Abstract
This work examines the role of the Arabidopsis thaliana RING E3 ligase, HISTONE MONOUBIQUITINATION1 (HUB1) in disease resistance. Loss-of-function alleles of HUB1 show increased susceptibility to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola, whereas HUB1 overexpression conferred resistance to B. cinerea. By contrast, responses to the bacterial pathogen Pseudomonas syringae are unaltered in hub1 plants. hub1 mutants have thinner cell walls but increased callose around an infection site. HUB1 acts independently of jasmonate, but ethylene (ET) responses and salicylate modulate the resistance of hub1 mutants to necrotrophic fungi. The ET response factor ETHYLENE INSENSITIVE2 is epistatic to HUB1 for A. brassicicola resistance but additive to HUB1 for B. cinerea resistance. HUB1 interacts with MED21, a subunit of the Arabidopsis Mediator, a conserved complex that regulates RNA polymerase II. RNA interference lines with reduced MED21 expression are highly susceptible to A. brassicicola and B. cinerea, whereas T-DNA insertion alleles are embryonic lethal, suggesting an essential role for MED21. However, HUB1-mediated histone H2B modification is independent of histone H3 and DNA methylation. In sum, histone H2B monoubiquitination is an important chromatin modification with regulatory roles in plant defense against necrotrophic fungi most likely through modulation of gene expression.
INTRODUCTION
The first line of defense against pathogen attack is the structural barrier provided by the plant cuticle and cell wall. The degree of susceptibility of the plant cell wall to degradation by cell wall–hydrolyzing enzymes can affect the severity of disease caused by the necrotrophic fungus Botrytis cinerea. Fungal polygalacturonases (PGs) hydrolyze the homogalacturonan of plant cell wall pectin and are important virulence factors for some necrotrophic fungi, whereas the plant polygalacturonase-inhibiting proteins contribute to resistance by counteracting fungal PGs (Powell et al., 2000; Ferrari et al., 2003). Furthermore, attacked plants build up papillae on the inner side of epidermal cell walls at the point of attempted pathogen entry. Papillae are largely composed of callose (a β-1,3-glucan) but also contain polysaccharides, phenolic compounds, and reactive oxygen intermediates (Flors et al., 2005). Contrary to intuitive expectations, defects in the plant secondary cell wall caused by a mutation in cellulose synthesis resulted in resistance to necrotrophic pathogens (Hernandez-Blanco et al., 2007). Similarly, the plant cuticle protects against pathogen penetration and hence prevents the pathogen from establishing infections. However, unexpectedly, Arabidopsis thaliana mutants defective in components of the cuticle were resistant to B. cinerea (Chassot et al., 2007), which was attributed to loss of virulence in B. cinerea in the absence of cuticle-derived signals. Thus, cell wall–based defense mechanisms can decrease or enhance pathogen resistance.
Plants activate defense responses based on recognition of race-specific or race nonspecific pathogen derived elicitors. Race-specific resistance has been extensively described for biotrophic plant interactions (Jones and Dangl, 2006). Resistance that counteracts toxins occurs for a number of necrotrophic fungal pathogens (Johal and Briggs, 1992; Brandwagt et al., 2000). Regardless of the lifestyle of the pathogen and its infection strategies, plants recognize pathogen-associated molecular patterns (PAMPs) to activate defense. PAMPs are evolutionarily conserved, and essential components of pathogens that are absent in host plants and serve as nonself recognition mechanisms. In the case of fungal attacks, chitins and glucans are PAMPs that may be recognized by pattern recognition receptors. Recognition of PAMPs activates basal defenses often without the hypersensitive response (HR), whereas recognition of effectors generally trigger the HR. Recognition of effectors and PAMPs trigger systemic acquired resistance (Mishina and Zeier, 2007). Systemic and local defenses mediated by ethylene (ET) and jasmonate (JA) are required for resistance to necrotrophic pathogens (Penninckx et al., 1996; Thomma et al., 1998; Thomma et al., 1999). These defense responses interact synergistically or antagonistically to fine-tune responses to pathogens (Schenk et al., 2000; Kunkel and Brooks, 2002; Anderson et al., 2004). Thus, complex networks of interacting defense mechanisms modulate responses to necrotrophic and biotrophic pathogens.
Resistance to microbial infections requires transcription of a wide range of genes encoding regulatory and antimicrobial proteins. This transcriptional control is likely to impact the state of chromatin and DNA modifications (Kouzarides, 2007), like other plant responses to the environment and many physiological processes. Changes in higher-order chromatin structure, such as chromatin condensation, occur during plant cell death caused by fungal toxins (Navarre and Wolpert, 1999; Liang et al., 2003). The fungal toxin victorin induces apoptosis-like responses, such as DNA laddering and heterochromatin condensation (Navarre and Wolpert, 1999). In rice (Oryza sativa), changes in histone H3K4 methylation and H3 acetylation occur at promoters of specific stress-inducible genes in response to submergence (Tsuji et al., 2006). A mutation in Arabidopsis histone deacetylase 19 compromises resistance to Alternaria brassicicola, suggesting a role of chromatin acetylation in disease resistance (Zhou et al., 2005). In animal cells, histone methylation occurs at chromatin of the lipopolysaccharide (LP)-inducible inflammatory genes in response to the LPs acting as a bacterial PAMP (Saccani and Natoli, 2002). Monoubiquitination of histone H2B is a less-studied chromatin modification in plants and other organisms. In human and Schizosaccharomyces pombe, a pair of RING E3 ligases act as a complex to monoubiquitinate histone H2B (Zhu et al., 2005; Tanny et al., 2007). In Saccharomyces cerivisae, a single E3 ligase, BRE1, ubiquitinates histone H2B (Wood et al., 2003b). Histone H2B ubiquitination regulates H3 methylation, gene silencing (Sun and Allis, 2002), and transcriptional elongation (Osley, 2004; Zhu et al., 2005; Laribee et al., 2007; Tanny et al., 2007).
In Arabidopsis, histone H2B is monoubiquitinated by two RING E3 ligases, HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2, which have recently been implicated in the control of the cell cycle and seed dormancy (Fleury et al., 2007; Liu et al., 2007). Here, we show that Arabidopsis HUB1 is a regulatory component of plant defense against necrotrophic fungal pathogens. Loss-of-function HUB1 alleles display extreme susceptibility to B. cinerea and A. brassicicola. Epistasis analyses between HUB1 and defense regulatory genes SA-INDUCTION DEFICIENT2 (SID2), PHYTOALEXIN DEFICIENT4 (PAD4), ETHYLENE INSENSITIVE2 (EIN2), and CORONATINE INSENSITIVE1 (COI1) suggest that both SA and ET affect the severity of hub1 disease susceptibility at the point of infection. Interestingly, the hub1 mutation reduces the thickness of epidermal cell walls, which may account for the disease resistance role of HUB1 consistent with the role of the plant cell wall in resistance to necrotrophic fungi. HUB1 interacts with MED21, a subunit of an evolutionarily conserved multisubunit Mediator complex, regulating the function of RNA polymerase II. Arabidopsis MED21 couples critical roles in disease resistance and embryo development based on the disease susceptibility and embryo-lethal phenotypes of plant lines with reduced MED21 gene expression. Thus, MED21, together with HUB1, controls critical components of defense against necrotrophic pathogens. The interaction between HUB1 and MED21 and their induction by chitin, a fungal PAMP, further supports their role in defense against necrotrophic fungi. These data suggest that MED21 relays signals from upstream regulators and chromatin modifications to the RNA polymerase.
RESULTS
Identification of HUB1 as a Regulator of Resistance to Necrotrophic Pathogens
Previously, we described the Arabidopsis BOTRYTIS INDUCED KINASE1 (BIK1) gene required for resistance to necrotrophic pathogens (Veronese et al., 2006). To identify genes acting downstream of BIK1, we compared expression profiles of Botrytis-infected bik1 and wild-type plants using Arabidopsis whole-genome microarrays (Affymetrix ATH1). Microarray analysis is described in Supplemental Figure 1 online. HUB1 was identified as a potential target of BIK1 because its expression was limited in noninfected wild-type plants, induced fivefold by Botrytis in wild-type plants and constitutive at higher levels in the bik1 mutant. The induction of HUB1 gene expression by Botrytis and in bik1 mutants was confirmed nonquantitatively by RT-PCR (see Supplemental Figure 1A online). HUB1 failed to interact with BIK1 in yeast cells when tested in a yeast two-hybrid assay, and the regulatory relationship between HUB1 and BIK1 remains unclear (see Supplemental Figure 2 online). HUB1 encodes a C3HC4- type RING E3 ligase that monoubiquitinates histone H2B, regulating seed dormancy and the plant cell cycle (Fleury et al., 2007; Liu et al., 2007).
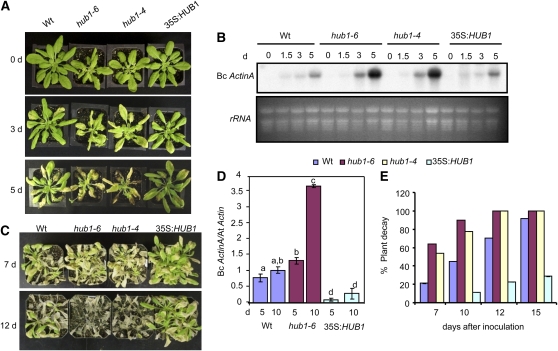
The responses of HUB1 T-DNA insertion alleles and 35S:HUB1 lines to different plant pathogens were tested to determine the role of HUB1 in plant defense. hub1-6 is a loss-of-function allele (see Supplemental Figures 1B and 1C online), and hub1-4 is a functional null allele described previously (Liu et al., 2007). We also selected two Arabidopsis 35S:HUB1 lines (35S:HUB1 #5 and #6) that constitutively express HUB1 at a higher level (see Supplemental Figure 1D online). Plants grown under 12:12-h dark-light cycles were tested for their responses to B. cinerea and A. brassicicola. The mutant plants had increased chlorosis leading to tissue damage 3 d after inoculation (DAI) with B. cinerea (2.5 × 105 spores/mL) (Figure 1A). The infection continued to spread and macerated the leaf tissue at 5 DAI in the mutant plants but caused relatively limited disease symptoms in the wild type. At 3 and 5 DAI, hub1-4 and hub1-6 plants harbor more fungal biomass than the wild type (Figure 1B) as assessed by the accumulation of the B. cinerea ActinA mRNA (Veronese et al., 2006). The 35S:HUB1 plants showed reduced disease symptoms compared with the wild-type plants, suggesting increased resistance to B. cinerea (Figure 1A). The 35S:HUB1 plants were indistinguishable from the wild type in terms of fungal growth at this level of inoculation (Figure 1B).
Figure 1.
HUB1 Contributes to B. cinerea Resistance.
(A) and (B) Disease symptoms (A) and RNA gel blot (B) showing fungal growth after inoculation with 2.5 × 105 Botrytis spores/mL.
(C) to (E) Disease symptoms (C), fungal growth (D), and percentage of decayed plants (E) after inoculation with 3.5 × 105 Botrytis spores/mL.
