Abstract
This work examines the involvement of chromatin looping in the transcriptional regulation of two epialleles of the maize (Zea mays) b1 gene, B-I and B'. These two epialleles are tissue-specifically regulated and are involved in paramutation. B-I and B' are expressed at high and low levels, respectively. A hepta-repeat ∼100 kb upstream of the transcription start site (TSS) is required for both paramutation and high b1 expression. Using chromosome conformation capture, we show that the hepta-repeat physically interacts with the TSS region in a tissue- and expression level–specific manner. Multiple repeats are required to stabilize this interaction. High b1 expression is mediated by a multiloop structure; besides the hepta-repeat, other sequence regions physically interact with the TSS as well, and these interactions are epiallele- and expression level–specific. Formaldehyde-assisted isolation of regulatory elements uncovered multiple interacting regions as potentially regulatory.
INTRODUCTION
Epigenetic regulation plays a crucial role in the control of eukaryotic gene expression (Henderson and Jacobsen, 2007; Martin and Zhang, 2007; Suzuki and Bird, 2008). Epigenetic mechanisms include DNA methylation and posttranslational modifications of histones and mediate heritable changes in gene expression without changing the DNA sequence. Our research focuses on an epigenetic phenomenon at the maize (Zea mays) booster 1 (b1) locus. The b1 gene is tissue-specifically expressed and encodes a transcription factor regulating the biosynthesis of flavonoid pigments (Coe, 1959). There are many different b1 alleles known (Selinger and Chandler, 1999). Two b1 alleles, B-I and B', are epialleles. They have an identical DNA sequence up to at least ∼110 kb upstream of the transcription start site (TSS; Stam et al., 2002a) but are nevertheless expressed at a different level. B-I is transcribed at a 10- to 20-fold higher level than B', and as a consequence, plants carrying B-I have high and plants carrying B' low pigment levels (Coe, 1966; Patterson et al., 1993). B-I and B' participate in paramutation, an epigenetic phenomenon involving a trans-interaction between alleles that results in a mitotically and meiotically heritable change in expression of one of the alleles (reviewed in Chandler, 2004; Louwers et al., 2005; Stam and Mittelsten Scheid, 2005). When B-I plants are crossed to B' plants, the low expressed B' epiallele changes the high expressed B-I epiallele into B' at a 100% frequency. Multiple tandem 853-bp repeats ∼100 kb upstream of the TSS are required for paramutation and for high b1 expression (Stam et al., 2002a, 2002b). B-I and B' contain seven copies of this 853-bp sequence, together called the hepta-repeat. Alleles with one copy of the repeat do not participate in paramutation (Stam et al., 2002a). The difference in expression level between B-I and B' is associated with differences in chromatin structure, particularly at the hepta-repeat (Stam et al., 2002a).
To investigate how the hepta-repeat functions, we applied chromosome conformation capture (3C) technology (Dekker et al., 2002; Tolhuis et al., 2002; Lanzuolo et al., 2007; Palstra et al., 2008). This technique, which has been previously applied to yeast, mammals, and Drosophila melanogaster, allowed the identification of physical interactions between DNA sequence regions at the b1 locus. Here, we show evidence for epiallele-specific physical chromatin interactions at the maize b1 locus, providing insight into the regulation of b1 expression. Tissue-specific as well as epiallele- and expression level–dependent interactions are observed between specific regions throughout the locus. The hepta-repeat interacts mainly in a tissue-specific manner, whereas regions other than the hepta-repeat interact in an epiallele- and expression level–specific manner. Formaldehyde-assisted isolation of regulatory elements (FAIRE; Hogan et al., 2006; Giresi et al., 2007) suggests that the latter regions have a regulatory function. The examination of chromatin looping adds a new dimension to the study of gene regulation in plants.
