Abstract
In rice (Oryza sativa), the shoot-borne crown roots are the major root type and are initiated at lower stem nodes as part of normal plant development. However, the regulatory mechanism of crown root development is poorly understood. In this work, we show that a WUSCHEL-related Homeobox (WOX) gene, WOX11, is involved in the activation of crown root emergence and growth. WOX11 was found to be expressed in emerging crown roots and later in cell division regions of the root meristem. The expression could be induced by exogenous auxin or cytokinin. Loss-of-function mutation or downregulation of the gene reduced the number and the growth rate of crown roots, whereas overexpression of the gene induced precocious crown root growth and dramatically increased the root biomass by producing crown roots at the upper stem nodes and the base of florets. The expressions of auxin- and cytokinin-responsive genes were affected in WOX11 overexpression and RNA interference transgenic plants. Further analysis showed that WOX11 directly repressed RR2, a type-A cytokinin-responsive regulator gene that was found to be expressed in crown root primordia. The results suggest that WOX11 may be an integrator of auxin and cytokinin signaling that feeds into RR2 to regulate cell proliferation during crown root development.
INTRODUCTION
The monocot cereals, such as rice (Oryza sativa) and maize (Zea mays), display a complex root structure with several root types, including embryonic primary roots, lateral roots, and shoot-borne roots (also known as adventitious roots). In rice, the embryonic primary root develops shortly after germination, whereas shoot-borne roots are initiated from stem nodes and are called crown roots (Hochholdinger et al., 2004). The fundamental difference between cereals and the dicot model plant Arabidopsis thaliana is the presence of an extensive postembryonic shoot-borne root system in cereals, which is missing in Arabidopsis. For example, a field-grown rice plant usually produces hundreds of crown roots, which form a so-called fibrous root system (Kawata and Harada, 1975).
To date, organization and cell differentiation processes in root development have been well characterized in Arabidopsis (reviewed in Scheres, 2002). Whereas mechanisms of radial root patterning are shared between cereals and eudicots (Cui et al., 2007), the regulation of root architecture in cereals involves some novel mechanisms. For instance, the rice LATERAL ORGAN BOUNDARIES domain gene, CROWN ROOTLESS1 (CRL1)/ADVENTITIOUS ROOTLESS1 (ARL1), is a key regulator of the initiation of crown roots and lateral roots (Inukai et al., 2005; Liu et al., 2005). A related maize gene is also shown to be required for shoot-borne root formation (Taramino et al., 2007).
Plant hormones, such as auxin and cytokinin, play important roles in regulating root development (reviewed in Dello Ioio et al., 2007a; De Smet and Jurgens, 2007). Auxin signaling has been shown to be required for the initiation of rice crown roots (Inukai et al., 2005). By contrast, cytokinin is known to have inhibitory effects on root growth. Transgenic plants overexpressing genes for cytokinin oxidases, which are involved in cytokinin degradation, have increased lateral root formation and enlarged root meristems leading to more rapidly growing roots in Arabidopsis (Werner et al., 2001, 2003). Cytokinin is shown to affect specifically the cell differentiation rate at the transition area between the cell division and elongation/differentiation zones but does not affect the rate of proliferation of cells in the meristem (Dello Ioio et al., 2007b). A recent study reveals that cytokinin acts directly on lateral root founder cells to inhibit root initiation in Arabidopsis (Laplaze et al., 2007). However, the role of cytokinin signaling in short-borne root development is not known.
WUSCHEL (WUS)-related Homeobox (WOX) genes have been shown to play an important role to coordinate gene transcription involved in both shoot and root meristem function and organogenesis (Haecker et al., 2004). In this work, we studied the expression and developmental function of a rice WOX gene, WOX11. We show that WOX11 is an auxin- and cytokinin-responsive gene that is expressed mainly in the cell division regions of both root and shoot meristems. Loss of function of the gene inhibited crown root development, while its overexpression accelerated crown root cell division and lead to precocious crown root growth and crown root production in upper stem nodes. The expression of both auxin- and cytokinin-responsive genes was affected by the mutation or overexpression of WOX11. Importantly, WOX11 is found to repress directly a cytokinin type-A responsive regulator (RR) gene, RR2, which is expressed in crown root primordia. These data together suggest that WOX11 may integrate both auxin and cytokinin signaling to stimulate cell division during crown root development.
RESULTS
WOX11 Is Expressed in Emerging Crown Roots and in Cell Division Regions of Root and Shoot Meristems
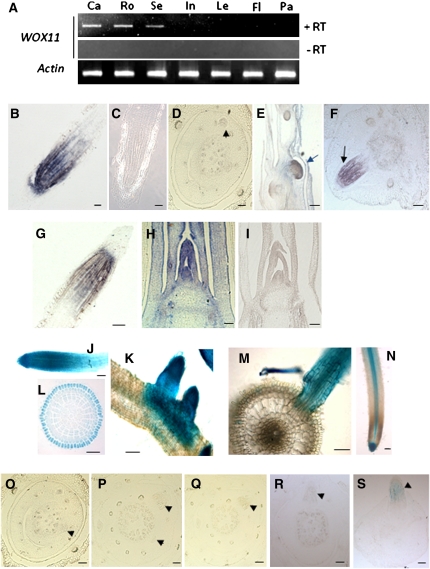
The rice genome contains at least 11 WOX genes. Phylogenetic analysis revealed that three rice (Os-WOX6, Os-WOX10, and Os-WOX11) and two Arabidopsis (At-WOX11 and At-WOX12) genes are found in the same clade (Dai et al., 2007). The function of this subgroup was unknown, and we chose to analyze Os-WOX11. RT-PCR analysis detected Os-WOX11 mRNA in calli, roots, and 7-d-old seedlings, but not in the other tested organs/tissues, including internodes, leaves, and panicles (Figure 1A). In situ hybridization analysis revealed that the gene was mainly expressed in the primary root meristematic (cell division) region (Figure 1B). The mRNA was hardly detectable in crown root initials (Figure 1D), but clearly detected in emerging crown roots (Figures 1E and 1F). In the mature crown roots, the expression pattern was similar to that in the primary root (Figure 1G). In the shoot, the mRNA was found in the shoot apical meristem (SAM), leaf primordia, and young leaves (Figure 1H).
Figure 1.
WOX11 Is Expressed in Cell Division Regions in Roots and Shoots.
(A) Detection of WOX11 transcript by RT-PCR in callus (Ca), roots (Ro), seedlings (Se), internodes (In), mature leaves (Le), flag leafs (Fl), and panicles (Pa). PCRs without prior reverse transcription (−RT) are included. Actin transcripts were detected as controls.
(B) to (I) In situ hybridization detection of WOX11 transcripts in the primary root with an antisense (B) or a sense probe (C), in crown root initials (D), in growing crown primordia ([E], longitudinal section; [F], transverse section), in a mature crown root (G), and in the SAM with an antisense (H) or a sense probe (I). Bars = 25 μm in (B), (C), (G), (H), and (I) and 50 μm in (D) to (F). Arrows indicate emerging crown roots.
(J) to (M) GUS activity in a 7-d-old WOX11p-GUS transgenic seedling: a primary root tip (J); lateral roots (K); and a transverse section of the root meristematic zone (L). (M) shows a transverse section through a lateral root.
(N) DR5-GUS expression profile in root. Bars = 25 μm in (J), (L), and (N) and 50 μm in (K) and (M).
