the most flexible area, judging from its temperature factors (Figure 8). Both β3 and β4 of PMC-2 are only slightly longer than those of stefin A and B and consequently form a similarly sized β-hairpin loop to those of stefin A and B.
By contrast, L2 of PMC-2, which connects β4 and β5, was found to have a superimposable backbone conformation with CEW cystatin, and it was rather different than those of stefin A and B, which have two more residues (Figure 10). The critical residue in L2, Trp-104, of CEW cystain, which corresponds to residue Trp-77 in PMC-2, has been previously shown to form a herring bone–like hydrophobic interaction with Trp-177 and Trp-181 of the target Cys protease (Wang et al., 2006). Trp-77 is conserved among all repeating units of PMC, CEW cystatin, oryzacystatin, cystatin F, and monellin, but not in type 1 cystatins, such as stefin 1 and stefin 2.
The C terminus of PMC-2 ends with β5 (Figure 3), similar to the structure of CEW cystatin and oryzacystatin; thus, it is again different from stefin A and B, which have a loop conformation extending more to the convex side of the β-sheet.
In addition to the similarity in the secondary structures discussed above, there are several residues that are highly conserved among the compared structures. For example, Tyr-53 is completely conserved in each protein (Figure 3). Tyr-53 is located near the interface with the target protease in the complex structure, as observed in the complex structures of stefin B (1STE) and stefin A (1NB5). In all cases except CEW cystatin, the hydroxyl group of Tyr-53 is hydrogen bonded to one of the residues in L2, Glu-77 in the case of PMC-2, which probably fixes its conformation. The next residue is also Tyr, Tyr-54, in all PMC repeat units, oryzacystatin-1, and stefin A. The hydroxyl group of Tyr-54 is within a hydrogen-bond distance with the side chain of Gln-46 in the QXVXG motif of L1. Therefore, two Tyr residues, Tyr-53 and Tyr-54, in PMC-2 repeat units, oryzacystatin-1 and stefin A, might be critical in maintaining the conformation of both the L1 and L2 sites.
The side chain of the other highly conserved residue, Asn-28, which is substituted to Glu in stefin A and B, is hydrogen bonded to the backbone of Leu-35, the first residue of the β2-strand. All other compared cystatins have a similar type of hydrogen bond in the identical area, which probably contributes to the bend in the β2-strand. Therefore, together with the three bulges mentioned earlier, this interaction contributes to wrapping the whole β-sheet around the α-helix.
Comparison with Monellin
The Dali search showed that the structure of PMC-2 is most similar to that of the single-chain monellin. Monellin is an intensely sweet-tasting protein isolated from the berries of the African plant D. cumminisii. The loop corresponding to the L1 position in PMC-2 has been engineered to form a single-chain monellin, while the natural monellin molecule is cleaved at this site, forming two chains, A and B. Naturally, this synthetic L1 becomes the most flexible area in the single-chain monellin (Ogata et al., 1897; Morris et al., 1973; Somoza et al., 1993; Lee et al., 1999; Spadaccini et al., 2001). The loop connecting β3 and β4 has been proposed to be a possible sweet receptor binding site because it cross-reacts with an antibody raised against another sweet-tasting protein, thaumatin, which is obtained from a plant (Thaumatococcus daniellii) native in West Africa (Kim et al., 1991; Spadaccini et al., 2001). Interestingly, except for the artificial L1 and the short loop between β3 and β4, the backbone of PMC-2 is superimposable on the single-chain monellin. As shown in Figure 3, many residues are conserved; thus, the previous notion that monellin and cystatin are descended from a common ancestor (Murzin, 1993) is clearly confirmed by the structure of PMC-2, a plant cystatin. Considering the close resemblance among the tertiary structures of the three plant proteins, PMC-2, oryzacystatin-1, and monellin, it is likely that divergence of monellin occurred after the split of the plant and animal cystatins.
