Abstract
As an important agronomic trait, rice (Oryza sativa L.) leaf rolling has attracted much attention from plant biologists and breeders. Moderate leaf rolling increases the photosynthesis of cultivars and hence raises grain yield. However, the relevant molecular mechanism remains unclear. Here, we show the isolation and functional characterization of SHALLOT-LIKE1 (SLL1), a key gene controlling rice leaf rolling. sll1 mutant plants have extremely incurved leaves due to the defective development of sclerenchymatous cells on the abaxial side. Defective development can be functionally rescued by expression of SLL1. SLL1 is transcribed in various tissues and accumulates in the abaxial epidermis throughout leaf development. SLL1 encodes a SHAQKYF class MYB family transcription factor belonging to the KANADI family. SLL1 deficiency leads to defective programmed cell death of abaxial mesophyll cells and suppresses the development of abaxial features. By contrast, enhanced SLL1 expression stimulates phloem development on the abaxial side and suppresses bulliform cell and sclerenchyma development on the adaxial side. Additionally, SLL1 deficiency results in increased chlorophyll and photosynthesis. Our findings identify the role of SLL1 in the modulation of leaf abaxial cell development and in sustaining abaxial characteristics during leaf development. These results should facilitate attempts to use molecular breeding to increase the photosynthetic capacity of rice, as well as other crops, by modulating leaf development and rolling.
INTRODUCTION
The three-dimensional structure of the plant leaf is crucial for its functions, including light capture, carbon fixation, and gas exchange for photosynthesis (Govaerts et al., 1996). Appropriate leaf shape is an important characteristic associated with the super-rice (a hybrid rice [Oryza sativa L.] with desirable characteristics) ideotype; the rolled leaf is regarded as a critical element for the ideal rice phenotype (Yuan, 1997; Chen et al., 2001). Moderate leaf rolling helps to maintain the erectness of leaves, which can increase the light transmission rate and light saturation point without affecting the light compensation point, resulting in a well-proportioned leaf area for photosynthesis (Duncan, 1971). Moderate leaf rolling improves photosynthetic efficiency, accelerates dry-matter accumulation, increases yield, reduces the solar radiation on leaves, and decreases leaf transpiration under drought stress (Lang et al., 2004). Therefore, the isolation of genes that control leaf rolling will be beneficial for breeding crops with the desired architecture and stress tolerance.
As a polymorphic crop, rice varieties and mutants exhibit several types of leaf rolling, including inward or outward rolling, and these various phenotypes are regulated by complicated developmental processes, including pattern formation, polarity establishment, and cell differentiation (Micol and Hake, 2003; Lang et al., 2004). Hsiao et al. (1984) reported that leaf rolling is induced by altered osmotic pressure between internal and external tissues. Moulia (1994) proposed that leaf rolling may result from varying degrees of dehydration in different cross sections of the rolled leaf. Price et al. (1997) suggested that leaf rolling results from decreased turgidity of bulliform cells. However, the mechanism underlying leaf rolling in monocots remains to be elucidated.
Due to its importance, many studies have been performed to characterize the genes controlling rice leaf rolling. Up to now, 11 rice mutants with rolled leaves (rl) have been characterized, for which six recessive genes (rl1-rl6) were mapped on rice chromosomes 1, 4, 12, 3, or 7 by morphological markers (Kinoshita, 1984; Khush and Kinoshita, 1991; http://www.gramene.org/). rl7-rl10 were mapped to chromosomes 5 (rl7; Li et al., 1998; rl8, 542 kb; Shao et al., 2005a) and 9 (rl9, 42 kb; Yan et al., 2006; rl10, 194 kb; Luo et al., 2007) by molecular markers. Rl(t), an incomplete recessive rolled leaf mutant, was fine-mapped to a 137-kb region of the long arm of chromosome 2 (Shao et al., 2005b). In addition, recent studies showed that overexpression of OsAGO7 results in the upward curling of rice leaves (Shi et al., 2007).
In Arabidopsis thaliana, mutants with differently curved leaves have been extensively described, including hyponastic leaves1 (Lu and Fedoroff, 2000), curly leaf (Goodrich et al., 1997), and incurvata (Berna et al., 1999). Aside from the effects of hormones and environmental cues, the defective establishment of leaf polarity is one important factor that results in rolled leaves (Byrne et al., 2000; Semiarti et al., 2001). Several adaxial-abaxial asymmetry-related genes have been characterized in Arabidopsis. Members of the class III homeodomain-leucine zipper (HD-ZIP III) family PHABULOSA (PHB), PHAVOLUTA (PHV) (McConnell and Barton, 1998; McConnell et al., 2001), and REVOLUTA (REV; Talbert et al., 1995; Otsuga et al., 2001) promote adaxial organ identity. On the other hand, KANADI (KAN; Kerstetter et al., 2001) and YABBY (YAB) family genes, including FILAMENTOUS FLOWER (Sawa et al., 1999a, 1999b), YAB2, and YAB3 (Eshed et al., 2001), are required to establish abaxial identity.
Other key regulators of leaf polarity include a group of functional homologs, including PHANTASTICA (PHAN) in Antirrhinum majus, ROUGH SHEATH2 in maize (Zea mays), and ASYMMETRIC LEAVES1 (AS1) in Arabidopsis. All are MYB domain–containing transcription factors (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Sun et al., 2002). ASYMMETRIC LEAVES2 (AS2), which encodes a protein containing a leucine-zipper motif associated with AS1, is important in promoting leaf adaxial fate (Iwakawa et al., 2002; Xu et al., 2002). The maize gene ROLLED LEAF1 (RLD1) encodes a HD-ZIP III family transcription factor and is involved in adaxial specification (Juarez et al., 2004b). Recent studies showed that the maize MILKWEED POD1 (MWP1) gene encodes a KANADI protein that is involved in the abaxial-adaxial patterning of leaves (Candela et al., 2008).
Posttranscriptional and posttranslational regulation are essential for proper leaf patterning (Xu et al., 2007). Transcripts of PHB, PHV, and REV contain complementary sites for microRNAs miR165 and miR166, which can direct their cleavage in vitro (Reinhart et al., 2002; Tang et al., 2003). The maize gene RLD1 has an miRNA166 complementarity site; its defective mutant, rld1, showed adaxialized or partially reversed axis patterns (Nelson et al., 2002). Additionally, ASYMMETRIC LEAVES ENHANCER3 (AE3) encodes the 26S proteasome subunit RPN8a and is involved in the AS1/AS2 pathway that regulates the development of leaf polarity (Huang et al., 2006).
The adaxial-abaxial pattern of the monocot leaf is different from that of dicot plants. In dicots, the internal mesophyll cells are polarized; the closely packed palisade cells are adjacent to the adaxial epidermis, and loosely packed spongy cells border the abaxial epidermis (McConnell and Barton, 1998). In monocots such as maize, the leaf is divided into five tissue layers along the adaxial-abaxial axis (Freeling and Lane, 1993). Its polarity characteristics include the macrohair and bulliform cells in the adaxial epidermis and ligule tissue on the adaxial surface of the blade sheath boundary (Nelson et al., 2002). Additionally, the middle vein on the maize leaf contains polarized xylem and phloem. In contrast with those of dicots, the mesophyll cells of some grasses, including maize and rice, have a uniform configuration without polarization (there is only spongy parenchyma in leaf), termed isobilateral mesophyll (Fahn, 1990); the development and regulation of polarity in isobilateral leaves remain to be elucidated. In addition, there is a distinct difference between the C3 plant rice and the C4 plant maize with regard to sclerenchymatous tissues. In rice, the sclerenchymatous tissues display excessive differentiation with a characteristic distribution, and these tissues are ubiquitous around the vascular tissues of both adaxial and abaxial epidermis. The hypodermal sclerenchyma of maize is rare, and, when found, it is predominantly associated with the abaxial epidermis (Nelson et al., 2002).
In the rice lamina, programmed cell death (PCD) occurs throughout the formation of sclerenchymatous cells, including tracheid and vessel elements in xylem and sclerenchyma tissues surrounding the vascular bundle (Lu et al., 1982). Along with the formation of conducting or mechanical tissues, the tracheary elements (TEs) or sclerenchyma tissue undergo protoplast degradation and are lignified with thickened secondary cell walls, forming the specialized cells and ultimately the nonliving cells responsible for water transport and support (Fahn, 1990). On the cellular level, the DNases, RNases, and proteases accumulate in the vacuoles, regulated by specific signals. The vacuoles collapse, and the cell contents of the protoplasts are degraded to form continuous canals (Fukuda, 2000).
The sclerenchymatous cells are lignified dead cells with thickened secondary cell walls. PCD is involved in the transdifferentiation of mesophyll cells to sclerenchymatous cells. This differentiation consists of three stages: dedifferentiation (stage I), conversion of mesophyll cells to TE precursor cells (procambium cells and immature xylem cells) (stage II), and TE-specific secondary wall thickening followed by lignification and cell death–related events (stage III) (Fukuda, 2000). Multiple genes are involved in this process. Expression of tracheary element dehydrogenase2 (TED2), a vascular cell-specific gene, increases in stage II and marks the initiation of stage III (Demura and Fukuda, 1993, 1994). Cinnamate 4-hydroxylase (C4H), which is transcribed in stages I and III, is associated with the lignification of secondary cell walls (Ye, 1996). Cys protease (CP), which is transcribed at stage III, degrades the cell contents and is critical for TE-PCD (Ye and Varner, 1996). Protease inhibitor (PI) associates with CP to function in the TE-PCD process (Solomon et al., 1999). Phe ammonia-lyase (PAL) and 4-coumarate-CoA ligase (4CL) are involved in secondary wall thickening during TE-PCD stage III (Lin and Northcote, 1990; Voo et al., 1995). However, little is known about the roles of PCD in the formation of TEs and sclerenchymatous cells during rice leaf development.