(O) to (S) Detection of WOX11p-GUS expression during the course of crown root initiation. GUS activity was detected in emerging crown roots ([R] and [S]) but not at the early stages during crown root initiation ([O] to [Q]). Arrowheads indicate crown root initials. Bars = 25 μm.
To complement the in situ hybridization results, a 2.4-kb promoter region of Os-WOX11 was used to direct β-glucuronidase (GUS) expression in transgenic rice. The GUS activity was mainly detected in the tips and cell division zones of the primary and lateral roots (Figures 1J and 1K). The expression was strongest in the epidermal cell layer (Figure 1L). However, no GUS activity was detected in the pericycle where the lateral roots were initiated (Figure 1M). The root expression pattern of the WOX11 promoter was different from the auxin-induced DR5 promoter activity that was mainly detected in the root meristem and the vascular cylinder (Figure 1N). During crown root development, the GUS activity was detectable only after the crown root was initiated (Figures 1O to 1S), confirming the in situ hybridization results (Figures 1D to 1F). Similarly, the promoter was found to be active only after the lateral root primordium was formed (see Supplemental Figure 1A online). The WOX11 protein fused with the green fluorescent protein (GFP) is localized in the nucleus (see Supplemental Figure 1B online).
Loss of WOX11 Function Affects Root Development
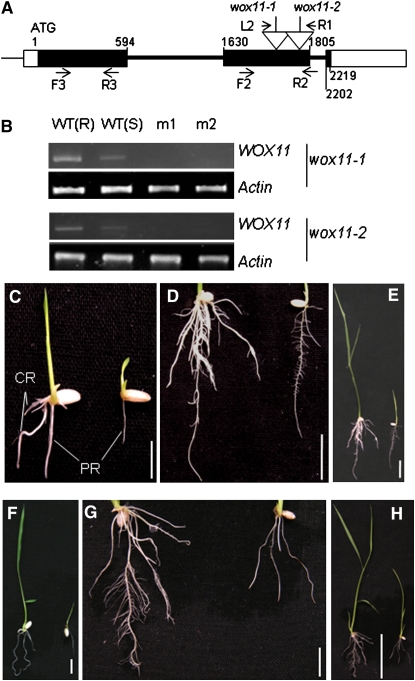
Searching in the rice insertion mutant databases led to the identification of two lines (2A00597 and 2D40272 from the Pohang databases) in Hwa and Dongjing backgrounds. These lines were renamed as wox11-1 and wox11-2, respectively. Homozygous plants for T-DNA insertions were identified by PCR (Figure 2A; see Supplemental Figure 2 online). RT-PCR using a primer set corresponding to the 5′ region relative to the insertion sites detected no transcripts of the gene in the mutants (Figure 2B). The homozygous seedlings of both insertion lines showed severe growth defects. The primary embryonic root was smaller than that of the wild-type and heterozygous seedlings (Figures 2C to 2H, Table 1). In particular, there was little or no crown root production in the mutants 2 weeks after germination (Figures 2C to 2H, Table 1). The mutants were adult-lethal.
Figure 2.
Characterization of Two T-DNA Insertion Lines of WOX11.
(A) Schematic representation of the WOX11 locus. Relative nucleotide positions of the coding region are indicated (with the initiation ATG codon assessed as 1). The T-DNA insertion positions in the second exon are indicated by open arrows. The positions of the primer L2 and R1 (corresponding to the T-DNA left and right borders, respectively) and the forward (F2) and reverse (R2) WOX11 primers are indicated by arrows. Positions of primers (F3 and R3) used to detect WOX11 expression are indicated. Filled boxes, exons; open boxes, untranslated exons; thick lines, introns.
(B) Detection of WOX11 transcripts in the root [WT(R)] or seedlings [WT(S)] of the wild type or seedlings of wox11-1 and wox11-2. Two different samples (m1 and m2) of each mutant line were used for the RT-PCR analysis. Actin transcripts were amplified as controls.
(C) to (E) Phenotypes of wox11-1 (on the right of each panel) compared with the wild type (left) at 7 d (C) and 14 d ([D] and [E]) after germination.
(F) to (H) Phenotype of wox11-2 (on the right of each panel) compared with the wild type (left) at 7 d (F) and 14 d ([G] and [H]) after germination.
PR, primary root; CR, crown roots. Bars = 1 cm in (C), (D), (F), and (G), 5 cm in (E), and 2 cm in (H).
[See online article for color version of this figure.]
Table 1.
Comparison of Primary Root Length and Crown Root Number in 2-Week-Old Seedlings of Wild-Type (Hwa), Heterozygous, and Homozygous wox11-1 Plants
| Genotype (Plant Number) | Primary Root Length | Crown-Borne Root Number |
|---|---|---|
| Hwa (20) | 2.7 ± 0.14 | 9.3 ± 0.37 |
| Heterozygote (20) | 2.5 ± 0.25 | 9.4 ± 0.21 |
| Homozygote (20) | 1.68 ± 0.18** | 0.69 ± 0.17** |
Data represent the means ± se of 20 plants in each line. Significant differences are indicated at the 5% (*) and 1% (**) of probability levels (Dunnett's two-tailed tests).
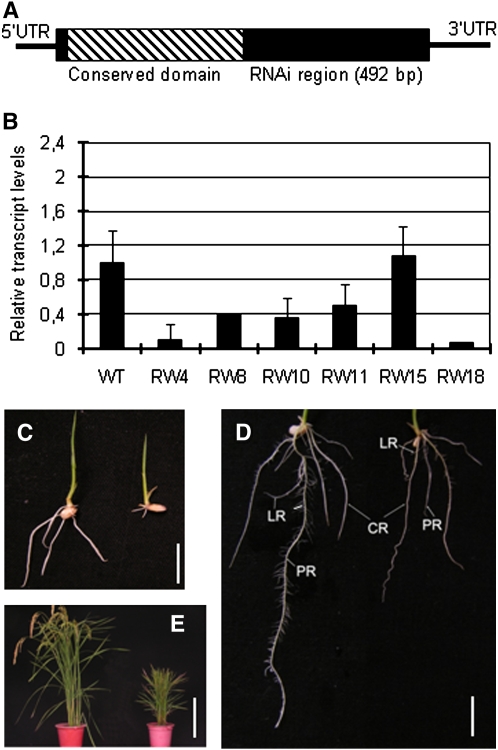
To confirm the observation, transgenic plants expressing double-strand RNA of the 3′ region of the gene (not including the conserved homeobox and WUS domains) were produced (Figure 3A). Expression analysis of the transgenic population identified five lines with reduced WOX11 mRNA levels (Figure 3B). Three of the RNA interference (RNAi) lines (RW4, RW10, and RW18) and a transgenic sibling that showed no effect on WOX11 mRNA accumulation (RW15) were selected for subsequent analysis. Two weeks after germination, the RNAi seedlings developed fewer crown roots. The length of the primary roots was about half that of the wild type (Figures 3C and 3D, Table 2). The plant height was reduced at mature stage (Figure 3E). Albeit less severe, the RNAi phenotypes were similar to the defects found in the T-DNA insertion mutants, indicating that WOX11 is required for root development in rice.
Figure 3.
Analysis of WOX11 RNAi Transgenic Plants.