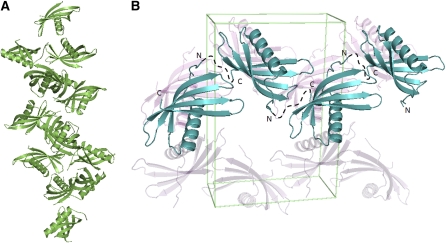
Structural Rationalization of Crystalline PMC
In spite of numerous attempts, the crystals of PMC isolated from potato did not diffract even at low resolution, reflecting apparent disorder in its lattice packing. Having identified the structural features of the individual repeating unit of PMC and the identity of the ∼35-kD fragment, it is now tempting to rationalize the unique natural existence of crystalline PMC. Careful observation of the interactions between the 14 molecules in the asymmetric unit in the C2 crystal lattice (Figure 11A) revealed that there are many different modes of interaction among PMC-2 molecules and possibly among the individual repeating units of the native PMC molecule. In addition, in the crystal lattice of the P212121 crystal, the individual PMC-2 molecules are packed in a head-to-tail manner, forming a linearly tethered arrangement (Figure 11B). Therefore, combining all the observations, including the trypsin digestion experiment, it is likely that native PMC has its domains 5 to 7 packed in a compact conformation, with domain 6 at the core, while domains 1 to 4 and 8 are in a more flexible, extended form. Using the versatile interactions among those exposed domains, PMC can precipitate into a crystalline form, but with only long-range order, in which a large amount of potent inhibitor can be stored in an inaccessible and inactive form. Crystalline PMC can easily be converted into its active 85-kD monomeric form through modulation of the levels of cytosolic PO4− and pH, which thus has the potential to dynamically alter tuber physiology and the interactions of the potato tuber with its environment.
Figure 11.
Arrangement of the PMC-2 Molecules in the Crystal Lattices.
(A) Diagram illustrating the arrangement of PMC-2 in the asymmetric unit of the C2 crystal lattice.
(B) Diagram illustrating the arrangement of the PMC-2 molecules in the unit cell of the P212121 crystal lattice. PMC-2 molecules are packed in a front-to-end interaction. Both Figures 10 and 11 were prepared using PyMol.
In its active monomer form, PMC has eight cystatin domains with the same global shape. It is interesting to speculate that the sequence heterogeneity among the eight domains, especially in their N-terminal trunk, L1, and L2 sites, may confer a wider range of binding affinity and inhibitory activity and thus efficacy toward a diversity of Cys proteases, including those in the midgut of insects (Murdock et al., 1987). It is also tempting to speculate that PMC likely has more than one target protease in vivo, thus enabling PMC to regulate myriad physiological events central to tuber ontogeny. As previously indicated, PMC appears to be involved in regulating protein deposition in tubers during development (Weeda et al., 2008) and protein mobilization from seed tubers during sprouting by regulating protease activity (Weeda et al., 2009). For example, during tuber development, active monomeric PMC accumulation precedes deposition of patatin, a major storage protein in the potato tuber (Weeda et al., 2008). In vitro studies show that proteolysis of patatin by Cys proteases can be inhibited by PMC (Kumar et al., 1999). As the tubers begin to sprout, PMC levels decline to effect mobilization of protein reserves (Weeda et al., 2009).
PMC Affinity Protein from S. tuberosum
The major protein band in the PMC pull-down assay was identified as a putative Cys protease (Avrova et al., 1999). However, the significance of the other faint bands in Figure 9 cannot be ignored, and their identity is under investigation. A search for similar amino acid sequences in the nonredundant National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/Genbank) using BLAST revealed that Cys protease TDI-65 from tomato (Solanum lycopersicum) showed the highest score (851 bits), with 87% amino acid identity to the identified putative Cys protease, followed by low temperature–induced Cys protease also from tomato, with a score of 642 bits and 89% identity. Overall, the level of sequence identity between the putative Cys protease and two tomato Cys proteases was high and the alignments were significant. However, the identified potato Cys protease has a molecular mass of ∼51 kD, which differs from the observed molecular mass of ∼45 kD (Figure 9), suggesting the protein is posttranslationally modified or is the product of another gene not identified due to incomplete sequencing of the potato genome. In a previous report (Avrova et al., 1999), the putative Cys protease identified here was found to be tightly regulated and to be induced by Phytophthora infestans in potato leaves. Considering a high level of sequence similarity to cathepsin K and the existence of a granulin binding domain in the C terminus, the putative Cys protease has been proposed to have caspase-like regulatory function through a specific target mechanism of its granulin binding domain (Avrova et al., 1999). Therefore, it is tempting to speculate that in tubers the same Cys protease or an isoform performs similar roles in programmed cell death during prolonged storage, mobilization of tuber reserves during sprouting, senescence, and other processes where the activity of this enzyme is tightly regulated by PMC, in contrast with the transcriptional regulation apparent in leaves. The observed pH- and/or PO4−-dependent monomer-polymer transition of PMC is likely the activation or deactivation mechanism.
