Abstract
Systemic acquired resistance (SAR) develops in response to local microbial leaf inoculation and renders the whole plant more resistant to subsequent pathogen infection. Accumulation of salicylic acid (SA) in noninfected plant parts is required for SAR, and methyl salicylate (MeSA) and jasmonate (JA) are proposed to have critical roles during SAR long-distance signaling from inoculated to distant leaves. Here, we address the significance of MeSA and JA during SAR development in Arabidopsis thaliana. MeSA production increases in leaves inoculated with the SAR-inducing bacterial pathogen Pseudomonas syringae; however, most MeSA is emitted into the atmosphere, and only small amounts are retained. We show that in several Arabidopsis defense mutants, the abilities to produce MeSA and to establish SAR do not coincide. T-DNA insertion lines defective in expression of a pathogen-responsive SA methyltransferase gene are completely devoid of induced MeSA production but increase systemic SA levels and develop SAR upon local P. syringae inoculation. Therefore, MeSA is dispensable for SAR in Arabidopsis, and SA accumulation in distant leaves appears to occur by de novo synthesis via isochorismate synthase. We show that MeSA production induced by P. syringae depends on the JA pathway but that JA biosynthesis or downstream signaling is not required for SAR. In compatible interactions, MeSA production depends on the P. syringae virulence factor coronatine, suggesting that the phytopathogen uses coronatine-mediated volatilization of MeSA from leaves to attenuate the SA-based defense pathway.
INTRODUCTION
Systemic acquired resistance (SAR) is an enhanced state of broad-spectrum disease resistance that develops in the whole plant in response to a locally restricted leaf inoculation with microbial pathogens (Métraux et al., 2002; Durrant and Dong, 2004). Induction of SAR occurs at the site of pathogen inoculation where presumed mobile long-distance signals are generated. The latter are thought to be subsequently transferred to and perceived in distant, noninfected plant parts. Therein, they are supposed to initiate signaling and amplification processes that lead to an increase of systemic defense responses to boost whole-plant resistance (Mishina and Zeier, 2006).
Induction of SAR is not restricted to hypersensitive response (HR)-inducing or necrotizing pathogens but also takes place upon leaf contact with high inoculi of nonpathogenic microbes or after local treatment with bacterial pathogen-associated molecular patterns, such as flagellin or lipopolysaccharides (Mishina and Zeier, 2007). Irrespective of the eliciting stimulus, the molecular events set in motion in inoculated leaves to initiate SAR in distant leaves are only partially understood. The recent finding that ectopic expression of Arabidopsis thaliana mitogen-activated protein kinase kinase7 in local tissue induces pathogenesis-related (PR) gene expression and resistance to Pseuodmonas syringae in systemic tissue indicates that mitogen-activated protein kinase-based signaling cascades are involved in the initiation of SAR long-distance signaling (Zhang et al., 2007). However, the chemical nature of putative mobile SAR signals remains elusive (Vlot et al., 2008a).
Mutational analyses in Arabidopsis suggest that peptide and lipid derivatives participate in signal transduction from inoculated to distant leaves (Grant and Lamb, 2006; Chaturvedi et al., 2008). A peptide signal might be generated by the apoplastic aspartic protease CONSTITUTIVE DISEASE RESISTANCE1, which is required for the execution of both local and systemic resistance responses (Xia et al., 2004). Moreover, DEFECTIVE IN INDUCED RESISTANCE1 (DIR1) bears homology to lipid transfer proteins and is involved in local generation or subsequent translocation of a mobile systemic signal, possibly by acting as a chaperone for a lipid-related signal (Maldonado et al., 2002). A glycerolipid-derivative might be a DIR1-interacting partner because the dihydroxyacetone phosphate reductase SUPPRESSOR OF FATTY ACID DESATURASE ACTIVITY1 (Nandi et al., 2004) and the fatty acid desaturase FAD7, both components of plastid glycerolipid biosynthesis, are necessary for SAR establishment and, together with DIR1, are required for the accumulation of a SAR-inducing activity in Arabidopsis petiole exudates (Chaturvedi et al., 2008). Moreover, the plant defense hormone jasmonic acid (JA) or a JA pathway-related oxylipin was proposed as the signal mediating long-distance information transmission during SAR (Truman et al., 2007). JA-mediated signaling is well established to participate in induced plant resistance against both insect herbivory and attack by necrotrophic pathogens, but its role in defense against biotrophic microbial pathogens is less well defined (Li et al., 2002; Glazebrook, 2005).
It has been known for more than a decade that salicylic acid (SA) acts as a major player during the establishment of SAR. SA accumulates both at inoculation sites and in distant leaves concomitant with the onset of SAR, and transgenic, SA hydroxylase (NahG) expressing plants not capable of SA accumulation are SAR deficient (Malamy et al., 1990; Métraux et al., 1990; Gaffney et al., 1993). The requirement for intact SA signaling during SAR is underlined by the failure of the Arabidopsis mutants salicylic acid induction-deficient1 (sid1) and sid2, which are both defective in induced SA production, to enhance systemic resistance after pathogen infection. SID1 and SID2 code for a multidrug and toxic compound extrusion transporter protein and isochorismate synthase1 (ICS1), respectively (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Nawrath et al., 2002). Grafting experiments using root stocks and scions from wild-type and NahG-expressing tobacco (Nicotiana tabacum) have indicated that SA itself is not a long-distance signal but that SA accumulation in distant leaves is critical for SAR (Vernooij et al., 1994).
SA can be biochemically modified to derivatives with altered physicochemical properties and bioactivity (Wildermuth, 2006). UDP-dependent SA-glucosyl-transferases transfer a glucose moiety to either the phenolic hydroxyl group or to the carboxyl group of SA, yielding the hydrophilic SA derivatives SA 2-O-β-d-glucose (SA glucoside [SAG]) or SA glucose ester (Lee and Raskin, 1999; Lim et al., 2002; Dean and Delaney, 2008). SAG, the most prominent glucosylated form of SA in many plant species, is produced from accumulating SA after pathogen infection (Malamy et al., 1992; Mishina et al., 2008). Furthermore, methylation of the free carboxyl group of SA yields the nonpolar and volatile SA methyl ester (methyl salicylate [MeSA]; Wildermuth, 2006). This reaction is catalyzed by SA methyl transferase (SAMT), which uses S-adenosine-l-methionine as methyl donor (Ross et al., 1999). In Arabidopsis, the BSMT1 gene codes for a protein with both benzoic acid and SA methylating activities (Chen et al., 2003). BSMT1 is highly expressed in flowers, and expression in leaves is upregulated by treatment with the antibiotic alamethicin, by methyl jasmonate application, and by herbivory. MeSA is a significant constituent of floral scents from various plant species and of volatile blends from herbivore-attacked vegetative plant parts, and it functions in pollinator attraction and defense against insects (Van Poecke et al., 2001; Effmert et al., 2005; Zhu and Park, 2005). Concomitant with SA biosynthesis, MeSA is produced in pathogen-infected tobacco and Arabidopsis leaves and emitted to significant amounts into the environment (Shulaev et al., 1997; Koo et al., 2007; Attaran et al., 2008).
Pathogen-elicited MeSA has been previously proposed as being an airborne signal involved in plant-to-plant communication (Shulaev et al., 1997). More recently, grafting experiments suggested that MeSA is a critical, phloem-mobile SAR long-distance signal in tobacco (Park et al., 2007). A model has been proposed in which the SA accumulating after tobacco mosaic virus (TMV) infection is converted to MeSA by SA methyl transferase (SAMT1) in inoculated tobacco leaves, and MeSA subsequently travels through the phloem to distant leaves. Here, by the methyl esterase activity of SA binding protein2 (Forouhar et al., 2005), MeSA is reconverted to active SA, which in turn triggers SAR in systemic tissue (Park et al., 2007). In addition to its movement through the phloem, MeSA has been suggested to act as a volatile intraplant signal that is capable of activating SAR in distant leaves of the same plant (Shulaev et al., 1997). Another recent study extended this putative signaling function of MeSA to SAR in Arabidopsis (Vlot et al., 2008b). In this species, 18 potentially functional methyl esterase genes exist, out of which five encode proteins with MeSA demethylase activity (Yang et al., 2008; Vlot et al., 2008b). Attempts to silence these five redundant methyl esterase genes by a combination of T-DNA knockout and RNA interference silencing strategies resulted in different transgenic lines with partial but not complete abrogation of SA methyl esterase expression. The failure of some of these lines to mount P. syringae–induced SAR was taken as supportive evidence for the notion that MeSA represents a universal mobile SAR signal in plants (Vlot et al., 2008a, 2008b).
In this study, we address the significance of MeSA during biologically induced SAR in Arabidopsis. We show that MeSA production strongly increases in leaves inoculated with SAR-inducing strains of P. syringae and that most of the generated MeSA is directly emitted into the atmosphere. Moreover, the SAR-deficient phenotype of several Arabidopsis defense mutants is not caused by a failure of MeSA production. Significantly, mutational defects in the Arabidopsis SA methyl transferase gene BSMT1 completely abolish pathogen-induced MeSA production but do not affect SAR. Together, these data show that MeSA production is dispensable for SAR in Arabidopsis and that the systemic increase in SA, which is crucial for SAR, is not based on translocation of MeSA from inoculated to distant leaves. Instead, our findings support the hypothesis that the systemic rises in SA occur via de novo synthesis in distant leaves. Our data also show that MeSA biosynthesis is largely regulated via the JA pathway but exclude a role for JA signaling in SAR establishment. Since MeSA production in compatible interactions largely depends on the capability of P. syringae to produce the bacterial phytotoxin coronatine, a possible virulence mechanism of this phytopathogen includes volatilization of MeSA from leaves to negatively interfere with SA-associated defense responses.
RESULTS
The bacterial plant pathogen P. syringae pv maculicola ES4326 (Psm) is able to rapidly multiply in apoplastic spaces of Arabidopsis leaves, thereby causing yellowish disease symptoms (Dong et al., 1991). Leaf inoculation of accession Columbia-0 (Col-0), which carries the Rpm1 resistance gene with Psm expressing the avirulence gene AvrRpm1 (Psm avrRpm1), by contrast, elicits an HR associated with rapid cell death at inoculation sites (Bisgrove et al., 1994; Delledonne et al., 1998). Early defense responses associated with the HR do not fully abrogate but significantly restrict bacterial multiplication. Both virulent Psm and avirulent Psm avrRpm1 trigger a robust SAR response in Col-0 plants (Mishina and Zeier, 2006; 2007).