(A) Schematic representation of the WOX11 cDNA. The coding region is boxed. The slash region in the box corresponds to the conserved homeodomain. The cDNA segment used to construct the RNAi vector is indicated. UTR, untranslated region.
(B) Real-time RT-PCR analysis of WOX11 transcripts in the wild type and six RNAi transgenic lines. The PCR signals were normalized with actin transcripts. Transcript levels from the wild type were set at 1. Data are means ± sd (n = 3)
(C) Comparison of 1-week-old seedlings between the wild type (left) and the RNAi line RW4 (right).
(D) Comparison of 2-week-old seedlings between the wild type (left) and the RNAi line RW4 (right).
(E) Comparison of mature plants between the wild type (left) and the RNAi line RW4 (right).
LR, lateral root; CR, crown root; PR, primary root. Bar = 1 cm in (C) and (D) and 30 cm in (E).
[See online article for color version of this figure.]
Table 2.
Comparison of Primary Root Lengths and Crown Root Numbers between Wild-Type and WOX11 Transgenic Plants
| Days after Germination | Genotype (Plant Number) | Primary Root Length | Crown-Borne Root Number |
|---|---|---|---|
| 1 Week | Wild type (16) | 4.5 ± 0.20 | 2.3 ± 0.11 |
| RW− (20) | 4.7 ± 0.20 | 2.2 ± 0.15 | |
| RW4 (18) | 2.3 ± 0.17** | 1.7 ± 0.26* | |
| RW10 (16) | 2.4 ± 0.20** | 1.5 ± 0.25** | |
| RW18 (15) | 2.0 ± 0.11** | 1.7 ± 0.23** | |
| OW− (18) | 4.9 ± 0.21 | 1.9 ± 0.17 | |
| OW1 (15) | 3.8 ± 0.16** | 3.5 ± 0.13** | |
| OW2 (15) | 3.1 ± 0.18** | 3.3 ± 0.12** | |
| OW7 (16) | 3.1 ± 0.15** | 3.5 ± 0.14** | |
| 2 Weeks | Wild type (16) | 9.4 ± 0.20 | 4.8 ± 0.21 |
| RW− (20) | 9.8 ± 0.28 | 4.7 ± 0.20 | |
| RW4 (13) | 5.0 ± 0.28** | 3.8 ± 0.28** | |
| RW10 (12) | 5.4 ± 0.22** | 4.3 ± 0.20* | |
| RW18 (14) | 5.4 ± 0.17** | 4.1 ± 0.23* | |
| OW− (18) | 9.6 ± 0.33 | 4.39 ± 0.19 | |
| OW1 (14) | 4.0 ± 0.16** | 8.3 ± 0.39** | |
| OW2 (15) | 5.2 ± 0.24** | 10.1 ± 0.36** | |
| OW7 (14) | 5.6 ± 0.19** | 11.8 ± 0.48** |
Data were collected from the wild type, WOX11 overexpression (OW1, OW2, and OW7), and RNAi transgenic positive (RW4, RW10, and RW18) and negative segregants (RW− and OW−) in the T1 generation lines. Data represent the means ± se of plants in each line. Significant differences are indicated at the 5% (*) and 1% (**) probability levels (Dunnett's two-tailed tests).
Overexpression of WOX11 Induces Ectopic Production of Shoot-Borne Roots
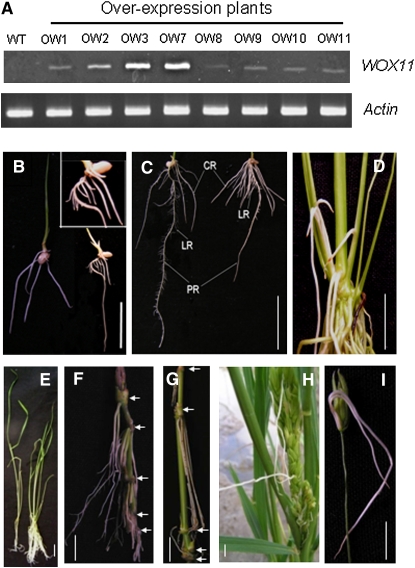
To further study the function of WOX11, a vector containing the WOX11 cDNA under the control of the maize ubiquitin promoter was transformed into rice plants. The T0 transgenic population contained many aberrant plants showing a large root system but poorly developed shoots (see Supplemental Figure 3 online). These plants died before maturity. The T0 plants showing less severe phenotypes produced seeds. The latter plants showed overexpression of WOX11 (Figure 4A). T2 seeds of three single-copy transgenic lines (OW1, OW2, and OW7) were selected for further analysis. One week after germination, the root system of the transgenic plants was more developed than the wild type, presenting a high number of crown roots and precocious lateral root development (Figure 4B, Table 2). Two weeks after germination, these plants developed at least twice as many crown roots as the controls (wild type and a transgenic sibling without overexpression of WOX11) (Figure 4C, Table 2). During vegetative growth, the plants overexpressing WOX11 produced an extensive root system with a large number of shoot-borne roots (Figures 4D and 4E). Later during development, shoot-borne roots were developed at every node of the culms (stems), including the base of florets (Figures 4F to 4I).
Figure 4.
Overexpression of WOX11-Induced Ectopic Crown Roots in Transgenic Rice.
(A) Detection of WOX11 transcripts by RT-PCR in eight phenotypic transgenic lines (OW). Actin transcripts were detected as controls.
(B) Comparison of 1-week-old seedlings of the wild type (left) with the overexpression plant (right). Inset: enlarged view of the transgenic root showing precocious production of lateral roots on the crown roots.
(C) Comparison of 2-week-old roots between the wild type (left) and the overexpression plants (right).
(D) An overexpression plant producing a large number of crown roots, showing roots growing out from the shoot.
(E) Comparison of a wild type (left) and an overexpression plant (right) at the four-leaf stage.
(F) Ectopic crown roots produced on the lower nodes of the overexpression plants (indicated by arrows).
(G) Ectopic crown roots produced on the upper nodes (indicated by arrows).
(H) Ectopic crown roots produced in the panicles.
(I) Two ectopic crown roots produced at the base of a floret in an overexpression plant. Pictures were taken from lines OW1, OW2, or OW7, which showed similar phenotypes.
LR, lateral root; CR, crown-borne root; PR, primary root. Bars = 2 cm.
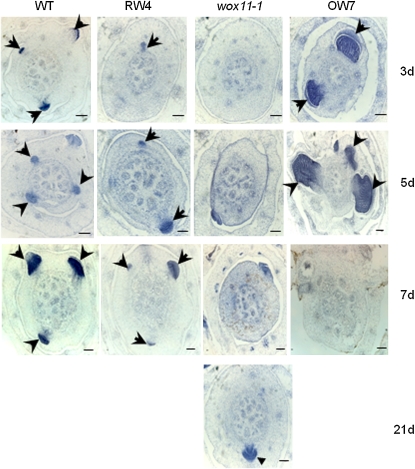
To check whether WOX11 affects the initiation of crown root primordia, serial cross sections of the coleoptilar nodal region of 3-, 5-, or 7-d-old seedlings were stained with hematoxylin. One set of the pictures is shown in Figure 5. In the wild type, the growth of crown roots started 3 d after germination, which was retarded or not initiated in the RNAi and wox11-1 plants. In fact, the initiation of a crown root in wox11-1 was observed only 21 d after germination (Figure 5). By contrast, the crown roots of overexpression plants grew much earlier and more rapidly, as they were fully developed only 7 d after germination. The increase was primarily due to production of crown roots on the third and subsequent nodes, although slight increases of root number on the first two nodes were observed (Table 3). The observations indicated that elevated WOX11 expression not only stimulated the growth rate of crown roots in lower stem nodes, but also activated ectopic crown root production on the upper stem nodes.