In summary, we have performed the comprehensive structural and functional characterization for both PMC protein and a single, representative domain of this protein. The single domain of PMC had structural similarity to both type I and type II cystatins and maintained its inhibitory activity, but lost the unique pH- and PO4−-dependent polymerization behavior of PMC. We also identified a Cys protease from potato, which has significant affinity for PMC. The observed heterogeneity among the eight cystatin domains, along with the pH/PO4−-dependent transition capability between active monomer and inactive crystalline forms, provides a structural regulation mechanism that would facilitate roles for PMC in self defense and the regulation of key physiological processes important during tuber development and sprouting.
METHODS
Tryptic Digestion of PMC
PMC was isolated from potato (Solanum tuberosum) tubers (Rodis and Hoff, 1984) and subjected to tryptic digestion at 37°C for 24 h. The purified PMC (43.5 μg) was incubated in 0.2 M Tris buffer, pH 7.4, with or without 959 units of bovine pancreatic trypsin (type I; Sigma-Aldrich) in a 120-μL total reaction volume. Following incubation, the sample was mixed with loading buffer and heated for 10 min at 100°C. Proteins resulting from tryptic digestion were resolved by SDS-PAGE (10% gel, 8.4 μg protein per lane) by following the methods of Laemmli (1970). The gel was stained with Coomassie Brilliant Blue, and the 35-kD PMC fragment was excised for MS analysis. In addition, the proteins resolved by SDS-PAGE were transferred electrophoretically to a polyvinylidene difluoride membrane and visualized by staining with 0.1% (w/v) Ponceau red S in 5% acetic acid. The 35-kD fragment of PMC was excised from the polyvinylidene difluoride membrane for N-terminal sequencing.
Expression and Purification of PMC-2
The PMC-2 gene (second cystatin domain-coding gene of potato multicystatin) was amplified from S. tuberosum genomic DNA by PCR (forward primer: 5′-AGTACCATGGGGGGCATTGTCAATGTTCCA-3′, reverse primer: 5′-CACACTCTCGAGAACAAGCTTGAATTCTACAAC-3′). The PCR product was cloned into the NcoI/XhoI recognition sites of the pET28a vector (Novagen), and the PMC-2 protein was expressed from this clone as a C-terminal His-tagged recombinant in Escherichia coli ER2566 (New England Biolabs) cells using the following procedure. An overnight culture in Luria-Bertani broth was used to inoculate 1 liter of Luria-Bertani in a 4-liter flask. The culture was incubated with shaking at 37°C to an A600 of 0.6. The temperature was then reduced to 20°C, expression of the protein was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for an additional 15 h. The cells were harvested by centrifugation and resuspended in 50 mM Na phosphate, pH 8.0, 300 mM NaCl, and 20 mM imidazole (lysis/binding buffer), and lysed by sonication (Model 450 Sonifier; Branson Ultrasonics). The lysates were cleared by centrifugation at 38,000g for 45 min. The cleared lysate was applied to a Ni2+-NTA column, and the column was extensively washed with lysis/binding buffer. The protein was eluted with lysis/binding buffer containing 300 mM imidazole. Protein-containing fractions were pooled, concentrated, and exchanged into 20 mM Na acetate buffer, pH 6.5, containing 100 mM NaCl and then applied to a Sephacryl S-100 (GE Healthcare) size exclusion column equilibrated in the same buffer. Fractions containing purified PMC-2 were pooled, concentrated, and exchanged into 20 mM Na acetate buffer, pH 5.0, containing 1 mM DTT.