Production and Fate of MeSA after Pathogen Attack
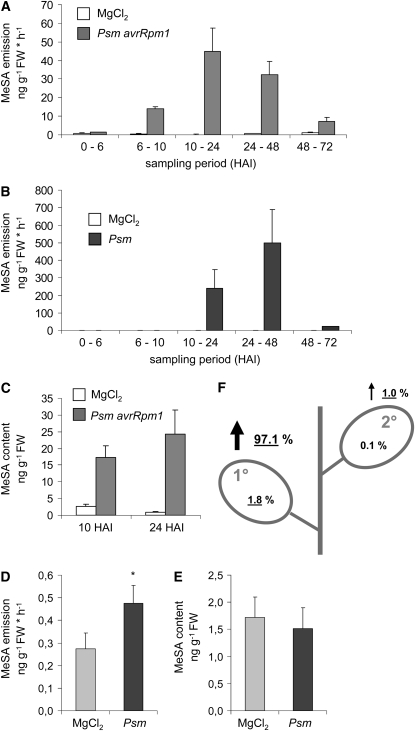
To assess the significance of MeSA during local and systemic resistance induction in Arabidopsis and its role in long-distance transport, we first determined leaf MeSA production upon P. syringae inoculation. Because of the volatile nature of MeSA, leaf emission of volatile organic compounds (VOCs) was determined from intact plants (Attaran et al., 2008). Following leaf inoculation with the avirulent Psm avrRpm1 strain, MeSA emission of Col-0 plants was not elevated before 6 h after inoculation (HAI) but strongly increased to ∼15 ng g−1 leaf fresh weight (FW) h−1 between 6 and 10 HAI compared with MgCl2-infiltrated control plants (Figure 1A). The release of MeSA further increased to 45 ng g−1 h−1 from between 10 and 24 HAI and then gradually decreased during the next 48 h of sampling. Comparatively, when plants were infected with virulent Psm, MeSA emission was delayed and not detectable before 10 HAI (Figure 1B). However, the quantity of emitted MeSA between 10 and 48 HAI was about one order of magnitude higher in the compatible than in the incompatible interaction, reaching values between 240 and 500 ng g−1 h−1. This strong MeSA release markedly declined after 2 d after inoculation (DAI). Emission of MeSA in mock-infiltrated control plants was low throughout the entire sampling period (0.2 to 0.9 ng g−1 h−1; Figures 1A and 1B). MeSA was the major Arabidopsis VOC induced after P. syringae infection. In addition, a significant amount of the volatile homoterpene (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene (TMTT) was emitted upon inoculation with both Psm and Psm avrRpm1, and lower increases in the amounts of the terpenes β-ionone and α-farnesene as well as of methyl benzoate were detected in the VOC blends during later stages of the compatible interaction (Attaran et al., 2008).
Figure 1.
Leaf MeSA Production in Arabidopsis Col-0 Plants upon P. syringae Inoculation.
(A) and (B) Time course of MeSA emission after inoculation with HR-inducing Psm avrRpm1 (gray bars) (A), inoculation with compatible Psm (black bars) (B), or infiltration with 10 mM MgCl2 (white bars). Mean values of ng emitted substance g−1 leaf FW h−1 (±sd) from three independent plants are given. The time periods during which volatiles were collected are indicated. HAI, h after inoculation.
(C) Leaf MeSA contents in response to inoculation with Psm avrRpm1 (gray bars) or infiltration with 10 mM MgCl2 (white bars) at 10 and 24 HAI (means ± sd, n = 3).
(D) Emission of MeSA from nontreated, distant leaves of Psm-inoculated or MgCl2-infiltrated Col-0 plants. Treated leaves were removed at the onset of SAR (at 2 DAI), and emission of the remainder of the plant was sampled from 2 to 3 DAI. Mean values of ng emitted MeSA g−1 leaf FW h−1 (±sd, n = 5) are given. Asterisk denotes statistically significant differences between Psm and MgCl2 treatments (P < 0.05).
(E) MeSA content in nontreated, distant leaves of Psm-inoculated or MgCl2-infiltrated Col-0 plants at 2 DAI (means ± sd, n = 5).
(F) Fate of MeSA after its production during SAR in a symbolized Col-0 plant. Percentages of total MeSA produced after a localized P. syringae inoculation are indicated. An underlined value indicates a significant increase after pathogen treatment. 1°, inoculated leaf; 2°, noninoculated, systemic leaf. Numbers given next to vertical arrows represent emission; numbers inside leaves represent leaf content.
In addition to analyzing the MeSA vaporizing from leaves, we also determined its actual content in control and pathogen-inoculated leaf tissue through solvent extraction followed by gas chromatography–mass spectrometry (GC-MS) analysis (Figure 1C). While mock-treated leaves contained between 0.8 and 2.5 ng MeSA g−1, the MeSA content was significantly higher in leaves inoculated with Psm avrRpm1, amounting to 17 and 24 ng g−1 at 10 and 24 HAI, respectively. Accordingly, the absolute value of MeSA retained in leaves after Psm avrRpm1 inoculation equaled the amount emitted from leaves within ∼30 min (Figure 1A).
An important requirement for SAR development is the accumulation of SA in distant, noninoculated leaves (Vernooij et al., 1994). Since systemic SA accumulation was proposed to be associated with phloem-based MeSA translocation from inoculated to distant leaves and subsequent MeSA to SA conversion (Park et al., 2007), we assessed MeSA emission and content systemically (i.e., in nontreated, distant leaves of pathogen-inoculated plants). A modest but statistically significant increase in emission of MeSA was observed in distant leaves after a remote Psm attack compared with a respective mock treatment (Figure 1D). However, emission rates from distant leaves were two to three orders of magnitude lower than the rates detected in pathogen-treated leaves and fell in the same range as those measured from MgCl2-infiltrated control leaves (Figures 1A and 1B). Moreover, the leaf contents of MeSA in nontreated, distant leaves of remotely Psm-inoculated plants (Figure 1E) were similar to those of MgCl2-infiltrated leaves (Figure 1C), and no significant differences in MeSA contents of systemic leaves existed between mock- and Psm-pretreated plants (Figure 1E).
In addition, we analyzed MeSA contents in petiole exudates collected from 6 to 48 HAI in mock- and pathogen-inoculated leaves. During this time period, a marked SAR response develops in Col-0 plants upon inoculation with the used inoculation density of Psm (OD 0.01), which is accompanied with systemic rises of 1 to 2 μg g−1 SA (Mishina and Zeier, 2007; Mishina et al., 2008). With 1.2 ng MeSA g−1 h−1, Psm-inoculated leaves exhibited a threefold higher exudation of MeSA from petioles than control leaves (see Supplemental Figure 1A online). However, these values might underestimate the actual MeSA exudation, as a fraction of the volatile could have escaped into the atmosphere during the exudate collection period. Nevertheless, these values are in the same order of magnitude as the MeSA levels estimated in exudates from tobacco leaves (Park et al., 2007). We also detected and quantified free and glucosidic SA in the collected petiole exudates, and both SA forms were found in similar scales in the exudates as MeSA. Whereas exudation of SAG from petioles increased from 1.1 to 4.0 ng g−1 h−1 upon Psm inoculation (see Supplemental Figure 1B online), leaf pathogen treatment did not significantly alter the levels of exuded free SA. The latter was released to ∼1 ng g−1 h−1 from both mock- and Psm-treated leaves (see Supplemental Figure 1C online).
In summary, these quantitative analyses show that MeSA production strongly increases in P. syringae–inoculated Arabidopis leaves. During the first 24 HAI, ∼0.75 μg g−1 MeSA are produced in the incompatible interaction, whereas 3.5 μg g−1 are generated in the compatible interaction. However, most (97%) of the MeSA is directly emitted into the atmosphere, and only minor amounts are retained in leaves (Figure 1F). Lower amounts of MeSA and SAG but not of free SA also accumulate in petiole exudates after pathogen infection. The calculated sum of estimated MeSA and detected SAG exuded during a 48-h SAR induction period (∼0.15 μg g−1) falls well below the usually observed systemic rises in SA (1 to 2 μg g−1; Mishina and Zeier, 2007; Mishina et al., 2008). Moreover, in leaves distant from pathogen attack, the content of MeSA is not elevated and its emission increases only marginally.
SA and MeSA Production in SAR-Deficient Arabidopsis Lines
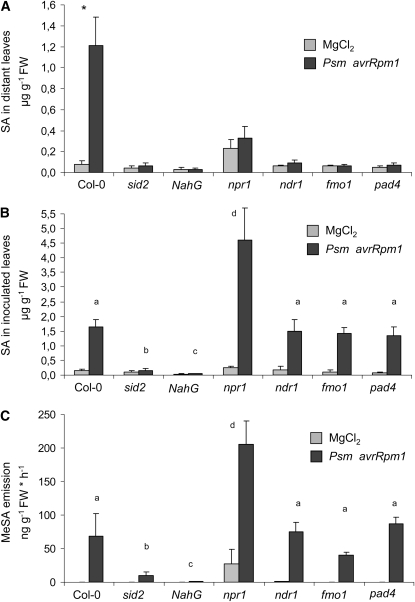
SAR is fully compromised in the Arabidopsis SA biosynthesis mutant ics1 (sid2), in the SA degrading NahG line, and in mutants of NON-EXPRESSOR OF PR1 (NPR1), which encodes a regulatory protein acting downstream of SA (Cao et al., 1994; Delaney et al., 1995; Lawton et al., 1995; Nawrath and Métraux, 1999). Moreover, mutants defective in NON RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1), and PHYTOALEXIN-DEFICIENT4 (PAD4) are also SAR deficient (Shapiro and Zhang, 2001; Mishina and Zeier, 2006, 2007). A general hallmark of these SAR-defective lines is that, unlike SAR-competent Col-0 plants, they do not accumulate SA in distant leaves after a local inoculation with P. syringae (Figure 2A). However, except for the SA biosynthesis-defective sid2 mutant and the SA nonaccumulating NahG line, these lines do produce SA in Psm avrRpm1-inoculated leaves to wild-type-like levels, or in the case of npr1, to levels even exceeding those of wild-type Col-0 (Figure 2B). These findings reflect the requirement of systemic but not local SA accumulation for SAR development, and they might be explained in two ways. The first scenario is that the systemic rises in SA that normally occur during SAR in wild-type plants are generated by de novo synthesis in distant leaves. The second possibility is that the SA accumulating in inoculated leaves is transported to distant leaves in free or derivatized form in the wild type but that this translocation is blocked in the different SAR-defective mutants. If MeSA were the translocated SA derivative (Park et al., 2007), a failure of the SAR-deficient lines to produce MeSA would explain the lack of systemic SA accumulation in these mutants (Figure 2A). We therefore tested whether the SAR-defective lines under investigation were defective in MeSA production after Psm avrRpm1 inoculation. However, except for sid2 plants, which emitted low but still increased levels of MeSA after pathogen treatment and the NahG line in which MeSA emission was nearly abolished, all the other SAR-defective lines emitted considerable amounts of MeSA after Psm avrRpm1 inoculation (Figure 2C). These data support the hypothesis that the majority of MeSA produced after pathogen inoculation is derived from SA synthesized by ICS1 and, more significantly for this study, indicate that the biosynthesis of MeSA is not impaired in several independent SAR-defective mutants.
