Abstract
Proteins of the regulators of G protein signaling (RGS) family modulate the duration of intracellular signaling by stimulating the GTPase activity of G protein α subunits. It has been established that the ninth member of the RGS family (RGS9) participates in accelerating the GTPase activity of the photoreceptor-specific G protein, transducin. This process is essential for timely inactivation of the phototransduction cascade during the recovery from a photoresponse. Here we report that functionally active RGS9 from vertebrate photoreceptors exists as a tight complex with the long splice variant of the G protein β subunit (Gβ5L). RGS9 and Gβ5L also form a complex when coexpressed in cell culture. Our data are consistent with the recent observation that several RGS proteins, including RGS9, contain G protein γ-subunit like domain that can mediate their association with Gβ5 (Snow, B. E., Krumins, A. M., Brothers, G. M., Lee, S. F., Wall, M. A., Chung, S., Mangion, J., Arya, S., Gilman, A. G. & Siderovski, D. P. (1998) Proc. Natl. Acad. Sci. USA 95, 13307–13312). We report an example of such a complex whose cellular localization and function are clearly defined.
Heterotrimeric G proteins act as molecular switches that relay excitation from activated receptors to effector molecules, such as enzymes or ion channels. A G protein becomes activated upon the receptor-stimulated binding of GTP to its α subunit and continues to modulate the activity of the effector until bound GTP is hydrolyzed (reviewed in refs. 1 and 2). In many signaling pathways, the duration of the signal under physiological conditions is much shorter than would be predicted from the intrinsic rate of α subunits of G proteins (Gα) GTPase activity. This is because GTPase activities of many Gαs are dramatically accelerated by RGS (regulators of G protein signaling) proteins or by the G protein effectors (reviewed in refs. 3–6). The phototransduction cascade of vertebrate photoreceptors represents one of the most sophisticated examples of such regulation where the GTPase activity of the G protein, transducin, is substantially enhanced by the cooperative action of RGS9 and the γ subunit of the effector of transducin, cGMP phosphodiesterase (PDEγ) (7–10). The role of RGS9 is to provide transducin with the RGS homology domain, which acts catalytically in stimulating the rate of transducin GTPase. PDEγ itself does not activate transducin GTPase but it enhances the catalytic action of RGS9. The degree of this potentiation observed in physiologically intact photoreceptors is ≈7-fold (10). We believe that this ability of PDEγ to potentiate RGS9 action is essential for photoreceptor function. When a rod photoreceptor is hit by photon of light it has to perform two tasks. First, it has to transmit the signal from excited rhodopsin to PDE with high efficiency. Second, it has to inactivate all activated proteins in the cascade, including transducin, within a fraction of 1 s. If transducin is allowed to be discharged by RGS9 before it forms a complex with PDE, then some transducin molecules would never activate PDE and signal amplification would be diminished. Therefore, making the GTPase activation contingent on transducin association with PDEγ ensures both high efficiency of signal transmission between transducin and PDE and timely photoresponse recovery.
Although the general scheme for transducin GTPase regulation, outlined above is well supported by experimental data, several fundamental mechanistic questions are not resolved. It remains unclear why the effect of PDEγ observed both with RGS9-containing photoreceptor membranes (7, 8) and intact photoreceptors (10) is substantially more pronounced than its effect in reconstituted system with recombinant catalytic domain of RGS9 (9). It is also not understood why the RGS9 catalytic domain is able to support the rate of transducin GTPase at physiologically fast rate, but intact RGS9 in the photoreceptor requires cooperation with PDEγ. A major difficulty in addressing these questions is that functionally active full-length RGS9 has never been purified or expressed. One possibility is that RGS9 in photoreceptors exists as a multiple subunit complex and that some of its properties could be manifested only within such a complex.
In this study we explored the hypothesis that RGS9 in photoreceptors exists as a multiple subunit complex and found that indeed it is present as a tight complex with the long splice variant of the type 5 β subunit of G proteins (Gβ5L) that cannot be separated from RGS9 under nondenaturing conditions. The existence of this complex has been shown by three complimentary approaches. First, RGS9 and Gβ5L comigrate when photoreceptor membrane proteins were subjected to various chromatographic procedures. Second, antibodies raised against each of these proteins quantitatively precipitated both RGS9 and Gβ5L. Finally, these proteins were coimmunoprecipitated after expression in cell culture.