The selenomethionine derivative of PMC-2 was prepared from pET28aPMC-2 expressed in the Met auxotroph B834(DE3) and cultured in minimal media supplemented with selenomethionine. The expression and purification of selenomethionyl-PMC-2 followed the protocol for the native protein, except that 1 mM DTT was included in every buffer to prevent oxidation of the selenomethionyl residue. The purification of both native and selenomethionyl-PMC-2 was followed at each step by SDS-PAGE on 15% gels. Protein concentrations were determined by the Bradford assay (Bio-Rad).
Papain Inhibitory Activity of Recombinant PMC-2
The ability of recombinant PMC-2 to inhibit catabolism of BAPNA by papain (Cys-type protease) was determined by modifying the methods of Walsh and Strickland (1993). The assay (1.0 mL) consisted of 894 μL of MES buffer (0.1 M, pH 6.2) containing 8 mM DTT, 4 mM EDTA, and 0.383 mM BAPNA (Sigma-Aldrich). The reaction was initiated by adding 2.1 units of papain (Sigma-Aldrich). Protease activity was monitored spectrometrically (Varian Cary 100 biospectrophotometer) at 23°C by following the increase in p-nitroanilide production at A395 for 10 min. Papain activity in the presence and absence of 0.5 and 1.0 μg PMC-2 was expressed as change in A395 min−1 over the 10-min interval.
Determination of Molecular Mass
Static Light Scattering
The weight averaged molecular mass of PMC-2 was determined using combined size exclusion chromatography and multiple angle laser light scattering. In brief, PMC-2 (0.2 mg) was injected onto a BioSep SEC S2000 column (Phenomenex) and eluted with PBS. The eluate at a rate of 1 mL/min was passed through a tandem UV detector (Gilson), laser light scattering detector (Optilab DSP; Wyatt Technology), and interferometric refractometer (Dawn EOS; Wyatt Technology). The light scattering data were analyzed using Astra software (Wyatt Technology).
Dynamic Light Scattering
The radius and molecular mass of PMC and PMC-2 were estimated using a DynaPro-Titan instrument (Wyatt Technology) at 22°C. Purified PMC or PMC-2 (2 mg mL−1) in a freshly prepared buffer were filtered through a polyvinylidene difluoride filter (0.2 μm; Millipore). Sodium acetate buffer, cacodylate buffer, and HEPES buffers were used for pH 5.5, 6.5, and 7.5, respectively. Scattering data were acquired through accumulation (five times) of 10 scans with 10 s/scan, with the laser intensity set to a range of 50 to 60% (30 to 36 mW). The corresponding molecular mass and radius were calculated using the software package DYNAMICS V6, which was supplied with the instrument.
PMC-2 Pull-Down Experiment
Potato tuber extracts were prepared as previously described (Kumar et al., 1999). Five milliliters of tuber extract was adjusted to pH 8.0 with 1 M Tris. Purified His-tagged PMC-2 (250 μg) was added, and the mixture was kept on ice for 30 min. The extract was then passed three times through a 1-mL Ni2+-NTA column (HisTrap HP; Pharmacia), which was equilibrated with buffer (50 mM Tris, pH 8.0, 300 mM NaCl, and 20 mM imidazole), followed by extensive washing with the same buffer. The bound potato proteins were eluted from the column with 5 mL of buffer containing 8 M urea. The column was washed again and eluted a second time with buffer containing 300 mM imidazole. Proteins in the eluates were precipitated with 10% trichloroacetic acid and separated by 12% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue R 250, and, following destaining, bands of interest were excised. Trypsin digests were performed in gel as previously described (Lopez et al., 2005). The digested samples were analyzed by LC-MS/MS using an LC Packings Ultimate Nano liquid chromatography system with a PS-DVB monolithic column, 0.1% formic acid/acetonitrile buffers for development, and an Esquire HCT electrospray ion trap (Bruker Daltonics) (Lopez et al., 2005; MacMillan et al., 2006). A local MASCOT server (www.matrixscience.com) was used to compare the MS/MS generated ion fragments to the Viridiplantae database (http://www.ncbi.nlm.nih.gov/).