Figure 2.
SA Accumulation and MeSA Production in P. syringae–Treated Wild-Type and SAR-Defective Mutant Plants.
(A) SA levels in nontreated, distant leaves of Psm avrRpm1–inoculated or MgCl2-infiltrated plants at 2 DAI (means ± sd, n = 4). Asterisk denotes statistically significant differences between Psm avrRpm1- and MgCl2-treated plants (P < 0.01).
(B) SA levels in Psm avrRpm1–inoculated leaves at 24 HAI (means ± sd, n = 4). Different characters symbolize statistically significant differences between Psm avrRpm1–treated plants from distinct lines (P < 0.05).
(C) MeSA emission from Psm avrRpm1- or mock-inoculated plants from 0 to 24 HAI (means ± sd, n = 4). Different characters symbolize statistically significant differences between Psm avrRpm1–treated plants from distinct lines (P < 0.05).
Arabidopsis bsmt1 Mutants Do Not Elevate MeSA after Pathogen Inoculation but Are SAR Competent
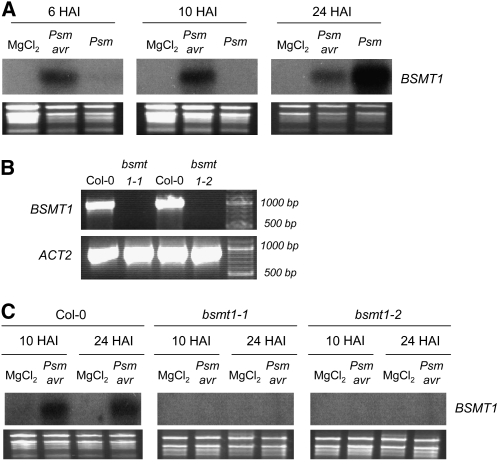
Arabidopsis BSMT1 has been previously identified as a methyl transferase with in vitro activity for SA to MeSA conversion (Chen et al., 2003). Expression of the BSMT1 gene in Col-0 leaves is virtually absent in mock-treated plants but is upregulated in response to P. syringae infection (Figure 3A). Whereas leaves inoculated with the incompatible Psm avrRpm1 strain induce expression of BSMT1 from 6 HAI onwards, expression of the gene in response to compatible Psm was slower but reached high values at 24 HAI. Thus, the temporal pattern and strength of leaf BSMT1 expression during the incompatible and the compatible P. syringae–Col-0 interaction closely resemble the relative timing and magnitude of MeSA emission (Figures 1A and 1B). This suggests that BSMT1 is directly involved in P. syringae–induced MeSA production.
Figure 3.
P. syringae–Induced Leaf Expression of the BSMT1 Methyl Transferase Gene and Identification of Nonexpressing T-DNA Insertion Lines.
(A) Expression of BSMT1 in Col-0 leaves inoculated with Psm avrRpm1 (Psm avr) or Psm. Control samples were infiltrated with 10 mM MgCl2. Leaf samples were taken at 6, 10, and 24 HAI for RNA gel blot analysis.
(B) PCR analyses using genomic DNA from Col-0, bsmt1-1 (SALK_140496), and bsmt1-2 (WiscDSLox430E05) mutant plants as templates and primers specific for the BSMT1 gene sequence. The actin gene ACT2 was amplified as a control.
(C) Expression patterns of BSMT1 in Col-0 and bsmt1 leaves infiltrated with 10 mM MgCl2 or Psm avrRpm1 (Psm avr) as assessed by gel blot analysis. Leaf samples were taken at 10 and 24 HAI.
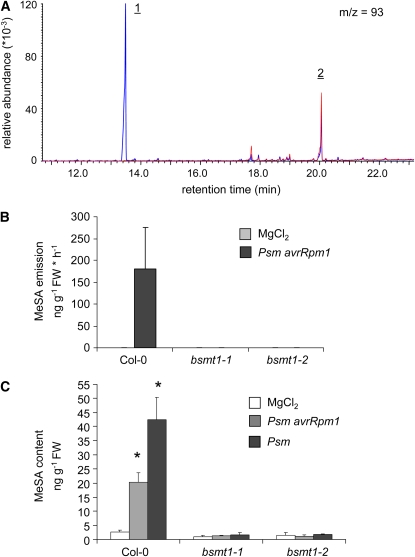
The T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress) predicts several lines with putative T-DNA insertions in the BSMT1 gene. We applied the PCR-based protocol described by Alonso et al. (2003) to confirm the predicted insertions and identified two lines, SALK_140496 and WiscDSLox430E05, which indeed harbor the T-DNA insert within the BSMT1 gene (Figure 3B). Lines homozygous for the insert, from now on designated as bsmt1-1 and bsmt1-2, do not exhibit any basal or pathogen-induced expression of BSMT1 (Figure 3C). Analyses of VOC emission from mock- and Psm avrRpm1–treated Col-0 or bsmt1 mutant plants revealed that MeSA was absent in blends of both bsmt1-1 and bsmt1-2 (Figures 4A and 4B). Moreover, the significant increase in leaf MeSA content that was detected in Col-0 upon P. syringae inoculation was not observed in bsmt1 mutant plants. The latter showed marginal basal leaf contents of MeSA, which were lower than those of noninoculated Col-0 controls and close to the analytical detection limit of ∼0.5 to 1 ng g−1 FW. These data demonstrate that BSMT1 is exclusively responsible for pathogen-induced MeSA production in Col-0 and suggest that a fraction of the already low basal MeSA levels might be produced independently from BSMT1. Compared with the wild type, neither bsmt1-1 nor bsmt1-2 plants had any obvious distinguishing morphological phenotype. Additionally, induced production of TMTT, the second most common volatile emitted from P. syringae–treated Arabidopsis leaves, was not affected in bsmt1 mutants (Figure 4A; see Supplemental Figure 2 online).
Figure 4.
bsmt1 Mutant Plants Are Completely Devoid of P. syringae–Induced MeSA Production.
(A) Ion chromatogram at m/z 93 of volatile samples from Col-0 plants (blue) and bsmt1-1 plants (red), illustrating MeSA (1) and TMTT (2) emission.
(B) Quantification of MeSA emitted from wild-type Col-0 and bsmt1 mutant plants inoculated with Psm avrRpm1 or infiltrated with MgCl2. Volatiles were collected from 0 to 24 HAI. Bars represent mean emission values (±sd, n = 4). MeSA emission was not detected in either bsmt1 mutant line (detection limit ∼0.05 ng g−1 FW h−1).
(C) Leaf MeSA contents of Col-0 and bsmt1 mutant plants in response to inoculation with Psm avrRpm1 (gray bars), Psm (black bars), or infiltration with 10 mM MgCl2 (white bars) at 24 HAI (means ± sd, n = 3). Asterisks denote statistically significant differences between P. syringae- and MgCl2-treated plants of a particular line (P < 0.003).
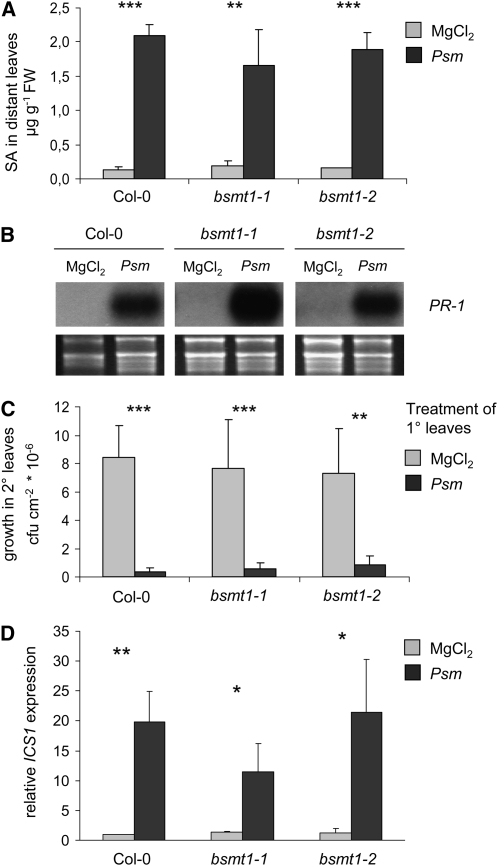
Although our data collected so far argued against a role of MeSA as a critical mobile SAR signal in Arabidopsis, a direct genetic examination of this putative function was still missing. With the availability of bsmt1 mutant plants lacking the ability to produce any pathogen-induced MeSA, the significance of MeSA during SAR could now be tested unequivocally. When plants of the different genotypes were inoculated with Psm in lower leaves to induce SAR, both bsmt1-1 and bsmt1-2 accumulated SA in upper, nontreated leaves, like the wild type, at day 2 after pathogen treatment (Figure 5A). Similarly, systemic expression of the SAR marker gene PATHOGENESIS-RELATED1 (PR-1) was increased in all the lines under investigation upon Psm but not after a mock pretreatment (Figure 5B). To test the enhancement of systemic resistance directly, we challenge-inoculated upper leaves with Psm 2 d after the primary MgCl2 or Psm treatment in lower leaves and assessed bacterial growth in upper leaves another 3 d later. When the primary, SAR-inducing Psm treatment in lower leaves was compared with the mock pretreatment, Col-0, bsmt1-1, and bsmt1-2 plants exhibited a similar, statistically highly significant containment of bacterial multiplication during the challenge infection in upper leaves (Figure 5C). These findings show that bsmt1 mutant plants are not affected in their abilities to enhance systemic SA levels, to systemically increase expression of the SAR gene PR-1, or to acquire resistance at the systemic plant level. Thus, MeSA is not required during SAR development and is not used as a long-distance signal ensuring systemic SA accumulation in Arabidopsis. As indicated by a strong upregulation of the SA biosynthesis gene ICS1 in systemic tissue upon primary Psm infection in the three investigated lines, the systemic accumulation of SA might rather be accomplished by de novo synthesis of SA in distant leaves (Figure 5D).
Figure 5.
P. syringae Induces SAR in bsmt1 Mutant Plants.
(A) Accumulation of SA in untreated, upper (2°) leaves after Psm inoculation, or MgCl2 infiltration of lower (1°) leaves. Treatments of 1° leaves were performed as described in (C). 2° leaves were harvested 2 d later for analyses. Bars represent mean values (±sd) of three independent samples, each sample consisting of six leaves from two different plants. Asterisks denote statistically significant differences in systemic SA levels between Psm and MgCl2 pretreated plants of a particular line (***P < 0.001; **P < 0.01).