MATERIALS AND METHODS
Purification and Washing of Rod Outer Segment (ROS) Membranes.
ROS were purified from frozen retinas (TA & WL Lowson, Lincoln, NE) under IR illumination as described (11). To obtain membranes lacking most peripheral proteins but retaining an active RGS9–Gβ5L complex, ROS were washed under IR illumination twice with isotonic buffer containing 100 mM KCl, 2 mM MgCl2, 1 mM DTT, and 10 mM Tris⋅HCl (pH 7.5) and three times by a hypotonic buffer containing 0.5 mM EDTA and 5 mM Tris⋅HCl (pH 7.5). Urea-washed photoreceptor discs lacking the activity of RGS9 were obtained as described (12). Rhodopsin concentration in all membrane preparations was determined spectrophotometrically (13).
Preparation of Proteins.
Transducin was purified from ROS as described (14). The only modification was that we bleached rhodopsin in the retinas before ROS purification to increase transducin yield by forming a tight complex between bleached rhodopsin and transducin, thus preventing transducin loss from ROS membranes during ROS isolation. Transducin concentration was first estimated by the Bradford assay (15), and then the exact concentration of active transducin in each preparation was determined by measuring the maximal amount of rhodopsin-catalyzed guanosine 5′[γ-thio]triphosphase (GTP[γS]) binding (16).
Recombinant PDEγ was purified by a combination of cation-exchange and reverse-phase chromatography (17) from Escherichia coli strain BL21 DE3 transformed with an expression plasmid containing a cDNA encoding PDEγ (18). The PDEγ concentration was determined spectrophotometrically at 280 nm by using a molar extinction coefficient of 7,100.
GTPase Measurements.
Transducin GTPase activity was determined by the single-turnover ([GTP] < [transducin]) technique described in detail previously (7, 19, 20). The measurements were conducted at room temperature (22–24°C) in a buffer containing 10 mM Tris⋅HCl (pH 7.8), 100 mM NaCl, 8 mM MgCl2, and 1 mM DTT. The urea-treated photoreceptor discs used as a source of rhodopsin were bleached on ice immediately before the experiments. For measuring the activity in the gel-filtration chromatography fractions, 10 μl of the fraction was mixed with 10 μl of the test system containing 20 μM rhodopsin and 2 μM transducin with or without 2 μM PDEγ (all concentrations final). The reaction was started by adding 5 μl of [γ-32P]GTP (≈105 dpm/sample) and conducted for 5 s. The reaction was stopped by the addition of 100 μl of 6% perchloric acid. 32Pi formation was measured by the activated charcoal binding assay (20).
Antibody Production and Protein Immunoprecipitation.
Sheep antibodies were produced by Elmira Biologicals (Iowa City, IA) against Gβ5L peptides DKCFKQRALRPVFKKS (N-terminal peptide, NTL), TLRVSPDGTAFCS (C-terminal peptide, CT), and MATDGLHENETLASLK (N terminus of the Gβ5 short splice variant, NTS). Each of the peptides contained an additional cysteine residue at the N terminus. Sheep antibodies were also produced against the His-tagged RGS9 fragment 226–484 called RGS9c by He et al. (9). The pET14b plasmid encoding the His-tagged RGS9c was a gift from T. G. Wensel (Baylor College of Medicine, Houston, TX). Rabbit antibodies were produced against the His-tagged RGS9c and Gβ5L N-terminal peptide. Rabbit anti-RGS9c antibodies used in some experiments were a gift from T. G. Wensel. Rabbit NTS antibodies were a gift from V. Z. Slepak (University of Miami). Each antibody was affinity purified on columns where antigens were covalently attached to SulfoLink medium (Pierce) according to instructions provided by Pierce. These antibodies were used for a standard Western blot analysis by using Amersham ECL kit for developing. No cross-reactivity with other ROS proteins on Western blots was observed with any of the affinity-purified antibodies. Some of the purified antibodies were also used for protein immunoprecipitation assays. In these cases antibodies were covalently attached to AminoLink medium (Pierce) according to instructions provided by Pierce. Five milligrams of the antibodies was attached to 1 ml of the beads. Pre-immune IgG were used as controls. All immunoprecipitation assays were performed in 200 μl of PBS buffer containing 0.5% lauryl sucrose. Fifty microliters of AminoLink beads was typically used in the assay. The samples were incubated for 2.5 h at room temperature upon mixing on the vortex shaker. The beads were separated from the supernatant by a brief centrifugation and washed twice with the same buffer. Bound proteins were extracted from the beads by 1-min boiling in the standard sample buffer for SDS/PAGE. Antibodies against Gγ2 were obtained from Santa Cruz Biotechnology.