RESULTS
3C at the b1 Locus: Experimental Approach
To test if physical, long-distance interactions are involved in gene regulation at the b1 locus, we applied the 3C technology (Dekker et al., 2002; Tolhuis et al., 2002; Hagege et al., 2007). 3C uses formaldehyde to cross-link DNA and proteins, conserving the three-dimensional chromosomal conformation. The cross-linked chromatin is then subjected to restriction digestion followed by intramolecular ligation. After reversing the cross-links, the DNA is purified and analyzed by quantitative PCR (qPCR), in our case using TaqMan probes (Splinter et al., 2006; Hagege et al., 2007). The quantity of specific ligation fragments is a measure of the interaction frequencies between chromatin regions in vivo. In all 3C experiments discussed in this article, BglII was used as a restriction enzyme. The BglII restriction fragments analyzed are indicated with a Roman numeral in the schematic representation of the b1 locus in Figure 1A. The extensive presence of transposon and other repeated sequences at the b1 locus (gray boxes in Figure 1A) prevented analysis of additional BglII fragments.
Figure 1.
Features of the b1 and Sam Loci.
(A) Schematic representation of the b1 (top panel) and Sam (bottom panel) loci. The b1 TSS is indicated at 0 kb with a hooked arrow; the open box represents the b1 coding region, and the hepta-repeat is indicated with arrowheads. Distances are indicated in kilobases relative to the TSS. The BglII fragments analyzed by 3C are indicated with Roman numerals. Gray bars represent transposon and other repetitive sequences (Stam et al., 2002b). Dr1 and Dr2, direct repeats 1 and 2; IR, inverted repeat; Uni, Unigene Zm.5756. The Sam locus was used to normalize the 3C, FAIRE, and ChIP data obtained for b1. The hooked arrow indicates the TSS (BT042811.1). Black box, Sam probe used for RNA gel blot shown in (B); B, BglII sites; triangles, 3C primers.
(B) RNA gel blot analyses of RNA isolated from B-I, B', and B-615 inner tissue and husk. Pictures from each type of tissue are shown above the respective lane; inner tissue consists of young tissue present inside the stem (see Methods). The blots were hybridized with probes recognizing the coding region of b1 and Sam. The arrow indicates the full-length b1 transcript. The green color of B-615 plant tissue is due to the presence of a very weak or null Pl1 allele and does not reflect the b1 expression level (Stam et al., 2002b). The bottom panel shows the corresponding ethidium bromide–stained gel. I, inner tissue; H, husk tissue.
(C) The band intensities of the full-length b1 and Sam transcripts of two independent experiments (gray and black bars) were quantified, the background signals subtracted, and the b1/Sam ratio calculated and depicted in the bar graph. The gray bars correspond to the blot shown in (B).
The necessary controls were included in the 3C experiments. To determine the PCR amplification efficiency of a given primer set, the pBACB'1 clone was used. This clone, which contains an ∼107-kb chromosomal region including the hepta-repeat and part of the b1 coding region (exons 1 and 2; Stam et al., 2002b), was digested and ligated, yielding a 3C control template that contains all possible ligation products in equimolar amounts. To account for differences in quality and quantity of the various DNA templates, we measured interaction frequencies between two BglII fragments at an unrelated, constitutively expressed locus, S-adenosyl-methionine decarboxylase (Sam; Figure 1A), and used these interaction frequencies to normalize the 3C data obtained for b1. The Sam gene is expressed at a similar level in the tissues examined (Figure 1B), and it is therefore reasonable to assume it adopts a similar conformation in these tissues.
To distinguish expression level–specific interactions from tissue-specific interactions, 3C was performed on two types of tissues of B-I and B' plants: inner stem and husk tissue. In inner stem tissue, which consists of young sheaths and leaves surrounding the shoot meristem, full-length b1 transcripts were not detected in B-I and B' (Figure 1B). In husk tissue, which consists of the leaves surrounding the corncob, expression of b1 is transcriptionally activated, resulting in a high expression level in B-I and a low expression level in B' husk tissue (Figure 1B). Comparison of the 3C data obtained with these four tissues allowed us to separate tissue-specific interactions (occurring primarily in either inner or husk tissue) from interactions that correlate with the b1 expression level.