Here, we report the isolation and functional characterization of a rice gene, SHALLOT-LIKE1 (SLL1), that regulates rice leaf rolling. SLL1 deficiency leads to the flawed formation of sclerenchymatous cells on the abaxial side of the leaf and results in extremely inwardly rolled leaves. SLL1 is crucial in polarity formation and helps to direct the development of the leaf abaxial cell layer. In addition, it is involved in the transdifferentiation of mesophyll cells to sclerenchymatous cells through the modulation of PCD, highlighting a possible mechanism underlying leaf rolling in monocot plants.
RESULTS
sll1, a Rice Mutant with Shallot-Like Leaves, Displays Abnormal Sclerenchymatous Cell Development in the Abaxial Cell Layers, Altered Mesophyll Cell Distribution, Increased Amounts of Chlorophyll, and Enhanced Photosynthesis
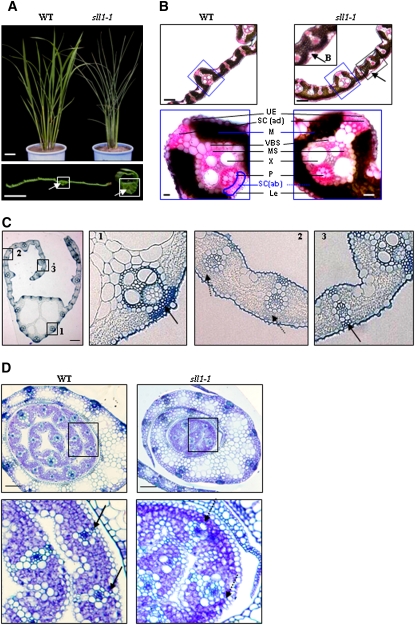
To investigate the molecular mechanisms of rice leaf rolling, a rice mutant population (Oryza sativa L. ssp. japonica variety Nipponbare) generated by ethyl methanesulphonate mutagenesis was screened. Two allelic mutants with extremely incurved leaves were identified, designated as sll1-1 and sll1-2 (Figure 1A). Phenotypic observation showed that sll1 mutant plants had narrow, extremely rolled and dark-green leaves, which appeared during the seedling stage. These leaves became more evident during plant growth. Observation of the cross section revealed enlarged clear cells in the midrib of sll1 plants (Figure 1A, bottom panel).
Figure 1.
sll1 Has Extremely Incurved Leaves, with Deficiency of Sclerenchymatous Cells at the Abaxial Side.
(A) Morphology of wild-type and sll1-1 plants and leaves. Mature wild-type and sll1-1 mutant plants were observed. The clear cells of the midrib (white box) are highlighted with arrows. Bars = 5 cm (top panel) or 2 mm (bottom panel).
(B) sll1-1 leaf blades display altered cellular organization. Bulliform cells on the abaxial surfaces of sll1-1 leaves are highlighted (top panel, arrows). The altered differentiation and distribution of mesophyll cells are highlighted (blue box) in the bottom panel. LE, lower epidermis; SC (ab), abaxial sclerenchyma; B, bulliform cells; P, phloem; X, xylem; MS, mestome sheath; VBS, vascular bundle sheath; M, mesophyll cells; SC (ad), adaxial sclerenchyma; UE, upper epidermis. Bars = 100 μm (top panel) or 20 μm (bottom panel). The broken line indicates the defective sclerenchyma at the abaxial epidermis. Sections (∼20 μm) of rice leaf blade were stained in Ruthenium red solution.
(C) The defective sclerenchymatous cells on the abaxial side of sll1-1 leaves. The defect is mainly in the small veins where the curl occurs (2; broken lines). Cellular organization at the midrib region (1) and the margin of the blade (3) in sll1-1 is normal. Bar = 50 μm.
(D) Transverse section of young seedlings at 5 d after germination. Differentiated sclerenchymatous cells on the wild-type abaxial epidermis are highlighted (left panel, arrows) but still lack a thickened secondary wall. These are deficient in sll1-1 (broken arrows). Bars = 100 μm.
In contrast with the wild type, mature sll1 leaves display altered mesophyll cell differentiation and distribution. sll1 does not form abaxial sclerenchymatous cells in the small veins of lateral region (Figure 1B), where the leaf starts to roll; however, the midrib region and the margin of the blade demonstrate cellular organization similar to the wild type (Figure 1C), indicating the modulated differentiation of sclerenchymatous cells in sll1. Bulliform cells, which are normally found in the adaxial epidermis, were found in abaxial surfaces of sll1-1 leaves (Figure 1B). Furthermore, the differences in abaxial sclerenchymatous specification can be observed in young leaves with curved leaf sheathes (plastochron 5), appearing after the mesophyll cells begin to differentiate. The mesophyll cells in abaxial layers do not differentiate to become sclerenchymatous cells in sll1-1 (Figure 1D).
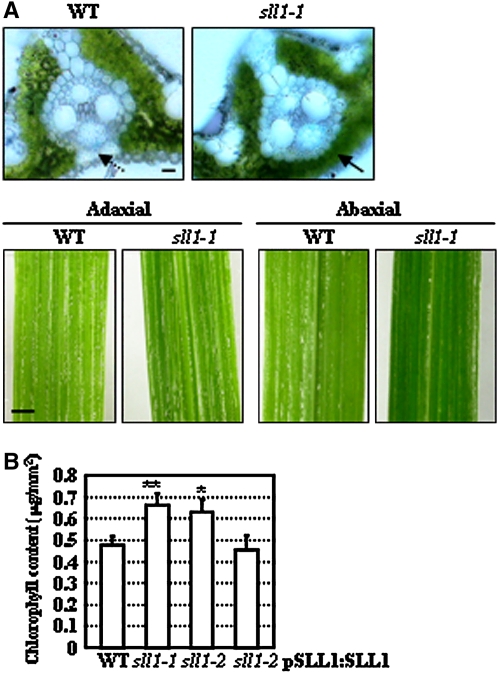
In wild-type leaf blades, the mesophyll cells on both adaxial and abaxial sides are interrupted by sclerenchymatous cells connecting vascular bundles and epidermis, while those on the abaxial side of sll1-1 leaves were replaced by mesophyll cells (Figure 2A, top panel). Leaves were a deeper green on the abaxial side (Figure 2A, bottom panel), indicating increased numbers of mesophyll cells. We also measured the chlorophyll in wild-type, sll11-1, sll1-2, or transgenic sll1-2 plants with complemented expression of SLL1. Measurements were performed on the same area of the plant (at the middle part of the second leaf from the top of the plant) at same growth stage. Chlorophyll measurements indicated that the increased numbers of mesophyll cells resulted in a corresponding increase in the amount of chlorophyll in sll1-1 and sll1-2 plants, and the complemented expression of SLL1 in sll1-2 plants restores normal chlorophyll levels (Figure 2B).
Figure 2.
SLL1 Deficiency Results in Increased Chlorophyll Content.
(A) Distribution of mesophyll cells on the abaxial side of wild-type and sll1-1 leaves. The broader distribution of mesophyll cells on the abaxial side of sll1-1 leaves is highlighted (arrows), rendering the deeper green color of sll1-1 abaxial epidermis. Sections (∼20 μm) of rice leaf blades were stained in aniline blue solution. Bars = 20 μm (top panel) or 2 mm (bottom panel).
(B) Chlorophyll levels in wild-type, sll1-1, sll1-2, and sll1-2 plants with complemented expression of SLL1. To measure chlorophyll levels, we collected samples from an area from the middle part of the second leaf from the top of wild-type, sll11-1, sll1-2, or transgenic sll1-2 plants with complemented expression of SLL1 (at the same growth stage). Experiments are biological replicates. Error bar represents sd (n > 30 in each measurement), and statistical analysis by heteroscedastic t test indicates the significant differences (* P < 0.05; ** P < 0.01).
Measurement of the quantum yield of photosystem II (PSII) and the maximal quantum yield of PSII (Fv/Fm) showed that both were slightly increased in sll1-1 or sll1-2 (Table 1), indicating increased photosynthesis due to the increased numbers of mesophyll cells and elevated chlorophyll content of the leaves.
Table 1.
The Photosynthetic Efficiency of Wild-Type, sll1-1, and sll1-2 Plants
| Wild Type | sll1-1 | sll1-2 | |
|---|---|---|---|
| Yield | 0.739 ± 0.017 | 0.761 ± 0.02** | 0.747 ± 0.018 |
| Fv/Fm | 0.799 ± 0.017 | 0.817 ± 0.006* | 0.808 ± 0.019 |
The quantum yield of PSII and maximal quantum yield of PSII (Fv/Fm) were measured and statistically analyzed. For measurements, the middle parts of the 8th to 10th rice leaves (at the same developmental stage) were used. Data are presented as means ± se (n > 10). Heteroscedastic t test indicates the significant differences in sll1-1 (* P < 0.05; ** P < 0.01) compared with the wild type.