Mass Spectrometric Fingerprinting.
Mass spectrometric fingerprinting analysis was performed by J. Lee at the Molecular Biology Core Facilities at the Dana–Farber Cancer Institute (Boston). Protein mixtures were subjected to SDS/PAGE, gels were stained by Coomassie blue G-250 (Sigma), and bands containing proteins of interest were cut from the gel and proteolyzed by trypsin as described (21). The molecular masses of proteolytic peptide fragments were determined by MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) spectrometry on Voyager-DE STR instrument (Perceptive Biosystems, Cambridge, MA). The values for molecular masses in digests were compared with the values from theoretical digests by using proteinprospector MS-Fit software (available via internet at http://prospector.ucsf.edu). The probability of the positive protein identification was assessed with the molecular weight search score described (22). Because the amount of protein in the bands originated from the gel filtration fractions was too small to obtain reliable data, we performed the analysis with the Gβ5L immunoreactive band obtained from the concentrated alkaline extract of washed ROS membranes. The extraction was performed as described (23) by 100 mM Na2CO3 with the pH adjusted to 12.0 by NaOH. Before the SDS/PAGE separation, proteins in the extract were concentrated by precipitation with 3% trichloroacetic acid.
cDNA Cloning and in Vitro Expression.
A mouse retinal cDNA library (provided by D. Roof, Harvard Medical School) was screened with a rat RGS9 cDNA probe (24) provided by M. Koelle (Yale University). Among 10 positive clones, 1 contained the complete coding sequence as concluded by its comparison with the published mouse RGS9 sequence (9). The mouse Gβ5L cDNA containing a point mutation that inactivates an alternative downstream translational initiation site (25) was provided by V. Z. Slepak. The bovine Gγ2 cDNA cloned in the expression vector pEV1 (26) was provided by Dr. A. Pronin (T. Jefferson University, Philadelphia).
For in vitro expression, the mouse RGS9 and Gβ5L coding regions were subcloned into the mammalian expression vector pcDNA3 (Invitrogen). The purified expression plasmids were transfected either singly or in combination into the HEK293 cell line by using the LipofectAmine Plus reagent (Life Technologies, Gaithersburg, MD) following the manufacturer’s protocols. Cells were harvested 48 hr posttransfection, and their membranes were purified as described (27).
RESULTS AND DISCUSSION
Comigration of RGS9 and Gβ5L upon Gel Filtration of ROS Membrane Proteins.
Our strategy in identifying ROS proteins that potentially associate with RGS9 was to look for proteins that comigrate with both RGS9 and the GTPase activating protein (GAP) activity upon chromatographic separation of solubilized ROS membranes. ROS membranes were washed as described in Materials and Methods to minimize the total amount of non-rhodopsin protein bands (but retaining RGS9) in the starting material. A nonionic detergent, lauryl sucrose, was used for membrane solubilization because we have found that it does not substantially influence the basal rate of transducin GTPase activity or the ability of RGS9 to stimulate this activity in the PDEγ-dependent manner at concentrations <0.25% (data not shown). Thus, this choice of detergent provided us with an advantage to analyze the GAP activity of RGS9 directly in the chromatography fractions.