The disease assays were done by spray inoculation of Botrytis spores. In (B), total RNA (10 μg) was loaded per lane. In (D), the data show the qPCR amplification of B. cinerea ActinA relative to the Arabidopsis Actin2 gene. In (E), plants were considered decayed when they were completely rotten due to infection. The data represent the mean ± se from a minimum of 26 plants. The experiments were repeated at least three times with similar results. Analysis of variance and Duncan's Multiple Range Test were performed to determine the statistical significance of the differences between the mean values using SAS software (SAS Institute, 1999). The mean values followed by different letters are significantly different from each other (P = 0.05). These experiments were repeated at least three times with similar results. Bc ActinA, Botrytis cinerea ActinA gene; At Actin, Arabidopsis Actin2 gene; d, days after inoculation.
To clearly establish whether HUB1 was sufficient for resistance to Botrytis, the 35S:HUB1 plants were challenged with a higher inoculum (3.5×105 spore/mL). The 35S:HUB1 line showed delayed chlorosis and necrosis and significantly lowered B. cinerea growth as determined by the amount of fungal DNA (Figures 1C and 1D). When infection was allowed to spread by extended incubation under conditions that promote infection, disease continued to spread and macerated the wild-type plants, but progressed slower in 35S:HUB1 plants (Figures 1C and 1E). Most of the 35S:HUB1 plants (75%) survived 2 weeks after inoculation, whereas most wild-type and hub1 plants were completely decayed between 10 and 12 DAI, indicating that HUB1 expression determines the level of resistance under increased pathogen pressure.
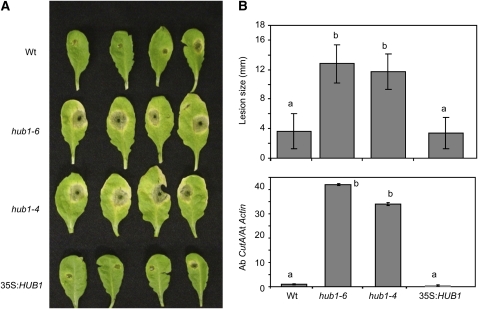
Resistance to A. brassisicola was determined by evaluating disease symptoms and fungal growth in inoculated plants. The relative amounts of A. brassicicola cutinase DNA normalized to Arabidopsis Actin2 DNA, based on real-time quantitative PCR (qPCR) amplification of DNA from inoculated leaves, was used to measure fungal growth (Brouwer et al., 2003). The mutant plants showed a threefold increase in the size of the disease lesions, and fungal growth was 42-fold higher in hub1-6 and 34-fold higher in hub1-4 plants compared with wild-type plants (Figures 2A and 2B). Interestingly, no statistically significant reduction in pathogen growth was observed in 35S:HUB1 plants relative to the wild type. This is most likely due to the high level of resistance bordering an incompatible interaction exhibited by Arabidopsis Columbia-0 (Col-0) wild-type plants. Thus, HUB1 is required for resistance to both necrotrophic fungi and its ectopic expression is sufficient for increased resistance to B. cinerea.
Figure 2.
HUB1 Is Required for Resistance to A. brassicicola.
Disease assays on hub1 and 35S:HUB1 plants showing disease symptoms (A), disease lesion size ([B]; top panel) and fungal growth (bottom panel) 6 DAI with A. brassicicola. The disease assays were done by drop inoculation of 5 × 105 A. brassicicola spore/mL. Data points represent average ± se from a minimum of 30 disease lesions. A. brassicicola growth was determined using qPCR amplification of the fungal Cutinase DNA (Ab CutA). The relative DNA levels were calculated by the comparative cycle threshold method (Applied Biosystems) with Arabidopsis Actin2 as the endogenous reference for normalization as described (Bluhm and Woloshuk, 2005). The statistical significance of the differences in the mean values was analyzed as described in the legend for Figure 1. These experiments were repeated at least three times with similar results. Ab CutA, A. brassicicola cutinase gene; At Actin, Arabidopsis Actin2 gene.
There was no difference in bacterial growth between hub1 and wild-type plants when infiltrated with the virulent (PstDC3000) or avirulent (PstDC3000AvrRpm1) strains of the bacterial pathogen Pseudomonas syringae (see Supplemental Figures 3A and 3B online). The sensitive assay of electrolyte leakage from inoculated leaves was evaluated to detect altered responses to bacterial infection and increased or decreased HR, but no difference was observed between wild-type and mutant plants (see Supplemental Figures 3C and 3D online). Similarly, hub1 plants did not show an altered response to the obligate fungal pathogen Erysiphe cichoracearum, the causal agent of powdery mildew disease (see Supplemental Figure 4 online). Sensitivity to the plant hormones abscisic acid, 1-aminocyclopropane-1-carboxylic acid (ACC), and Methyl jasmonate (MeJA) and high salinity were also unaltered in the hub1 plants (see Supplemental Figures 5 and 6 online). Thus, HUB1 is not involved in general stress tolerance and resistance to obligate pathogens but is specifically required for resistance to necrotrophic fungi, independently of hormone responses.
HUB1 Controls Flowering Time Independent of Photoperiod
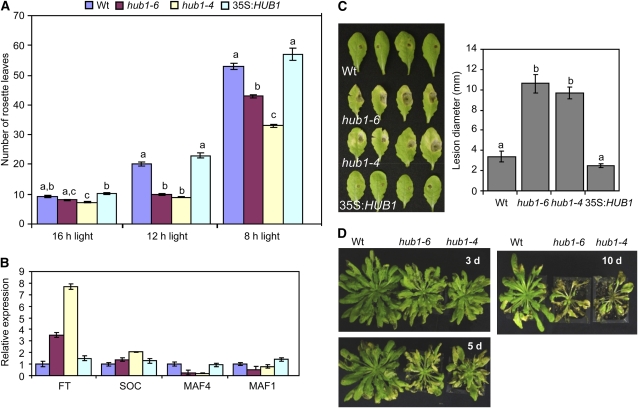
The hub1 plants grown for disease assays flowered significantly earlier than wild-type Col-0 plants. We examined flowering time by growing plants under long (LD1, 12 h light; LD2, 16 h light) and short (SD, 8 h light) day conditions. In LD1, hub1-4 and hub1-6 plants flowered within 3 weeks, producing an average of 9 ± 1.5 and 10 ± 1.4 rosette leaves, respectively, compared with 20 ± 3.6 leaves in Col-0 plants (Figure 3A; see Supplemental Figure 7A online). The 35S:HUB1 plants flowered later than the wild type, producing an average of 23 ± 4 leaves. Under LD2, intriguingly, the hub1-4 functionally null allele showed significant early flowering, while hub1-6 was comparable to the wild type. In SD, the wild type produced an average of 53 ± 3.2 leaves compared with 43 ± 1.7 in hub1-6, 33 ± 1.5 in hub1-4, and 59 ± 6.88 in the 35S:HUB1 plants. Thus, HUB1 levels regulate flowering time in the Col-0 background. To investigate the molecular basis of HUB1 function in flowering, we studied expression of MADS AFFECTING FLOWERING1 (MAF1), MAF4, FLOWERING LOCUS T (FT), and SUPPRESSOR OF CONSTANS OVEREXPRESSION1 (SOC1) genes that regulate flowering in Arabidopsis (Figure 3B). Quantitative RT-PCR results reveal that the expression of MAF1 and MAF4 genes is reduced in hub1. MAF1 and MAF4 are members of the FLC gene family that can act as floral repressors when expressed constitutively to high levels in transgenic plants, while they are downregulated in vernalized plants (Ratcliffe et al., 2001, 2003). On the other hand, the FT gene in hub1 was expressed at a higher level than in the wild-type and 35S:HUB1 plants, consistent with the role of FT as a promoter of flowering in Arabidopsis (Turck et al., 2008). The early flowering of hub1 is not due to increased endogenous SA levels, as removal of SA through genetic crosses to SID2/ICS1, required for the synthesis of SA, failed to alter its flowering time (see next section). Thus, loss of HUB1 alters a bona fide regulatory mechanism for the control of flowering time.
Figure 3.
HUB1 Regulates Flowering Time in Arabidopsis.
(A) Number of rosette leaves on plants grown under different photoperiods indicative of early flowering in hub1.
(B) Expression of flowering genes.
(C) and (D) Susceptibility of hub1 plants grown under short days to A. brassicicola (C) and B. cinerea (D).
In (A), analysis of variance was performed to determine the statistical significance of the differences between mean numbers of rosette leaves using SAS software (SAS Institute, 1999). Means with different letters are significantly different from each other (P = 0.05). In (B), quantitative RT-PCR was used to determine the expression of flowering genes relative to Actin2 gene. Experiments were repeated at least three times with similar results. d, days after inoculation.
The Susceptibility of hub1 to Necrotrophic Fungi Is Not Due to Its Early Flowering or Senescence
Since the hub1 disease phenotype was initially observed under 12-h daylength where the mutants flower significantly early, we investigated whether the disease susceptibility of the mutant to necrotrophic fungi was connected with early flowering and eventual early senescence of leaves. Necrotrophic fungi can take advantage of senescent tissue to colonize host plants. hub1 plants grown under SD grow very robustly and do not flower until 8 weeks of growth (see Supplemental Figure 7B online). SD-grown plants were therefore tested for disease resistance at week 6, before the onset of flowering. When challenged with A. brassisicola, SD-grown hub1 mutant plants exhibited increased susceptibility with enhanced disease symptoms similar to plants grown under LD conditions (Figure 3C). Similarly, SD-grown hub1 plants showed increased susceptibility to B. cinerea with enhanced chlorosis and necrosis, like plants in LD (Figure 3D). Thus, the disease phenotype of hub1 is not due to the very early flowering observed in LD. In a detached leaf senescence assay, the hub1 alleles did not show an enhanced senescence phenotype relative to the wild-type plants, suggesting that the disease responses are not due to easier colonization of aging tissue (see Supplemental Figure 7C online).
hub1 Has Altered Cell Wall Thickness and Callose Accumulation
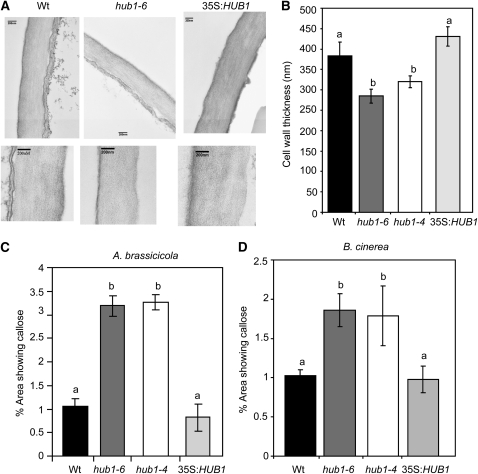
Mutants in the S.pombe homolog of HUB1 show enhanced septation (cell separation) (Tanny et al., 2007), a process closely linked to cell wall restructuring (Humbel et al., 2001). To explore whether altered cell wall functions are conserved and could explain the HUB1 role in plant defense, the cell wall of hub1 plants was studied by transmission electron microscopy. Interestingly, the thickness of the cell wall in hub1 plants was significantly reduced (Figures 4A and 4B). The average thickness of the epidermal cell wall of 4-week-old plants was 285.5 ± 33.7 nm for hub1-6, 320.8 ± 14.6 for hub1-4, and 430.8 ± 23.57 for 35S:HUB1 compared with 383 ± 33.7 nm in the wild type (Figure 4B). The 35S:HUB1 plants were comparable to the wild type in cell wall thickness, suggesting that the HUB1 disease resistance cannot be accounted for by cell wall thickness alone. Interestingly, the thickness of the main stem of hub1 plants was reduced, whereas that of 35S:HUB1 plants increased (see Supplemental Figure 8 online).