SLL1 Encodes a SHAQKYF Class MYB Transcription Factor
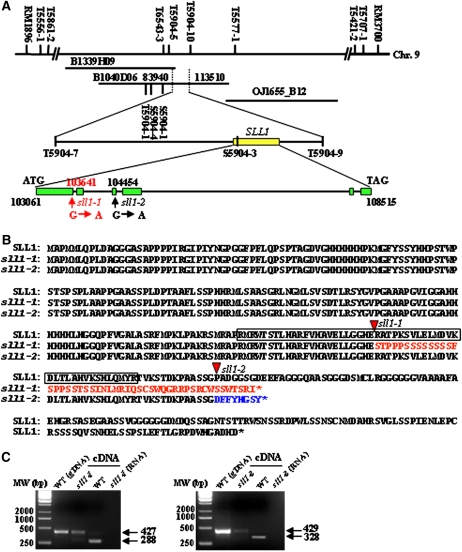
Genetic analysis of reciprocal crosses between the sll1-1, sll1-2, and wild-type plants revealed that the abnormal character of sll1 was controlled by a single recessive gene. To isolate the relevant mutant gene, SLL1 was mapped to the long arm of rice chromosome 9 between markers RM1896 and RM3700. This conclusion is based on analysis of the F2 population from a cross between sll1-2 and Nanjing 6 (Figure 3A). A large F2 mapping population was then generated, allowing the fine-mapping of SLL1 to a 29.57-kb region, using the sequence tagged site and simple sequence repeat markers (Figure 3A; see Supplemental Table 1 online). Three annotated candidate genes, encoding a hypothetical protein, an En/Spm-like transposon, and a transcription factor containing a MYB-like domain, respectively, were located in this region (The Institute for Genomic Research; http://rice.plantbiology.msu.edu/). Further amplification of the relevant DNA fragments and sequence comparison revealed differences in sll1-1 and sll1-2 alleles in the gene encoding the transcription factor containing the MYB-like domain. The sll1 allele carried a single base substitution (G to A) in the intron splicing site of both sll1-1 (the first intron) and sll1-2 (the third intron), resulting in altered mRNA splicing and premature termination of the encoded protein (Figure 3B). To verify this, primers located in the two open reading frames (ORFs) flanking the mutation site were used to amplify the relevant fragments. The longer mRNA molecules were indeed amplified in sll1-1 and sll1-2 (Figure 3C), confirming the altered splicing of sll1 transcripts.
Figure 3.
Map-Based Cloning of SLL1, Which Encodes an MYB Transcription Factor.
(A) Mapping of SLL1. SLL1 was mapped primarily to the long arm of rice chromosome 9 between markers RM1896 and RM3700 and then to a 29.57-kb region in a large F2 mapping population. Amplification of relevant DNA fragments and sequence comparison revealed that the sll1 alleles resulted from a single base substitution (G to A) in the intron-splicing site of both sll1-1 (the first intron) and sll1-2 (the third intron).
(B) SLL1 encodes an MYB transcription factor. Nucleotide mutations result in altered splicing of mRNA and premature termination of the protein. The boxed region in the amino acid sequence represents the MYB domain. The red triangles demarcate the start point of nucleotide mutations and altered transcription. The red or blue markings indicate the mutated amino acid sequences of sll1-1 (red) or sll1-2 (blue).
(C) Altered splicing of sll1-1 (left panel) and sll1-2 (right panel) transcripts. mRNA molecules were amplified with primers located in two ORFs flanking the mutation site.
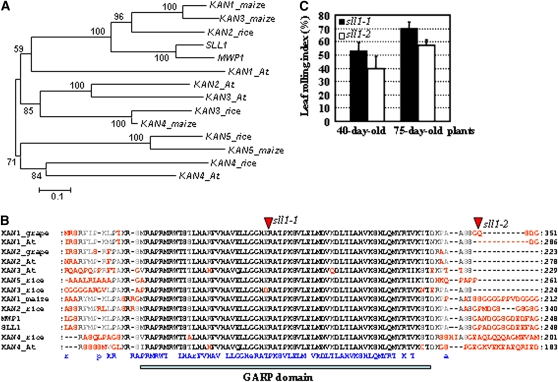
Comparison with the corresponding genomic sequence revealed that the SLL1 gene consists of six exons and five introns. The gene encodes a 377–amino acid MYB family transcription factor (Figure 3B). Homologous analysis showed that SLL1 shares high similarity with the KAN family members in Arabidopsis (Table 2). The Arabidopsis KAN1 is involved in the polarity regulation of lateral organs, and three other KAN genes (KAN2, KAN3, and KAN4) also redundantly specify abaxial fate (Eshed et al., 1999, 2001; Kerstetter et al., 2001; Hawker and Bowman 2004). This finding suggested the possible effects of SLL1 in polarity identity. Neighbor-joining phylogenetic tree analysis showed that among the KANADI members in rice, maize, and Arabidopsis, SLL1 is closest to maize MWP1 (MILKWEED POD1, which has been shown to be involved in adaxial/abaxial patterning) (Candela et al., 2008), two other KANADI members (KAN1 and KAN3), and rice KANADI member KAN2 (Figure 4A). SLL1 is closest to At KAN1 among the Arabidopsis KANADI members (Figure 4A, Table 2), and structural organization analysis showed that all KANADI proteins share a highly conserved GARP domain (Figure 4B). The GARP domain is a plant-specific DNA binding domain of the helix-loop-helix superfamily that is distantly related to the MYB domain (GARP is an acronym for the founding members of the family: maize Golden 2, ARR [Arabidopsis response regulators], and Psr1 [phosphorous stress response1 from Chlamydomonas]). Studies have shown that the GARP domain is crucial for regulating the transcription of downstream genes (Kerstetter et al., 2001). The shared identity for this GARP domain in the KANADI family is higher than 85% (the overall protein identity is lower than 43%; Table 2).
Table 2.
Amino Acid Identities (%) among Arabidopsis and Rice KANADI Proteins and Maize MWP1
| SLL1 | MWP1 | KAN2_rice | KAN3_rice | KAN4_rice | KAN1_At | KAN2_At | KAN3_At | KAN4_At | |
|---|---|---|---|---|---|---|---|---|---|
| SLL1 | – | 58 | 42 | 26 | 19 | 23 | 24 | 20 | 17 |
| MWP1 | 100 | – | 49 | 27 | 18 | 24 | 25 | 19 | 16 |
| KAN2_rice | 96 | 96 | – | 28 | 18 | 22 | 22 | 18 | 16 |
| KAN3_rice | 93 | 93 | 90 | – | 20 | 22 | 26 | 25 | 20 |
| KAN4_rice | 93 | 93 | 92 | 89 | – | 18 | 19 | 16 | 27 |
| KAN1_At | 96 | 96 | 96 | 92 | 92 | – | 29 | 24 | 19 |
| KAN2_At | 96 | 96 | 93 | 92 | 93 | 96 | – | 41 | 18 |
| KAN3_At | 89 | 89 | 85 | 85 | 89 | 85 | 89 | – | 19 |
| KAN4_At | 92 | 92 | 89 | 89 | 90 | 90 | 90 | 89 | – |
The upper numbers (above the dashes) indicate identities among the entire proteins and the lower numbers indicate the identities among GARP domains.
Figure 4.
SLL1 Has a Close Relationship with KANADI Family Members.
(A) Phylogenetic relationships among the KANADI family members. KANADI proteins from Arabidopsis (KAN1, 2, 3, and 4), rice (SLL1, KAN2, 3, 4, and 5), and Z. mays (MWP1, KAN1, 3, 4, and 5) are analyzed. The numbers at each node represent the bootstrap support (percentage). The scale bar indicates genetic distance based on branch length.
(B) Alignment analysis of KANADI proteins. This analysis revealed the conservation of a GARP domain among the KANADI proteins. The conserved amino acid residues are highlighted in the bottom line in blue.
(C) LRI of sll1 mutant plants. sll1-1 has a more evident LRI than sll1-2 at different developmental stages (40 or 75 d). Error bars represent sd (n > 30).
Although both mutant alleles show rolled leaves, leaf rolling index (LRI) analysis showed that sll1-1 has more severe rolling (Figure 4C), which is consistent with the fact that mutation of sll1-1 affects the GARP domain, while that of sll1-2 does not (Figure 4B). These phenotypic differences indicate the importance of the GARP domain to the function of SLL1, probably because of its central role regulating gene transcription.
The Expression Pattern of SLL1
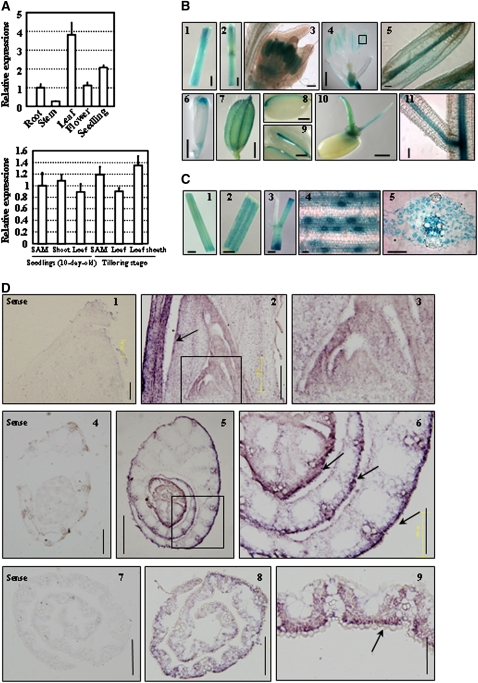
Quantitative real-time RT-PCR (qRT-PCR) analysis revealed the expression of SLL1 in various tissues, including roots, stems, leaves, flowers, and seedlings, with relatively high expression in leaf and seedlings (Figure 5A, top panel). With an emphasis on the leaf, analysis by qRT-PCR showed that SLL1 is ubiquitously expressed in shoot apical meristem (SAM), leaf blade, or leaf sheath at different developmental stages (Figure 5A, bottom panel).
Figure 5.
SLL1 Expression Pattern Analysis.
(A) qRT-PCR analysis of SLL1 expression in various tissues. Expression of SLL1 in roots (from 10-d-old seedlings), stems, leaves (middle part of 4th leaf), flowers, and 10-d-old seedlings (whole plants including roots and shoots) (top panel) in the SAM, shoot (whole plant without roots), and the 4th leaf of 10-d-old seedlings and in the SAM, 10th leaf, and leaf sheath of plants at the tillering stage (bottom panel) are analyzed. For SAM samples, the precisely defined intact SAM and young leaf primordia (without leaf sheath) were collected. Experiments are biological replicates.