Figure 5.
Crown root growth rates in wild-type and WOX11 RNAi, mutant, and over-expression plants.
Hematoxylin-stained cross sections of the coleoptilar node of wild-type, WOX11 RNAi (RW4), wox11-1 mutant, and WOX11-overexpressing (OW7) seedlings at 3, 5, 7, or 21 d after germination as indicated. Arrows indicate emerging crown root initials. Bars = 50 μm.
Table 3.
Comparison of Crown Root Numbers at the Different Stem Nodes between Wild-Type and WOX11 Overexpression (OW7) Plants
| Node | Root Number Overexpression | Wild Type |
|---|---|---|
| 1st | 26.6 ± 1.3 | 24.6 ± 1.1 |
| 2nd | 22.6 ± 1.2 | 18.4 ± 1.0 |
| 3rd | 17.6 ± 1.8 | 0.7 ± 0.4 |
| 4th | 11.2 ± 1.7 | 0 |
| 5th | 10.5 ± 0.9 | 0 |
| 6th | 6.9 ± 1.3 | 0 |
| 7th | 3.5 ± 0.7 | 0 |
| 8th | 2.4 ± 0.8 | 0 |
| ≥9th | 0.7 ± 1.2 | 0 |
The crown root numbers were surveyed from 10 plants (tillers) for each genotype. Average numbers with sd are shown.
Examination of the anatomy of crown roots revealed a disorganization of the cortical cell layers and altered cell structure of the cells adjacent to the epidermis in the mutants, while the overexpression of WOX11 led to an increase of cortical and parenchyma cell layers (see Supplemental Figure 4 online).
WOX11 Expression Is Regulated by Auxin and Cytokinin
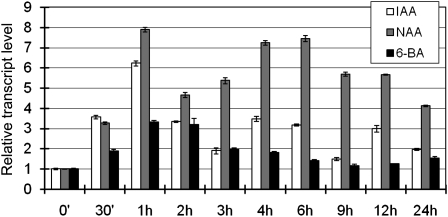
As auxin and cytokinin are known to regulate root growth, we checked whether WOX11 was regulated by those hormones. Wild-type seedlings (10 d after germination) were transferred to liquid media containing indole-3-acetic acid (IAA), naphthaleneacetic acid (NAA), or 6-benzylaminopurine (6-BA). The seedlings were harvested for RNA extraction at different time points during the treatment, and the WOX11 transcripts were quantified by real-time RT-PCR. The analysis revealed that the WOX11 expression could be induced by IAA and NAA after 30 min of treatment, with the highest induction (6 to 8 times) at 1 h of treatment (Figure 6). To a lesser extent, cytokinin (6-BA) could also induce the WOX11 expression (Figure 6).
Figure 6.
Kinetics of Induction of WOX11 in Response to Plant Hormones IAA, NAA, and 6-BA.
The transcript levels of WOX11 in 10-d-old light-grown wild-type seedlings treated with IAA, NAA, or 6-BA for the indicated times were plotted as the relative expression (fold) of water-treated seedlings during the same durations. The PCR signals were normalized with those of the actin transcripts. Data are means ± sd (n = 3).
Expression of Hormone-Responsive Genes in WOX11 Transgenic Plants
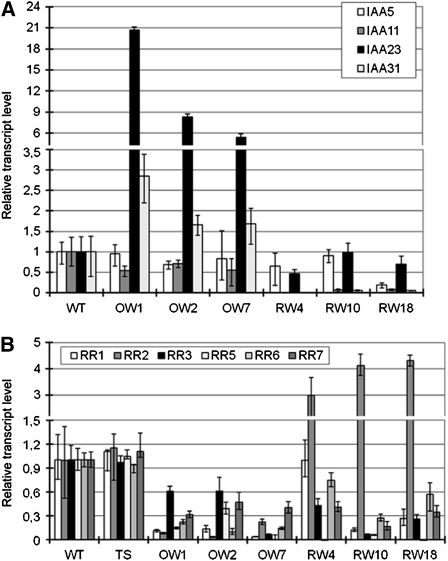
To check whether auxin and cytokinin signaling was affected in the transgenic plants, the expressions of four putative auxin-responsive Aux/IAA and six putative cytokinin type-A RR genes in 1-week-old seedlings were analyzed. These genes have been shown to be expressed in rice roots (Jain et al., 2006; Du et al., 2007). IAA23 and IAA31 were more highly expressed in WOX11 overexpression seedlings than in the wild type, while IAA11 and IAA31 were repressed in all three RNAi lines (Figure 7A). All of the six tested RR genes (RR1, RR2, RR3, RR5, RR6, and RR7) were repressed in WOX11-overexpressing plants. However, these genes, except for RR2, were also repressed in the RNAi lines. RR2 showed approximately three- to fourfold induction in the RNAi plants compared with a nonphenotypic transgenic sibling and the wild-type plants (Figure 7B). This result suggests that RR2 might be regulated by WOX11.
Figure 7.
Expression of Auxin- and Cytokinin-Responsive Genes in Wild-Type, WOX11 RNAi, and Overexpression Plants.
(A) Relative expression levels determined by real-time RT-PCR of four rice Aux/IAA genes (IAA5, IAA11, IAA23, and IAA31) in three WOX11 overexpression (OW) or three RNAi (RW) lines compared with the wild type.
(B) Relative expression levels of six rice type-A RR genes. A nonphenotypic RNAi transgenic sibling (TS) was included for comparison. The PCR signals were normalized with those of the actin transcripts. Transcript levels from the wild type were set at 1. Data are means ± sd of three biological replicates.
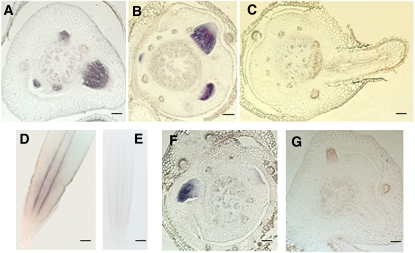
In situ hybridization revealed that RR2 is expressed in crown root initials and during the early stages of the crown root development on the coleoptilar node (Figures 8A and 8B). The expression of RR2 was not detected in more developed crown roots protruding out of the node (Figure 8C). In developed crown roots, the hybridization signals were weak and limited to the periphery of the vascular cylinder (Figure 8D). However, RR2 transcripts were detected in crown root primordia of 21-d-old wox11-1 plants (Figure 8F), which were at comparable levels to those in the wild type (Figure 8A). The signals were reduced in WOX11 overexpression plants (Figure 8G). The observations suggest that WOX11 might negatively regulate RR2 expression during crown root development.
Figure 8.
In Situ Hybridization Detection of RR2 Transcripts in Crown Roots.
(A) to (C) In coleoptilar node sections of a 5- (A), 7- (B), or 12-d-old (C) seedlings.
(D) and (E) In mature crown roots with the antisense (D) or sense (E) probes.
(F) In a coleoptilar node section of a 21-d-old wox11-1 seedling.
(G) In a coleoptilar node section of a 5-d-old WOX11 overexpression seedling (OW7).