Figure 4.
hub1 Plants Show Reduced Cell Wall Thickness in Epidermal Tissues but Increased Callose Accumulation at the Site of Fungal Infections.
(A) Representative pictures showing the size of epidermal cell wall.
(B) Mean thickness of epidermal cell walls.
(C) and (D) hub1 plants show increased callose accumulation at the site of A. brassiciola (C) and B. cinerea (D) inoculation.
In (A), the bars = 200 nm. In (B), the data represent mean ± se from 20 samples. The experiment was repeated twice. The statistical significance of the differences in the mean thickness of the cell wall was analyzed as described in the legend for Figure 1. Means followed by different letters are significantly different from each other (P = 0.05). In (C) and (D), the callose staining assays were from plants 2 d after drop inoculation with 5 × 105 A. brassicicola spores/mL (middle) or 5 × 104 Botrytis spores/mL. The callose data shown in (C) and (D) were quantified using an image analysis program as described in Methods.
To determine callose levels in the cell wall, tissues inoculated with A. brassicicola and B. cinerea were stained with aniline blue. Mock-inoculated plants accumulated no detectable callose, while wild-type plants responded to pathogen inoculation with significant callose deposition in cells surrounding the site of inoculation (Figures 4C and 4D). hub1 plants accumulated even more callose than the wild type after inoculation with A. brassicicola (Figure 4C). Furthermore, inoculation with B. cinerea, performed with a low spore titer of 5×104 spores/mL, to avoid early tissue maceration, induced more callose production in hub1 mutants than in the wild type (Figure 4D). Callose deposition in the 35S:HUB1 plants was comparable to that in wild-type plants. Consistent with the increased callose, hub1 had increased callose synthase1 (CALS1, 1,3-β-Glucan synthase 1; At1g05570) gene expression relative to the wild-type plants (Fleury et al., 2007).
To elucidate the role of callose in resistance to necrotrophic fungi and the phenotypes of hub1, we tested two loss-of-function alleles of the callose synthase gene PMR4, the point mutation pmr4-1 (Nishimura et al., 2003), and the T-DNA insertion allele pmr4-2 (SAIL-294-D06) for resistance to A. brassicicola and B. cinerea (see Supplemental Figure 9 online). The pmr4 alleles showed increased susceptibility to A. brassicicola, suggesting that the inability to accumulate callose can also correlate with susceptibility to necrotrophic fungi (see Supplemental Figures 9A and 9B online). The pmr4-1 mutation is connected with an activation of salicylic acid (SA)-dependent disease resistance (Nishimura et al., 2003), resulting in downregulation of resistance against A. brassicicola as recently observed (Flors et al., 2008). In spite of these observations, the pmr4 mutant alleles did not show altered responses to B. cinerea (see Supplemental Figure 9C online). Therefore, reduced or increased amounts of callose deposition can be associated with increased susceptibility to A. brassicicola, but loss of callose had no effect on B. cinerea resistance, suggesting that additional factors determine the outcome of plant responses to necrotrophic infection.
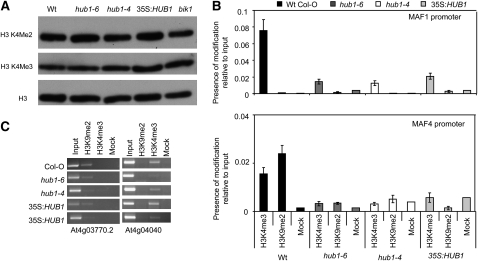
Global Histone H3 Lysine 4 Di- and Trimethylation and DNA Methylation Are Not Changed in hub1 Mutant Plants
In yeast, Drosophila, and human cells, H2B ubiquitination serves as a signal for methylation of histone H3, which in turn regulates transcription (Shilatifard, 2006). In Arabidopsis, mutations in the deubiquitination enzyme SUP32/UBP26 decrease the dimethylation of lysine 9 in histone H3 and release heterochromatic silencing (Sridhar et al., 2007). Thus, the status of global histone methylation was studied in hub1 and 35S:HUB1 plants using H3K4-specific antibodies. Immunoblots of histone-enriched protein revealed no significant changes in global histone methylation at these residues (Figure 5A), indicating that histone monubiquitination may not serve as a signal for global histone H3 methylation in plants. Alternatively, it is possible that changes would only affect chromatin at promoters of specific HUB1 target genes. A candidate would be the flowering control gene MAF1, a MADS domain containing gene whose expression depends on HUB1 (Figure 3C). Therefore, we analyzed H3K4 trimethylation and H3K9 dimethylation at the MAF1 and MAF4 promoters by chromatin immunoprecipitation (ChIP) in wild-type, hub1 mutants, and HUB1-overexpressing lines. No consistent difference was observed in histone H3 methylation at the MAF1 promoter (Figure 5B). The variation between the samples is attributed to the overall low level of histone modification as only slightly higher levels than background could be detected. Since deubiquitination of Arabidopsis H2B has been further linked to reduction of DNA methylation (Sridhar et al., 2007), we analyzed the global level of methylcytosine by HPLC after hydrolysis of genomic DNA from wild-type, hub1 mutants, HUB1-overexpressing lines, and ddm1 (decreased DNA methylation; Vongs et al., 1993). Besides ddm1 control, there was no significant difference between the lines (see Supplemental Figure 10A online). To exclude that changes in DNA methylation would only affect specific targets, we performed DNA gel blot analysis with methylation-sensitive restriction enzymes and hybridization to a TSI (for transcriptionally silent information) probe (Steimer et al., 2000). TSI represents repeats from heterochromatic pericentromeric regions of Arabidopsis chromosomes that are heavily methylated (Steimer et al., 2000). Genomic DNA of wild-type, hub1 mutants, HUB1-overexpressing lines, and ddm1 was digested with HpaII and MspI. Both enzymes recognize the sequence CCGG. HpaII digestion is inhibited by methylation of either of the two cytosines of the recognition site, whereas MspI is inhibited only by methylation of the first cytosine. While DNA of ddm1 is clearly hypomethylated, changes of HUB1 levels do not affect cytosine methylation at TSI loci, neither at CpG nor at CpHpG sequences (see Supplemental Figure 10B online). While the deubiquitinating enzyme seems to be required for methylation-based heterochromatin maintenance (Sridhar et al., 2007), the ubiquitinating enzymes do not seem to be connected with the DNA modification.
Figure 5.
Global and Locus-Specific Histone H3 Methylation Is Not Altered in hub1 Plants.
(A) Immunoblot showing global H3K4 methylation. Histone-enriched protein was extracted and immunoblotted using antibodies specific to methylated histones H3K4 methylation. Histone H3 total protein was used as a loading control.
(B) ChIP qPCR analysis of histone H3K4 trimethylation and H3K9 dimethylation at the MAF1 and MAF4 promoters. ChIP results for histone H3K4 trimethylation and H3K9 dimethylation at the MAF1 and MAF4 promoters are represented relative to input; the error bars indicate the se. The variation between the samples is very likely due to the overall low level of histone modification as only slightly higher levels than background could be detected.
(C) ChIP analysis of control sequences.
A heterochromatin control (At4g03770.2, a Gypsy-like retrotransposon) reacts with antibody to H3K9me2, whereas a control for euchromatin (At4g04040, a putative phosphofructokinase beta subunit) reacts with antibody to H3K4me3. Mock, no-antibody control.
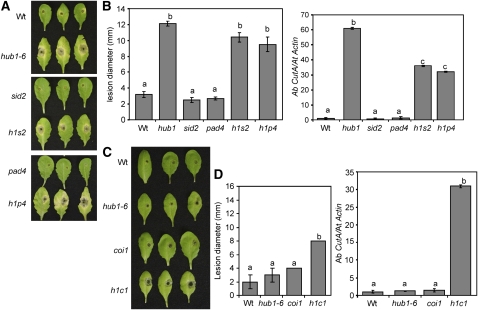
SA Promotes Development of a Disease Lesion at the Site of A. brassicicola Infection in the hub1 Mutant
SA is a defense molecule that modulates plant resistance to diverse pathogens. In the case of necrotrophs, increased SA has been associated with susceptibility (Veronese et al., 2006). The role of SA in hub1 was examined by analyzing the disease responses of hub1 sid2 and hub1 pad4 double mutants. The SID2 gene encodes an isochorismate synthase involved in the synthesis of SA in Arabidopsis (Wildermuth et al., 2001). Arabidopsis PAD4 encodes a lipase-like protein that regulates SA accumulation in response to some pathogens (Jirage et al., 1999). Both hub1 sid2 and hub1 pad4 plants had comparable A. brassicicola disease lesions to hub1-6 (Figures 6A and 6B). However, hub1 sid2 and hub1 pad4 plants support significantly less A. brassicicola growth than the hub1 single mutant. Thus, SA promotes fungal growth in hub1 possibly through the suppression of defense against necrotrophic pathogens or by promoting cell death at the point of inoculation. In the case of B. cinerea, hub1 sid2 and hub1 pad4 all show comparable levels of disease symptoms and fungal growth to hub1 in spray-inoculated plants (see Supplemental Figure 11 online), suggesting that B. cinerea susceptibility of hub1 is largely independent of SA. The results indicate the subtle differences in the role of SA in regulating disease severity to A. brassicicola and B. cinerea infections.
Figure 6.
HUB1 Functions Independently of JA-Mediated Defense, but SA Modulates Pathogen Growth in the hub1 Mutant.
(A) and (B) A. brassicicola disease assays showing disease symptoms (A), size of disease lesion ([B]; left), and pathogen growth (right) on hub1, sid2, pad4, and double mutants.
(C) and (D) Disease symptoms (C), disease lesion size ([D]; left), and pathogen growth on hub1, coi1, and hub1 coi1 plants.
All disease assays were repeated at least twice. The disease response data in (A) and (B) are from 5 DAI, whereas those in (C) and (D) are from 2 DAI. The values in (B) and (D) represent mean ± se from at least 30 lesions. h1, hub1-6; h1s2, hub1 sid2; h1p4, hub1 pad4; h1c1, hub1 coi1. Fungal growth was assessed using qPCR amplification of the A. brassicicola cutinase DNA as described in the legend for Figure 1 and in Methods.