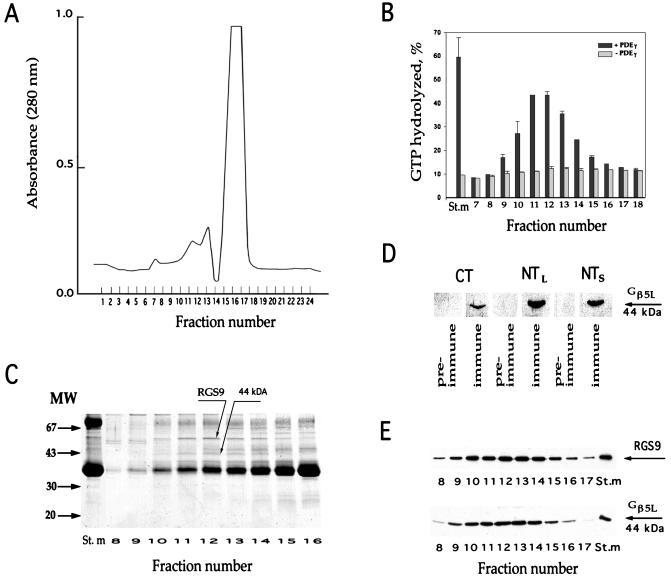
Gel filtration of solubilized washed membranes on the Superose-6 column is illustrated in Fig. 1A. The ability of the fractions to stimulate transducin GTPase in a PDEγ-dependent manner (Fig. 1B) correlated with the presence of the RGS9 band (Fig. 1C). The only other band whose density profile precisely followed the activity profile was the band with an apparent molecular mass of 44 kDa (Fig. 1C). This molecular mass corresponds to that of Gβ5L (43,566 Da in mouse), which was previously reported to be a ROS-specific, membrane-associated protein of unknown function (25). To test the hypothesis that the 44-kDa protein is Gβ5L, we generated three different antibodies against peptides from various regions of Gβ5L, including the N-terminal sequence which is absent in the Gβ5 short splice variant. Fig. 1D shows that all three antibodies provided strong immunostaining of the 44-kDa band on Western blots. The data from Fig. 1E, where both RGS9 and Gβ5L are immunostained by corresponding antibodies, provide clear evidence that these proteins comigrate during the gel filtration.
Figure 1.
Comigration of RGS9 and Gβ5L during gel filtration. Washed ROS membranes (containing 3 mg rhodopsin) were solubilized in 400 μl of buffer (20 mM Hepes adjusted to pH 7.4 by KOH, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, and 2% lauryl sucrose) and loaded on the Superose 6 column attached to the fast protein liquid chromatography (FPLC) system (Pharmacia). The column was equilibrated by the same buffer containing 0.5% lauryl sucrose, 5% glycerol, and 2 mg/ml soybean l-α-phosphatidylcholine (Sigma product P-5638). The elution rate was 0.4 ml/min; the fraction size was 0.4 ml. (A) Protein elution profile monitored at 280 nm. The major source of the UV absorbance is rhodopsin. (B) GAP activity of RGS9 in chromatography fractions. Single-turnover transducin GTPase measurements were performed in duplicate with and without PDEγ (see Materials and Methods). The y-axis value represents the percentage of GTP hydrolysis over the 5-s period; error bars indicate the range of determined values. The St.m bars represent the activity in the starting material after it was diluted to achieve the same lauryl sucrose, glycerol, and phosphatidylcholine concentration as in the fractions. (C) Coomassie staining of proteins in fractions surrounding the peak of RGS9 activity. (D) Western blot immunostaining of the 44-kDa protein band by three immune and pre-immune serums raised in sheep against the Gβ5L C-terminal peptide (CT), Gβ5L N-terminal peptide (NTL), and N-terminal peptide of the Gβ5 short splice variant (NTS). (E) Comigration of RGS9 and Gβ5L in the chromatography fractions. Western blots were probed with rabbit anti-RGS9c and anti-NTS antibodies.
Mass spectrometric fingerprinting analysis was used to obtain an independent confirmation that the 44-kDa protein is Gβ5L (see Materials and Methods for details). Nineteen major peptides were identified in the tryptic digest of the putative Gβ5L band. Protein database search analysis indicated that 10 peptides in the digest potentially originate from mouse Gβ5L with the molecular weight search (MOWSE) score of 70,500. Because the amino acid sequence of bovine Gβ5 is not currently known, this score should be considered as the lowest estimate. The MOWSE score for the next most likely candidate among all known sequenced mammalian proteins was only 601 (human carboxylesterase hCE-2). Another way to stress the uniqueness of the Gβ5L identification is to note that not a single sequenced mammalian protein with molecular mass between 40 and 50 kDa could contribute more than three of the 19 peptides to this digest (assuming the same stringency of the search parameters).