Bars = 50 μm.
WOX11 Directly Represses RR2 Expression
Nuclear localization of WOX11-GFP (see Supplemental Figure 1B online) suggests that WOX11 functions in the nucleus. To study whether WOX11 directly regulates the expression of RR2, WOX11 was fused with the glucocorticoid receptor (GR). The latter sequestrates the fusion protein in the cytoplasm but allows entry to the nucleus in the presence of dexamethasone (DEX). The system has been previously used in rice (Inukai et al., 2005; Dai et al., 2007). The WOX11-GR fusion under the control of the double enhancers of the cauliflower mosaic virus 35S promoter was introduced into rice plants by Agrobacterium tumefaciens–mediated transformation.
RT-PCR analysis showed that several transgenic lines expressed the fusion mRNA (see Supplemental Figure 5A online). These lines did not show any root phenotype in the absence of DEX, but in the presence of DEX showed a similar root phenotype as in the overexpression lines (see Supplemental Figure 5B online). Wild-type plants and siblings from each of three selected transgenic lines (WG1-WG3) were divided into four groups for treatment as described previously (Dai et al., 2007). One group was treated with DEX and a second one was treated with cycloheximide (CHX) to inhibit protein synthesis. The third group was treated first with CHX for 1 h, then with DEX for 3 h, and the fourth one received no treatment. Whole seedlings were harvested for RNA extraction and analyzed by real-time PCR to examine the transcript levels of RR2. The DEX treatment resulted in a two- to fourfold decrease of RR2 transcripts in the presence or absence of CHX, while the expression of RR2 in wild-type plants was not affected by the treatments (see Supplemental Figure 5C online). The data suggest that WOX11 is a direct repressor of RR2.
WOX11 Interacts with the RR2 Promoter in Vitro and in Vivo
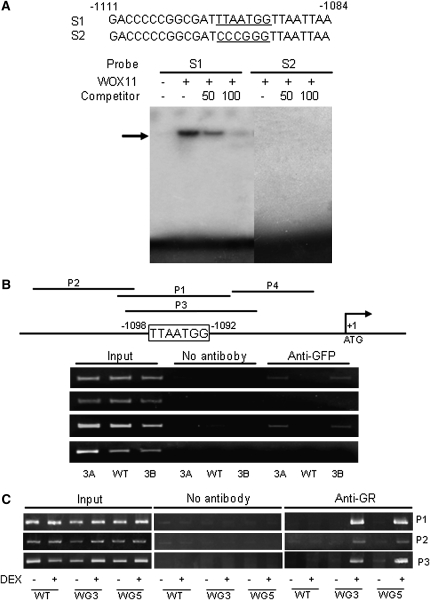
It has been shown that WUS directly represses some of the type-A RR genes in Arabidopsis SAMs (Leibfried et al., 2005). WUS interacts with the TTAATGG sequence within AGAMOUS and RR genes (Lohmann et al., 2001; Leibfried et al., 2005). In addition, rice WOX proteins QHB (for quiescent center-specific homeobox) and WOX3 have been shown to bind to the same DNA sequence in different contexts (Kamiya et al., 2003; Dai et al., 2007). Sequence analysis revealed that the Os-RR2 promoter region contains a potential WOX binding site (Figure 9A). WOX11 protein was produced in Escherichia coli and used for in vitro binding assays. As shown in Figure 9A, WOX11 could bind to the RR2 promoter fragment containing the TTAATGG motif, but not to a mutated version.
Figure 9.
WOX11 Interacts with the RR2 Gene in Vitro and in Vivo.
(A) Gel shift assays of WOX11 protein binding to a promoter sequence (S1) of RR2 containing the WOX binding site (underlined) or a mutant version of the promoter (S2). E. coli–produced WOX11 protein was incubated with 32P-labeled S1 or S2 in the absence or presence of 50 or 100 M excess of the corresponding cold probes and analyzed by electrophoresis. The shifted band is indicated by an arrow.
(B) ChIP analysis of transgenic plants expressing a WOX11-GFP fusion protein. Nuclei from two WOX11-GFP expression lines (3A and 3B) and the wild-type plants were immunoprecipitated by anti-GFP or without antibody. The precipitated chromatin fragments were analyzed by 26 cycles of PCR using four primer sets (P1 to P4) amplifying four RR2 promoter regions as indicated. The relative nucleotide positions of the putative WOX binding site are indicated (with the initiation ATG codon assessed as +1). One-tenth of the input chromatin was analyzed as controls.
(C) ChIP analysis of WOX11-GR transgenic plants using antibodies against mouse glucocorticoid receptor. Nuclei from two WOX11-GR expression lines (WG3 and WG5) and wild-type plants treated with (+) or without (–) DEX were immunoprecipitated by anti-GR or without antibody. The precipitated chromatin fragments were analyzed by 28 cycles of PCR using three primer sets (P1 to P3) as indicated. One-tenth of the input chromatin was analyzed as controls.
To study whether WOX11 interacts with RR2 in planta, we produced transgenic rice plants expressing a WOX11-GFP fusion. Chromatin of wild type and two lines of the transgenic plants were isolated and used for chromatin immunoprecipitation (ChIP) with antibodies against GFP. The precipitated products in the presence or absence of the antibody as well as input materials were analyzed by PCR using four primer sets corresponding to different regions of the RR2 promoter (Figure 9B). The results showed that only the P1 and P3 regions covering the TTAATGG sequence were amplified. To confirm the observations, transgenic lines WG3 and WG5 that express the WOX11-GR fusion shown in Supplemental Figure 5A online were assayed for ChIP using antibody against mouse GR. As shown in Figure 9C, treatment with DEX highly enriched the promoter regions P1 and P3 precipitated by the GR antibody. These data together indicate that WOX11 was associated to the RR2 promoter in vivo.
DISCUSSION
WOX11 May Stimulate Cell Division in the Crown Root Meristem
In this work, we presented evidence that WOX11 is required for the development of rice crown roots. During rice crown root formation, different developmental stages can be distinguished: initiation, early development, and emergence of the root primordia (Itoh et al., 2005). The rice gene CRL1/ARL1 is expressed during the initiation of crown root primordia and is shown to be essential for the crown root initiation (Inukai et al., 2005; Liu et al., 2005). Our data suggest that WOX11 is a distinct regulator of crown root development, as the expression of WOX11 was mainly detected in emerging crown root primordia and later in the active cell division region of the root meristem (Figures 1D to 1G and 1Q to 1S), suggesting that WOX11 may have a function to regulate the emergence and the growth of crown roots.
Crown roots are initiated in the stem nodes as part of normal plant development in rice, and crown root initials are detected in upper stem nodes (Liu et al., 2005). However, the root initials on the upper nodes do not emerge through the nodal epidermis (Bleecker et al., 1987; Lorbiecke and Sauter, 1999; Liu et al., 2005), except in deeper-water rice when treated with submergence (Lorbiecke and Sauter, 1999). The inhibition or delay of crown root growth in loss-of-function mutants and downregulation plants of WOX11 supports the hypothesis that WOX11 is required for the emergence and elongation of crown root primordia. Interestingly, elevated WOX11 expression induced not only precocious growth of the crown root from the lower stem nodes (Figures 4 and 5), but also crown root production from the upper stem nodes (Figure 4, Table 3), suggesting that WOX11 may be sufficient to activate the growth of crown root initials. WOX11 may regulate the development of the crown root system by stimulating cell division. This is supported by accelerated growth of crown root primordia and extensive production of lateral roots from the crown roots in the overexpression plants (Figures 4 and 5). However, a function of WOX11 in crown root initiation is not excluded, as wox11 mutations reduced crown root initials, and the overexpression of the gene induced supernumerary crown roots on the basal nodes.