Light Microscopy
Localization of PMC crystals in situ was examined by light microscopy. Potato tuber tissue was fixed in 4% (w/v) paraformaldehyde and 3% (v/v) glutaraldehyde in 0.2 M phosphate buffer, pH 8.0, overnight at 4°C. The tissue was then fixed in 2% (v/v) osmium tetroxide for 2 h at room temperature prior to dehydration in a graded ethanol-propylene oxide series. Samples were embedded in Spurr's epoxy resin, sectioned at 500 nm, and stained with 1% toluidine blue O with 1% borax.
Crystallization and Data Collection
Crystals of native PMC-2 were grown using the hanging drop vapor diffusion method. A solution of the purified protein in 20 mM Na acetate, pH 5.0, was mixed with an equal volume of 0.1 M TrisHCl , pH 8.5, and 8% polyethylene glycol 8000 and incubated with the same solution as a reservoir at 4°C. Crystals of diffraction quality appeared after 7 to 10 d. Crystals of the selenomethionyl derivative were prepared in the same fashion, with the inclusion of 0.1 mM DTT. Native and selenomethionyl crystals were determined as belonging to the monoclinic C2 space group, with a = 210.85, b= 85.71, c = 96.86, and β = 100.6°, and a = 211.56, b = 85.92, c = 96.97, and β = 100.6°, respectively. The crystals diffracted to ∼2.7 and 2.8 Å, respectively, and there were 14 PMC-2 molecules in the asymmetric unit. In addition, during the continuous crystallization trails of native PMC-2, new cuboidal shaped crystals were produced when the volume ratio of reservoir solution to protein solution was changed to 2:1 instead of 1:1, as with the above-mentioned monoclinic crystal. These crystals belonged to the orthorhombic crystal system, with unit cell dimensions of a = 25.46 Å, b = 54.53 Å, and c = 79.15 Å. The space group of these crystals was determined to be P212121, and there was one PMC-2 molecule in the asymmetric unit.
Phasing and Refinement
Initial phases of the monoclinic PMC-2 crystal structure were determined by the MAD phasing method (Hendrickson, 1991) using SOLVE software (Terwilliger and Berendzen, 1999) after prior approaches with the molecular replacement method were unsuccessful. Data collected at the remote wavelength were treated as the reference data set, and resolution limits of 40 to 2.7 Å were imposed. Experimental values of f ' and f"' estimated from fluorescence spectra were used. The selenium site was located, and the resulting phases have a figure of merit of 0.52. A density modification process using the maximum likelihood method was performed with RESOLVE software (Terwilliger and Berendzen, 1999), which eventually resulted in a clearly interpretable electron density map with many well-defined secondary structural elements. The corresponding amino acids were assigned and manually fitted into this map using the software O (Jones et al., 1991). The resulting rough coordinates of the 14 PMC-2 structures were refined using X-PLOR (Brunger et al., 1998), with the simulated annealing protocol resulting in a crystallographic R value of 28%. Several rounds of manual adjustment followed cycles of refinement, and picking solvent molecules led to an R factor of 21.2% (Rfree 24.6%, for the random 5% data). The structure of PMC-2 in the P212121 space group was then solved using the refined coordinates of one molecule in the asymmetric unit. The final R factor (Table 1) for the P212121 crystal data was 19.3% (Rfree = 23.2%, for the random 5% data). The number of reflections above the 2σ level for C2 and in the P212121 space group was 3837 (94.5% completeness, 10 to 2.5 Å) and 13,629 (75.6%, 10 to 2.7 Å), respectively. The rmsd from ideal geometry of the final coordinates corresponding to C2 and P212121 crystal data were 0.014 and 0.016 Å for bonds and 3.18 and 3.82° for angles, respectively. Both PMC-2 coordinates have been deposited in the Protein Data Bank (2W9P and 2W9Q).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AAD48496.1 for Cys protease TDI-65 from tomato, CAB53515 for putative Cys protease from S. tuberosum, and P20721 for low temperature–induced Cys protease from tomato.