RGS9 and Gβ5L Could Not Be Separated upon Ion Exchange Chromatography and Immunoprecipitation by Specific Antibodies.
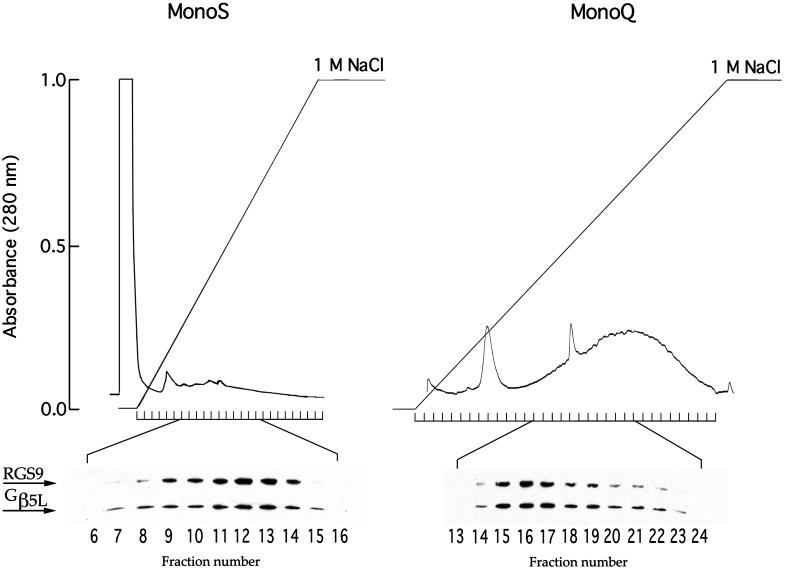
Our first approach to demonstrate that RGS9 and Gβ5L indeed form a tight complex in ROS was to show that these proteins comigrate upon various chromatographic separations of the ROS membrane proteins. As seen clearly in Fig. 2, a precise comigration of these proteins is observed when solubilized ROS membranes were subjected to both cation exchange chromatography on Mono-S column and anion exchange chromatography on Mono-Q column. We did not observe any chromatography fractions where either protein was present alone or where RGS9 and Gβ5L relative abundance was noticeably mismatched.
Figure 2.
Comigration of RGS9 and Gβ5L during cation-exchange and anion-exchange chromatography. Washed ROS membranes (containing 3 mg rhodopsin) were solubilized in 500 μl of 2% lauryl sucrose either in 20 mM Hepes-KOH (pH 6.0) with 2 mM MgCl2 (MonoS) or in 50 mM Tris⋅HCl (pH 7.8) with 2 mM MgCl2 (MonoQ). The columns were equilibrated by the corresponding buffers containing 0.5% lauryl sucrose. A 0–1 M gradient of NaCl was used to elute the bound proteins. The flow rate was 1 ml/min; the fraction size was 1 ml. Western blots of the chromatography fractions were probed by a mixture of rabbit anti-RGS9c and anti-NTS antibodies against Gβ5 at dilutions yielding similar intensities of immunostaining. No GAP activity in corresponding fractions was present in these cases, consistent with previous reports that this activity is extremely unstable in detergent solutions (32).
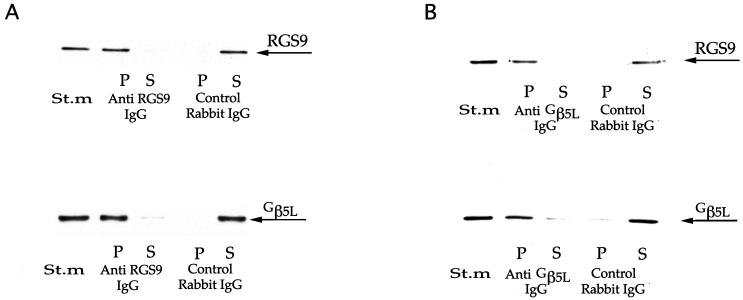
Our second approach to establish the existence of the RGS9–Gβ5L complex was to show that both of these proteins could be immunoprecipitated by antibodies raised against each individual protein. As reported by Cowan et al. (23), polyclonal antibodies raised against the RGS9c domain have an ability to interact with RGS9 in its native conformation and to deplete its GAP activity from solubilized membranes. A similar observation for the anti-RGS9c antibodies obtained in our study is illustrated in Fig. 3A. RGS9 precipitation was also accompanied by a practically complete precipitation of Gβ5L. No precipitation of either protein was observed with control pre-immune IgG. Similarly, an antibody against the N-terminal domain of Gβ5L quantitatively precipitated both Gβ5L and RGS9 (Fig. 3B). Consistently with the data by Cowan et al. (23), the precipitation of RGS9 by either antibody resulted in the depletion of the PDEγ-stimulated GAP activity for transducin from the solubilized ROS membranes (data not shown).