WOX11 is also expressed in the shoot meristem. Loss-of-function mutation or downregulation of WOX11 reduced the shoot growth, but did not affect the morphology of the SAM (see Supplemental Figure 6 online). The reduced shoot growth may be an indirect effect of disrupted root function. Alternatively, the loss of function of WOX11 in the SAM may be compensated by functional redundancy of other SAM-expressed WOX genes. Unlike crown roots, the shoot growth in WOX11 overexpression plants was not stimulated (Figure 4), suggesting that either WOX11 is not limiting or WOX11 may function with a different mechanism to regulate cell division in the shoot.
WOX11 Functions in Auxin and Cytokinin Signaling to Promote Crown Root Development
Auxin is essential for root development, including root meristem function, lateral root formation, and root elongation. Auxin signaling is shown to be required for crown root initiation in rice (Inukai et al., 2005). Auxin treatment of wox11 mutants failed to induce crown root production as in wild-type plants (see Supplemental Figure 7A online). This suggests that WOX11 may be required for auxin induction of crown root development. Conversely, treatment with the auxin inhibitor N-1-naphthylphthalamic acid reduced crown root production in WOX11-overexpressing plants (see Supplemental Figure 7B online), indicating a role of auxin in WOX11-mediated regulation of crown root growth. Although the expression pattern of WOX11 did not overlap with the accumulation of auxin in roots (Figure 1), the gene was rapidly induced by exogenous treatment of auxin (Figure 6), suggesting that WOX11 is an auxin-responsive gene. These data suggest that the action of exogenous auxins on crown roots may be at least partly mediated by an effect on WOX11 expression. At the same time, because WOX11 expression correlates with the expression of auxin-responsive Aux/IAA genes (Figure 7A), it is possible that WOX11 is part of a positive feedback loop of auxin signaling.
We showed that the expression of WOX11 was induced by cytokinin treatment, and downregulation or gain-of-function of WOX11 altered the expression of type-A RR genes. Type-A RR genes have been shown to be transcriptionally upregulated by cytokinin and to function as redundant negative regulators of cytokinin signaling (To et al., 2004). In this work, we showed that WOX11 directly represses one of the type-A genes, RR2 (Figures 8 and 9; see Supplemental Figure 5 online). RR2 is rapidly induced by cytokinin (Jain et al., 2006, Du et al., 2007). Activation of RR2 may have a direct or indirect negative effect on the expression of other type-A RR genes. Reduction of WOX11 activity in the mutants or RNAi plants derepresses RR2, which then represses other type-A RR genes (Figure 7B), thus enhancing cytokinin signaling and inhibiting crown root development. In the overexpression lines, the expression of RR2 is turned off, but the other RR genes are also repressed (Figure 7B), possibly due to a repressive effect of activation of the auxin pathway. Therefore, WOX11 is likely to function in a part of a homeostatic feedback loop of both cytokinin and auxin signaling to promote crown root development.
The direct repression of RR2 by WOX11 in crown root primordia is reminiscent of the regulation of type-A ARR genes by WUS in Arabidopsis SAMs (Leibfried et al., 2005). WUS, a positive regulator of meristem stem cells, directly represses the transcription of several ARR genes (ARR5, ARR6, ARR7, and ARR15). ARR7 is expressed in a specific domain of the Arabidopsis SAM. WUS was shown to bind directly to sequences in the ARR7 promoter. Overexpression of an active form of ARR7 caused a meristem determination phenotype, similar to that of wus mutants (Leibfried et al., 2005), indicating that ARR genes negatively regulate meristem size and that their repression by WUS is required for meristem activity. Similarly, the mutation of a maize type-A RR gene, ABPHL1 (ABPH1), induces an increased meristem size (Giulini et al., 2004), suggesting that ABPH1 may restrict the meristem size by inhibiting cell division. RR2 was expressed in crown root primordia, but downregulated in emerging and developed crown roots (Figure 5), suggesting that RR2 may have a similar function as the above mentioned Arabidopsis and maize type-A genes, negatively regulating cell proliferation in crown root meristem, and its repression by WOX11 may be necessary to promote the emergence of crown roots. Recent results show that overexpression of RR6 also results in a poorly developed root system as well as a dwarf phenotype in rice (Hirose et al., 2007).
Developmental Functions of WOX Genes
WOX genes are suggested to play an important role in region-specific transcription programming during embryogenesis and lateral organ development (Haecker et al., 2004). In this work, we showed that WOX11 is expressed in rapid cell division regions of both root and shoot meristems. The expression pattern and the growth defects induced by WOX11 mutation and downregulation suggest that WOX11 may be involved in establishing a transcriptional program by coordinating both auxin and cytokinin signals to stimulate cell division in root and shoot meristems in rice. In addition, the root epidermal expression pattern of WOX11 and the alteration of cell structure in the root epidermis and in cells adjacent to the epidermis suggest that WOX11 may function in specification of cell identity in roots (Figure 1; see Supplemental Figure 5 online). This expression pattern is distinct from the other studied rice WOX genes. For instance, the expression of QHB is confined in the quiescent center of the root meristem (Kamiya et al., 2003), whereas WOX3 transcript is detected in leaf primordia but is excluded from the SAM (Dai et al., 2007). QHB is required for the root apical meristem function (Kamiya et al., 2003). Ectopic expression of QHB affects normal shoot and leaf development in rice (Kamiya et al., 2003). The malformed leaves induced by QHB overexpression are much like WOX3 overexpression leaves (Dai et al., 2007), suggesting that basic regulatory functions, such as DNA binding and transcriptional repression, of the two WOX proteins might have been conserved during evolution, while their natural developmental functions may diverge partially as a result of their differential expression patterns. Similarly, Arabidopsis WOX5 and WUS are interchangeable in stem cell regulation (Sarkar et al., 2007). Our data show that overexpression of Os-WOX11 induced an extensive crown root production, suggesting that this protein may have a different biochemical or transcriptional function than QHB or WOX3. Similarly, Arabidopsis WOX8 or WOX9 could not rescue wus mutant defects (Sarkar et al., 2007). These observations suggest that subgroups of WOX proteins may have distinct transcriptional and cellular functions. Our data showed a direct repression of a type-A RR gene by WOX11 required for crown root cell proliferation. This is similar to the repression of several type-A ARR genes by WUS required for Arabidopsis meristem function, suggesting a conservation of some of the regulatory properties among the two WOX proteins.
METHODS
Materials
The rice variety Zhonghua11 (Oryza sativa spp Japonica) was used for transformation in this study. Rice seeds were surface-sterilized and germinated in media containing 0.8% agar supplemented with 3% (w/v) sucrose at 28°C (in light) and 24°C (in dark) with a 14-h-light/10-h-dark cycle. The full-length cDNA of WOX11 was isolated from a normalized Zhonghua11 cDNA library (Zhang et al., 2005).