(B) Expression of the SAR marker gene PR-1 in untreated, upper (2°) leaves after Psm inoculation or MgCl2 infiltration of lower (1°) leaves, as assessed by gel blot analyses. 2° leaves were harvested 2 d after the 1° treatment for analyses.
(C) Bacterial growth quantification to directly assess enhancement of systemic resistance. Plants were pretreated with either 10 mM MgCl2 or Psm (OD = 0.01) in three lower (1°) leaves. Two days later, three upper leaves (2°) were challenge infected with Psm (OD = 0.002). Bacterial growth in upper leaves was assessed 3 d after the 2° leaf inoculation. Bars represent mean values (±sd) of colony-forming units (cfu) per square centimeter from at least seven parallel samples each consisting of three leaf disks. Asterisks denote statistically significant differences of bacterial growth in 2° leaves between Psm and MgCl2 pretreated plants of a particular line (***P < 0.001; **P < 0.01).
(D) Relative expression levels of ICS1, as assessed by quantitative real-time PCR analysis. ICS1 expression values were normalized to those for the reference gene (At1g62930) and expressed relative to the wild-type MgCl2 sample. For each expression value of one sample, three PCR replicates were performed and averaged. The depicted bars represent mean values (±sd) of three biologically independent samples. Asterisks denote statistically significant differences in systemic SA levels between Psm and MgCl2 pretreated plants of a particular line (**P < 0.01; *P < 0.05).
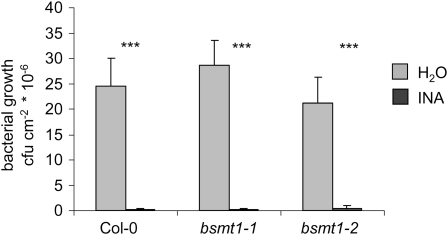
The SAR process is often investigated by whole-plant treatment of resistance-enhancing chemical agents such as 2,6-dichloroisonicotinic acid (INA), benzothiadiazole, or SA itself (Cao et al., 1994; Lawton et al., 1996), although such studies do not properly reflect the distinct spatial processes occurring after a localized induction of SAR with microbial pathogens. To test whether the chemical enhancement of resistance through SA analogs is dependent on functional BSMT1, we assayed leaf resistance against Psm of plants previously sprayed with a solution of 0.65 mM INA. Compared with water-sprayed control plants, a strong and highly significant enhancement of resistance by a factor of ∼50 was detected in INA-treated Col-0, bsmt1-1, and bsmt1-2 plants, indicating that INA-induced resistance is not affected by defects in BSMT1 (Figure 6).
Figure 6.
INA-Induced Resistance in Col-0 and bsmt1 Mutant Plants.
Plants were sprayed with 0.65 mM INA or water, and three leaves per plant infected 2 d later with Psm (OD = 0.002). Bacterial growth was assessed 3 d after inoculation (***P < 0.001).
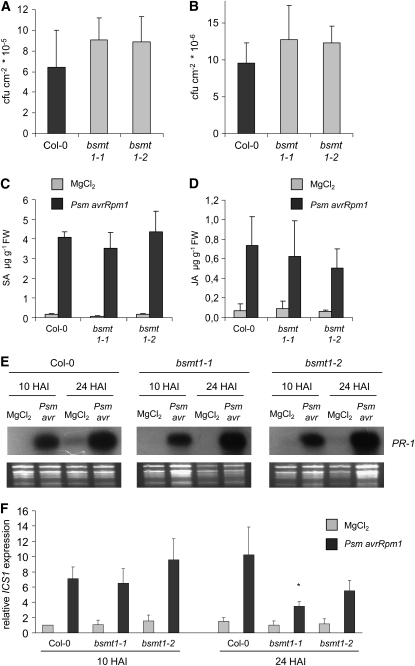
The bsmt1 mutants also allowed us to test whether disease resistance at inoculation sites and associated local defense responses would be influenced by MeSA production. Local resistance against both the incompatible Psm avrRpm1 strain and the compatible Psm strain were similar in wild-type and bsmt1 mutant plants (Figures 7A and 7B). Moreover, local accumulation of the defense signals SA and JA, and PR-1 expression patterns at infection sites were not impaired in the bsmt1 lines (Figures 7C to 7E). This indicates that, like SAR, induced resistance toward P. syringae at the site of pathogen inoculation is established independently of MeSA production.
Figure 7.
Local Defense Responses in bsmt1 Plants Are Similar to Those in the Wild Type.
(A) and (B) Bacterial growth quantification of Psm avrRpm1 (OD = 0.005) (A) and Psm (OD = 0.002) (B) in leaves of wild-type and bsmt1 mutant plants 3 DAI. Bars represent means (±sd) of cfu per cm2 from at least six parallel samples from different plants, each sample consisting of three leaf disks. No significant differences in bacterial numbers were detected at 3 DAI and 1 HAI (data not shown) for samples from different lines.
(C) and (D) Accumulation of the defense hormones SA (C) and JA (D) at sites of Psm avrRpm1 inoculation (10 HAI). Control samples were infiltrated with 10 mM MgCl2.
(E) RNA gel blot analysis of PR-1 expression in Col-0 and bsmt1 leaves infiltrated with 10 mM MgCl2 or Psm avrRpm1 (Psm avr). Leaf samples were taken at 10 and 24 HAI.
(F) Relative ICS1 expression in Col-0 and bsmt1 leaves infiltrated with 10 mM MgCl2 or Psm avrRpm1, as assessed by quantitative real-time PCR analyses (see Figure 5D for details). Leaf samples were taken at 10 and 24 HAI. Asterisk indicates statistically significant differences between Psm avrRpm1–treated wild-type and mutant samples (P < 0.05).
JA Signaling Regulates MeSA Production but Not SAR
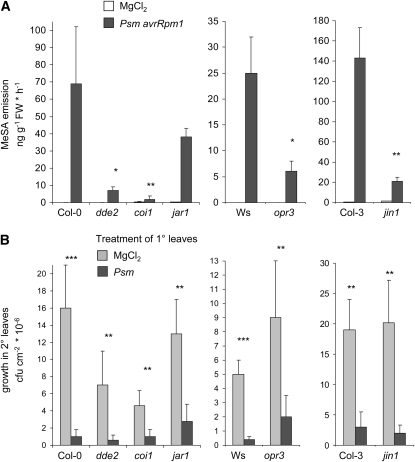
Induced biosynthesis of terpenoid volatiles in Arabidopsis and other plant species is dependent on JA signaling (Ament et al., 2006; Arimura et al., 2008; Attaran et al., 2008; Herde et al., 2008). By determining pathogen-induced MeSA emission from different Arabidopsis JA pathway mutants, we tested whether P. syringae–induced MeSA production would also require JA biosynthesis or associated downstream signaling events. The Arabidopsis DDE2 and OPR3 genes code for allene oxide synthase and 12-oxophytodienoic acid (OPDA) reductase, respectively (Stintzi and Browse, 2000; von Malek et al., 2002). The dde2 mutant is therefore defective in the synthesis of both JA and its signaling competent precursor OPDA (Mueller et al., 2008), whereas opr3 is compromised in JA but not in OPDA synthesis. Although Psm avrRpm1 inoculation enhanced MeSA emission in dde2 and opr3, the amounts of released MeSA were significantly lower in these mutants than the amounts emitted from the corresponding wild-type background lines Col-0 and Wassilewskija (Ws) after pathogen treatment (Figure 8A). The COI1 ubiquitin ligase is required for jasmonate-regulated defense responses (Xie et al., 1998), and coi1 mutant plants displayed a strongly attenuated emission of MeSA after Psm avrRpm1 inoculation (Figure 8A). Similarly, compared with the Col-3 wild type, induced MeSA production was markedly reduced in the jin1 mutant carrying a defect in the transcription factor MYC2, which also acts downstream of JA (Lorenzo et al., 2004). By contrast, mutational defects in the JAR1 gene, encoding jasmonate amino acid synthetase (Staswick and Tiryaki, 2004), only moderately affected Psm avrRpm1–induced MeSA production (Figure 8A). These data indicate that MeSA production induced by avirulent P. syringae partially requires JA biosynthesis and depends on COI1- and MYC2-mediated downstream signaling.
Figure 8.
MeSA Production but Not SAR Is Regulated by JA Signaling.
(A) Leaf MeSA emission from Psm avrRpm1- or mock-inoculated JA pathway mutants and their corresponding wild-type lines (dde2, coi1, and jar1 are in Col-0, opr3 is in Ws, and jin1 is in Col-3 background). Volatiles were sampled from 0 to 24 HAI, and mean values (±sd, n = 4) are given. Asterisks indicate whether statistically significant differences exist between Psm avrRpm1–treated JA mutant plants and the corresponding wild type (**P < 0.01; *P < 0.05). Note the different scales of the y axes.
(B) SAR assessment via bacterial growth quantification in challenge-infected upper (2°) leaves of pretreated (1°) JA pathway mutants and respective wild-type plants. For experimental details, see legend to Figure 5C. Bars represent means (±sd) of cfu per cm2 from at least seven parallel samples. Asterisks denote statistically significant differences of bacterial growth in 2° leaves between Psm and MgCl2 pretreated plants of a particular line (***P < 0.001; **P < 0.01). No statistically significant differences (P > 0.05) exist between Psm-treated wild-type and mutant samples with respect to a particular background, indicating a similar strength of SAR induction for the different lines. Note the different scales of the y axes.
As part of the hypothesis that MeSA functions as a SAR signal (Park et al., 2007), JA was suggested to strengthen the MeSA component of SAR signaling (Vlot et al., 2008a, 2008b). Moreover, JA or related oxylipins were postulated to act as critical SAR long-distance signals in their own right (Truman et al., 2007), although the significance of JA for SAR long-distance signaling has recently been questioned (Chaturvedi et al., 2008). To clarify the importance of JA signaling during SAR, we examined whether biological induction of SAR occurs in Arabidopsis mutants defective in distinct steps of JA signaling. Compared with MgCl2 pretreated control plants, Psm preinoculated plants of opr3, jar1, and jin1 mutant lines were all able to significantly increase their resistance toward subsequent challenge infections in distant leaves (Figure 8B). Similarly, a statistically significant enhancement of resistance upon Psm pretreatment was observed for dde2 and coi1 mutant plants, which already exhibit a somewhat higher degree of basal resistance toward P. syringae than the Col-0 background line (Figure 8B; Kloek et al., 2001; Raake et al., 2006). These increases in whole-plant resistance upon localized Psm infection of the different JA-related mutants indicate that SAR can be established without a functional JA signaling pathway and thus rule out a function of JA or OPDA derivatives in SAR long-distance signaling. Together with our previous data (Figure 5), these findings also exclude a mechanism in which JA signaling strengthens SAR establishment through MeSA production.