(B) Promoter-reporter gene (GUS) fusion studies on the expression of SLL1. Expression of SLL1 in stem (1 and 2), anthers (3 to 5), pistil tip (6), seed vascular tissues (7 to 9), coleoptile and embryonic roots of germinating seeds (10), and root vascular tissues (11) is analyzed. Bars = 1 mm (4 and 6), 2 mm (1, 2, and 7 to 10), or 100 μm (3, 5, and 11).
(C) Promoter-GUS fusion studies of expression of SLL1 in leaf. Expression of SLL1 in young leaf (middle part of 2nd leaf of 10-d-old seedling; 1), mature leaf and leaf vein (2), leaf sheath (3), guard cells (4), and vascular tissues (5) is analyzed. Bars = 1 mm (1), 5 mm (2 and 3), or 50 μm (4 and 5).
(D) In situ hybridization analysis of expression of SLL1 in the abaxial cell layer and in tracheal elements throughout leaf development. The SLL1 signal was detected at the shoot apical meristem and leaf primordia (2; enlarged in 3, arrows). Cross section of the shoot apex showed higher expression of SLL1 in the abaxial cell layer, including the epidermis and vascular system of early leaf blades (5; enlarged in 6, arrows). In the mature leaf, there is an accumulation of SLL1 in the region of the developing abaxial sclerenchymatous cells and tracheal elements (8 and 9, arrow). Arrows highlight the more intense accumulation of SLL1 at the abaxial cell layer. The sense probe was hybridized and used as control (1, 4, and 7). Bars = 200 μm (1 to 4 and 6 to 8), 505 μm (5), or 100 μm (9).
To assess the expression pattern comprehensively, β-glucuronidase (GUS) activity was examined histochemically in transgenic plants carrying an SLL1 promoter-GUS reporter gene. Results showed that SLL1 was transcribed in stem (Figure 5B, panels 1 and 2), anthers of young or mature flowers (Figure 5B, panels 3 to 5), pistil tip (Figure 5B, panel 6), glume (Figure 5B, panel 7), vascular tissues of mature seeds (Figure 5B, panels 8 and 9), coleoptile and embryonic root of germinating seedlings (Figure 5B, panel 10), and root vascular tissues (Figure 5B, panel 11). In addition, SLL1 is highly transcribed in leaf veins and leaf sheath (Figure 5C, panels 1 to 3), guard cells, and tracheal elements (Figure 5C, panels 4 and 5).
We further examined the spatial and temporal localization of SLL1 during leaf development by in situ hybridization analysis. The mRNA expression of SLL1 was detected throughout the young leaf primordium (plastochrons 1 to 3) and was more intense in abaxial cell layer through leaf development (plastochrons 4 and 5) (Figure 5D, panel 2). However, the SLL1 transcript did not demonstrate apical/basal polarity and did not accumulate at the apex of the meristem (Figure 5D, panel 3). Cross-section analysis of the shoot apex showed that SLL1 was more highly expressed in the abaxial cell layer, including the epidermis and vasculature of the early leaf blade (Figure 5D, panels 5 and 6). In the mature leaf, SLL1 mainly accumulated at the abaxial epidermis, abaxial mesophyll cells, and vasculature (Figure 5D, panels 8 and 9). In contrast with the sense probe (Figure 5D, panels 4 and 7), SLL1 was transcribed at a relatively low level throughout other leaf positions including the adaxial epidermis (Figure 5D, panels 5 and 6).
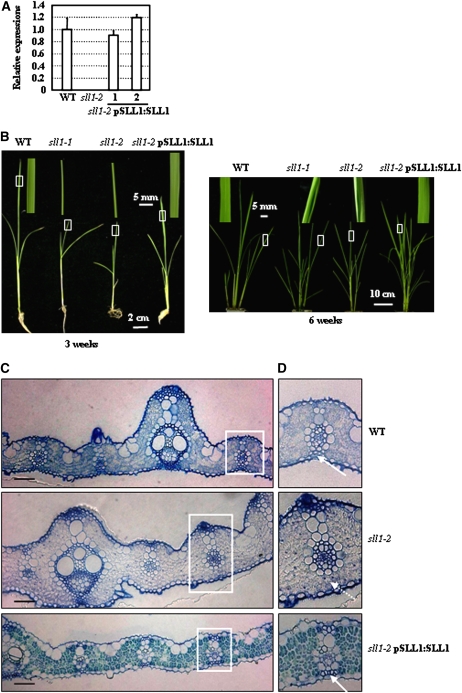
Complemented Expression of SLL1 Rescued the Rolled Leaves of sll1
The physiological effect of SLL1 in leaf rolling was further confirmed by genetic complementation analysis. A construct carrying the 7.8-kb genomic fragment containing the entire promoter region and SLL1 ORF (excised from BAC B1040D06 and subcloned into vector pCAMBIA1300) was transformed into sll1-2 plants (sll1-1 plants are almost sterile). Independent transgenic lines were identified through qRT-PCR analysis of SLL1 expression using primers matching the sequence of normal transcript after correct splicing. The transcript in the sll1-2 mutant could not be amplified with these primers due to the altered splicing (Figure 6A). Phenotypic observation confirmed that complementary SLL1 expression of the normal splicing product rescued leaf rolling, with restoration of normal leaf shape at different developmental stages (3 or 6 weeks; Figure 6B). In addition, observations of leaf cross sections indicated the normal formation of sclerenchymatous cells at the abaxial epidermis (Figures 6C and 6D).
Figure 6.
SLL1 Functionally Complements the Defective Sclerenchymatous Cells on the Abaxial Side of sll1-2.
(A) qRT-PCR analysis on the expression of SLL1. Normal splicing of SLL1 in wild-type or transgenic sll1-2 plants with complemented expression of SLL1 following expression of the SLL1 genomic fragment is detected, but cannot be detected in sll1-2. Experiments are biological replicates.
(B) Morphology of leaves of wild-type, sll1-1, sll1-2, and transgenic sll1-2 plants with complemented expression of SLL1. Leaf shape of these plants at different developmental stages (3 or 6 weeks, respectively) is observed. The squared leaf regions (white box) are enlarged and shown above the plants to highlight the leaf shape.
(C) Recovered formation of sclerenchymatous cells in sll1-2 with complemented expression of SLL1. Cross-sectional analysis is performed using the 5th leaf from 6-week-old seedlings, and formation of sclerenchyma at the abaxial epidermis of sll1-2 after complemented expression of SLL1 is observed. Bars = 50 μm.
(D) Enlarged squared regions from (C). Arrows highlight the positions of sclerenchymatous cells on the abaxial side, lacking in sll1-2.
SLL1 Modulates Leaf Rolling by Regulating Abaxial Mesophyll Cell PCD
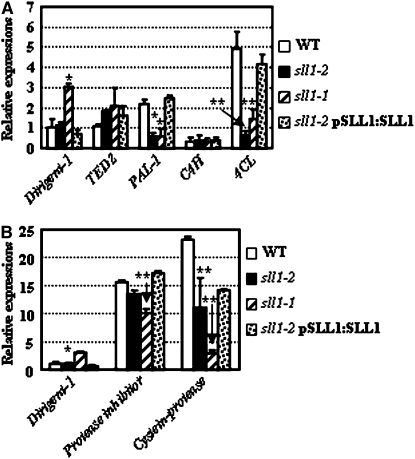
Defective formation of sclerenchymatous cells in the curve region of sll1 leaves (Figure 1C) suggested that loss of mechanical strength in abaxial sclerenchymatous cells leads to inward rolling and that SLL1 is required for the normal formation of sclerenchymatous cells. Previous studies have shown the involvement of PCD in the differentiation of sclerenchymatous cells and that multiple genes are involved in this process. We performed qRT-PCR analysis to determine the expression of rice genes that share high homology with TE-PCD marker genes in Zinnia or other species. These genes include TED2, C4H, CP, PI, PAL, 4CL, and Dirigent-1 (involved in lignin assembly rather than as a marker gene of the TE-PCD process, as determined by the finding that proteins harboring arrays of monolignol radical binding sites are involved in macromolecular lignin assembly; Davin and Lewis, 2005). The expression of CP, PI, PAL, and 4CL genes were suppressed in sll1-2, while TED2, C4H, and Dirigent-1 were not obviously altered (Figure 7). The only known marker gene of stage II is TED2, which did not show obvious changes in the presence of SLL1 deficiency. Several genes marking stage III, such as CP, PI, PAL and 4CL, are evidently altered. This indicates that SLL1 mainly modulates the stage III events of PCD during the transdifferentiation of mesophyll cells to sclerenchymatous cells. The absence of an alteration in Dirigent-1 expression further confirmed that SLL1 specifically regulated the PCD in abaxial sclerenchyma development.
Figure 7.