Characterization of WOX11 Insertion Mutants
The T-DNA insertion mutant lines 2A00597 and 2D40272 and the respective wild-type varieties (Hwa and Dongjing) were obtained from the Pohang University of Science and Technology, South Korea. The insertions were confirmed by PCR using WOX11-specific primers F2 and R2 and a T-DNA left side primer L2 or right side primer R1. For RT-PCR analysis of WOX11 transcripts in the mutants, the primer set F3 and R3 was used. Actin transcript was amplified as controls using the primer set Actin-F and Actin-R. Nucleotide sequences of the primers are listed in Supplemental Table 1 online.
Vector Construction and Rice Transformation
For the construction of the fusion between the WOX11 promoter and the GUS coding sequence (WOX11p-GUS), the 2.4-kb WOX11 promoter was amplified from Zhonghua11 genomic DNA and inserted into pCAMBIA1381Xb (obtained from the Centre for the Application of Molecular Biology to International Agriculture, CAMBIA, Australia) at the SalI and BamHI sites. The primers used in PCR amplification were WOX11gus-F and WOX11gus-R. The DR5-GUS construct is described by Scarpella et al. (2003).
To construct the RNAi vector, a 492-bp cDNA fragment of WOX11 was amplified from the cDNA clone using the primer set WOX11RNAi-F and WOX11RNAi-R and was inserted into the KpnI and BamHI sites (for forward insert) and the SacI and SpeI sites (for the reverse insert) of the pDS1301 vector (Chu et al., 2006).
For overexpression, the full-length cDNA of WOX11 amplified with the primer set OXWOX11-F and OXWOX11-R was inserted into the KpnI and BamHI sites of pU1301, previously known as p1301DS (Dai et al., 2007).
To construct the translational WOX11-GFP fusion, the GUS fragment of pCAMBIA1391Xb was replaced by a maize (Zea mays) ubiquitin promoter-GFP cassette from pU1301.The coding region of WOX11 was amplified using the primers WOX11gfp-F and WOX11gfp-R. The amplified fragment was inserted into the KpnI and BamHI sites of modified pCAMBIA1391Xb to create pU1391-WOX11-GFP for nuclear localization assays and for rice transformation.
To create the WOX11-GR fusion construct, the full-length coding sequence of WOX11 was amplified by PCR. The stop codon was removed and replaced with a SalI site. A KpnI site was added to the end of the forward primer. The primers used in PCR were WOX11gr-F and WOX11gr-R. The amplified fragment was inserted into the plasmid pSport1 (Clontech). In the same way, the DNA fragment encoding the steroid binding domain of the mouse GR was amplified using the plasmid pBI-GR as template. The PCR primers were GR-F with a SalI adaptor and GR-R with a BamHI adaptor. The amplified GR fragment was inserted downstream to and in frame with WOX11 in pSport1. The fused WOX11-GR sequence was then cut from pSport1 and inserted into pU1301 digested with KpnI and BamHI.
The sequences of the primers used in vector construction are listed in Supplemental Table 1 online.
Agrobacterium tumefaciens (strain EHA105)–mediated transformation of Zhonghua11 plants was conducted according to Dai et al. (2007).
RNA Isolation and RT-PCR
For gene expression analysis, total RNA was extracted from wild-type and transgenic plants using TRIzol (Invitrogen). For RT-PCR analysis, total RNA (2 μg) was reverse transcribed in a total volume of 20 μL with 0.5 μg oligo(dT)15, 0.75 mM deoxynucleotide triphosphate, 10 mM DTT, and 100 unites of SuperScript II RNase H− reverse transcriptase (Invitrogen). PCR was performed in a total volume of 20 μL with 1 μL of the reverse transcription reactions, 0.2 μM gene-specific primers, and 1 unite of rTaq (TaKaRa). Twenty-five to 30 reaction cycles were performed. Rice actin gene transcripts were detected by PCR as internal controls. Real-time PCR was performed using gene-specific primers in a total volume of 25 μL with 1.5 μL of the RT reactions, 0.25 μM gene-specific primers, and 12.5 μL SYBR Green Master mix (Applied Biosystems) on a 7500 real-time PCR machine (Applied Biosystems) according to the manufacturer's instructions. The rice actin gene was used as the internal control. All primers were annealed at 58°C. The relative expression level of each gene in transgenic plants was compared with the wild type, after normalization with actin transcript and averaged from three biological replicates. The sequences of the primers used are listed in Supplemental Table 1 online.
In Situ Hybridization
The hybridization and immunological detection were performed as described by Dai et al. (2007). The WOX11 probe was amplified using the gene-specific primers WOX11RNAI-F and WOX11RNAi-R. The RR2 probe was amplified using the gene-specific primers RR2situ-F and RR2situ-R (see Supplemental Table 1 online). The PCR fragments were inserted into the SpeI and BamHI sites of pGEM-T (Promega) and transcribed in vitro from either T7 or SP6 promoter for sense or antisense strand synthesis using the Digoxigenin RNA labeling kit (Roche).
Promoter Activity Detection and Nuclear Localization
Transgenic plants harvested at different developmental stages were incubated with X-gluc buffer overnight at 37°C (Jefferson et al., 1987). After being stained, the tissues were rinsed and fixed in formalin acetic acid alcohol fixation solution (50% ethanol, 5% acetic acid, and 3.7% formaldehyde) at 4°C overnight, embedded in paraffin, sectioned, mounted on slides, and photographed using a LEICA MZFL111 stereomicroscope with a color CCD camera.
For the nuclear localization analysis, plasmids (5 μg) were coprecipitated with 3 mg of gold particles. The particles were resuspended in ethanol in a total volume of 60 μL and divided into five aliquots for bombarding onion epidermal cells using the PDS-1000 System (Bio-Rad) at 1100 p.s.i. helium pressure. The expression of the fusion protein of WOX11-GFP in the onion epidermal cells was observed by a confocal microscope (Leica) 36 h after bombardment.
Exogenous IAA, NAA, NPA, and 6-BA Treatment
Seeds were sowed and germinated on agar medium. After 10 d, the seedlings were transferred to media with or without 10−6 M NAA, 10−6 M IAA, or 10−5 M 6-BA. Total RNA was extracted after 0, 0.5, 1, 3, 6, 9, 12, and 24 h of treatment and analyzed by real-time RT-PCR. For root growth tests, mutant, transgenic, and wild-type seeds were germinated on agar medium containing 10−6 M NAA or 10−6 M NPA. Four weeks after treatment with NPA and 9 d with NAA, crown root numbers were recorded.
Histological Observation
Root, shoot, and stem node sections from mutant, wild-type, or transgenic plants were stained with hematoxylin. The procedures of staining, dehydration, clearing, infiltration, and embedding were performed according to Liu et al. (2005). The microtome sections (8 to 10 μm) were mounted on glass slides for imaging.
Electrophoretic Mobility Shift Assay
To produce the WOX11 protein, the full-length cDNA amplified with primers WOX1132a-F and WOX1132a-R was inserted into the KpnI and BamHI sites of the pET-32a expression vector (Novagen) and expressed in Escherichia coli DE3 cells (Novagen). The target protein was purified with B-PER 6X His Spin Purification kit (Pierce). The RR2 promoter DNA S1 (including the putative WUS binding site: TTAATGG) and S2 (with nucleotide substitutions in the WUS binding site) were produced by annealing of oligonucleotides S1-F and S1-R, or S2-F and S2-R, respectively. The double-stranded oligonucleotides S1 and S2 were labeled with 32P-dCTP using Klenow fragment. DNA binding reactions were performed in the presence or absence of unlabeled S1 fragment at 50 or 100 molar excess at room temperature for 20 min in 10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM MgCl2, 5% glycerol, and 50 mg/L poly(dI-dC) poly(dI-dC) (Amersham Pharmacia Biotech). The reactions were resolved on 6% polyacrylamide gels in Tris-glycine (0.3% Tris and 1.88% glycine) buffer and visualized by autoradiography. The sequences of the primers used are listed in Supplemental Table 1 online.