Acknowledgments
We thank C. Ralston (Berkeley Advanced Light Source, beamline 8.2.1), G. Munske (Washington State University, LBB1), and V. Lynch-Holm (Washington State University, Franceschi Microscopy and Imaging Center). This work was partially supported by grants from the USDA-Cooperative State Research, Education and Extension Service (CSREES) National Research Initiative, the USDA-Agricultural Research Service, the American Heart Association, and Murdock Charitable Trust.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: ChulHee Kang (chkang@wsu.edu) and N. Richard Knowles (rknowles@wsu.edu).
Open Access articles can be viewed online without a subscription.
References
- Aagaard, A., Listwan, P., Cowieson, N., Huber, T., Ravasi, T., Wells, C., Flanagan, J., Kellie, S., Hume, D., Kobe, B., and Martin, J. (2005). An inflammatory role for the mammalian carboxypeptidase inhibitor latexin: relationship to cystatins and the tumor suppressor TIG1. Structure 13 309–317. [DOI] [PubMed] [Google Scholar]
- Abe, K., Emori, Y., Kondo, H., Arai, S., and Suzuki, K. (1988). The NH2-terminal 21 amino acid residues are not essential for the papain-inhibitory activity of oryzacystatin, a member of the cystatin superfamily. Expression of oryzacystatin cDNA and its truncated fragments in Escherichia coli. J. Biol. Chem. 263 7655–7659. [PubMed] [Google Scholar]
- Avrova, A., Stewart, H., De Jong, W., Heilbronn, J., Lyon, G., and Birch, P. (1999). A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol. Plant Microbe Interact. 12 1114–1119. [DOI] [PubMed] [Google Scholar]
- Bode, W., Engh, R., Musil, D., Thiele, U., Huber, R., Karshikov, A., Brzin, J., Kos, J., and Turk, V. (1988). The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 7 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W., and Dziegielewska, K. (1997). Friends and relations of the cystatin superfamily–new members and their evolution. Protein Sci. 6 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Dieckmann, T., Mitschang, L., Hofmann, M., Kos, J., Turk, V., Auerswald, E., Jaenicke, R., and Oschkinat, H. (1993). The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution. J. Mol. Biol. 234 1048–1059. [DOI] [PubMed] [Google Scholar]
- Garcia-Olmedo, F., Salcedo, G., Sanchez-Monge, R., Gomez, L., Royo, J., and Carbonero, P. (1987). Plant proteinaceous inhibitors of proteinases and α-amylases. Oxf. Surv. Plant Mol. Cell Biol. 4 275–334. [Google Scholar]
- Hendrickson, W. (1991). Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254 51–58. [DOI] [PubMed] [Google Scholar]
- Hoff, J., Jones, C., Sosa, M., and Rodis, P. (1972). Naturally occurring crystals in the potato: Isolation and identification as a protein. Biochem. Biophys. Res. Commun. 49 1525–1529. [DOI] [PubMed] [Google Scholar]
- Holm, L., and Sander, C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138. [DOI] [PubMed] [Google Scholar]
- Irie, K., Hosoyama, H., Takeuchi, T., Iwabuchi, K., Watanabe, H., Abe, M., Abe, K., and Arai, S. (1996). Transgenic rice established to express corn cystatin exhibits strong inhibitory activity against insect gut proteinases. Plant Mol. Biol. 30 149–157. [DOI] [PubMed] [Google Scholar]
- Janin, J., and Chothia, C. (1980). Packing of alpha-helices onto beta-pleated sheets and the anatomy of alpha/beta proteins. J. Mol. Biol. 143 95–128. [DOI] [PubMed] [Google Scholar]
- Jenko, S., Dolenc, I., Guncar, G., Dobersek, A., Podobnik, M., and Turk, D. (2003). Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J. Mol. Biol. 326 875–885. [DOI] [PubMed] [Google Scholar]
- Jones, T., Zou, J., Cowan, S., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Jongsma, M., and Bolter, C. (1997). The adaptation of insects to plant proteinase inhibitors. J. Insect Physiol. 43 885–895. [DOI] [PubMed] [Google Scholar]
- Kim, S., Kang, C., and Cho, J. (1991). Sweet proteins: biochemical studies and genetic engineering. In Sweetners, Discovery, Molecular Design and Chemoreception, D.E. Waters, F.T. Orthofer, and G.E. Dubois, eds (Washington, DC: ACS), pp. 28–40.