Altered Expression of Genes Encoding Proteins Related to TE-PCD in sll1-1 and sll1-2.
qRT-PCR analysis of rice gene expression. These genes include (A) TED2, C4H, PAL, dirigent-1 (macromolecular lignin assembly protein), and 4CL; and (B) dirigent-1, PI, and CP in sll1-1, sll1-2, or transgenic sll1-2 plants with complemented expression of SLL1. The experiments were repeated three times using biological replicates; the data were statistically analyzed and presented as means ± sd. Heteroscedastic t test indicated the significant differences (* P < 0.05; ** P < 0.01).
sll1 Leaves Showed Adaxialized Trends in Abaxial Epidermis, while Enhanced SLL1 Expression Suppresses the Adaxial Characteristics and Stimulates Abaxial Leaf Features
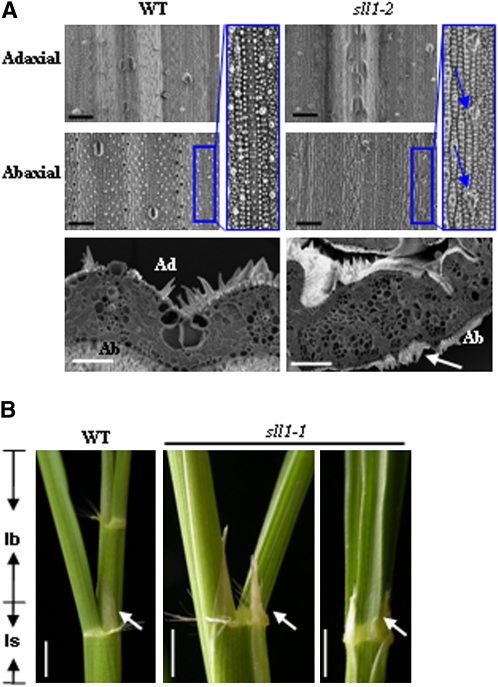
As mentioned above, a distinguishing characteristic of the monocot leaf is that microhairs or ligules are present only at the adaxial surface. In sll1-1, the microhairs can be observed on both epidermal surfaces of some plants (Figure 8A), and the ligules exist on the abaxial side of the joint (Figure 8B). This suggests an adaxialized trend in the abaxial epidermis of sll1-1 and indicates the involvement of SLL1 in the development of polarity throughout the leaf abaxial epidermis.
Figure 8.
Sll1 Alters Adaxial-Abaxial Pattern Formation.
(A) Altered epidermal features of sll1-2 leaves. Scanning electron microscopy analysis showed that the microhairs can be observed at the abaxial epidermis of sll1-2, highlighted in the bottom panel (arrows) or enlarged (blue box, arrows). Bars = 50 μm.
(B) The ectopic ligules in sll1-1 mutants. The ligules, which are normally found on the adaxial side of wild-type leaves (left), can be observed on the abaxial side at the junction of the leaf blade (lb) and leaf sheath (ls) of sll1-1 mutants. Arrows indicate the positions of ligules. Bars = 1 cm.
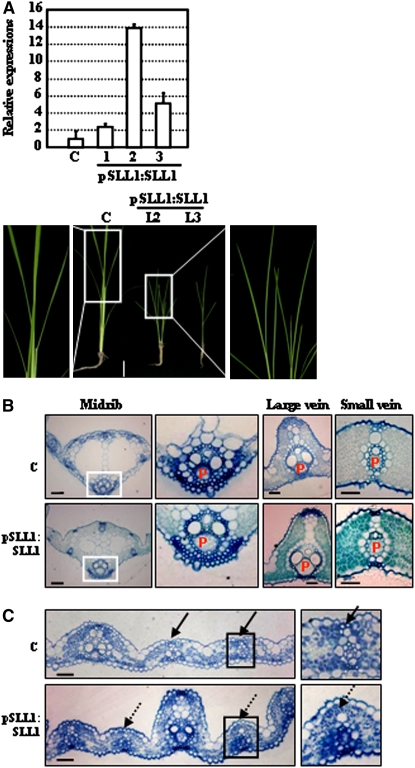
Furthermore, transgenic rice lines overexpressing SLL1 genomic DNA (driven by its own native promoter) were generated on a wild-type background (Figure 9A, top panel). Observations of leaf growth showed that SLL1 overexpression resulted in dwarf plants with twisted and abnormal inner rolled leaves (Figure 9A, bottom panel). We also observed enlarged phloem in the midrib as well as large and small veins (Figure 9B, Table 3), revealing the enhanced abaxial features of leaves following SLL1 overexpression.
Figure 9.
Enhanced Expression of SLL1 Stimulated Phloem Development in the Abaxial Cell Layer and Suppressed the Formation of Sclerenchymatous Cells in the Adaxial Cell Layer.
(A) qRT-PCR analysis confirmed the enhanced expression of SLL1 (top panel). The transgenic plants (3-week-old seedlings) revealed a phenotype with upper-rolled leaves (bottom panel, bar = 5 cm). The enlarged squared regions highlight leaf rolling compared with control. Experiments are biological replicates.
(B) Enhanced development of phloem of SLL1-overexpressing plants. Cross-section analyses were performed to analyze the development of phloem in the midrib (the squared regions are enlarged) and the large and small veins of wild-type and SLL1-overexpressing plants. P, phloem. Bars = 50 μm.
(C) Defective formation of sclerenchymatous cells in the adaxial cell layer of SLL1-overexpressing plants. Broken arrows indicate the regions where sclerenchymatous cells would have formed in normal leaf (bottom panel). Bars = 100 μm.
Table 3.
Measurement of the Vascular Elements in Wild-Type and SLL1-Overexpressing Plants
| Phloem (μm2) | Phloem/Xylem | Phloem/Vascular Bundle | |
|---|---|---|---|
| Wild Type | 495 ± 148 | 1.25 ± 0.24 | 0.18 ± 0.02 |
| pSLL1:SLL1 | 1700 ± 477** | 2.18 ± 0.39** | 0.39 ± 0.03** |
The areas of phloem, xylem, and whole vascular bundles in large veins were measured, as well as the ratios of phloem/xylem and phloem/vascular bundle. The large veins for measurement were selected from 10 rolled leaves of three independent transgenic lines. The data are statistically analyzed and presented as means ± sd (n > 30). Heteroscedastic t test indicated the significant differences (** P < 0.01) compared with the wild type.
Our observations of the bulliform cells formed on the adaxial epidermis revealed the loss of the normal fan-shaped anatomical structure. The adaxial sclerenchymatous cells proximal to the vascular bundle were defective in some transgenic lines (Figure 9C), indicating the suppression of adaxial characteristics and demonstrating that SLL1 is important in controlling the specification of abaxial epidermis.
SLL1 Controls Multiple Developmental Processes
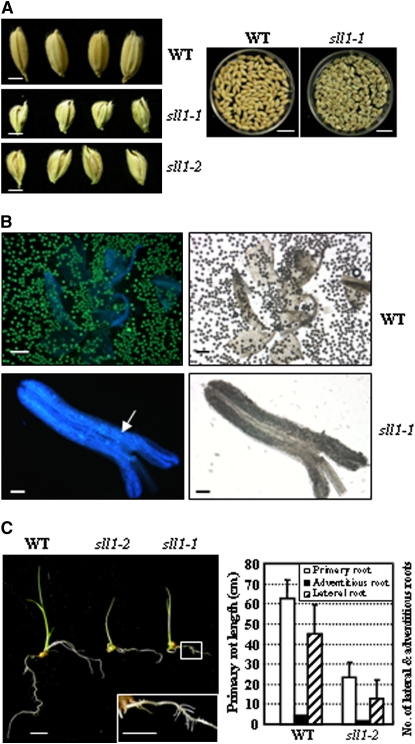
SLL1 affected developmental processes in multiple developmental stages, in accordance with its expression patterns. Aside from altered leaf morphology, other tissues, including seeds, anthers, and roots, displayed abnormal development in sll1 mutants. In seeds, both the palea and outer glume had a defective shape and failed to close; the seeds were smaller and exposed (Figure 10A). With regard to anther development, negligible pollen was generated and the generated pollen fell in a scattered fashion. Extremely few pollen grains were active when analyzed by fluorescein diacetate (FDA) staining (Figure 10B). In addition, abnormal root growth was observed in SLL1-deficient plants. In comparison with the wild type, the lengths of primary roots and lateral roots were shortened and the number of adventitious roots was significantly decreased (Figure 10C).
Figure 10.
SLL1 Affected Multiple Developmental Processes in Rice.
(A) Altered seed development and maturation of sll1. Bars = 2 mm (left panel) or 1.5 cm (right panel).
(B) Reduced pollen grains of sll1-1 mutant. Bars = 200 μm (wild type) or 100 μm (sll1-1).
(C) Abnormal root development of sll1 seedlings. Phenotypic observation (left panel, bars = 1 cm) and measurement analysis (right panel, n > 30 in each experiment, and experiments are biological replicates) showed that sll1 had shortened primary roots and reduced numbers of lateral and adventitious roots. The boxed region was enlarged to highlight the abnormal roots of sll1-1 seedlings (left panel, bars = 1 cm).
DISCUSSION
Sclerenchymatous tissues present different characteristic distributions in C3 and C4 plants. This study expands our knowledge of rice leaf development, especially the roles of leaf sclerenchymatous cells. Although the rice plant has an equifacial leaf, our results support independent regulation of differentiation in the adaxial and abaxial mesophyll cell layers. This modulates the shape of rice leaves as well as photosynthesis efficiency by regulating the development of polarity.
SLL1 Arrests Normal Sclerenchymatous Cell Formation and Controls Leaf Rolling
Multiple factors are involved in the control of leaf form, including axis-determining leaf genes (Eshed et al., 2001; Bowman et al., 2002), miRNAs and phytohormones (Juarez et al., 2004a; Kinder and Martienssen, 2004), as well as factors controlling the establishment of leaf polarity. The form of the dicot leaf is determined strongly by the mesophyll cell type; altered polarity in spongy tissue or palisade tissues will result in leaf rolling. In the monocot leaves, alteration in the distribution of leaf polarity characteristics, such as ligules and epidermal hairs (microhairs), does not lead to leaf rolling. Indeed, the cell types on the adaxial and abaxial epidermal surfaces are very similar in some grasses (but not in dicots). Although the sll1 mutants showed altered leaf polarity characteristics in the abaxial epidermis (presence of bulliform cells in the abaxial epidermis [Figure 1B] and microhairs at the blade sheath boundary of the abaxial epidermis [Figure 6A]), this change in epidermal markers of leaf polarity is not observed in all rolled leaf mutants. By contrast, because sclerenchymatous cells contribute continuous mechanical strength to maintain normal leaf shape, the lack of sclerenchymatous cells on the abaxial surface in regions of leaf curving in sll1 mutants indicates the critical role of sclerenchyma in controlling leaf rolling in monocots and the role of SLL1 in schlerenchyma formation that influences leaf rolling.