In Vivo Binding Assay of WOX11
For ChIP assays, wild-type, WOX11-GFP transgenic lines, and WOX11-GR trangenic plants treated with or without DEX were used for chromatin extraction and immunoprecipitation as described by Huang et al. (2007). Briefly, roots were treated with formaldehyde and the nuclei were isolated and sonicated using an Ultrasonic Crasher Noise Isolating Chamber (SCIENTZ). The soluble chromatin fragments were isolated and preabsorbed with sheared salmon sperm DNA/protein A-agarose (Sigma-Aldrich) to remove nonspecific binding. Immunoprecipitations with anti-GFP (Abcam; ab290), anti-GR (Santa Cruz Biotechnology; sc-1002), or without any serum were performed as described. The precipitated DNA was analyzed by PCR using specific primer sets (see Supplemental Table 1 online).Typically, 26 to 28 cycles of PCR were performed, and the products were analyzed by agarose gel electrophoresis.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: WOX11, AK073232, LOC_Os07g48560; RR1, AK072736, LOC_Os04g36070; RR2, AK070645, LOC_Os02g35180; RR3, AP004043, LOC_Os02g58350; RR5, AK106426, LOC_Os04g44280; RR6, AK059734, LOC_Os04g57720; RR7, AP003802, LOC_Os07g26720; IAA5, AK106121, LOC_Os01g48450; IAA11, LOC_Os03g43400; IAA23, AK069376, LOC_Os06g39590; and IAA31, AK073361, LOC_Os12g40900.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of WOX11p-GUS and Nuclear localization of WOX11-GFP Protein.
Supplemental Figure 2. Genotyping of the WOX11 Locus in the Two Insertion Lines and the Respective Wild Type (Hwa and Dongjing).
Supplemental Figure 3. Severe Phenotypes Found in the T0 Population of WOX11 Overexpression Plants.
Supplemental Figure 4. Comparison of Crown Root Cross and Longitudinal Sections of Wild-Type, wox11-1 Mutant, and Overexpression Plants.
Supplemental Figure 5. Production of Transgenic Lines Expressing the WOX11-GR Fusion.
Supplemental Figure 6. Comparison of Shoot Longitudinal Sections from Wild Type and wox11 Mutants.
Supplemental Figure 7. Effect of Auxin and an Auxin Inhibitor on Crown Root Growth.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Gynheung An for proving the T-DNA insertion mutant line and P.B. Ouwerkerk for providing the binary vector pCAMBIA containing the DR5-GUS construct. This work was supported by grants from the National Special Key Program of Rice Functional Genomics and the National Natural Science Foundation of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dao-Xiu Zhou (dao-xiu.zhou@u-psud.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Bleecker, A.B., Rose-John, S., and Kende, H. (1987). An evaluation of 2,5-norbornadiene as a reversible inhibitor of ethylene action in deepwater rice. Plant Physiol. 84 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., Li, X., Fu, B., Li, Z., Bennetzen, J.L., Zhang, Q., and Wang, S. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H., Levesque, M.P., Vernoux, T., Jung, J.W., Paquette, A.J., Gallagher, K.L., Wang, J.Y., Blilou, I., Scheres, B., and Benfey, P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316 421–425. [DOI] [PubMed] [Google Scholar]
- Dai, M., Hu, Y., Zhao, Y., Liu, H., and Zhou, D.X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio, R., Linhares, F.S., and Sabatini, S. (2007. a). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11 23–27. [DOI] [PubMed] [Google Scholar]
- Dello Ioio, R., Linhares, F.S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., and Sabatini, S. (2007. b). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17 678–682. [DOI] [PubMed] [Google Scholar]
- De Smet, I., and Jurgens, G. (2007). Patterning the axis in plants–Auxin in control. Curr. Opin. Genet. Dev. 17 337–343. [DOI] [PubMed] [Google Scholar]
- Du, L., Jiao, F., Chu, J., Jin, G., Chen, M., and Wu, P. (2007). The two-component signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics 89 697–707. [DOI] [PubMed] [Google Scholar]
- Giulini, A., Wang, J., and Jackson, D. (2004). Control of phyllotaxy in maize by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034. [DOI] [PubMed] [Google Scholar]
- Haecker, A., Gross-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., and Laux, T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668. [DOI] [PubMed] [Google Scholar]
- Hirose, N., Nobue, N., Kojima, M., Kamada-Nobusada, T., and Sakakibara, H. (2007). Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48 523–539. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F., Park, W.J., Sauer, M., and Woll, K. (2004). From weeds to crops: Genetic analysis of root development in cereals. Trends Plant Sci. 9 42–48. [DOI] [PubMed] [Google Scholar]
- Huang, L., Sun, Q., Qin, F., Li, C., Zhao, Y., and Zhou, D.-X. (2007). Down-regulation of a Silent Information Regulator2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., Hasegawa, Y., Ashikari, M., Kitano, H., and Matsuoka, M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Jain, M., Kaur, N., Garg, R., Thakur, J.K., Tyagi, A.K., and Khurana, J.P. (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 47–59. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: b-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, N., Nagasaki, H., Morikami, A., Sato, Y., and Matsuoka, M. (2003). Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 35 429–441. [DOI] [PubMed] [Google Scholar]
- Kawata, S., and Harada, J. (1975). On the development of the crown root primordial in rice plants. Proc. Crop Sci. Soc. Jpn. 45 438–457. [Google Scholar]
- Laplaze, L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried, A., To, J.P., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175. [DOI] [PubMed] [Google Scholar]
- Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., Shou, H., and Wu, P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43 47–56. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Lorbiecke, R., and Sauter, M. (1999). Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol. 119 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, A.K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., Scheres, B., Heidstra, R., and Laux, T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. [DOI] [PubMed] [Google Scholar]
- Scarpella, E., Rueb, S., and Meijer, A.H. (2003). The RADICLELESS1 gene is required for vascular pattern formation in rice. Development 130 645–658. [DOI] [PubMed] [Google Scholar]
- Scheres, B. (2002). Plant patterning: TRY to inhibit your neighbors. Curr. Biol. 12 R804–R806. [DOI] [PubMed] [Google Scholar]
- Taramino, G., Sauer, M., Stauffer, J.L., Jr., Multani, D., Niu, X., Sakai, H., and Hochholdinger, F. (2007). The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50 649–659. [DOI] [PubMed] [Google Scholar]
- To, J.P., Haberer, G., Ferreira, F.J., Deruère, J., Masson, M.G., Shaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A ARRs are partially redundant negative regulators of cytokinin signaling in Arabidopsis. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmulling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Strnad, M., and Schmulling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Feng, Q., Jin, C., Qiu, D., Zhang, L., Xie, K., Yuan, D., Han, B., Zhang, Q., and Wang, S. (2005). Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J. 42 772–780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.