Figure 3.
Reciprocal coimmunoprecipitation of RGS9 and Gβ5L by rabbit anti-RGS9c (A) and anti-Gβ5L NTL (B) antibodies. Washed ROS membranes, containing either 180 (A) or 8 (B) μg rhodopsin, were solubilized in 0.5% lauryl sucrose and subjected to the immunoprecipitation. The difference in the amounts of membranes used in A and B reflects ≈20-fold difference in precipitating capacities of the antibodies used in the assay. Western blots from the samples originated from the starting material (St.m), unbound proteins in the supernatant (S) and proteins bound to the pelleted beads (P) were probed with purified sheep anti-RGS9c and anti-NTS antibodies.
The data from Figs. 1–3 thus provide independent evidence that RGS9 exists as a tight complex with Gβ5L in photoreceptors and that neither protein exists in any significant amount free from the other.
RGS9 and Gβ5L Form a Complex upon Their Expression in the HEK293 Cells.
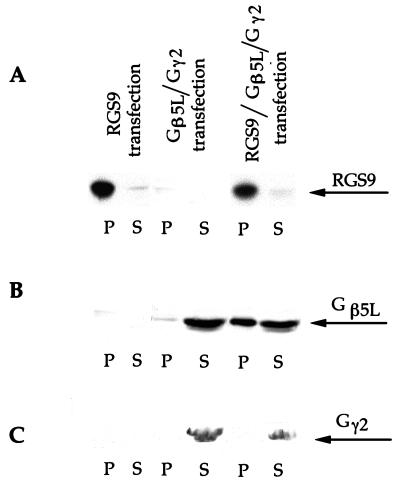
The data from Fig. 4 indicate that RGS9 and Gβ5L also form a complex after being coexpressed in human embryonic kidney cells, HEK293. When RGS9 cDNA was expressed alone, anti-RGS9c antibodies immunoprecipitated the entire pool of RGS9 from the membrane fraction of HEK293 cells. As expected, when Gβ5L was expressed in the absence of RGS9 it was not precipitated by the same antibodies. However, when RGS9 and Gβ5L were coexpressed, a substantial fraction of Gβ5L was precipitated. In spite of the fact that the ability of expressed proteins to form complex was preserved, no detectable GAP activity of RGS9 was observed in these experiments. We coexpressed Gβ5L with Gγ2 because this resulted in a significant increase in the levels of Gβ5L expression, as reported by others (28, 29). This led to the observation illustrated in Fig. 4C. Gγ2 was completely absent from the Gβ5L fraction precipitated by the anti-RGS9 antibodies. Instead, the entire pool of Gγ2 was found in the Gβ5L fraction that did not contain RGS9. These data suggest that Gβ5L binding to RGS9 occurs as an alternative to the Gβ5L binding to Gγ2. This finding is consistent with the idea that RGS9 contains the G protein γ subunit-like domain (GGL domain), which should make it possible for Gβ5 to form a complex with RGS9 instead of forming a complex with a Gγ (30). Interestingly, we did not observe coprecipitation of Gβ5L with Gγ2 by the NTL anti-Gβ5L antibodies, in spite of the fact that the level of Gβ5L expression with Gγ2 was significantly higher than without Gγ2. This is similar to the lack of Gβ5 coprecipitation with Gγ2 in COS-7 cells reported by Snow et al. (30). This apparent discrepancy between the ability of Gγ2 to enhance Gβ5 expression and the lack of their coimmunoprecipitation might be explained if the Gβ5–Gγ2 is of low affinity, or if their association is transient, but necessary for the proper Gβ5 processing.