In wild-type rice plants, the specification of mesophyll cells occurs during the early stages of leaf development, when the young leaf remains wrapped inside the sheath in a crimped state. At plastochron 5, the differentiation of sclerenchymatous cells is visible; however, the mechanical strength is not yet developed, requiring the initiation of PCD and thickening of the secondary cell walls. These processes progress gradually, which is crucial for leaf flattening. The defective differentiation of sclerenchymatous cells in sll1 results in deficient mechanical strength and, hence, the rolled leaf. Throughout tissue differentiation, SLL1 accumulates in the abaxial cell layer (plastochrons 4 and 5). This pattern of expression is more distinct during the early stages of leaf development (plastochrons 1 to 3), consistent with the role of SLL1 in the differentiation of sclerenchymatous cells.
The structure, number, and distribution of sclerenchymatous cells differ between C3 and C4 plants, and sclerenchyma is critical to determining leaf form in both C3 and C4 plants. The maize leaf shows polarization of a few sclerenchymatous cells on the abaxial side, while rice has sclerenchymatous cells on both the abaxial and adaxial sides (corresponding to vascular positions). Notably, mutants with defective abaxial/adaxial patterning show varying sclerenchyma specification. We compared two abaxial specification defective mutants (rld1 and sll1). Maize RLD1 promotes adaxial identity, and the abaxial misexpression of which results in ectopic switch sclerenchyma cells extending from the abaxial side to the adaxial side along a subset of veins (due to the partially reversed D/V patterning). By contrast, sll1 forms sclerenchyma cells on the adaxial sides, but these plants exhibit defective abaxial sclerenchyma development, indicating defective abaxial specification or partially abaxialized development without reverse D/V patterning. These findings suggest the possibility of differential roles for sclerenchymatous cells in the control of leaf development in C3 compared with C4 plants.
When the effects of SLL1 are compared with the effects of the maize MWP1 (the closest KANADI protein to SLL1, involved in the abaxial-adaxial patterning of leaves), both sll1 and mwp1 show adaxialized sectors of cells in the sheath. However, it is not yet clear whether the phenotypic differences between sll1 and mwp1 are due to the difference between the influence of KANADI proteins on C3 and C4 leaf anatomies or because different KANADI proteins have different functions in determining abaxial polarity of different cell types in different species. There are five KANADI proteins in rice, and it is possible that other KANADI proteins contribute to the polarity determination of rice leaves, and SLL1 has a specialized role in schlerenchyma formation.
SLL1 Controls the Establishment of Rice Leaf Polarity
Arabidopsis AS1, which encodes an R2-R3 MYB transcription factor, is closely related to PHAN in Antirrhinum and ROUGH SHEATH2 in maize. This gene negatively regulates the expression of KNOX, a marker of founder cell recruitment into promordium (Smith et al., 1992; Jackson et al., 1994), thus modulating the establishment of leaf polarity (Waites et al., 1998; Byrne et al., 2000; Semiarti et al., 2001). However, our studies showed that the function of SLL1 is not similar to the functions of these factors in regulating founder cell identity. SLL1 contains a SHAQKYF class Myb-like DNA binding domain, and analysis of homologs showed that SLL1 was similar to Arabidopsis KANADI1 family members sharing a highly conserved GARP domains (Kerstetter et al., 2001). SLL1 was involved in the establishment of leaf polarity, and recently studies showed that maize mwp1 (the gene most homologous to SLL1) displayed adaxialized sectors in the sheath and proximal part of the leaf (Candela et al., 2008), confirming the conserved function of KANADI family members in monocots.
In dicots, KANADI and YABBY are two primary determinants in establishing abaxial identity (Sawa et al., 1999a, 1999b; Siegfried et al., 1999; Kerstetter et al., 2001). Eshed et al. (2004) proposed that, in Arabidopsis, initial asymmetric leaf development is regulated primarily by KANADI activity. KANADI translation modulates the polar expression of YAB, contributing to abaxial cell fate. By contrast, in monocots, Yamaguchi et al. (2004) observed that YAB genes may direct midrib cell division but are not sufficient for the control of dorsoventrality. YAB1 is expressed throughout the rice leaf and functions in gibberellic acid regulation (Dai et al., 2007), and YAB4 is expressed in developing vascular tissue and may regulate the development of vasculature in rice (Liu et al., 2007). Indeed, the unaltered expression of YAB in sll1 (see Supplemental Figure 1 online) supports the idea that the rice YAB genes have different functions to the dicot genes, underpinning a view that the molecular mechanisms for the establishment of leaf polarity differ between monocots and dicots.
Although rice YAB1 and YAB2 are both expressed in precursor cells of certain specific cell types, including abaxial sclerenchyma, which suggests that these genes may be involved in leaf cell differentiation (Toriba et al., 2007), there is no direct evidence that SLL1 regulates abaxial sclerenchyma formation and functions in the PCD process through sustaining the normal expression of rice YAB in abaxial sclerenchyma.
Although the PCD process during sclerenchyma differentiation is altered in sll1, we suggest that SLL1 functions primarily as a key factor in specifying polarity during adaxial/abaxial patterning. Abnormal sclerenchyma formation resulted in defective PCD, affecting adaxial/abaxial patterning; thus, SLL1 probably regulates PCD indirectly.
In addition, the monocot mesophyll cells show a uniform configuration without polarization, in contrast with the adaxial-palisade cells or abaxial-spongy mesophyll cells in dicots, indicating differential controls for the establishment of leaf polarity. In dicots, altered adaxial-abaxial pattern formation often results in rolled leaves; while the leaf rolling of sll1 is due to the defective differentiation of sclerenchymatous cells at the abaxial surface. However, the differentiation of sclerenchymatous cells on the adaxial surface is unaffected in sll1. These findings not only indicate the specific role of SLL1 in controlling abaxial identity and determining abaxial mesophyll cell fate in rice but also suggest that the mesophyll cells of monocot plants do not really have isobilateral mesophyll and, in fact, have polarity. Alternatively, the specification of mesophyll cells on either the adaxial or the abaxial side may rely on distinct signals.
SLL1 Modulates Photosynthesis by Regulating Cell Fate
The rolled leaf is an important agronomic trait, directly related to photosynthesis and crop yield. The erect, half-rolled rice leaf results in enhanced light capture capacity and increased chlorophyll contents and, hence, increased photosynthesic efficiency and yield. Previous studies in maize, sugarcane, and grain sorghum showed that the arrangement of leaf vascular bundles and mesophyll cells is closely related to photosynthesic efficiency. In maize, the developed vascular bundle contains more chlorophyll and compact mesophyll cells, resulting in a higher photosynthetic capacity. SLL1 deficiency suppresses the specification of sclerenchymatous cells in the abaxial mesophyll and may thus help to increase the chlorophyll content through increasing the proportion of mesophyll cells, consequently enhancing photosynthesis and providing a useful tool for rice breeders. Accordingly, the photosynthetic efficiency of sll1-2 mutants was slightly increased (∼3%), consistent with the increased amounts of chlorophyll. However, photosynthetic efficiency is affected by multiple factors, and other photosynthesis-related processes are suppressed in sll1-2, which may explain why the increased chlorophyll content does not result in a marked improvement of photosynthetic efficiency as compared with sll1.
Because leaf rolling may be subject to mechanical regulation (the maintenance of longitudinal rolling requires a continuous mechanical force), the modulation of sclerenchymatous cell formation will provide new insights into the regulation of leaf rolling. Our studies elucidate the control of leaf polarity establishment. This information may help to increase area yield by increasing the number of photosynthetic cells in the leaf and thus enhancing overall photosynthesis.
METHODS
Plant Material and Growth Conditions
The sll1 rice mutant was isolated from the population of Oryza sativa L. ssp. japonica cultivar Nipponbare treated with a 1% ethyl methanesulfonate solution. Nipponbare plants represent the wild type. Mutant sll1 was crossed with an indica rice variety, Nanjing6 (with flat leaf); the resultant F1 plants were selfed to produce the F2 seeds for constructing the F2 mapping population. Rice plants were cultivated in an experimental field at the China National Rice Research Institute under natural growing conditions; field management adhered to normal agricultural practice. For transgenic rice growth, rice seeds were germinated in sterilized water and then grown in pots in a phytotron with a 12-h-light (26°C) and 12-h-dark (18°C) cycle.
Map-Based Cloning of SLL1
SLL1 was mapped primarily with simple sequence repeat (http://www.gramene.org/microsat/ssr.html) and sequence tagged site (see Supplemental Table 1 online) markers using 152 F2 mutant plants. The SLL1 locus was further localized within a 29.57-kb region between markers T5904-7 and T5904-9 using 711 F2 mutant plants. New molecular markers (see Supplemental Table 1 online) were developed by comparing original or cleaved amplified polymorphic sequences between indica variety 9311 and Nipponbare, according to data published at http://www.ncbi.nlm.nih.gov.
To define molecular lesions, 29.57-kb genomic DNA from the sll1 and relevant wild-type variety (Nipponbare) were amplified by PCR. All PCR products were sequenced and the candidate gene was amplified from both sll1 and Nipponbare genomic DNAs using different primers (see Supplemental Table 2 online). Obtained sequences were analyzed with DNAMAN software (version 5.2.2).