Because most of the produced MeSA is emitted from leaves (Figure 1F), JA could negatively affect SA levels in plant pathogen interactions by promoting the conversion of SA to MeSA. However, considering this mechanism, the bsmt1 mutants should exhibit higher SA levels after pathogen infection than wild-type plants and show increased PR-1 gene expression, which is not the case (Figures 7C and 7E). To explain these unexpected results, we determined expression of ICS1 after pathogen infection in bsmt1 mutants and detected a slightly attenuated induction of the SA biosynthesis gene at 24 HAI compared with Col-0 (Figure 7F). Thus, although MeSA is not produced and emitted from bsmt1 plants after pathogen infection, induced SA levels might remain at a wild-type-like level in the mutants because transcription of SA biosynthesis is alleviated to a certain extent.
Virulent P. syringae Mediate Leaf MeSA Release but Not SAR via Coronatine
Coronatine is a phytotoxin produced by several P. syringae pathovars, including Psm and P. syringae pv tomato DC3000 (Pst; Bender et al., 1999). It acts as a bacterial virulence factor that counteracts SA-dependent plant defense reactions by acting as a structural and functional mimic of bioactive jasmonates, most notably JA-Ile (Brooks et al., 2005; Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008). The availability of coronatine-deficient (cor−) Pst mutants (Brooks et al., 2004) allowed us to test whether P. syringae–induced MeSA production would require the action of coronatine. Infection of Col-0 leaves with the coronatine-producing Pst wild-type strain evoked a strong emission of MeSA, which was similar in magnitude to the MeSA released after Psm infection (Figures 1B and 9A). By contrast, leaf MeSA emission from plants infected with the Pst cor− strain DB29 (Brooks et al., 2004) was only marginally elevated, falling by a factor of 60 below the amounts induced by wild-type Pst (Figure 9A). Because coronatine functions as a virulence factor to promote bacterial multiplication in planta (Brooks et al., 2005), we comparatively determined the growth of wild-type Pst and of Pst cor− at 24 HAI, the endpoint of MeSA sampling in the above experiment (Figure 9A). Leaf bacterial numbers were about twofold lower for Pst cor− than for Pst (see Supplemental Figure 3 online). However, this relatively small growth difference is not likely to account for the large differences in leaf MeSA emission observed after treatments of plants with Pst and Pst cor−, respectively. Thus, MeSA release from Pst-infected leaves is mainly triggered by the action of the phytotoxin coronatine. Since MeSA is produced from SA by BSMT1 and predominantly lost into the atmosphere (Figures 1 and 4; Chen et al., 2003), coronatine-mediated MeSA volatilization has the potential to decrease SA levels at infection sites and thus to constitute a bacterial virulence mechanism that negatively influences SA-based plant defenses.
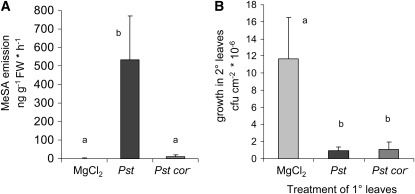
Figure 9.
P. syringae–Induced MeSA Formation but Not SAR Is Dependent on Bacterial Production of the Phytotoxin Coronatine.
(A) MeSA emission from Col-0 leaves after inoculation with coronatine-producing Pst, coronatine-deficient Pst cor−, and MgCl2 infiltration. Volatiles were sampled from 0 to 24 HAI, and mean values of ng emitted substance g−1 leaf FW h−1 (±sd, n = 7) are given. Different letters symbolize statistically significant differences between treatments (P < 0.002).
(B) SAR induction by Pst and Pst cor− in Col-0 plants. 1° leaves were infiltrated with MgCl2, Pst, or Pst cor− (OD 0.01 each), 2° leaves were challenge-infected 2 d later with Psm (OD 0.002), and quantities of Psm in 2° leaves were determined another 3 d later (see Figure 5C for details). Bars represent means (±sd) of cfu per cm2 from at least six parallel samples. Different characters symbolize statistically significant differences between treatments (P < 0.01).
Finally, to test whether bacterial induction of SAR is affected by the ability of Pst to produce coronatine, we comparatively analyzed the systemic resistance of Col-0 plants after a remote infection with Pst and with Pst cor−. Since the primary infection with Pst cor− triggered SAR to the same extent as infection with Pst (Figure 9B), SAR is established independently of coronatine in the Arabidopsis–Pseudomonas interaction. Because of the large discrepancies between MeSA production in Pst- and Pst cor−-infected plants, this result further corroborates our findings that MeSA formation is dispensable for SAR establishment in Arabidopsis.
DISCUSSION
The state of increased systemic disease resistance that develops during SAR requires elevated levels of SA and the mobilization of SA-dependent defenses in leaves distant from pathogen inoculation (Vernooij et al., 1994). The earliest candidate for a mobile long-distance signal traveling from inoculated to systemic tissue was SA itself. SA accumulates both at inoculation sites and in distant leaves concomitant with the onset of SAR, is found in phloem exudates of infected cucumber leaves, is distributed inside an Arabidopsis plant when applied externally to a single leaf, and its exogenous application increases whole-plant resistance in many species (Malamy et al., 1990; Métraux et al., 1990; Kiefer and Slusarenko, 2003). However, evidence from detailed physiological and grafting experiments has essentially excluded a function of SA as the phloem-mobile long-distance signal (Rasmussen et al., 1991; Vernooij et al., 1994). Still, instead of SA itself, modified forms, such as MeSA or SAG, are candidate molecules that might travel from inoculated to distant leaves. MeSA was recently proposed as being a critical, phloem-mobile SAR signal in tobacco. The respective model includes SA to MeSA conversion by SAMT in inoculated leaves, transport of MeSA to distant leaves, and subsequent reconversion to active SA by SA methyl esterase (Park et al., 2007). From SAR phenotypes of Arabidopsis lines in which different SA methyl esterase isoforms were concomitantly silenced, it was further concluded that MeSA functions as a conserved SAR signal in Arabidopsis and possibly other species (Vlot et al., 2008a, 2008b).
Our approach has tackled the problem from the side of MeSA production. BSMT1 belongs to a group of Arabidopsis methyl transferases and bears in vitro SA to MeSA converting activity (Chen et al., 2003). The BSMT1 gene is strongly upregulated in response to P. syringae leaf inoculation (Figure 3A), and its expression kinetics closely correlates with the timing of MeSA production (Figures 1A and 1B). Two independent Arabidopsis lines, bsmt1-1 and bsmt1-2, both with predicted T-DNA insertions in the BSMT1 coding region, not only fail to express the gene but also lack any pathogen-induced elevation of MeSA production (Figures 3C and 4). This demonstrates that BSMT1 is the single methyl transferase that catalyzes induced production of MeSA in Arabidopsis leaves. If MeSA were critical for SAR in Arabidopsis, the bsmt1 mutants would exhibit a SAR-compromised phenotype. Our findings that both bsmt1-1 and bsmt1-2 are able to mount a wild-type-like SAR response associated with conventional systemic SA elevation and PR gene expression shows that MeSA is dispensable for systemic SA accumulation and SAR in Arabidopsis (Figure 5). Thus, in this species, MeSA neither functions as a critical long-distance signal nor in any other SAR relevant process, including systemic SA accumulation. MeSA production is also not required for chemical induction of Arabidopsis resistance by the SA analog INA (Figure 6).
Our findings in Arabidopsis contradict the events described for TMV-induced SAR in tobacco (Park et al., 2007) and indicate the existence of species differences in the molecular nature of SAR long-distance signals. This is surprising because the SAR phenomenon has been observed in many plant species, and the associated responses, such as systemic SA accumulation, increased PR gene expression, or the timing of SAR induction, are well-conserved between species (Sticher et al., 1997). Nevertheless, we provide direct evidence that MeSA is not a conserved SAR signal in all species, and this is in sharp contrast with the previously proposed generalized model (Park et al., 2007; Vlot et al., 2008a, 2008b).
Mere physicochemical considerations and the experimentally determined in planta properties of MeSA also argue against a function of the molecule as an effective phloem-directed long-distance signal. Methylation of SA to MeSA does strongly increase membrane permeability and volatility, and this is reflected by our finding that the predominant part of the produced MeSA is lost into the atmosphere by emission, and only a small portion is retained in leaves or is detectable in petiole exudates (Figure 1; see Supplemental Figure 1 online). A directed and efficient mass flow of this volatile SA derivative through the phloem or other conductive parts of the stem therefore does not seem realistic. Moreover, the amount of MeSA accumulating after bacterial inoculation in leaf exudates during a 48-h SAR induction period is modest and falls well below the usually observed systemic elevation of SA levels observed during P. syringae–induced SAR in Arabidopsis (1 to 2 μg g−1; Mishina and Zeier, 2007; Mishina et al., 2008). Finally, we did not observe increases in MeSA content and detected only a small elevation of MeSA emission in noninoculated leaves after pathogen treatment (Figures 1D to 1F), indicating that a flow of MeSA from inoculated to systemic leaves, if present at all, is only marginal. This is consistent with the minor and statistically barely significant elevations of systemic MeSA reported previously (Park et al., 2007; Vlot et al., 2008b).
The major part of MeSA produced in P. syringae–inoculated Arabidopsis leaves is released into the atmosphere. For the incompatible Psm avrRpm1–Arabidopsis interaction, emission rates of 50 ng g−1 h−1 are accompanied by leaf contents of 20 to 25 ng g−1, meaning that the amounts retained in leaves equal the value emitted during ∼30 min (Figure 1). Although MeSA production starts later in the compatible Psm–Arabidopsis interaction, the values emitted around 24 HAI are about one order of magnitude higher than in the incompatible one. In total, ∼0.75 and 3.5 μg g−1 MeSA are volatilized during the first 24 HAI from leaves inoculated with Psm avrRpm1 and Psm, respectively (Figures 1A and 1B). Considering that in those interactions, SA and SAG accumulate in leaves at 24 HAI to ∼1 to 1.5 μg g−1 and 4 to 6 μg g−1, respectively (Figure 4B; Mishina et al., 2008), a marked percentage of the totally produced SA is lost as volatilized MeSA. The MeSA amounts emitted from pathogen-treated tobacco plants are of the same order of magnitude as those emitted from Arabidopsis. Shulaev et al. (1997) detected emission rates from TMV-infected tobacco leaves of ∼20 to 300 ng h−1 per plant.