- Kitamura, N., and Maruyama, Y. (1985). Cysteine endopeptidase activity in sprouting potato tubers. Agric. Biol. Chem. 49 1591–1597. [Google Scholar]
- Kumar, G.N.M., Houtz, R., and Knowles, N. (1999). Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 119 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, G.N.M., and Knowles, N.R. (2003). Wound-induced superoxide production and PAL activity decline with potato tuber age and wound healing ability. Physiol. Plant. 117 108–117. [Google Scholar]
- Kumar, G.N.M., and Knowles, N.R. (2007). Strboh A homologue of NADPH oxidase regulates wound-induced oxidative burst and facilitates wound-healing in potato tubers. Planta 227 25–36. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, S., Lee, J., Chang, H., Cho, J., Jung, J., and Lee, W. (1999). Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations. Biochemistry 38 2340–2346. [DOI] [PubMed] [Google Scholar]
- Lopez, J., Siems, W., Palmer, G., Brayton, K., McGuire, T., Norimine, J., and Brown, W. (2005). Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73 8109–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan, H., Brayton, K., Palmer, G., McGuire, T., Munske, G., Siems, W., and Brown, W. (2006). Analysis of the Anaplasma marginalemagor surface protein 1 complex protein composition by tandem mass spectrometry. J. Bacteriol. 18 4983–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J., Craven, C., Jerala, R., Kroon-Zitko, L., Zerovnik, E., Turk, V., and Waltho, J. (1995). The three-dimensional solution structure of human stefin A. J. Mol. Biol. 246 331–343. [DOI] [PubMed] [Google Scholar]
- Michaud, D., Nguyen-Quoc, B., Bernier-Vadnais, N., Faye, L., and Yelle, S. (2006). Cysteine proteinase forms in sprouting potato tuber. Physiol. Plant. 90 497–503. [Google Scholar]
- Morris, J., Martenson, R., Deibler, G., and Cagan, R. (1973). Characterization of monellin, a protein that tastes sweet. J. Biol. Chem. 248 534–539. [PubMed] [Google Scholar]
- Murdock, L., Brookhart, G., Dun, P., Foard, D., Kelley, S., Kitch, L., Shade, R., Shukle, R., and Wolfson, J. (1987). Cysteine digestive proteinases in coleoptera. Comp. Biochem. Physiol. 87B 783–787. [Google Scholar]
- Murzin, A. (1993). Sweet-tasting protein monellin is related to the cystatin family of thiol proteinase inhibitors. J. Mol. Biol. 230 689–694. [DOI] [PubMed] [Google Scholar]
- Nagata, K., Kudo, N., Abe, K., Arai, S., and Tanokura, M. (2000). Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 39 14753–14760. [DOI] [PubMed] [Google Scholar]
- Ogata, C., Hatada, M., Tomlinson, G., Shin, W., and Kim, S. (1897). Crystal structure of the intensely sweet protein monellin. Nature 328 739–742. [DOI] [PubMed] [Google Scholar]
- Orr, G., Strickland, J., and Walsh, T. (1994). Inhibition of Diabrotica larval growth by a multicystatin from potato tubers. J. Insect Physiol. 40 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau, L., Gruppen, H., Piersma, S., van den Broek, L., van Koningsveld, G., and Voragen, A. (2001). Relative abundance and inhibitory distribution of protease inhibitors in potato juice from cv. Elkana. J. Agric. Food Chem. 49 2864–2874. [DOI] [PubMed] [Google Scholar]
- Rawlings, N., and Barrett, A. (1990). Evolution of proteins of the cystatin superfamily. J. Mol. Evol. 30 60–71. [DOI] [PubMed] [Google Scholar]
- Rodis, P., and Hoff, J. (1984). Naturally occurring protein crystals in the potato: Inhibitor of papain, chymopapain, and ficin. Plant Physiol. 74 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C. (1990). Proteinase inhibitors in plants: Genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 28 425–449. [Google Scholar]