Histology and Microscopy Observation
Fresh hand-cut sections (∼20 μm) of rice leaf blade were stained in Ruthenium red solution (0.2 mg/mL) or in aniline blue solution (5 mg aniline blue in 50% ethanol). Treated samples were transferred into water. The sections were microscopically examined (Leica DMR) and photographed.
Leaves at certain developmental stages were sampled for resin sectioning, intenerated by hydrofluoric acid, fixed in 2.5% glutaraldehyde (16 to 48 h, 48°C), and dehydrated through a graded ethanol series. The samples were embedded in Epon812 resin (Fluka) and polymerized at 60°C. Sections (3 μm) were cut and stained with filtered 1% toluidine blue, microscopically examined (Leica DMR), and photographed. Area measurements of vascular elements were performed using Leica Qwin software.
FDA was used to determine the viability of pollen grains. Mature flowers before opening were harvested, and picked anthers were crushed into a fine powder and treated with FDA solution (10 mg FDA dissolved in 1 mL acetone as stock, diluted with 7% sucrose to a concentration of 100 μg/mL for use) for 5 min and then observed under UV light (Leica DMR). Pollen grains emitting green fluorescence were regarded as viable pollen.
Measurement of Chlorophyll Content in Leaves, Photosynthetic Efficiency, and Leaf Rolling Index
The second leaf from the top of wild-type, sll1-1, sll1-2, or transgenic sll1-2 plants with complemented expression of SLL1 (at the same growth stage) was collected using a hole-puncher to cut three circles in the center area of the leaf beside the midvein. Before sampling, the paper circle was cut using the same hole-puncher to calculate the areas of leaf samples. The collected leaf samples were incubated in 5 mL 80% acetone (v/v) for 24 h under darkness. Chlorophyll contents were determined by measuring the absorbance at 652 nm according to Arnon (1949).
The middle part of the 8th to 10th rice leaves in wild-type, sll1-1, and sll1-2 plants at the tilling stage were collected and used to measure the photosynthetic efficiency. The TEACHIG-PAM-2100 chlorophyll fluorometer was used to detect the quantum yield of PSII and the maximal quantum yield of PSII, Fv/Fm, through the RUN1 and RUN2 programs (Schreiber, 1997).
The widths of 4th to 5th or 10th to 11th leaves in 40-d-old or 75-d-old sll1-1 and sll1-2 plants were measured under either the natural (Ln) or unfolding state (Lw). The LRI was calculated as LRI = (Lw − Ln)/Lw.
Phylogenetic Analysis
The BLAST search program (http://www.ncbi.nlm.gov/BLASTp/) was used to identify the homologous sequence of SLL1 using its amino acid sequence as query in searches of the National Center for Biotechnology Information and The Institute for Genomics Research. All the resulting sequences were aligned using ClustalX1.83 multiple alignment mode, and the neighbor-joining phylogenetic tree was generated using MEGA 4.0 (Tamura et al., 2007). The bootstrap values for nodes in the phylogenetic tree are from 1000 replications. The handing gap option was pairwise deletion, and the numbers at the branching points indicated the bootstrap values. The peptide identities among the different proteins were calculated using Genedoc software.
Promoter-Reporter Gene Fusion Studies
For promoter-GUS fusion studies, a 2.1-kb genomic DNA fragment containing the promoter region of the SLL1 gene was amplified by PCR using primers 5′-GCGTCGACTTTTGGCTCTGGCTAACTTA-3′ and 5′-GCTCTAGACACCTCCTTCTCCACCTC-3′ and then subcloned into vector pCAMBIA1301 (Cambia), resulting in fusion of the promoter and the GUS reporter gene. The positive construct was transformed into wild-type plants as described above. About 20 independent transgenic lines were obtained after screening, and GUS activities were histochemically detected as described (Jefferson et al., 1987).
In Situ Hybridization Analysis
Wild-type leaves from two developmental stages (4th seedling leaf and 10th mature leaf) were fixed in a formaldehyde solution (4%), dehydrated through an ethanol series, embedded in paraffin (Sigma-Aldrich), and sectioned at 8 μm using an YL3-A rotary microtome (Shanghai Instrument Factory). A 343-bp gene-specific region of SLL1 cDNA, amplified by PCR reaction (primers 5′-CCGTGAAGAGCACTGACAAGCC-3′ and 5′-GAAGACAATCCTACGGAGCGATG-3′) and a 528-bp gene-specific region of rice YAB1 cDNA, amplified by PCR reaction (primers 5′-GCCAAAAGGGTCAAGAGG-3′ and 5′-GGACGTATAGGTGACAC-3′), were used as the probes. The amplified DNA fragments were subcloned into a pGEM-T easy vector (Promega) in two orientations; the sense and antisense probes were synthesized and used to generate the RNA probe. In situ hybridization was performed as described (Luo et al., 1996).
Confirmation of SLL1 Mutation in sll1-1 and sll1-2 Plants
Total RNA was extracted from the 4th or 5th leaf blade of wild-type and sll1 mutant lines using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The isolated RNA was reverse-transcribed (SuperScript preamplification system) and used as templates for analysis. The SLL1 transcripts in sll1-1 or sll1-2 mutants were amplified through RT-PCR with primers for sll1-1 (5′-AGGCTCAACGGCATGCTCTC-3′ and 5′-GGCTCTTGACATGCGCTAGC-3′) or for sll1-2 (5′-TGTATCGCACCGTGAAGAGCA-3′ and 5′-TTGAGGAGTTGCTCCACCTGG-3′).
Constructs and Rice Transformation
A 7.8-Kb genomic DNA fragment containing the entire SLL1 gene (coding region, 1600-bp upstream sequence and 900-bp downstream sequence) was cut from BAC B1040D06 and subcloned into vector pCAMBIA1300. The construct obtained was introduced into the Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by electroporation; positive clones were used for transformation of the wild type and sll1-2 using immature embryos as materials.
Real-Time qRT-PCR Analysis
Real-time qRT-PCR analysis was performed to examine the expression pattern of SLL1 in various tissues and different stages of leaf growth to identify the transgenic lines with enhanced or complemented SLL1 expression as well as to identify altered transcripts of PCD-related genes in the wild type compared with sll1-1 or sll1-2 plants.
Total RNA was extracted using TRIzol solution and reverse-transcribed according to the manufacturer's instructions (SuperScript preamplification system) and then quantitatively analyzed on the Rotor-Gene real-time thermocycler R3000 (Corbett Research) with real-time PCR Master Mix (Toyobo). For the analysis, a linear standard curve was generated using a series of dilutions for each PCR product; the levels of the transcript in all unknown samples were determined according to the standard curve. The rice ACTIN gene (Os03g50890, amplified by primers 5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCAGGCATCTG-3′) was amplified and used as an internal standard to normalize the expression of SLL1 and the other tested genes.
To test the expressions of the PCD-related genes and rice YAB genes, leaf samples were collected from the middle part of the 7th leaf from different lines. Each sample was duplicated independently to ensure validity with biological replicates. All experiments were repeated three times; the data were statistically analyzed and presented as means plus sd.
The primers used to test the expression of SLL1 were as follows: in various tissues, 5′-CAGGTGTCCAACCATGAGC-3′ and 5′-GCCTCTGTGATTGCCATCTAAT-3′; in transgenic plants, 5′-CTCTTCAGGGCCGGCGGA-3′ and 5′-TGAGGAGTTGCTCCACCT-3′. To test the expression of PCD-related genes, the following genes were examined: Dirigent1 (Os11g42500, 5′-CCTACAGTTGTACAGATGCAATCG-3′ and 5′-GATGGAGGAATGGATTGATGGT-3′), PI (Os05g06780, 5′-ATCAAGCCGGAGGTCGCCATC-3′ and 5′-CAAGGCAGCGTGTAATCTCC-3′), TED2 (Os10g41170, 5′-AAAGCAATCCAAGCAGCCGAAGACG-3′ and 5′-CCACGCGGCATCAGACACTCCATTG-3′), PAL (Os05g35290, 5′-TACACCGACCACCTCACCCACA-3′ and 5′-CGAGCCTCTTCGCCTCCTTCAT-3′), C4H (Os05g25640, 5′-GGCGAGATCAACCACGACAACG-3′ and 5′-GCAACCGCAGCGTCTCCTTCA-3′), 4CL (Os06g44620, 5′-GTCGTCGCCCTCCCTTACTCCT-3′ and 5′-AACAGCGGCAGCAAGCACAGC-3′), and CP (Os08g44270, 5′-CCAACAGGTTCGCCGACCTCAC-3′ and 5′-CCGCACGAGCCTTGGTCCTTGA-3′). To test the expression of Os YAB genes, the primers used were as follows: Os YAB1 (Os07g06620, 5′-GCCAACCAGTCAGCAGCAAGTGTCA-3′ and 5′-CAAAATGGAGCCGGGGAAGATGAG-3′), Os YAB2 (Os03g44710, 5′-CGGCGCCGGAGCATGTGTG-3′ and 5′-TGGCCACAACGGACGGTCACGA-3′), and Os YAB6 (Os12g42610, 5′-GCCTCCTGCTGCTGCTGCCATGG-3′ and 5′-AATGGATGTTTGGATAATGT-3′).
Scanning Electron Microscopy
The 4th leaves of the wild type and sll1-2 were collected and fixed in formalin:glacial acetic acid:50% ethanol (2:1:17, v:v:v) overnight. After dehydration in a graded ethanol series, the sections were dried for 4 h (Hitachi critical point dryer), surface-sprayed with gold powder, and examined under a scanning electron microscope (Hitachi S-450).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: rice SLL1 (Os09g23200), ACTIN (Os03g50890), Dirigent1 (Os11g42500), PI (Os05g06780), TED2 (Os10g41170), PAL (Os05g35290), C4H (Os05g25640), 4CL (Os06g44620), CP2 (Os08g44270), KAN2 (Os08g33050), KAN3 (Os08g06370), KAN4 (Os03g55760), and KAN5 (Os02g46940); Arabidopsis KAN1 (At5g16560), KAN2 (At1g32240), KAN3 (At4g17695), KAN4 (At5g42630); maize KAN1 (ABB89931), KAN3 (ACF87413), KAN4 (ACG26785), KAN5 (NP_001132886), and MWP1 (ACG26785); and grape KAN1 (CAO38744) and KAN2 (CAO63351).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Unaltered Expression of rice YABBY Genes in sll1-1.