We excluded MeSA as a phloem-mobile long-distance signal during SAR in Arabidopsis. However, considering the substantial levels of MeSA emitted from leaves, does MeSA act as an airborne SAR signal, as proposed previously (Shulaev et al., 1997)? The answer for Arabidopsis is clearly no, and this negative statement again relies on the wild-type-like SAR phenotype of the bsmt1 mutant plants that fail to elevate production and emission of MeSA after inoculation (Figures 3 to 5). It is noteworthy in this context that bsmt1 mutants also develop SAR when wild-type plants, which are possible sources of MeSA, are absent from the experimental growth chamber. A second reasoning is that in our experimental setting for SAR assessments, mock-treated and pathogen-inoculated plants are routinely located in direct proximity, and several leaves of differently treated plants are often in close contact. Nevertheless, we observe statistically robust differences in acquired resistance between mock- and pathogen-treated plants (Figure 5), indicating that signaling processes within the plant but not airborne communication dominate during SAR. Further, SAR is suppressed in cucumber (Cucumis sativus) plants when petioles of inoculated leaves are girdled, suggesting an intraplant and more specifically a phloem-based signal transmission pathway (Guedes et al., 1980; van Bel and Gaupels, 2004).
This does not rule out that under certain artificially provoked and nonphysiological conditions, gaseous MeSA from external sources or from plants is able to heighten plant resistance, presumably by leaf uptake followed by conversion to bioactive SA (Shulaev et al., 1997; Koo et al., 2007; Park et al., 2007). The minimum concentration of externally applied gaseous MeSA at which tobacco plants start to significantly elevate resistance is ∼10 μg L−1 (Shulaev et al., 1997), and concentrations of up to 1 mg L−1 have been used for this purpose in other experiments (Park et al., 2007). Considering the measured Psm-induced volatile emission in Col-0 plants during the first 48 h after inoculation (Figure 1B), and the 500-liter volume of the experimental compartment, and assuming a total of 50 Psm-treated plants from which three leaves (∼0.1 g fresh weight) each have been inoculated, we calculate a concentration of 0.1 μg L−1 MeSA in our experimental chambers during a SAR experiment. Even with this relatively high plant density, the restricted volume, and the high inoculation frequency, the calculated value is about two orders of magnitude lower than the minimum concentration previously determined to be sufficient for resistance induction (Shulaev et al., 1997). By contrast, when MeSA produced by donor plants is pointedly directed into low volume vessels containing acceptor plants, plant resistance might be elevated in the acceptor plants. For instance, considerable amounts of MeSA that were emitted from 150 SA-treated Arabidopsis plants overexpressing the BSMT1 rice (Oryza sativa) homolog were conducted into sealed 0.4-liter vessels containing Col-0 acceptor plants. This treatment increased expression of PR-1 in the acceptor plants (Koo et al., 2007). However, this highly directed bulk flow of gaseous MeSA into a small-volume acceptor compartment is rather artificial and hardly reflects the physiological circumstances occurring during SAR.
As a relatively strong acid with a pKa value of 3, nonderivatized SA predominantly exists as an anion in most subcellular compartments (an exception might be the fairly acidic vacuole), and its membrane permeability should therefore be low in the absence of a specific transport protein (Chatton et al., 1990). MeSA might thus represent a membrane-permeable, mobile form of SA able to travel over shorter cellular distances by diffusion. Our finding that MeSA but not SA levels increase in Arabidopsis leaf exudates after pathogen inoculation supports this view. Interestingly, SA glycosylation also enhances petiole exudation (see Supplemental Figure 1 online). However, overall exudation rates of SAG are too low to markedly contribute to the systemic rises of SA occurring during SAR via phloem-based long-distance transport. Moreover, the SAR-deficient Arabidopsis mutants npr1, ndr1, fmo1, and pad4 are able to elevate local production of SA (Figure 2B), MeSA (Figure 2C), and SAG (see Supplemental Figure 4 online) but fail to increase SA levels in distant leaves (Figure 2A). The likewise SAR-deficient phytochrome photoreceptor double mutant phyA phyB exhibits a similar behavior (Griebel and Zeier, 2008). Because there is no obvious physiological reason why these different mutational defects should all block systemic translocation of locally accumulating SA derivatives, it seems reasonable to assume that neither SA itself nor a modified form of SA, such as MeSA or SAG, travels from inoculated to distant leaves during SAR. Together with the observation that the SA biosynthesis gene ICS1 is strongly upregulated in distant leaves after local pathogen inoculation (Figure 5D), the above results support the hypothesis that the systemic rises in SA during SAR are achieved via de novo synthesis in distant leaves. This view is consistent with the outcome of SAR experiments using tobacco grafts with SA hydroxylase-expressing root stocks and wild-type scions (Vernooij et al., 1994).
A significant early production of JA occurs in Arabidopsis leaves following recognition of avirulent P. syringae (Mishina et al., 2008). According to the analyses of JA biosynthesis mutants (Figure 8A), this transient JA accumulation must be the main driving force for Psm avrRpm1–triggered MeSA production. By contrast, virulent strains, such as Psm or Pst, do not evoke significant rises in leaf JA levels during the first 2 d after infection when modest inoculum concentrations are applied (see below; Mishina and Zeier, 2007; Mishina et al., 2008). According to our results, the compatible bacteria rather use the phytotoxin and JA-Ile mimic coronatine to provoke leaf MeSA emission (Figure 9A). Further downstream of the JA pathway, both COI1 and MYC2-mediated signaling events are required for induced MeSA production (Figure 8A). The JA pathway-dependent regulation of MeSA formation is thus similar to the regulation of TMTT biosynthesis, the second significant Arabidopsis leaf volatile induced upon P. syringae attack (Attaran et al., 2008; Herde et al., 2008). Although production of the homoterpene TMTT is more tightly dependent on JA than synthesis of the phenylpropanoid MeSA, a common regulatory mechanism of these biochemically unrelated, major Arabidopsis leaf volatiles is apparent. The regulation of MeSA synthesis through the JA pathway occurs at the transcriptional level because exogenous treatment with methyl jasmonate is sufficient to trigger BSMT1 expression (Chen et al., 2003; Koo et al., 2007). Despite this coregulation, production of TMTT is not influenced by MeSA generation and vice versa (see Supplemental Figure 2 online; Attaran et al., 2008).
The significance of the JA pathway during SAR has recently been debated. On the one hand, a major role for JAs during SAR has been suggested, with JA or a related oxylipin derivative possibly initiating or directly mediating systemic long-distance signaling (Grant and Lamb, 2006; Truman et al., 2007). Experimental support for this proposition includes the finding that several JA pathway mutants show attenuated SAR in response to Pst avrRpm1, that foliar JA application enhances systemic resistance, and that JA levels increase in Arabidopsis leaf petiole exudates as well as in distant leaves after inoculation with high inoculum density (OD 0.2) of Pst avrRpm1 (Truman et al., 2007). Other experiments, on the other hand, argue against a role for JA as a mobile SAR signal. Chaturvedi et al. (2008) have shown that a SAR-inducing activity collected from petiole exudates of Pst avrRpm1–inoculated leaves does not copurify with JA, and that neither JA nor MeJA reconstitute an inducer activity in SAR-inactive leaf exudates. Our presented results rule out a decisive role of the JA pathway during SAR because systemic resistance in the JA biosynthesis mutants dde2 and opr3, as well as in the downstream signaling mutants coi1, jar1, and jin1, is significantly enhanced in response to a local Psm inoculation (Figure 8B). A SAR-positive phenotype for coi1 mutants has also been reported by Cui et al. (2005). The correlation between SAR, JA petiole exudation, and systemic JA elevation reported by Truman et al. (2007) is questionable because it was not tested in this study whether the high inoculum (OD 0.2) used for analytical JA determinations indeed induces a SAR response. Instead, bacterial ODs that were several orders of magnitude lower than 0.2 were used by Truman et al. (2007) for SAR bioassays. Previous experiments with various bacterial inoculation densities conducted in our laboratory indicate that the magnitude of P. syringae–induced SAR is low for high inoculation densities (OD 0.2), although these ODs provoke, besides heavy tissue necrosis, strong JA elevation at inoculation sites. By contrast, modest inoculi (OD 0.005 to 0.02), which result in much lower or even no detectable rises of local JA, trigger a significantly stronger SAR response (Mishina and Zeier, 2007). In addition, we have never detected increased levels of JA or OPDA in distant tissue under these conditions (Mishina et al., 2008). Taken together, data from our and other laboratories (Cui et al., 2005; Chaturvedi et al., 2008) argue against a significant function of the JA pathway during SAR establishment and long-distance signaling. Moreover, the wild-type-like SAR-inducing capacity of Pst cor− mutants reveals that bacterial production of the JA-Ile-mimicking phytotoxin coronatine does not affect the SAR process, neither positively nor negatively (Figure 9B). SAR induction through Pst cor− is associated with a largely suppressed leaf MeSA production (Figure 9A), and this further corroborates the dispensability of MeSA during SAR in Arabidopsis.
In summary, our data exclude an essential function of both MeSA and JA signaling during systemic long-distance signaling and SAR in Arabidopsis. Other hitherto unidentified molecules are likely to travel from inoculated to distant tissue in this species to set in gear signal transduction and amplification mechanisms in distant leaves. The latter processes can then drive the systemic de novo biosynthesis of SA, which in turn is known to trigger expression of PR genes and SAR (Cao et al., 1994). A conceivable function of SA methylation in plant defense is to prevent SA levels from accumulating to toxic concentrations by vaporization of volatile MeSA into the atmosphere. JA may regulate this process because it promotes SA to MeSA conversion (Figure 8A). Analyses of bsmt1 mutants cannot definitively prove this statement because MeSA depletion in these plants seems to negatively affect SA biosynthesis at the transcriptional level (Figure 7F). In addition to MeSA volatilization, SAG formation and subsequent vacuolar storage is an alternative way to handle an excess of SA (Lee et al., 1995; Dean et al., 2005). MeSA formation might also influence the interplay between SA and JA, which trigger distinct sets of defense responses and thereby often behave in a counteractive manner (Traw et al., 2003; Koornneef et al., 2008). JA-mediated MeSA production and subsequent release of the volatile might thus be one means by which negative crosstalk between SA and JA signaling is realized. Moreover, the strong induced production of MeSA by coronatine suggests a bacterial virulence mechanism through negative interference with the SA defense pathway: coronatine triggers SA to MeSA conversion, and the subsequent emission of volatile MeSA from the plant results in a lowering of the leaf SA pool. In support of this, coronatine-mediated attenuation of plant SA accumulation and downstream defenses have been reported previously (Brooks et al., 2005; Uppalapati et al., 2007). In this context, it is interesting to note that overexpression of the rice homolog of BSMT1 in Arabidopsis resulted in constitutively enhanced MeSA emission and attenuated disease resistance due to SA depletion (Koo et al., 2007).
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown on an autoclaved mixture of soil (Klasmann), vermiculite, and sand (10:0.5:0.5) in a controlled environmental chamber (J-66LQ4; Percival) with a 9-h day (photon flux density 70 μmol m−2 s−1)/15-h night cycle and a relative humidity of 70%. Growth temperatures during the day and night period were 21 and 18°C, respectively. Experiments were performed with 6-week-old naïve and unstressed plants exhibiting a uniform appearance. If not otherwise stated, Arabidopsis accession Col-0 was used for experiments.