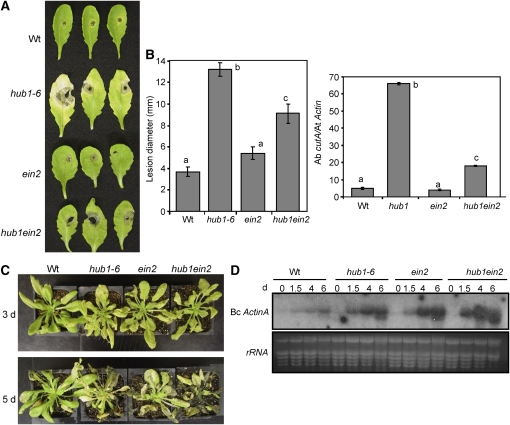
COI1 Is Additive to HUB1 for B. cinerea and A. brassicicola Resistance, whereas EIN2 Is Epistatic to HUB1 for A. brassicicola but Additive for B. cinerea Resistance
Arabidopsis resistance to B. cinerea requires both JA and ET signaling, whereas resistance to A. brassicicola requires only JA responses (Thomma et al., 1999; van Wees et al., 2003a). To investigate whether HUB1 functions independently of or synergistically with the ET and JA responses, we constructed hub1 coi1 and hub1 ein2 double mutants. COI1 functions in JA responses (Xie et al., 1998), and a coi1 mutant shows susceptibility to B. cinerea and A. brassicicola (van Wees et al., 2003a). The ein2 mutant is impaired in ET responses (Guzman and Ecker, 1990) and shows increased susceptibility to B. cinerea but wild-type resistance to A. brassicicola (Thomma et al., 1999; van Wees et al., 2003).
We evaluated the responses of hub1 coi1 to A. brassisicola at 2 DAI since our initial observations suggested that hub1 coi1 plants were extremely susceptible to this pathogen. At this time point, hub1 coi1 had developed significantly larger lesions than the hub1-6 and coi1 single mutant plants (Figures 6C and 6D). At 2 DAI, hub1-6 did not differ from the wild-type plants in the size of the disease lesions and pathogen growth. hub1 coi1 supported ∼30-fold more A. brassicicola growth compared with hub1-6 or coi1 single mutants. After inoculation with B. cinerea, more tissue maceration was observed in hub1 coi1 than in the wild type and the single mutants (see Supplemental Figure 12 online). The greater severity of disease in hub1 coi1 relative to the single mutants suggests that HUB1 and COI1 affect resistance in two independent pathways. Thus, COI1 is additive to HUB1 for resistance to both B. cinerea and A. brassicicola.
Consistent with previous reports, ein2 did not change resistance to A. brassicicola in an otherwise wild-type background (Figures 7A and 7B). Nevertheless, the hub1 ein2 double mutant was more resistant to A. brassicicola than hub1-6 alone, suggesting that the susceptibility of hub1 partially depends on functional ET responses. The ein2 mutation in a hub1 background completely eliminated the chlorosis surrounding the infection site and considerably reduced the extent of pathogen proliferation (Figures 7A and 7B). By contrast, after B. cinerea inoculation, ein2 hub1 plants were more susceptible than either ein2 or hub1 single mutants and supported significantly more fungal growth (Figures 7C and 7D). At 5 DAI with B. cinerea, ein2 hub1 plants were completely macerated compared with the single mutants. Taken together, our data suggest that ET responses enhance A. brassicicola disease symptoms and pathogen growth in the hub1 mutant, most likely by promoting tissue damage at the site of infection. Thus, the function of HUB1 is additive to EIN2 with respect to B. cinerea but partially epistatic to HUB1 for A. brassicicola resistance.
Figure 7.
Ethylene Signaling Promotes hub1 Susceptibility to A. brassicicola but Is Required for Resistance to Botrytis.
Disease assays on hub1, ein2, and hub1ein2 double mutant showing A. brassicicola disease symptoms (A), size of disease lesion ([B]; left) and fungal growth (right), B. cinerea disease symptoms (C), and fungal growth (D). In (D), total RNA (10 μg) was loaded per lane. All disease assays were repeated at least twice. Data in (B) represent mean ± se from at least 30 lesions. Bc ActinA, B. cinerea ActinA gene; d, days after inoculation.
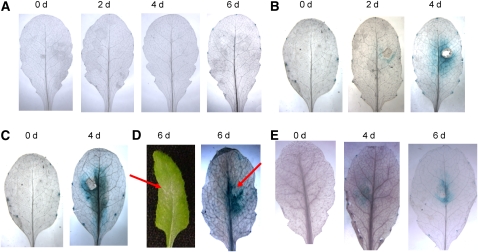
HUB1 Is Induced at the Site of Infection by Biotrophic and Necrotrophic Fungal Pathogens
To determine the expression of HUB1 during pathogen infection, we generated transgenic lines expressing the β-glucuronidase (GUS) reporter gene under the control of the HUB1 promoter (HUB1Pr:GUS). Homozygous transgenic lines with relatively low GUS activities were used to investigate the pathogen-induced HUB1 gene expression. HUB1 expression was not induced by mock inoculation but is induced at the site of A. brassicicola, B. cinerea, and E. cichoracearum infection consistent with its role in defense against these pathogens (Figures 8A to 8D). Inoculation with a Cochliobolus carbonum strain producing HC toxin (Tox+) also induced HUB1 gene expression at the infection site at 4 to 6 DAI (Figure 8E). Although hub1 plants do not show altered responses to this pathogen, the HC toxins produced by certain strains of the fungus are known to inhibit histone deacetylase activity and are likely to interfere with chromatin modifications (Brosch et al., 1995). HUB1 expression was not induced by treatments with MeJA, SA, ACC, and the herbicide paraquat (methyl viologen) (see Supplemental Figure 13 online). Thus, HUB1 is induced specifically by pathogen infection, consistent with the specificity of the hub1 mutant phenotypes.
Figure 8.
HUB1 Is Induced at the Site of Infection by Fungal Pathogens.
Expression of HUB1-GUS promoter fusion in responses to mock (A), Botrytis (B), A. brassicicola (C), E. cichoracearum (D), and C. carbonum (E) (Tox+) inoculation. d, days after inoculation.
HUB1 Interacts with the MED21 Subunit of the Arabidopsis Mediator Complex
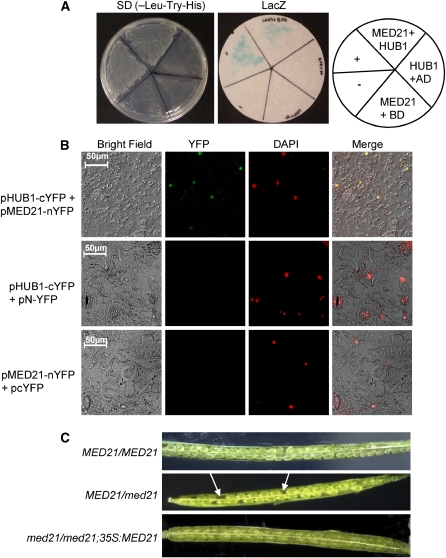
To gain further insights into the mechanisms of HUB1 function, we screened for HUB1 interacting proteins using a yeast two-hybrid screen. MED21 (At4g04780), a homolog of human and yeast MED21, constituents of the Mediator complex involved in transcriptional regulation (Bjorklund and Gustafsson, 2005), was identified as a strong interactor of HUB1. Figure 9A shows growth on selective media and β-gal activity when the HUB1 cDNA in the bait vector (pBD-HUB1) was coexpressed with MED21 in the prey vector (pAD-MED21), indicating their specific interaction in yeast. In combination with the pAD empty vector, the bait plasmid pBD-HUB1 failed to activate the transcription of the β-gal reporter gene and did not allow growth on selective media, suggesting that HUB1 does not autoactivate.
Figure 9.
The RING E3 Ligase HUB1 Interacts with the MED21 Subunit of Arabidopsis Mediator Complex.
(A) HUB1 interacts with MED21 in the yeast two-hybrid assay. Yeast strains containing the MED21 in the prey vector (pAD-MED21) and HUB1 in the bait prey (pBD-HUB1) were assayed for growth on selective medium (-Leu, -Trp, and -His) (left) and β-galactosidase activity (right) showing interaction between MED21 and HUB1 in yeast. The β-galactosidase activity was assayed from yeast cells grown on synthetic complete medium. The positive (+) and negative (−) controls from the Stratagene kit were also assayed in parallel.
(B) HUB1 interacts with MED21 in vivo. Bimolecular fluorescence complementation assay showing in vivo interaction between HUB1 and MED21. pHUB1-cYFP and pMED21-nYFP were transiently coexpressed or were coexpressed with the vector alone in N. benthamiana leaf cells. YFP fluorescence was detected when pHUB1-cYFP was coexpressed with pMED21-nYFP. Images were examined under the bright field (left column), fluorescence (YFP), and as a merged image (bottom) showing either no interaction or interaction in the nucleus. DAPI, 4',6-diamidino-2-phenylindole.
(C) Siliques from wild-type, MED21/med21, and med21/med21;35S:MED1 lines. Arrows indicate aborted embryos.
To confirm the specific interaction in planta, a bimolecular fluorescence complementation assay was performed using full-length HUB1 and MED21. HUB1 was translationally fused with the N-terminal 155–amino acid portion of the yellow fluorescent protein (YFP) (pHUB1-cYFP), and MED21 was fused with the C-terminal 86–amino acid portion of YFP (pMED21-nYFP). pHUB1-cYFP and pMED21-nYFP were cotransformed into Nicotiana benthamiana leaves through agroinfiltration. YFP fluorescence was observed only when the two constructs were coexpressed (Figure 9B, top row). Leaves from plants infiltrated with either of the constructs alone, or in combination with the empty vector, did not show any YFP fluorescence (Figure 9B, middle and bottom rows). Staining of cells with the fluorescent nuclear stain 4',6-diamidino-2-phenylindole (DAPI) revealed fluorescence in the nucleus of cells cotransformed with both constructs, indicating interactions in the nucleus. The interaction of HUB1 and MED21 in the nucleus is consistent with the function of HUB1 in histone monoubiquitination and the function of MED21 as a transcriptional coregulator (Boube et al., 2002; Takagi and Kornberg, 2006).
We cloned the full-length cDNA of MED21 and found it to be shorter than the predicted MED21 cDNA (www.Arabidopsis.org). The MED21 cDNA contains an open reading frame of 140 amino acids encoding a predicted protein of 15.8 kD. A comparison of MED21 protein sequences from various eukaryotes only shows a few conserved segments in the N-terminal, central, and C-terminal regions, suggesting the divergence of MED21 protein sequences in various species (see Supplemental Figure 14A online). Other plant MED21 sequences available from rice, Sorghum bicolor (sorghum), and the moss Physcomitrella patens are relatively closer related with Arabidopsis MED21 than those from other kingdoms, with the Arabidopsis MED21 sharing 57 and 52% overall identity with the P. patens and sorghum MED21 proteins, respectively. The MED21 sequences from the two monocot species, sorghum and rice, share even greater sequence similarity with each other (70% identity and 79% similarity). Phylogenetic analysis reveals that Arabidopsis MED21 is most closely related to MED21 from P. patens (see Supplemental Figure 14B online). The overall size is conserved for all MED21 proteins from various organisms and is shown or predicted to be around 15 kD (Backstrom et al., 2007).