Figure 4.
Coimmunoprecipitation of RGS9 and Gβ5L expressed in the HEK293 cells. HEK293 cells were transfected with pcDNA3-RGS9 plasmid, pcDNA3-Gβ5L and pEV1-Gγ2 plasmids, or all three plasmids. Membranes from each transfected line were solubilized in 0.5% lauryl sucrose and immunoprecipitated with rabbit anti-RGS9c antibodies. Three identical Western blots were performed with the aliquots of unbound proteins in the supernatant (S) and proteins bound to the pelleted beads (P). Each blot was probed with one of three antibodies: purified sheep anti-RGS9c (A), purified sheep anti-Gβ5L NTS antibodies (B), and commercial rabbit anti-Gγ2 antibodies (C).
Consistent with the idea that RGS9 substitutes Gγ in the complex with Gβ5L, we failed to identify any Gγ within the RGS9–Gβ5L complex obtained from ROS membranes. This result is evident from three lines of experiments. (i)The mass spectrometric fingerprinting analysis of the alkaline protein extract from washed ROS membranes was performed for the entire area of the SDS/PAGE gels where all known Gγs could be found (between the dye front and the 14-kDa molecular mass standard). It revealed a strong signal for Gγ1 and no hint for any other known Gγ. Further analysis with the Gγ1-specific antibodies (by both chromatography and immunoprecipitation) has indicated that Gγ1 is found only in fractions containing the β subunit of transducin, Gβ1, but not in the Gβ5L fractions. (ii)Immunostaining of both gel-filtration fractions and RGS9–Gβ5L immunoprecipitates by all commercially available antibodies against Gγ (Gγ1, Gγ2, Gγ3, Gγ5, and Gγ7) provided negative results. (iii) The sample from the RGS9–Gβ5L immunoprecipitate did not produce a detectable band on the SDS/PAGE gel in the area where a transducin sample with similar amount of Gβ1 produced a clear staining of Gγ1 (data not shown).
The Ability to Form Complexes with Gβ5 Is a General Property of Several RGS Proteins.
The observations presented in this study are consistent with the data from two recent publications obtained with RGS proteins other than RGS9. Snow et al. (30) have found that RGS11 contains the GGL domain which enables it to form a complex with both short and long splice variants of Gβ5 upon their coexpression in cell culture. This interaction is highly specific, and no other known type of G protein β subunit, besides Gβ5, can interact with RGS11. Similar domains are present in RGS6, RGS7, RGS9 and Caenorhabditis elegans protein EGL-10 (30). They have also observed that the RGS11–Gβ5 complex has a GAP activity with an unusually high selectivity toward the α subunit of the G protein, Go. In another study by Cabrera et al. (31), a short splice variant of Gβ5 was copurified with RGS7 from the retinal extract depleted of ROS, indicating that they might be present as a complex.
Taken together, these studies challenge one of the central paradigms in G protein signaling. It is generally accepted that Gβs always function as tight complexes with Gγs. However, it is now clear that Gβ5 is capable of forming complexes with RGS proteins instead of binding to a Gγ. Our experiments provide an example of such a complex whose cellular localization and physiological function are clearly defined. The RGS9–Gβ5L complex is responsible for activating transducin GTPase in photoreceptor outer segments, and it does this in a cooperative interaction with the effector of transducin, PDEγ. The challenge for future experiments is to define the role of each individual protein subunit in this task.
Acknowledgments
We thank Drs. M. D. Bownds, A. Bohm, C. L. Makino, M. Sokolov, and P. D. Calvert for critically reading the manuscript, and Mr. M. Goulston for technical assistance. This work was supported by National Institutes of Health Grant EY 10336, a grant from the Massachusetts Lions Eye Research Fund, and a Jules and Doris Stein Professorship from Research to Prevent Blindness (to V.Y.A.). T.L. is a recipient of a Career Development Award from Research to Prevent Blindness.