The bstm1-1 and bstm1-2 mutant lines represent the T-DNA insertion lines SALK_140496 and WiscDSLox430E05, respectively, which are both in the Col background. Homozygous insertion mutants were identified by PCR, using gene-specific (BSMT1-1-forward, 5′-GCAAAAACTTCAAATATATTATGCATG-3′; BSMT1-1-reverse, 5′-GAAATCATTTTCCGGGAGATC-3′; BSMT1-2-forward, 5′-ATAAAACGGCATGTTGAATGC-3′; BSMT1-2-reverse, 5′- GGTCCAGTATCACATTATCACGG -3′) and T-DNA-specific primers as described by Alonso et al. (2003). The JA pathway mutants opr3 (Stintzi and Browse, 2000) and jin1 (Berger et al., 1996) are in the Ws and Col-3 backgrounds, respectively. All other Arabidopsis lines used in this study (dde2-2 [von Malek et al., 2002], coi1-35 [Staswick and Tiryaki, 2004], jar1-1 [Staswick and Tiryaki, 2004], sid2-1 [Nawrath and Métraux, 1999], NahG [Lawton et al., 1995], npr1-2 [NASC line N3801], ndr1 [Century et al., 1995], fmo1 [Mishina and Zeier, 2006], and pad4-1 [Glazebrook et al., 1997]) have background Col-0.
Cultivation of Bacteria
Pseudomonas syringae pv maculicola strain ES4326 (Psm), Psm carrying the avrRpm1 avirulence gene (Psm avrRpm1), P. syringae pv tomato DC3000 (Pst; strain KP105; Brooks et al., 2004), and Pst cor− (strain DB 29; Brooks et al., 2004) were grown in King's B medium containing the appropriate antibiotics at 28°C. Overnight log phase cultures were washed three times with 10 mM MgCl2 and diluted to different final optical densities for leaf inoculations.
Assessment of SAR and Local Resistance Responses
For SAR experiments, plants were first infiltrated into three lower (1°) leaves with a suspension of Psm (OD = 0.01) or with 10 mM MgCl2 as a control treatment. Two days after the primary treatment, upper (2°) leaves were either harvested for SA determination and gene expression analysis or inoculated with Psm (OD 0.002). Growth of Psm in 2° leaves was scored another 3 d later by homogenizing discs originating from infiltrated areas of three different leaves in 1 mL 10 mM MgCl2, plating appropriate dilutions on King's B medium, and counting colony numbers after incubating the plates at 28°C for 2 d.
For the determination of local defense responses, bacterial suspensions of OD 0.005 (determination of gene expression, metabolite levels, and Psm avrRpm1 growth assay) or OD 0.002 (Psm growth assays) were infiltrated into three full-grown leaves per plant. Bacterial growth was assessed 3 d after infiltration as described above.
INA-induced resistance was assessed by spraying whole plants with a solution of 0.65 mM INA or water as a control, leaf inoculation of Psm (OD 0.002) 2 d later, and determination of bacterial growth as described above.
Determination of VOC Emission Including MeSA
To assess P. syringae–induced plant VOC emission, including emission of MeSA, bacterial suspensions of OD 0.01 were infiltrated from the abaxial side into seven full-grown rosette leaves per Arabidopsis plant using a 1-mL syringe without a needle. Control treatments were performed by infiltrating a 10 mM MgCl2 solution. To determine induced MeSA production in noninoculated systemic leaves, four lower leaves per plant were treated and removed at 2 DAI when SAR is just induced in the pathosystem (Mishina et al., 2008). The remainder plant was then sampled for VOC emission from day 2 to day 3 after inoculation.
Volatiles emitted by individual plants were collected in a push/pull apparatus as described by Attaran et al. (2008). Plants were placed in collection chambers ∼30 min after leaf infiltrations and trapping filters consisting of glass tubes packed with Super-Q absorbent (VCT-1/4X3-SPQ; Analytical Research Systems) were attached. Charcoal-filtered and humidified air was pushed into each sampling chamber at a rate of 1.2 L min−1. The air flow containing plant volatiles was pulled through the trapping filter with a vacuum pump (ME2; Vacuubrand), and volatiles were collected for 10 to 24 h.
After each collection, trapping filters were eluted with 1 mL CH2Cl2, and 200 ng of n-octane was added as internal standard. The mixture was concentrated to a volume of 25 μL under a gentle stream of nitrogen, strictly avoiding evaporation to dryness, and analyzed by GC-MS. Aliquots (3 μL) of the sample mixture were separated on a GC (6890N; Agilent Technologies) that was equipped with a split/splitless injector and a fused silica capillary column (HP-1; 30 m × 0.25 mm ID, 0.25 μm film thickness) and combined with a 5975 mass spectrometric detector (Agilent Technologies). Samples were injected in pulsed splitless mode, and helium was used as a carrier gas. The temperature of the oven was held at 50°C for 2 min and then increased at 8°C/min to 300°C. Mass spectra were recorded at 70 eV. Substances were identified by comparison of mass spectra with those of the National Institute of Standards and Technology (NIST 98) reference library. Compound identities were confirmed by comparison of mass spectra and retention times with those of standard substances. To allow sensitive quantification of VOCs, substance peaks originating from selected ion chromatograms were integrated (generally m/z 120 for MeSA and m/z 81 for TMTT). The resulting peak areas were related to the peak area of the n-octane standard (ion chromatogram m/z 114), whereby experimentally determined correction factors were considered for each substance.
Determination of Leaf MeSA Contents
Frozen leaf tissue (150 mg) was homogenized with 600 μL of extraction buffer (water:1-propanol:HCl = 1:2:0.005). After addition of 200 ng D3-methylsalicylate (Sigma-Aldrich) as internal standard and 1 mL of methylene chloride, the mixture was shaken thoroughly and centrifuged at 14,000 rpm for phase separation. The lower, organic phase was removed, dried over Na2SO4, and subject to a vapor phase extraction procedure using a Super-Q collector trap. The final evaporation temperature was set to 200°C, and samples were eluted from the collector trap with 1 mL methylene chloride. Finally, the sample volume was reduced to 25 μL in a stream of nitrogen, and GC-MS analysis was performed as described above.
Determination of Leaf SA, SAG, and JA Levels
Leaf SA, SAG, and JA contents were determined by vapor-phase extraction and subsequent GC-MS analysis according to Mishina and Zeier (2006).
Collection of Leaf Petiole Exudates and Exudate Analyses
Petiole exudates were collected essentially as described previously (Maldonado et al., 2002; Chaturvedi et al., 2008). Plant leaves were either infiltrated with a suspension of Psm (OD 0.01) or with 10 mM MgCl2 as a mock inoculation. Six hours after infiltration, leaves were cut at the base of their petioles and the cut surface sterilized by successive dipping for 10 s in 50% ethanol and in 0.0005% bleach. After rinsing petioles with sterile 1 mM EDTA, pH 8.0, they were submerged in fresh EDTA-solution for exudate collection. Twelve-well tissue culture plates were used for this purpose, whereas each well was filled with 2.5 mL of collection solution and equipped with 10 harvested leaves. Exudates were continuously collected in the period from 6 to 48 HAI.
For MeSA analyses, 10 mL of pooled exudate solution was extracted three times with 3 mL of CH2Cl2 after 200 ng D3-MeSA was added as internal standard. The combined organic extracts were analyzed by vapor phase extraction and GC-MS as described above.
For SA determination, the aqueous phase remaining after solvent extraction was acidified with 0.1 M HCl to a final pH of 3, supplemented with internal standard (200 ng of D6-SA; Sigma-Aldrich), and extracted three times with 3 mL of CH2Cl2/methanol (2:1, v/v). The combined organic phases were analyzed according to Mishina and Zeier (2006). For SAG analysis, the acidic aqueous phase remaining after solvent extraction was brought to pH 1.0 with HCl and heated for 30 min at 100°C, and the free SA liberated by hydrolysis was determined as described above.
Analysis of Gene Expression
Expression levels of PR-1 and BSMT1 were determined by RNA gel blot analysis as outlined by Mishina and Zeier (2006). ICS1 expression was analyzed by quantitative real-time PCR, essentially as described by Schlaeppi et al. (2008). Total RNA was isolated from frozen leaves using peqGOLD RNAPure reagent (PeqLab). RNA samples were reverse transcribed using an Omniscript Reverse Transcription kit (Qiagen) with 1 μg of total RNA. The resulting cDNA samples were diluted 10-fold with water, and quantitative real-time PCR was performed in triplicate using the SensiMixPlus SYBR kit (Quantace) in a Rotor-Gene 2000 apparatus (Corbett Research). In a 15-μL reaction volume, 5 μL of the cDNA sample was combined with 7.5 μL of 2 SYBR Green mix, 1.5 μL water, and 0.5 μL of each primer (both at 10 μM). The cycling included 95°C for 10 min, followed by 45 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and finally 72°C for 3 min. The following gene-specific primers were used: 5′-TTCTGGGCTCAAACACTAAA-AC-3′ (ICS1-forward) and 5′- GGCGTCTTGAAATCTCCATC-3′ (ICS1-reverse). The At1g62930 gene, which is no-responsive to P. syringae inoculation (Czechowski et al., 2005), was used as a reference gene and amplified with the primers 5′-GAGTTGCGGGTTTGTTGGAG-3′ (At1g62930-forward) and 5′-CAAGACAG-CATTTCCAGATAGCAT-3′ (At1g62930-reverse). The data were analyzed using the Rotor-Gene 6000 software, setting the threshold of the normalized fluorescence to 0.15, which corresponded to the exponential phase of the fluorescence signal. The resulting CT and E values were used to calculate the relative mRNA abundance according to the ΔΔCT method. The values were normalized to those for the reference gene and expressed relative to the MgCl2-treated wild-type control sample.
Reproducibility of Experiments and Statistical Analyses
All pathogen experiments and the respective bacterial growth analyses, metabolite determinations, and gene expression analyses depicted in the figures were conducted three times with similar results or tendencies. Statistical analyses were performed using Student's t test for comparison of two data sets and using analysis of variance (Fisher's Least Significant Difference test) to analyze multiple data sets from comparable treatments.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g11480 (BSMT1), At2g14610 (PR-1), and At1g74710 (ICS1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Petiole Exudation of SA Derivatives from P. syringae- and Mock-Inoculated Col-0 Leaves.
Supplemental Figure 2. TMTT Emission from Wild-Type Col-0 and bsmt1 Mutant Plants.
Supplemental Figure 3. Growth of Pst and Pst cor− in Col-0 Leaves.
Supplemental Figure 4. SAG Accumulation in P. syringae–Treated Wild-Type and SAR-Defective Mutant Plants.