Arabidopsis MED21 Is Required for Embryo Development and for Defense against Necrotrophic Fungi
To determine the function of MED21 in Arabidopsis, a population with a segregating loss-of-function T-DNA insertion allele of MED21 was screened for plants with the homozygous med21/med21 genotype. The mutant allele carries a T-DNA insertion in the first exon of MED21 and is predicted to be a null allele. Among 65 plants screened by PCR, 40 heterozygous (MED21/med21) and 25 wild types (MED21/MED21) but no homozygous T-DNA insertion genotypes were recovered. The siliques of MED21/med21 plants show aborted embryos at a rate of 25% (Figure 9C). Thus, MED21 is essential for embryo development in Arabidopsis. MED21 knockout mice are also not viable (Tudor et al., 1999). Hemizgous MED21/med21 plants were transformed with the 35S:MED21 construct. We recovered med21/med21;35S:MED21 plants that show no aborted embryos, suggesting that ectopic expression of MED21 rescued the embryonic lethal phenotype of the med21 mutant (Figure 9C, bottom panel).
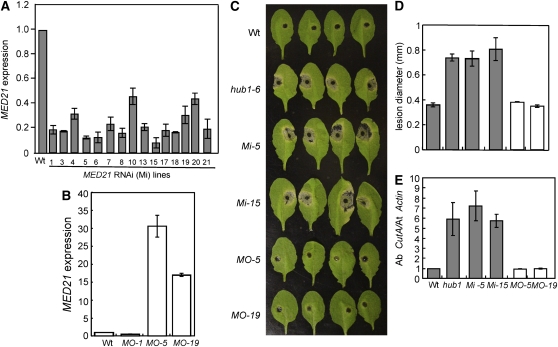
To determine the defense function of MED21 more directly, we generated MED21 RNA interference (RNAi) and 35S:MED21 lines. Most MED21 RNAi (Mi) transgenic lines generated show significantly reduced MED21 gene expression, whereas two 35S:MED21 lines overexpress MED21 (MO-5 and MO-15) relative to the wild-type plants (Figures 10A and 10B). The MED21 RNAi lines were more susceptible to A. brassicicola with enhanced disease symptoms and fungal growth similar to the disease response of the hub1 mutant (Figures 10C to 10E). The MO lines did not significantly differ from the wild-type plants. Similarly, the MED21 RNAi lines showed increased susceptibility to B. cinerea (see Supplemental Figure 15 online).
Figure 10.
Arabidopsis MED21 Is Required for Resistance to A. brassicicola.
(A) and (B) Quantitative RT-PCR showing the expression of MED21 in transgenic MED21 RNAi (A) and 35S:MED21 (B) lines.
(C) to (E) Disease symptoms (C), size of disease lesion (D), and fungal growth (E) in MED21 RNAi and overexpression lines after A. brassicicola inoculation.
The quantitative RT-PCR data represent the mean ± se from three replicates. In (C), the data are the mean ± se from a minimum of 20 disease lesions. In (E), fungal growth in inoculated plants was determined using the amplification levels of A. brassicicola CutinA (Ab cutA) relative to the Arabidopsis Actin2 gene (At Actin). MO, MED21 overexpression lines; Mi, MED21 RNAi lines. In all cases, the experiments were repeated at least twice.
Mediator is required for transcription of nearly all RNA polymerase II–dependent genes in S. cerevisiae and other organisms, and posttranslational modifications of specific Mediator subunits can affect global patterns of gene transcription (Kornberg, 2005; Takagi and Kornberg, 2006). Consistent with a requirement for embryo development, MED21 is strongly expressed in the later stages of embryo development and especially during cotyledon expansion (Arabidopsis eFP browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). It is also highly expressed in suspension culture cells (Gene Atlas, https://iii.genevestigator.ethz.ch). Significant expression in rosette leaves could correspond with a possible role in defense. Congruent with that, MED21 (ATH1 probe set: 256448_s_at) is induced more than twofold by infection with E. cichoracearum and by treatment with the ethylene precursor ACC, brassinolide, gibberellic acid (GA), auxin, zeatin, glucose, and cold stress, suggesting that MED21 may be activated by microbial infection and other factors involved in stress signaling (Genevestigator response viewer, http://iii.genevestigator.ethz.ch) (Zimmermann et al., 2004). A dual role of a protein connecting defense and embryo development has been documented in Drosophila (Lemaitre et al., 1996).
Defense Gene Expression in the hub1 Mutant
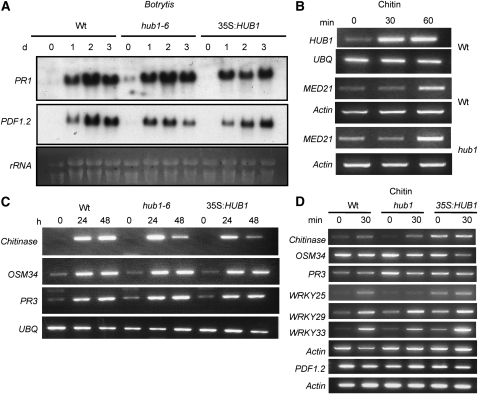
PDF1.2 and PR1 genes are widely used as markers for the SA- or JA/ET-regulated defense pathways in Arabidopsis. The hub1 and 35S:HUB1 plants showed no altered expression of PDF1.2 in responses to B. cinerea infection (Figure 11A); basal expression of PR1 was only slightly increased in hub1 plants compared with wild-type plants. The 35S:HUB1 plants were comparable to the wild type for expression of both genes. Thus, HUB1 functions independently of pathways leading to the expression of the PR1 and PDF1.2 genes, consistent with the results from the double mutants.
Figure 11.
Induction of Defense Genes, HUB1 and MED21, by Chitin and B. cinerea.
Expression of PR-1 and PDF1.2 genes during B. cinerea infection (A), HUB1 and MED21 upon exposure to chitin (B), Chitinase, osmotin like, and PR3 genes by B. cinerea infection (C), and Arabidopsis defense-related and WRKY transcription factor genes upon exposure to chitin (D). For the RNA blot in (A), total RNA (10 μg) was loaded per lane. In (B) to (D), RT-PCR was performed as described in Methods with 28 cycles, which was within the linear range of amplification. The experiments were repeated at least three times with similar results. Arabidopsis Actin2 and Ubiquitin genes were used as constitutive controls. UBQ, Arabidopsis ubiquitin; d, days after inoculation; h, h after treatment.
Interestingly, both HUB1 and MED21 are induced by chitin treatment of plants, indicating their involvement in basal defense (Figure 11B). The chitin-induced expression of MED21 was independent of HUB1, suggesting that their regulatory relationship is posttranscriptional. The hub1 and wild-type plants were comparable for B. cinerea- and chitin-induced expression of the JA-regulated genes chitinase, osmotin like (OSM34), and PR3 genes (Figures 11C and 11D). The chitin-induced expression of WRKY33, WRKY22, and WRKY29 genes, previously implicated in signaling of PAMP-mediated defense response in Arabidopsis (Asai et al., 2002; Miya et al., 2007; Wan et al., 2008), was also unaffected in the hub1 mutant (Figure 11D). By contrast, the chitin-induced expression of WRKY25 is reduced in the hub1 mutant. The 35S:HUB1 plants show increased basal expression of the three WRKY genes tested. Thus, HUB1 regulates resistance to necrotrophic fungi through a novel signaling pathway.
DISCUSSION
Two previous reports have implicated Arabidopsis H2B ubiquitination in the control of the plant cell cycle and seed dormancy (Fleury et al., 2007; Liu et al., 2007). This report adds two major aspects to our understanding of the role of HUB1, the E3 ligase for histone H2B. (1) HUB1 plays an important role in defense against necrotrophic fungal pathogens, as is obvious from the modified susceptibility to B. cinerea and A. brassicicola, two representatives for this perilous class of pathogens, upon modified HUB1 levels. The mechanisms could involve HUB1-mediated changes in the epidermal cell wall and gene expression. (2) HUB1 interacts specifically with the MED21 subunit of the middle module of Mediator, an evolutionarily conserved protein complex with a role in relaying signals from other regulators to RNA polymerase II (Boube et al., 2002; Lewis and Reinberg, 2003). Both aspects might be functionally connected since HUB1 and MED21 are transcriptionally induced by chitin, an elicitor derived from fungal cell walls. Significantly, reduced expression of MED21 in MED21 RNAi lines resulted in susceptibility to the two necrotrophic fungi, providing a strong link between HUB1 and MED21 functions in defense against necrotrophic fungi. The defense function of HUB1 and MED21 is likely linked to the regulation of gene expression. Beyond pathogen response, HUB1 affects flowering time, and MED21 is a gene essential for proper embryo development. Therefore, H2B ubiquitination by HUB1 seems to affect diverse physiological processes in plants, apparently through a mechanism conserved in other organisms.
In contrast with polyubiquitination, monoubiquitination does not lead to the destruction of ubiquitinated proteins by the 26S proteosome (Glickman and Ciechanover, 2002). Instead, monoubiquitination of proteins regulates transcription, receptor internalization, and endosomal sorting (Liu et al., 2005). In yeast and Drosophila, H2B ubiquitination is required for proper histone H3 methylation by other enzymes, suggesting a crosstalk between histone modifications (Shilatifard, 2006). In the case of Arabidopsis hub1, we observed no altered global H3K4 methylation; therefore, this dependence may not exist in plants. Our observation is consistent with data from fission yeast where the primary function of H2B ubiquitination is to stimulate transcription, independent of histone methylation (Tanny et al., 2007). However, we cannot exclude the possibility that HUB1-regulated histone ubiquitination could determine other chromatin modifications not analyzed or histone methylation at a minor and specific subset of target genes. In sum, HUB1 function in disease resistance seems to be attributed to multiple regulatory events that depend on histone H2B monoubiquitination.
The state of chromatin regulates many physiological processes, including flowering time and responses to environmental stresses (Bastow et al., 2004; Tsuji et al., 2006). Interestingly, some necrotrophs produce toxins that interfere with plant chromatin or the chromatin modification machinery as a virulence target to suppress expression of plant defense genes. The host-selective virulence factor HC toxin, produced by some strains of C. carbonum, inhibits host histone deacetylases and thus suppresses elicitor-activated defense in maize (Zea mays) (Brosch et al., 1995; Ransom and Walton, 1997). A. brassicicola produces depudecin, a toxin that also inhibits histone deacetylase (Privalsky, 1998). Consistent with this, the lack of HISTONE DEACETYLASE19 (HDA19) increased susceptibility to A. brassicicola in Arabidopsis (Zhou et al., 2005). Furthermore, the Arabidopsis HDA6 interacts with the JA response regulator COI1 that is also required for resistance to necrotrophic fungi (Devoto et al., 2002, 2003). Thus, several chromatin modifications, including H2B ubiquitination, as demonstrated in this report, have regulatory functions in plant responses to necrotrophic pathogens.
The yeast homolog of HUB1 and its E2 enzyme Rad6 associate with elongating RNA polymerase II and ubiquitinate histone H2B on the body of a transcribed gene (Xiao et al., 2005). An enrichment of ubiquitinated histones at transcriptionally active gene loci has been reported (Zhang, 2003), indicating a correlation between histone ubiquitination and transcription. The interaction between MED21 and HUB1 narrows this gap, identifies a specific subunit of the Mediator complex as a specific interacting molecule, and provides a novel link between H2B ubiquitination and RNA polymerase II functions in transcription. However, the order of events during the interaction remains to be analyzed.