- Samotus, B., and Schwimmer, S. (1962. a). Phytic acid as a phosphorus reservoir in the developing potato tuber. Nature 194 578–579.13911680 [Google Scholar]
- Samotus, B., and Schwimmer, S. (1962. b). Effect of maturity on distribution of phosphorus among starch and other components of potato tuber. Plant Physiol. 37 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, T., Poppy, G., Kerry, B., and Denholm, I. (1998). Insect-resistant transgenic plants. Trends Biotechnol. 16 168–175. [DOI] [PubMed] [Google Scholar]
- Somoza, J., Jiang, F., Tong, L., Kang, C., Cho, J., and Kim, S. (1993). Two crystal structures of a potently sweet protein. Natural monellin at 2.75 A resolution and single-chain monellin at 1.7 A resolution. J. Mol. Biol. 234 390–404. [DOI] [PubMed] [Google Scholar]
- Spadaccini, R., Crescenzi, O., Tancredi, T., De Casamassimi, N., Saviano, G., Scognamiglio, R., Di Donato, A., and Temussi, P. (2001). Solution structure of a sweet protein: NMR study of MNEI, a single chain monellin. J. Mol. Biol. 305 505–514. [DOI] [PubMed] [Google Scholar]
- Stubbs, M., Laber, B., Bode, W., Huber, R., Jerala, R., Lenarcic, B., and Turk, V. (1990). The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: A novel type of proteinase inhibitor interaction. EMBO J. 9 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, S., Ushioda, T., Utsunomiya-Tate, N., Shibuya, K., Ohyama, Y., Nakano, Y., Kaji, H., Inagaki, F., Samejima, T., and Kainosho, M. (1995). Solution structure of a human cystatin A variant, cystatin A2-98 M65L, by NMR spectroscopy. A possible role of the interactions between the N- and C-termini to maintain the inhibitory active form of cystatin A. Biochemistry 34 14637–14648. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele, U., Auerswald, E., Gebhard, W., Assfalg-Machleidt, I., Popović, T., and Machleidt, W. (1988). Inhibitorily active recombinant human stefin B. Gene synthesis, expression and isolation of an inhibitory active MS-2 pol-stefin B fusion protein and preparation of Des[Met1,2(2)]stefin B. Biol. Chem. Hoppe Seyler 369 1167–1178. [DOI] [PubMed] [Google Scholar]
- Turk, V., and Bode, W. (1991). The cystains: Protein inhibitors of cysteine proteinases. FEBS Lett. 285 213–219. [DOI] [PubMed] [Google Scholar]
- Waldron, C., Wegrich, L.M., Merlo, P.A., and Walsh, T.A. (1993). Characterization of a genomic sequence coding for potato multicystatin, an eight-domain cysteine proteinase inhibitor. Plant Mol. Biol. 23 801–812. [DOI] [PubMed] [Google Scholar]
- Walsh, T., and Strickland, J. (1993). Proteolysis of the 85-kilodalton crystalline cysteine proteinase inhibitor from potato releases functional cystatin domains. Plant Physiol. 103 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Pandey, K., Somoza, J., Sijwali, P., Kortemme, T., Brinen, L., Fletterick, R., Rosenthal, P., and McKerrow, J. (2006). Structural basis for unique mechanisms of folding and hemoglobin binding by a malarial protease. Proc. Natl. Acad. Sci. USA 103 11503–11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda, S.M., Kumar, G.N.M., and Knowles, N.R. (2008). Developmentally linked changes in proteases and protease inhibitors may regulate protein accumulation in potato tubers. Proceedings of the 91st Annual Meeting of the Potato Association of America. Am. J. Pot. Res. 85 33. [Google Scholar]
- Weeda, S.M., Kumar, G.N.M., and Knowles, N.R. (2009). Changes in protease inhibitors during protein mobilization from seed-tubers. Proceedings of the 92nd Annual Meeting of the Potato Association of America. Am. J. Pot. Res., in press.
- Zavala, J., Patankar, A., Gase, K., Hui, D., and Baldwin, I. (2004). Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol. 134 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]