Supplementary Material
Acknowledgments
E.A. is a fellow of the German Academic Exchange Service (DAAD). Financial support from the German Research Foundation (DFG Graduiertenkolleg 1342) is gratefully acknowledged. We are indebted to Barbara Kunkel for kindly providing the Pst strains DB29 and KP105 and thank Jean-Pierre Métraux and Felix Mauch for critically reading the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jürgen Zeier (juergen.zeier@unifr.ch).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ament, K., Van Schie, C.C., Bouwmeester, H.J., Haring, M.A., and Schuurink, R.C. (2006). Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224 1197–1208. [DOI] [PubMed] [Google Scholar]
- Arimura, G.I., Garms, S., Maffei, M., Bossi, S., Schulze, B., Leitner, M., Mithoefer, A., and Boland, W. (2008). Herbivore-induced terpenoid emission in Medicago truncatula: Concerted action of jasmonate, ethylene and calcium signaling. Planta 227 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran, E., Rostás, M., and Zeier, J. (2008). Pseudomonas syringae elicits emission of the terpenoid (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene in Arabidopsis leaves via jasmonate signaling and expression of the terpene synthase TPS4. Mol. Plant Microbe Interact. 21 1482–1497. [DOI] [PubMed] [Google Scholar]
- Bender, C.L., Alarcon-Chaidez, F., and Gross, D.C. (1999). Pseudomonas syringae phytotoxins: Mode of action, regulation and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of atvsp in response to methyl jasmonate and wounding. Plant Physiol. 111 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, A., and Innes, R.W. (1994). A disease resistance gene in Arabidopsis with specificity for 2 different pathogen avirulence genes. Plant Cell 6 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, D.M., Bender, C.L., and Kunkel, B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6 629–639. [DOI] [PubMed] [Google Scholar]
- Brooks, D.M., Hernández-Guzmán, G., Kloek, A.P., Alarcón-Chaidez, F., Sreedharan, A., Rangaswamy, V., Peñaloza-Vázquez, A., Bender, C.L., and Kunkel, B.N. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato strain DC3000. Mol. Plant Microbe Interact. 17 162–174. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X.N. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton, J.Y., Besseghir, K., and Roch-Ramel, F. (1990). Salicylic acid permeability properties of the rabbit cortical collecting duct. Am. J. Physiol. 259 F613–F618. [DOI] [PubMed] [Google Scholar]
- Chaturvedi, R., Krothapalli, K., Makandar, R., Nandi, A., Sparks, A.A., Roth, M.R., Welti, R., and Shah, J. (2008). Plastid omega 3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54 106–117. [DOI] [PubMed] [Google Scholar]
- Chen, F., D'Auria, J.C., Tholl, D., Ross, J.R., Gershenzon, J., Noel, J.P., and Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36 577–588. [DOI] [PubMed] [Google Scholar]
- Cui, J., Bahrami, A.K., Pringle, E.G., Hernandez-Guzman, G., Bender, C.L., Pierce, N.E., and Ausubel, F.M. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 102 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.V., and Delaney, S.P. (2008). Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132 417–425. [DOI] [PubMed] [Google Scholar]
- Dean, J.V., Mohammed, L.A., and Fitzpatrick, T. (2005). The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221 287–296. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal-transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y.J., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394 585–588. [DOI] [PubMed] [Google Scholar]
- Dong, X., Mindrinos, M., Davis, K.R., and Ausubel, F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. [DOI] [PubMed] [Google Scholar]
- Effmert, U., Saschenbrecker, S., Ross, J., Negre, F., Fraser, C.M., Noel, J.P., Dudareva, N., and Piechulla, B. (2005). Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 66 1211–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S.W., Chiang, Y., Acton, T.B., Montelione, G.T., Pichersky, E., Klessig, D.F., and Tong, L. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 102 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. [DOI] [PubMed] [Google Scholar]
- Grant, M., and Lamb, C. (2006). Systemic immunity. Curr. Opin. Plant Biol. 9 414–420. [DOI] [PubMed] [Google Scholar]
- Griebel, T., and Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes, M.E.M., Richmond, S., and Kuc, J. (1980). Induced systemic resistance to anthracnose in cucumber as influenced by the location of the inducer inoculation with Colletotrichum lagenarium and the onset of flowering and fruiting. Physiol. Plant Pathol. 17 229–233. [Google Scholar]
- Herde, M., Gartner, K., Kollner, T.G., Fode, B., Boland, W., Gershenzon, J., Gatz, C., and Tholl, D. (2008). Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C-16-homoterpene TMTT. Plant Cell 20 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir, L., Schilmiller, A.L., Staswick, P.E., He, S.Y., and Howe, G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer, I.W., and Slusarenko, A.J. (2003). The pattern of systemic acquired resistance induction within the Arabidopsis rosette in relation to the pattern of translocation. Plant Physiol. 132 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26 509–522. [DOI] [PubMed] [Google Scholar]
- Koo, Y.J., et al. (2007). Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol. Biol. 64 1–15. [DOI] [PubMed] [Google Scholar]
- Koornneef, A., Leon-Reyes, A., Ritsema, T., Verhage, A., Den Otter, F.C., Van Loon, L.C., and Pieterse, C.M.J. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8 863–870. [DOI] [PubMed] [Google Scholar]
- Lawton, K.A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Kessmann, H., Staub, T., and Ryals, J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10 71–82. [DOI] [PubMed] [Google Scholar]
- Lee, H.I., Leon, J., and Raskin, I. (1995). Biosynthesis and metabolism of salicylic-acid. Proc. Natl. Acad. Sci. USA 92 4076–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.I., and Raskin, I. (1999). Purification, cloning, and expression of a pathogen inducible UDP-glucose:salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 274 36637–36642. [DOI] [PubMed] [Google Scholar]
- Li, C., Williams, M.M., Loh, Y.-T., Lee, G.I., and Howe, G.A. (2002). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, E.K., Doucet, C.J., Li, Y., Elias, L., Worrall, D., Spencer, S.P., Ross, J., and Bowles, D.J. (2002). The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277 586–592. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sánchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J., Carr, J.P., Klessig, D.F., and Raskin, I. (1990). Salicylic acid - A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250 1002–1004. [DOI] [PubMed] [Google Scholar]
- Malamy, J., Hennig, J., and Klessig, D.F. (1992). Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic-virus infection. Plant Cell 4 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., and Cameron, R.K. (2002). A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419 399–403. [DOI] [PubMed] [Google Scholar]
- Melotto, M., Mecey, C., Niu, Y., Chung, H.S., Katsir, L., Yao, J., Zeng, W., Thines, B., Staswick, P., Browse, J., Howe, G.A., and He, S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux, J.P., Nawrath, C., and Genoud, T. (2002). Systemic acquired resistance. Euphytica 124 237–243. [Google Scholar]
- Métraux, J.P., Signer, H., Ryals, J., Ward, E., Wyssbenz, M., Gaudin, J., Raschdorf, K., Schmid, E., Blum, W., and Inverardi, B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250 1004–1006. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E., Griebel, T., Geuecke, M., Attaran, E., and Zeier, J. (2008). New insights into the molecular events underlying systemic acquired resistance. Paper 81 in: M. Lorito, S.L. Woo, F. Scala, eds, Biology of Plant-Microbe Interactions CD, Vol 6, International Society for Molecular Plant-Microbe Interactions, St. Paul, MN.
- Mishina, T.E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50 500–513. [DOI] [PubMed] [Google Scholar]
- Mueller, S., Hilbert, B., Dueckershoff, S., Roitsch, T., Krischke, M., Mueller, M.J., and Berger, S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, A., Welti, R., and Shah, J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D.F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318 113–116. [DOI] [PubMed] [Google Scholar]
- Raake, I., Mueller, M.J., and Berger, S. (2006). Defects in allene oxide synthase and OPDA reductase alter the resistance to Pseudomonas syringae and Botrytis cinerea. J. Phytopathol. 154 740–744. [Google Scholar]
- Rasmussen, J.B., Hammerschmidt, R., and Zook, M.N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 97 1342–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-L-methionine: Salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367 9–16. [DOI] [PubMed] [Google Scholar]
- Schlaeppi, K., Bodenhausen, N., Buchala, A., Mauch, F., and Reymond, P. (2008). The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55 774–786. [DOI] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signaling by methyl salicylate in plant pathogen resistance. Nature 385 718–721. [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Métraux, J.P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35 235–270. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Traw, M.B., Kim, J., Enright, S., Cipollini, D.F., and Bergelson, J. (2003). Negative cross-talk between salicylate- and jasmonate-mediated pathways in the Wassilewskija ecotype of Arabidopsis thaliana. Mol. Ecol. 12 1125–1135. [DOI] [PubMed] [Google Scholar]
- Truman, W., Bennett, M.H., Kubigsteltig, I., Turnbull, C., and Grant, M. (2007). Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 104 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati, S.R., Ishiga, Y., Wangdi, T., Kunkel, B.N., Anand, A., Mysore, K.S., and Bender, C.L. (2007). The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 20 955–965. [DOI] [PubMed] [Google Scholar]
- van Bel, A.J.E., and Gaupels, F. (2004). Pathogen resistance and alarm signals via the phloem. Mol. Plant Pathol. 5 495–504. [DOI] [PubMed] [Google Scholar]
- Van Poecke, R.M.P., Posthumus, M.A., and Dicke, M. (2001). Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene expression analysis. J. Chem. Ecol. 27 1911–1928. [DOI] [PubMed] [Google Scholar]
- Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditzjawhar, R., Ward, E., Uknes, S., Kessmann, H., and Ryals, J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C., Klessig, D.F., and Park, S.-W. (2008. a). Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11 436–442. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C., Liu, P.-P., Cameron, R.K., Park, S.-W., Yang, Y., Kumar, D., Zhou, F., Padukkavidana, T., Gustafsson, C., Pichersky, E., and Klessig, D.F. (2008. b). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56 445–456. [DOI] [PubMed] [Google Scholar]
- von Malek, B., van der Graaff, E., Schneitz, K., and Keller, B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216 187–192. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. (2006). Variations on a theme: Synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 9 288–296. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Xia, Y.J., Suzuki, H., Borevitz, J., Blount, J., Guo, Z.J., Patel, K., Dixon, R.A., and Lamb, C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Xu, R., Ma, C.J., Vlot, A.C., Klessig, D.F., and Pichersky, E. (2008). Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 147 1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Dai, Y., Xiong, Y., DeFraia, C., Li, J., Dong, X., and Mou, Z. (2007). Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 52 1066–1079. [DOI] [PubMed] [Google Scholar]
- Zhu, J.W., and Park, K.C. (2005). Methyl salicylate, a soybean aphid-induced plant volatile attractive to the predator Coccinella septempunctata. J. Chem. Ecol. 31 1733–1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.