Mediator is a large multisubunit protein complex conserved in eukaryotic cells and was implicated in transcriptional activation and repression (Bjorklund and Gustafsson, 2005). Recently, proteomic and database similarity searches identified 19 potential constituents of an Arabidopsis Mediator complex, confirming its conservation in plants (Backstrom et al., 2007; Gonzalez et al., 2007). Mediator consists of head, middle, and tail modules. MED21 is an evolutionarily conserved subunit of the middle module (Boube et al., 2002) that is thought to play a key role in regulating RNA polymerase II activity by direct interaction (Hallberg et al., 2006). Mediator relays regulatory information from enhancers and other control elements to the basal RNA polymerase II transcription machinery (Kim and Lis, 2005). Therefore, it was to be expected that Arabidopsis MED21 would also have a significant regulatory role. Consistent with this, MED21 RNAi lines show enhanced susceptibility to B. cinerea and A. brassicicola. The induced expression of HUB1 and MED21 in response to pathogen and/or pathogen-derived signals also suggests involvement in defense signaling. Mediator subunits regulate expression of innate immune response genes in Drosophila where most innate immune response genes require subunits of the Mediator complex to activate their target genes. LPs induce the expression of antimicrobial peptides, and subunits of Mediator act as coactivators of LP-induced transcriptional activation of antimicrobial peptides (Kim et al., 2004). The MED17 and MED16 subunits of Mediator are required for transcriptional activation of the antimicrobial peptides, drosomycin and Attacin A (Park et al., 2003). This observation suggests a conserved role in transcription of defense genes in Arabidopsis and Drosophila. Although PR1 and PDF1.2 do not appear to be under the regulation of HUB1 during Botrytis infection, it will be worth analyzing whether other annotated or yet unknown defense genes, including novel antimicrobial peptides, are under the control of HUB1.
Recently, Arabidopsis PHYTOCHROME AND FLOWERING TIME1 (PFT1) and STRUWWELPETER were isolated as the MED14 and MED25 subunits of the Arabidopsis Mediator complex (Backstrom et al., 2007). PFT1 mediates flowering time in response to light quality, suggesting that specific plant Mediator subunits are linked to the regulation of specialized developmental processes. In addition, subunits of Mediator link transcriptional regulators and chromatin modification enzymes to RNA polymerase II function in transcription activation and elongation. The Arabidopsis transcription corepressor LEUING interacts with the histone deacetylase (HDA19) and Mediator component MED14 and CDK8 to repress transcription (Gonzalez et al., 2007). Thus, specific components of the Mediator complex may also connect different environmental signals to transcriptional outputs.
hub1 plants show altered growth characteristics, including reduced cell wall thickness and increased inducible callose accumulation. Loss of H2B ubiquittination in S. pombe causes defects in cell growth, increased septation, altered nuclear structure, and impaired transcriptional elongation of target genes (Tanny et al., 2007). In yeast, septation is a complex process, and the septum is composed of 1,3 β-glucan layers. Interestingly, the yeast sep10+ and sep15+ genes that are involved in septation encode subunits of the Mediator complex. Consistent with the S. pombe data, hub1 has increased inducible callose accumulation. Loss of inducible callose has been linked to resistance to powdery mildew (Nishimura et al., 2003) but susceptibility to A. brassicicola (Flors et al., 2008). The hub1 mutant plants are susceptible to both B. cinerea and A. brassicicola. The susceptibility of pmr4 alleles to A. brassiciola has been linked to the increased SA levels that accompany loss of callose (Flors et al., 2008). In light of this data, it is unexpected that the pmr4 plants show wild-type resistance to B. cinerea. Thus, the role of callose accumulation in resistance to necrotrophic pathogens remains unclear. The reduced cell wall size in hub1 may be due to the altered expression of genes involved in cell wall biosynthesis (Fleury et al., 2007) and is consistent with the disease responses. Necrotrophic fungi target the plant cell wall for degradation, and the strength of the wall upon such an attack contributes directly to resistance against these pathogens. In tomato (Solanum lycopersicum), inhibition of expansin and PGs involved in the disassembly of the plant cell wall during fruit ripening resulted in reduced cell wall thickness and decreased the susceptibility of the fruits to Botrytis, which supports the hypothesis that the cell wall is an important virulence target for Botrytis (Cantu et al., 2008).
The studies on genetic interactions between HUB1 and key defense regulatory genes suggest that pathogen growth and symptom development in hub1 are modulated by endogenous SA levels and ET responses. ICS1/SID2-dependent SA promotes pathogen growth at the infection site but also has slight effects on symptom development in hub1. These results can be due to antagonism between SA- and JA/ET-dependent responses (Spoel et al., 2007). The hub1 ein2 double mutant highlights the differences in the role of ET responses in resistance to the two necrotrophic fungi. Loss of ET signaling in hub1 significantly reduced pathogen growth and disease symptoms caused by A. brassiciola. The ein2 mutation reduced the disease symptoms caused by P. syringae without affecting pathogen growth (Bent et al., 1992). Intriguingly, HUB1 and EIN2 act additively with respect to B. cinerea resistance. Despite a significant overlap in plant defense mechanisms against Botrytis and A. brassicicola, the role of ET varies. In contrast with ET, functional JA responses are required for the function of HUB1 in resistance to both B. cinerea and A. brassicicola.
HUB1 was initially identified due to its increased expression in the bik1 mutant. Closer examination of the hub1 mutant phenotypes, including growth-related traits, particularly leaf shape and early flowering, suggest that the two proteins act in the same pathway. The disease responses of the two mutants are also consistent with the two genes acting in Arabidopsis defense against necrotrophic fungi. However, there are distinct disease responses and molecular data that indicate meaningful differences, although the exact mechanisms of their relationships are not clear at this stage. The increased expression of HUB1 in the bik1 mutant may have to do with its increased susceptibility to pathogens rather than a direct regulation of HUB1 by BIK1. Future studies will address the relationship between BIK1 and HUB1. BIK1 is a membrane-associated protein kinase that may act early in defense, whereas HUB1 is localized in the nucleus. It will be interesting to find out how and across which signaling pathways pathogen recognition reach the nucleus and affect H2B monoubiquitination and cell wall formation. The findings described here open a new avenue for examining chromatin-based transcriptional regulation of plant resistance to microbial infections and further studies on the role of Mediator subunits in plant immune responses, growth, and development.
The function of HUB1 in the control of flowering and disease resistance is likely to be exerted through two independent mechanisms. In yeast, Mediator and Paf1 are two biochemically distinct large subunit complexes that are associated with histone monoubiquitination and also regulate the activities of RNA polymerase II. The yeast Paf1 complex is essential for H2B monoubiquitination by BRE1 (Wood et al., 2003a). Strains deleted for several components of the Paf1 complex are defective in monoubiquitination of histone H2B, which results in the loss of H3K4 and H3K79 methylation. The Arabidopsis EARLY FLOWERING (ELF7 and ELF8) and VERNALIZATION INDEPENDENCE (VIP) genes encode proteins that are homologs of the components of the budding yeast Paf1 complex (He et al., 2004; Oh et al., 2004). ELF7 and ELF8 are required for the enhancement of H3K4 trimethylation in FLC chromatin. On the other hand, VIP genes do not affect global histone H3 methylation. The elf and vip mutations result in early flowering phenotypes, and the elf mutants have pleiotropic developmental defects. HUB1 neither affects global methylation nor repeat-associated methylation in Arabidopsis, and H3K4 methylation is also not affected generally or at the chromatin of the upregulated MAF1 and MAF4 genes. The requirement of the PAF1 complex for H2B ubiquitination in Arabidopsis is not yet determined. Thus, we speculate that the PAF complex is required for the function of HUB1 in the control of flowering time, whereas interaction of HUB1 with the Mediator complex is required for its disease resistance functions.
METHODS
Plant Growth Conditions and Diseases Assays
Plant growth conditions and media were described previously (Zheng et al., 2006). Botrytis cinerea strain BO5-10 and Alternaria brassicicola strain MUCL20297 were cultured on 2 × V8 agars (36% V8 juice, 0.2% CaCO3, and 2% Bacto-agar) and incubated at 20 to 25°C. Collection of B. cinerea and A. brassicicola conidia and disease assays were as described (Veronese et al., 2006). B. cinerea disease assays were performed on soil-grown plants by spray inoculations. All A. brassicicola disease assays were done on detached leaves by drop inoculation of a single 5- to 7-μL droplet of spore suspension (5 × 105 spores/mL) in water on each leaf. In both cases, inoculated plants were kept under a transparent cover to maintain high humidity.
The culture and disease assay of the strains of Pseudomonas syringae pv tomato were done as described (Zheng et al., 2006). For measurements of electrolyte leakage, bacterial suspension (OD600 of 0.1) was infiltrated into leaves. Leaf disks (4 mm diameter) were collected from the infiltrated area and washed with water for 50 min and then placed in a tube containing 15 mL of water. Conductivity was measured from five replicates for each treatment using a conductivity meter (Model AB30, AccumetR BASIC; Fisher Scientific) following the procedure described (Kawasaki et al., 2005).
Determination of Fungal Growth in Inoculated Plants
As an indicator of B. cinerea growth in inoculated plants, the levels of the B. cinerea ActinA mRNA (Benito et al., 1998) were determined by RNA gel blots. The B. cinerea ActinA gene was amplified from the B. cinerea genomic DNA and used as a template for random prime labeling. RNA blots from infected tissues were hybridized to 32P-labeled ActinA gene. A. brassicicola growth was evaluated by qPCR analysis of the A. brassicicola CutinaseA (Ab cutA) DNA (van Wees et al., 2003). The relative amplifications of the A. brassicicola–specific Ab cutA relative to that of Arabidopsis thaliana–specific Actin2 DNA (At3G18780) were determined on the Stratagene Mx3000P quantitative PCR system. Three technical replicates of the qPCR assay were used for each sample, and there were at least two biological replicates. The DNA levels were calculated by the comparative cycle threshold method (Applied Biosystems) with Arabidopsis Actin2 as the endogenous reference for normalization as described (Bluhm and Woloshuk, 2005).
Generation of Transgenic Lines and Identification of the Mutant Alleles
The HUB1 cDNA (clone name U16885) was obtained from the ABRC. To generate overexpression plants, full-length HUB1 cDNA was cloned after the cauliflower mosaic virus 35S promoter into a modified version of binary vector pCAMBIA 1200, transformed into Agrobacterium strain GV3101, and transformed into plants by Arabidopsis floral dip transformation (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog (MS) medium containing hygromycin, and the lines overexpressing HUB1 were identified by RNA gel blots hybridized to the full-length HUB1 cDNA. Transgenic lines with the HUB1 promoter driving GUS gene expression were generated by cloning a 1500-bp fragment of the HUB1 promoter region into the binary vector pCAMBIA1391 carrying the GUS gene. Transformants were selected on MS medium supplemented with hygromycin. T3 homozygous lines were used to study the expression of HUB1 during infection. hub1-4 and hub1-6 were isolated from SALK-122512 and WiscDsLox433B10, respectively, and the med21 allele was identified from WISCDSLOX461-464K13 (stock number CS856922) obtained from ABRC. To generate MED21 RNAi lines, the first 250 bp of the MED21 cDNA, starting from the ATG, was amplified and cloned into the RNAi vector pGSA1252 (http://www.chromdb.org/rnai/order_vectors.html). The expression of MED21 in RNAi and 35S:MED21 lines was determined using real-time quantitative RT-PCR.