ABBREVIATIONS
- RGS
regulators of G protein signaling
- Gα
α subunits of G proteins
- Gβ
β subunits of G proteins
- Gγ
γ subunits of G proteins
- ROS
rod outer segments
- PDE
type 6 cGMP phosphodiesterase from ROS
- PDEγ
the γ subunit of PDE
- GGL domain
G protein γ subunit-like domain
- GAP
GTPase activating protein
- Gβ5L
long splice variant of Gβ
References
- 1.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 2.Hildebrandt J D. Biochem Pharmacol. 1997;54:325–339. doi: 10.1016/s0006-2952(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 3.Koelle M R. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 4.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 5.Berman D M, Gilman A G. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 6.Arshavsky V Y, Pugh E N. Neuron. 1998;20:11–14. doi: 10.1016/s0896-6273(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 7.Angleson J K, Wensel T G. J Biol Chem. 1994;269:16290–16296. [PubMed] [Google Scholar]
- 8.Arshavsky V Y, Dumke C L, Zhu Y, Artemyev N O, Skiba N P, Hamm H E, Bownds M D. J Biol Chem. 1994;269:19882–19887. [PubMed] [Google Scholar]
- 9.He W, Cowan C W, Wensel T G. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 10.Tsang S H, Burns M E, Calvert P D, Gouras P, Baylor D A, Goff S P, Arshavsky V Y. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 11.McDowell J H. Methods Neurosci. 1993;15:123–130. [Google Scholar]
- 12.Nekrasova E R, Berman D M, Rustandi R R, Hamm H E, Gilman A G, Arshavsky V Y. Biochemistry. 1997;36:7638–7643. doi: 10.1021/bi970427r. [DOI] [PubMed] [Google Scholar]
- 13.Bownds M D, Gordon-Walker A, Gaide-Huguenin A-C, Robinson W. J Gen Physiol. 1971;58:225–237. doi: 10.1085/jgp.58.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ting T D, Goldin S B, Ho Y-K. Methods Neurosci. 1993;15:180–195. [Google Scholar]
- 15.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Fung B B-K, Hurley J B, Stryer L. Proc Natl Acad Sci USA. 1981;78:152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown R L, Stryer L. Proc Natl Acad Sci USA. 1989;86:4922–4926. doi: 10.1073/pnas.86.13.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slepak V Z, Artemyev N O, Zhu Y, Dumke C L, Sabacan L, Sondek J, Hamm H E, Bownds M D, Arshavsky V Y. J Biol Chem. 1995;270:14319–14324. doi: 10.1074/jbc.270.24.14319. [DOI] [PubMed] [Google Scholar]
- 19.Arshavsky V Y, Antoch M P, Lukjanov K A, Philippov P P. FEBS Lett. 1989;250:353–356. doi: 10.1016/0014-5793(89)80754-9. [DOI] [PubMed] [Google Scholar]
- 20.Arshavsky V Y, Gray-Keller M P, Bownds M D. J Biol Chem. 1991;266:18530–18537. [PubMed] [Google Scholar]
- 21.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 22.Pappin D J C, Hojrup P, Bleasby A J. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 23.Cowan C W, Fariss R N, Sokal I, Palczewski K, Wensel T G. Proc Natl Acad Sci USA. 1998;95:5351–5356. doi: 10.1073/pnas.95.9.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 25.Watson A J, Aragay A M, Slepak V Z, Simon M I. J Biol Chem. 1996;271:28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
- 26.Pronin A N, Gautam N. Proc Natl Acad Sci USA. 1992;89:6220–6224. doi: 10.1073/pnas.89.13.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathans J, Weitz C J, Agarwal N, Nir I, Papermaster D S. Vision Res. 1989;29:907–914. doi: 10.1016/0042-6989(89)90105-3. [DOI] [PubMed] [Google Scholar]
- 28.Watson A J, Katz A, Simon M I. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 29.Zhang S, Coso O A, Lee C, Gutkind J S, Simonds W F. J Biol Chem. 1996;271:33575–33579. doi: 10.1074/jbc.271.52.33575. [DOI] [PubMed] [Google Scholar]
- 30.Snow B E, Krumins A M, Brothers G M, Lee S F, Wall M A, Chung S, Mangion J, Arya S, Gilman A G, Siderovski D P. Proc Natl Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabrera J L, De Freitas F, Satpaev D K, Slepak V Z. Biochem Biophys Res Commun. 1998;249:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- 32.Cowan C W, Angleson J K, Wensel T G. Invest Ophthalmol Vis Sci. 1996;37:S809. [Google Scholar]