Supplemental Table 1. New Developed Polymorphic STS and SSR Markers Used for Map-Based Cloning of SLL1.
Supplemental Table 2. Primers Used to Amplify the Genomic DNA Fragment of Candidate Gene from sll1 and Wild-Type Variety.
Supplemental Data Set 1. Alignment Analysis of the KANADI Proteins, Including Those from Arabidopsis (KAN1, 2, 3, and 4), rice (SLL1 and KAN2, 3, 4, and 5), and Zea mays (MWP1 and KAN1, 3, 4, and 5).
Supplementary Material
Acknowledgments
The study was supported by the State Key Project of Basic Research (2005CB120803 and 2006AA10A102), the National Science Foundation of China (30425034 and 30623006), and the National Key Laboratory of Plant Molecular Genetics. We thank Shu-Ping Xu for help with rice transformation, Wei Huang for in situ hybridization, and Hui-Qiong Zheng and Xiao-Yan Gao for the leaf cross-section technique.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Qian Qian (qianqian188@hotmail.com) and Hong-Wei Xue (hwxue@sibs.ac.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenol-oxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna, G., Robles, P., and Micol, J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Candela, H., Johnston, R., Gerhold, A., Foster, T., and Hake, S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell . [DOI] [PMC free article] [PubMed]
- Chen, Z.X., Pan, X.B., and Hu, J. (2001). Relationship between rolling leaf and ideal plant type of rice (Oryza sativa L.) (in Chinese). Jiangsu Agricultural Res. 22 88–91. [Google Scholar]
- Dai, M.Q., Zhao, Y., Ma, Q., Hu, Y.F., Hedden, P., Zhang, Q.F., and Zhou, D.X. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin, L.B., and Lewis, N.G. (2005). Lignin primary structures and dirigent sites. Curr. Opin. Biotechnol. 16 407–415. [DOI] [PubMed] [Google Scholar]
- Demura, T., and Fukuda, H. (1993). Molecular cloning and characterization of cDNAs associated with tracheary element differentiation in cultured Zinnia cells. Plant Physiol. 103 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura, T., and Fukuda, H. (1994). Novel vascular cell-specific genes whose expression is regulated temporally and spatially during vascular system development. Plant Cell 6 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, W.G. (1971). Leaf angle, leaf area, and canopy photosynthesis. Crop Sci. 11 482–485. [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fahn, A. (1990). Plant Anatomy, 4th ed. (New York: Pergramon Press).
- Freeling, M., and Lane, B. (1993). The maize leaf. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 17–28.
- Fukuda, H. (2000). Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 44 245–253. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–45. [DOI] [PubMed] [Google Scholar]
- Govaerts, Y.M., Jacquemoud, S., Verstraete, M.M., and Ustin, S.L. (1996). Three-dimensional radiation transfer modeling in a dicotyledon leaf. Appl. Opt. 35 6585–6598. [DOI] [PubMed] [Google Scholar]
- Hawker, N.P., and Bowman, J.L. (2004). Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2 208–218. [Google Scholar]
- Hsiao, T.C., O'Toole, J.C., Yambao, E.B., and Turner, N.C. (1984). Influence of osmotic adjustment on leaf rolling and tissue death in rice. Plant Physiol. 75 338–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.H., Pi, L.M., Liang, W.Q., Xu, B., Wang, H., Cai, R., and Huang, H. (2006). The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18 2479–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C.P. (2004. a). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Twigg, R.W., and Timmermans, M.C.P. (2004. b). Specification of adaxial cell fate during maize leaf development. Development 131 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethlg, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Khush, G.S., and Kinoshita, T. (1991). Rice karyotype, marker genes, and linkage group. In Rice Biotechnology, G.S. Khush and G.H. Toenniesen, eds (Wallingford, UK: CAB International), pp. 83–107.
- Kinder, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T. (1984). Gene analysis and linkage map. In Biology of Rice, S. Tsunoda and N. Takahashi, eds (Tokyo: JSSP/Elsevier), pp. 187–274.
- Lang, Y.Z., Zhang, Z.J., Gu, X.Y., Yang, J.C., and Zhu, Q.S. (2004). A physiological and ecological effect of crimpy leaf character in rice (Oryza sativa L.) II. Photosynthetic character, dry mass production and yield forming. Acta Agron. Sin. 30 883–887. [Google Scholar]
- Li, S.G., Ma, Y.Q., He, P., Li, H.Y., Chen, Y., Zhou, K.D., and Zhu, L.H. (1998). Genetics analysis and mapping the flag leaf roll in rice (Oryza sativa L.). J. Sichuan Agric. Uni. 16 391–393. [Google Scholar]
- Lin, Q., and Northcote, D.H. (1990). Expression of phenylalanine ammonia-lyase gene during tracheary-element differentiation from cultured mesophyll cells of Zinnia elegans L. Planta 182 591–598. [DOI] [PubMed] [Google Scholar]
- Liu, H.L., Xu, Y.Y., Xu, Z.H., and Chong, K. (2007). A rice YABBY gene, OsYABBY4, preferentially expresses in developing vascular tissue. Dev. Genes Evol. 217 629–637. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S.W., Xu, X.S., and Shen, M.J. (1982). Botany (in Chinese), 2nd ed. (Beijing: Higher Education Press).
- Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. [DOI] [PubMed] [Google Scholar]
- Luo, Z., Yang, Z., Zhong, B., Li, Y., Xie, R., Zhao, F., Ling, Y., and He, G. (2007). Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome 50 811–817. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Micol, J.L., and Hake, S. (2003). The development of plant leaves. Plant Physiol. 131 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulia, B. (1994). The biomechanics of leaf rolling. Biomimetics 2 267–281. [Google Scholar]
- Nelson, J.M., Lane, B., and Freeling, M. (2002). Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development 129 4581–4589. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 223–236. [DOI] [PubMed] [Google Scholar]
- Price, A.H., Young, E.M., and Tomos, A.D. (1997). Quantitative trait loci associated with stomatal conductance leaf rolling and heading date mapped in upland rice (Oryza sativa L.). New Phytol. 137 83–91. [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Ito, T., Shimura, Y., and Okada, K. (1999. a). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999. b). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, U. (1997). Chlorophyll Fluorescence and Photosynthetic Energy Conversion: Simple Introductory Experiments with the TEACHING-PAM Chlorophyll Fluorometer. (Effeltrich, Germany: Heinz Walz GmbH).
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shao, Y.J., Chen, Z.X., Zhang, Y.F., Chen, E.H., Qi, D.C., Miao, J., and Pan, X.B. (2005. a). One major QTL mapping and physical map construction for rolled leaf in rice (in Chinese). Acta Genet. Sin. 32 501–506. [PubMed] [Google Scholar]
- Shao, Y.J., Pan, C.H., Chen, Z.X., Zuo, S.M., Zhang, Y.F., and Pan, X.B. (2005. b). Fine mapping of an incomplete recessive gene for leaf rolling in rice (Oryza sativa L.). Chin. Sci. Bull. 50 2466–2472 (in Chinese). [Google Scholar]
- Shi, Z.Y., Wang, J., Wan, X.S., Shen, G.Z., Wang, X.Q., and Zhang, J.L. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226 99–108. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., Greene, B., Veit, B., and Hake, S. (1992). A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116 21–30. [DOI] [PubMed] [Google Scholar]
- Solomon, M., Belenghi, B., Delledonne, M., Menachem, E., and Levine, A. (1999). The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214 694–702. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, G.L., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151–153. [DOI] [PubMed] [Google Scholar]
- Toriba, T., Harada, K., Takamura, A., Nakamura, H., Ichikawa, H., Suzaki, T., and Hirano, H.Y. (2007). Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genomics 277 457–468. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 154–156. [DOI] [PubMed] [Google Scholar]
- Voo, K.S., Whetten, R.W., O'Malley, D.M., and Sederoff, R.R. (1995). 4-Coumarate: coenzyme A ligase from loblolly pine xylem: isolation, characterization, and complementary DNA cloning. Plant Physiol. 108 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789. [DOI] [PubMed] [Google Scholar]
- Xu, L., Yang, L., and Huang, H. (2007). Transcriptional, post-transcriptional and post-translational regulations of gene expression during leaf polarity formation. Cell Res. 17 512–519. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Sun, Y., Liang, W., and Huang, H. (2002). The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Bot. Sin. 44 1194–1202. [Google Scholar]
- Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C.J., Yan, S., Zhang, Z.Q., Liang, G.H., Lu, J.F., and Gu, M.H. (2006). Genetic analysis and gene fine mapping for a rice novel mutant rl9(t) with rolling leaf character. Chin. Sci. Bull. 51 63–69 (in Chinese). [Google Scholar]
- Ye, Z.H. (1996). Expression patterns of the cinnamic acid 4-hydroxylase gene during lignification in Zinnia elegans. Plant Sci. 121 133–141. [Google Scholar]
- Ye, Z.H., and Varner, J.E. (1996). Induction of cysteine and serine proteases during xylogenesis in Zinnia elegans. Plant Mol. Biol. 30 1233–1246. [DOI] [PubMed] [Google Scholar]
- Yuan, L.P. (1997). Super-high yield hybrid rice breeding. Hybrid Rice 12 1–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.