RNA Gel Blots and RT-PCR
Total RNA was isolated as described (Lagrimini et al., 1987) or with Trizol reagent according to the manufacturer's instructions (Invitrogen). For RNA gel blots, total RNA was separated on 1.2% agarose-formaldehyde gels and blotted to Hybond N+ nylon membranes (Amersham Pharmacia Biotech). Probes were labeled with 32P using the random labeling system (Redi Prime II; GE Healthcare). Hybridization of probe and subsequent washings were performed as described (Church and Gilbert, 1984). Membranes were exposed to film for 24 h at −80°C (Biomax XAR Film; Kodak). Ethidium bromide staining of rRNA was used as a loading control.
RT-PCR was performed after DNase treatment of RNA and first-strand cDNA synthesis according to the manufacturer's instructions (Promega). cDNA was synthesized from both control and treated samples using equal amounts of total RNA (2 μg), AMV reverse transcriptase (Promega), and oligo(dT15) primers according to standard protocols. The PCR was performed for 28 cycles using 2.5 μL of cDNA as a template and specific primer pairs (94°C for 30 s, 52°C for 30 s, and 72°C for 1 min). qPCR was performed essentially as described by Bluhm and Woloshuk (2005). Total RNA (1 μg) was reverse transcribed using oligo(dT) primers and superscript II reverse transcriptase (Invitrogen). The resulting cDNA was subjected to qPCR using specific primers. The expression levels were calculated by the comparative cycle threshold method (Applied Biosystems) with Arabidopsis Actin2 (At3G18780) as the endogenous reference for normalization. qRT-PCR expression of FLM, MAF4, MAF1, FLC, SOC, and FT was determined from 10-d-old seedlings grown with a 12-h photoperiod on MS media using gene-specific primers (see Supplemental Table 1 online) and previously described primers (Jin et al., 2008).
Induction experiments with various chemicals and pathogens were performed as described (Veronese et al., 2006). The induction with chitin was done by treating 2-week-old seedlings with crab shell chitin as described (Sigma-Aldrich; 100 mg/L) (Zhang et al., 2002).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed with the GAL4 system according to the manufacturer's instructions (Stratagene). The HUB1 coding sequence was amplified and cloned into pBD-GAL4 to generate DNA binding domain bait protein fusion and was verified for the absence of transcriptional activation of LacZ. We built a cDNA library from B. cinerea–infected Arabidopsis tissue in the HybriZAP-2.1 vector according to the manufacturer's instructions (Stratagene). At least 106 yeast colonies were screened by transformation into the YRG-2 yeast strain (Stratagene) expressing HUB1. Interacting proteins were initially selected for complementation of His auxotrophy on selective medium lacking His, Leu, and Tyr. The putative interactors were then tested by assaying for the lacZ reporter gene activation by performing the filter lift assay as described in the Stratagene protocol. Interactions were retested for His3+, Trp+, and Leu+ auxotrophy and LacZ reporter activity (β-galactosidase assay). The plasmids from the positive clones were then isolated using the Zymoprep kit, sequenced, and reintroduced into the original yeast bait and control bait strains to verify interaction.
Bimolecular Fluorescence Complementation Assays
DNA sequences for the N-terminal 173–amino acid YFP (N-YFP) and the C-terminal 64–amino acid (C-YFP) fragments were PCR amplified and cloned into a plant expression vector derived from pCAMBIA1300 to generate pCAMBIA-N-YFP and pCAMBIA-C-YFP, respectively. The MED21 full-length cDNA was inserted into pCAMBIA-N-YFP to generate the N-terminal in-frame fusions with N-YFP (pMED21-nYFP), whereas HUB1 was introduced into pCAMBIA-C-YFP to form C-terminal in-frame fusions with C-YFP (pHUB1-cYFP). The constructs were verified by sequencing. The binary plasmids were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. The Agrobacterium carrying the appropriate plasmids was expressed in Nicotiana benthamiana leaf tissue by agroinfiltration. In vivo interaction was observed under an epifluorescent microscope (Nikon Eclipse E800).
Construction of Double Mutants
We used the Arabidopsis mutants ein2 (Guzman and Ecker, 1990), coi1 (Xie et al., 1998), sid2 (Wildermuth et al., 2001), and pad4 (Jirage et al., 1999) for genetic crosses to the hub1-6 allele. The coi1 mutant plants are male sterile and fail to set seed upon self-fertilization. Homozygous coi1/coi1 plants were identified from the progeny of coi1/COI1 heterozygous plants by plating seeds on MS medium (Sigma-Aldrich) supplemented with 50 μM MeJA. To select the coi1 hub1 double mutant, we first identified plants homozygous for hub1 mutations from the segregating population. Homozygous hub1 coi1 double mutants were then identified by selecting on 50 μM MeJA. Seedlings that develop normally on MeJA were considered to be coi1 coi1 homozygous (Xie et al., 1998; Liu et al., 2007) and were transplanted into soil for disease assays. The hub1 ein2 mutation was identified by plating seeds on 100 μM ACC. ACC-insensitive plants were selected for the hub1 mutation using PCR with primers specific for the hub1-6 allele.
The sid2-5 mutant was characterized by Glazebrook's lab previously and was isolated from Syngenta's Arabidopsis Insertion Library as described (van Wees and Glazebrook, 2003). The primers used for identification of the double mutants are presented in the Supplemental Table 1 online. The pad4 and coi11 alleles carry point mutations and were identified using cleaved-amplified polymorphic sequence markers (Xie et al., 1998; Jirage et al., 1999).
Callose Staining and Quantification
Callose staining was done 48 h after drop inoculation with Botrytis (5×104 spores/mL) or A. brassisicola (5×105 spores/mL). The inoculated leaves were collected 48 h after inoculation, cleared with 96% ethanol, and incubated with sodium phosphate buffer (0.07 M, pH 9) for 30 min. The leaves were then immersed in 0.005% aniline blue solution prepared in sodium phosphate buffer for 1 to 2 h and were visualized using a Nikon Eclipse E800 microscope. The pictures were taken with an epifluorescent microscope equipped with a UV filter under ×100 magnification. The region surrounding the infection site was analyzed for callose deposition and compared with the control pictures from mock-inoculated leaves. The pictures were taken at a similar exposure for all the genotypes and treatments. The quantification of callose in inoculated tissue was done using ImageJ software (http://rsb.info.nih.gov/ij/download.html). The same threshold defining a fluorescent and a nonfluorescent area was used for all the infected samples and controls, respectively. The area (in percentage) showing fluorescence in the infected tissue above the mock-inoculated control was calculated.
GUS Staining
To determine GUS activity, homozygous HUB1 Prom:GUS plants were drop inoculated with 5 μL of B. cinerea (2.5×105 spores/mL) or A. brassisicola (5×105 spores/mL). The infection with Erysiphe cichoracearum was done on whole plants and samples for GUS staining taken at 6 DAI. Histochemical staining for GUS was done as described (Liu et al., 2007).
Electron Microscopy
The ninth leaves from 4-week-old plants were used for the study, and fixation was done using the microwave method. The primary fixation was done with 2% paraformaldehyde + 2.5% glutaraldehyde in 0.1 M potassium phosphate buffer, pH 6.8. Secondary fixation was done with reduced osmium [1% OsO4 +1.5% K3Fe(CN)6]. Dehydration was done using an ethanol series and propylene oxide. Embedding was done in the polymerized resin. and ultrathin sections were viewed under an FEI/Philips CM-10 transmission electron microscope.
Immunoblot Analysis
Histone-enriched nuclear protein extracts were separated on a SDS-PAGE and transferred to a nitrocellulose membrane. To determine global levels of histone modifications, the membranes were probed with H3K4Me2 and H3K4M3 antibodies (Abcam). For signal detection, the immunoblots were then hybridized to peroxidase-conjugated secondary antibodies and incubated with enhanced chemiluminescence reagents (Pierce Scientific). The signals were detected by BioMax XAR film (Eastman Kodak), with exposure times selected to be in the linear range of detection. Histone H3 total protein was used as a loading control.
ChIP
ChIP was performed as described in http://www.epigenome-noe.net/researchtools/protocol.php?protid=13 using 3-week-old seedlings. The chromatin was immunoprecipitated with antibodies to histone H3 trimethyl K4 (Upstate; 07-473) and dimethyl K9 (prepared in Jenuwein laboratory). Immunoprecipitated DNA was purified using the Qiagen PCR purification kit and eluted in 50 μL of EB buffer. The control PCR was performed in total reaction volume of 25 μL, and PCR conditions were as follows: 96°C, 3 min; 30 cycles of 94°C, 30 s; 51°C, 30 s; 68°C, 1 min; followed by 68°C, 6 min. The primer sets for control PCR are listed (Huettel et al., 2006). Quantitative real-time PCR ChIP data for MAF1 and MAF4 promoters were obtained using the 2x SensiMix Plus SYBR and Fluorescein Kit (Quantace) in a 20-μL qPCR reaction according to the manufacturer's instructions. The samples were amplified using an iQ5 real-time PCR system (Bio-Rad Laboratories). qPCR data were analyzed according to the percentage of input method described (Haring et al., 2007).
Cation-Exchange High-Pressure Liquid Chromatography
Total cytosine methylation was determined from 2 μg of genomic DNA (extracted from 3-week-old seedlings with DNeasy; Qiagen) as described (Rozhon et al., 2008).
DNA Gel Blot Analysis
DNA methylation analysis with methylation-sensitive restriction enzymes was performed using genomic DNA prepared from 3-week-old seedlings (Phytopure; Amersham). Five micrograms of DNA was digested overnight with 2 units of HpaII and MspI (MBI Fermentas). Subsequently, samples were electrophoretically separated on 1% TAE agarose gels, depurinated for 10 min in 250 mM HCl, denaturated for 30 min in denaturation solution containing 0.5 M NaOH and 1.5 M NaCl, and neutralized twice in 0.5 M Tris, 1.5 M NaCl, and 1 mM EDTA at pH 7.2 for 15 min. The DNA gel was blotted onto Hybond N+ (Amersham) membranes overnight with 20× SSC, washed, and UV-crosslinked using a Stratalinker (Stratagene). Hybridization was performed using the Amersham Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare) according to the manufacturer's protocol. A probe specific for TSI pericentromeric repeats was used for hybridization (Steimer et al., 2000). Signals were detected with Phosphorimager Screens (Bio-Rad) and scanned with a Molecular Imager FX (Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HUB1 (At2g44950), HUB2 (At1g55250 and At1g55255), MED21 (GenBank accession number FJ769239), PR1 (At2g14610), PDF1.2 (At5g44420), PR3 (AT3G12500), OSM (At4G11650), Chitinase (At2G43580), MAF4 (At5g65070), MAF1 (At1g77080), BIK1 (At2g39660), WRKY33 (At2g38470), WRKY25 (At2g30250), and WRKY29 (At4g23550).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of HUB1, Characterization of the hub1 Mutations, and the 35S:HUB1 Plants.
Supplemental Figure 2. HUB1 Does Not Interact with BIK1 in Yeast Two-Hybrid Assays.
Supplemental Figure 3. HUB1 Has No Role in Resistance to P. syringae.
Supplemental Figure 4. The hub1 Plants Show No Altered Responses to Erysiphe cichoracearum Infection.
Supplemental Figure 5. hub1 Shows Wild-Type Responses to Plant Hormones.
Supplemental Figure 6. hub1 Shows Wild-Type Responses to NaCl and ABA.
Supplemental Figure 7. Flowering, Growth, and Senescence of hub1.
Supplemental Figure 8. Reduced Stem Thickness in hub1 Plants.
Supplemental Figure 9. The PMR4 Gene Is Required for Resistance to A. brassicicola.
Supplemental Figure 10. Global and Locus-Specific DNA Methylation.
Supplemental Figure 11. SA Has Only a Marginal Effect on the Functions of HUB1 in Limiting Botrytis Growth and Disease Symptoms.
Supplemental Figure 12. COI1 Function in Botrytis Resistance Is Additive to HUB1.
Supplemental Figure 13. RT-PCR Showing the HUB1 Gene Expression in Response to Plant Hormones and Methyl Viologen (Paraquat).
Supplemental Figure 14. MED21 Is an Evolutionarily Conserved Eukaryotic Protein.
Supplemental Figure 15. MED21 Is Required for Resistance to B. cinerea.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Data Set 1. Multiple Sequence Alignment Used as Input for the Phylogenetic Tree Presented in Supplemental Figure 14.
Supplementary Material
Acknowledgments
We thank Kristin Laluk for comments on the manuscript and for the phylogenetic analysis and Maria Simmons for editing the manuscript. This research was funded by National Science Foundation Grants IOB-0618897 and IOB-0749865 to T.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tesfaye Mengiste (mengiste@Purdue.edu).
Online version contains Web-only data.
References
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Backstrom, S., Elfving, N., Nilsson, R., Wingsle, G., and Bjorklund, S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26 717–729. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. [DOI] [PubMed] [Google Scholar]
- Benito, E.P., ten Have, A., van 't Klooster, J.W., and van Kan, J.A.L. (1998). Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur. J. Plant Pathol. 104 207–220. [Google Scholar]
- Bent, A.F., Innes, R.W., Ecker, J.R., and Staskawicz, B.J. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant Microbe Interact. 5 372–378. [DOI] [PubMed] [Google Scholar]
- Bjorklund, S., and Gustafsson, C.M. (2005). The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30 240–244. [DOI] [PubMed] [Google Scholar]
- Bluhm, B.H., and Woloshuk, C.P. (2005). Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant Microbe Interact. 18 1333–1339. [DOI] [PubMed] [Google Scholar]
- Boube, M., Joulia, L., Cribbs, D.L., and Bourbon, H.M. (2002). Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110 143–151. [DOI] [PubMed] [Google Scholar]
- Brandwagt, B.F., Mesbah, L.A., Takken, F.L., Laurent, P.L., Kneppers, T.J., Hille, J., and Nijkamp, H.J. (2000). A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. USA 97 4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch, G., Ransom, R., Lechner, T., Walton, J.D., and Loidl, P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, M., Lievens, B., Van Hemelrijck, W., Van den Ackerveken, G., Cammue, B.P., and Thomma, B.P. (2003). Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiol. Lett. 228 241–248. [DOI] [PubMed] [Google Scholar]
- Cantu, D., Vicente, A.R., Greve, L.C., Dewey, F.M., Bennett, A.B., Labavitch, J.M., and Powell, A.L. (2008). The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 105 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot, C., Nawrath, C., and Metraux, J.P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49 972–980. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Muskett, P.R., and Shirasu, K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6 307–311. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32 457–466. [DOI] [PubMed] [Google Scholar]
- Ferrari, S., Ausubel, F.M., Cervone, F., and De Lorenzo, G. (2003). Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury, D., et al. (2007). The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors, V., Ton, J., Jakab, G., and Mauch-Mani, B. (2005). Abscisic acid and callose: Team players in defense againist pathogens? J. Phytopathol. 153 377–383. [Google Scholar]
- Flors, V., Ton, J., van Doorn, R., Jakab, G., Garcia-Agustin, P., and Mauch-Mani, B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54 81–92. [DOI] [PubMed] [Google Scholar]
- Glickman, M.H., and Ciechanover, A. (2002). The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82 373–428. [DOI] [PubMed] [Google Scholar]
- Gonzalez, D., Bowen, A.J., Carroll, T.S., and Conlan, R.S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, M., Hu, G.Z., Tronnersjo, S., Shaikhibrahim, Z., Balciunas, D., Bjorklund, S., and Ronne, H. (2006). Functional and physical interactions within the middle domain of the yeast mediator. Mol. Genet. Genomics 276 197–210. [DOI] [PubMed] [Google Scholar]
- Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., and Stam, M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Doyle, M.R., and Amasino, R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Blanco, C., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel, B., Kanno, T., Daxinger, L., Aufsatz, W., Matzke, A.J., and Matzke, M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25 2828–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbel, B.M., Konomi, M., Takagi, T., Kamasawa, N., Ishijima, S.A., and Osumi, M. (2001). In situ localization of β-glucans in the cell wall of Schizosaccharomyces pombe. Yeast 18 433–444. [DOI] [PubMed] [Google Scholar]
- Jin, J.B., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal, G.S., and Briggs, S.P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258 985–987. [DOI] [PubMed]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Nam, J., Boyes, D.C., Holt III, B.F., Hubert, D.A., Wiig, A., and Dangl, J.L. (2005). A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J. 44 258–270. [DOI] [PubMed] [Google Scholar]
- Kim, T.W., Kwon, Y.J., Kim, J.M., Song, Y.H., Kim, S.N., and Kim, Y.J. (2004). MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc. Natl. Acad. Sci. USA 101 12153–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.J., and Lis, J.T. (2005). Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem. Sci. 30 245–249. [DOI] [PubMed] [Google Scholar]
- Kornberg, R.D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30 235–239. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 325–331. [DOI] [PubMed] [Google Scholar]
- Lagrimini, L.M., Burkhart, W., Moyer, M., and Rothstein, S. (1987). Molecular cloning of complementary DNA encoding the liginin-forming peroxidasew from tobacco: Molecular analysis and tissue-specific expression. Proc. Natl. Acad. Sci. USA 84 7542–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee, R.N., Fuchs, S.M., and Strahl, B.D. (2007). H2B ubiquitylation in transcriptional control: A FACT-finding mission. Genes Dev. 21 737–743. [DOI] [PubMed] [Google Scholar]
- Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J.M., and Hoffmann, J.A. (1996). The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86 973–983. [DOI] [PubMed] [Google Scholar]
- Lewis, B.A., and Reinberg, D. (2003). The mediator coactivator complex: Functional and physical roles in transcriptional regulation. J. Cell Sci. 116 3667–3675. [DOI] [PubMed] [Google Scholar]
- Liang, H., Yao, N., Song, J.T., Luo, S., Lu, H., and Greenberg, J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Koornneef, M., and Soppe, W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.C., Penninger, J., and Karin, M. (2005). Immunity by ubiquitylation: A reversible process of modification. Nat. Rev. Immunol. 5 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50 500–513. [DOI] [PubMed] [Google Scholar]
- Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre, D.A., and Wolpert, T.J. (1999). Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell 11 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Oh, S., Zhang, H., Ludwig, P., and van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley, M.A. (2004). H2B ubiquitylation: The end is in sight. Biochim. Biophys. Acta 1677 74–78. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Kim, J.M., Kim, L.K., Kim, S.N., Kim-Ha, J., Kim, J.H., and Kim, Y.J. (2003). Signal-induced transcriptional activation by Dif requires the dTRAP80 mediator module. Mol. Cell. Biol. 23 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, A.L., van Kan, J., ten Have, A., Visser, J., Greve, L.C., Bennett, A.B., and Labavitch, J.M. (2000). Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol. Plant Microbe Interact. 13 942–50. [DOI] [PubMed] [Google Scholar]
- Privalsky, M.L. (1998). Depudecin makes a debut. Proc. Natl. Acad. Sci. USA 95 3335–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom, R.F., and Walton, J.D. (1997). Histone hyperacetylation in maize in response to treatment with HC-toxin or infection by the filamentous fungus Cochliobolus carbonum. Plant Physiol. 115 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., and Riechmann, J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon, W., Baubec, T., Mayerhofer, J., Mittelsten Scheid, O., and Jonak, C. (2008). Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 375 354–360. [DOI] [PubMed] [Google Scholar]
- Saccani, S., and Natoli, G. (2002). Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 16 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (1999). The SAS System for Windows, Release 8.0. (Carry, NC: SAS Institute).
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 75 243–269. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., Johnson, J.S., and Dong, X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar, V.V., Kapoor, A., Zhang, K., Zhu, J., Zhou, T., Hasegawa, P.M., Bressan, R.A., and Zhu, J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447 735–738. [DOI] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Mittelsten Scheid, O., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z.W., and Allis, C.D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 104–108. [DOI] [PubMed] [Google Scholar]
- Takagi, Y., and Kornberg, R.D. (2006). Mediator as a general transcription factor. J. Biol. Chem. 281 80–89. [DOI] [PubMed] [Google Scholar]
- Tanny, J.C., Erdjument-Bromage, H., Tempst, P., and Allis, C.D. (2007). Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H., Saika, H., Tsutsumi, N., Hirai, A., and Nakazono, M. (2006). Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 47 995–1003. [DOI] [PubMed] [Google Scholar]
- Tudor, M., Murray, P.J., Onufryk, C., Jaenisch, R., and Young, R.A. (1999). Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59 573–594. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., Chang, H.S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S.C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33 733–742. [DOI] [PubMed] [Google Scholar]
- Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., Dietrich, R.A., Hirt, H., and Mengiste, T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wan, J., Zhang, X.C., Neece, D., Ramonell, K.M., Clough, S., Kim, S.Y., Stacey, M.G., and Stacey, G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Wood, A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., Johnston, M., and Shilatifard, A. (2003. b). Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11 267–274. [DOI] [PubMed] [Google Scholar]
- Wood, A., Schneider, J., Dover, J., Johnston, M., and Shilatifard, A. (2003. a). The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278 34739–34742. [DOI] [PubMed] [Google Scholar]
- Xiao, T., Kao, C.F., Krogan, N.J., Sun, Z.W., Greenblatt, J.F., Osley, M.A., and Strahl, B.D. (2005). Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Ramonell, K., Somerville, S., and Stacey, G. (2002). Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 15 963–970. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. (2003). Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17 2733–2740. [DOI] [PubMed] [Google Scholar]
- Zheng, Z., Qamar, S.A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. [DOI] [PubMed] [Google Scholar]
- Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B., Zheng, Y., Pham, A.D., Mandal, S.S., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2005). Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 20 601–611. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.