Tissue- and Expression Level–Specific Physical Interactions throughout the b1 Locus
To obtain insight into the spatial organization of the b1 locus and to identify physical interactions associated with the transcriptional differences between B-I and B', 3C-qPCR technology was applied. The locus-wide cross-linking frequencies were determined for three fragments within the b1 locus (fixed fragments). To measure interaction frequencies with the TSS region, BglII fragment I, containing proximal b1 promoter sequences (Figure 1A), was used as a fixed fragment.
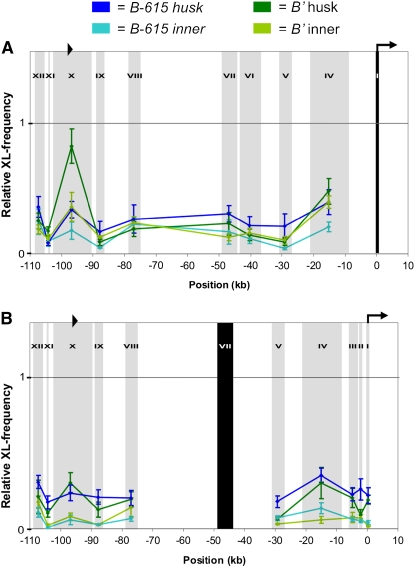
Previous work showed that the hepta-repeat is required for the high B-I expression level (Stam et al., 2002a), indicating it contains a transcriptional enhancer. Here, we show that in high expressing B-I husk tissue, high cross-linking frequencies were observed between fragment I and fragment X, which contain the TSS and the hepta-repeat, respectively (Figure 2A). Interestingly, elevated cross-linking frequencies between fragments I and X were also observed in low expressing B' husk tissue, although the height of the peak was significantly lower than in B-I husk (Student's t test, confidence interval [CI] = 90%, see Methods; Figure 2A). In contrast with husk tissue, in the B-I and B' inner stem tissues, low interaction frequencies were measured between fragments I and X (Figure 2A). These results demonstrate that the physical interaction between the TSS region and the hepta-repeat is tissue-specific: it occurs significantly more frequently in husk than in inner tissue (Student's t test, CI = 99% for B-I and 95% for B'). In addition, this interaction is in part expression level-specific: the interaction frequency is higher in the high expressing B-I than in the low expressing B' husk tissue, suggesting this interaction plays a role in enhancing b1 expression. The 3C data with fragment I as the fixed fragment indicated additional interactions within the b1 locus. Interestingly, these interactions correlate with high b1 expression. In high expressing B-I husk tissue, we detected high interaction frequencies between fragments I and VII (∼47 kb upstream of TSS) and between I and XII (∼107 kb upstream of TSS). These interactions were not observed in nonexpressing B-I and B' inner tissue nor in low expressing B' husk tissue. Our data suggest that in low expressing B' husk, a single loop is formed between the TSS region and the hepta-repeat, while in high expressing B-I husk a multiloop is formed involving the TSS, hepta-repeat, and the sequences ∼47 and ∼107 kb upstream.
Figure 2.
3C-qPCR Analyses Demonstrate Tissue-Specific and Expression Level–Dependent Chromatin Looping at the b1 Locus.
The b1 locus is shown at the top of each graph (see also Figure 1A). The x axis shows the position in kilobases relative to the transcription start site (hooked arrow). The hepta-repeat is indicated with arrowheads. The position and size of the BglII fragments analyzed is indicated by vertical gray shading and Roman numerals; black shading represents the fixed fragment for each experiment. The y axis depicts relative cross-linking frequencies. Data were normalized against cross-linking frequencies measured for the Sam locus. The data for B-I inner and husk tissue are indicated in pink and purple, respectively. Dark green represents B' husk tissue, and light green represents B' inner tissue. Error bars indicate the standard error of mean of four to eight different samples. Relative cross-linking frequencies are shown between fixed fragment I (TSS region) and the rest of the b1 locus (A); fixed fragment X (containing the hepta-repeat) and the rest of the b1 locus (B); and fixed fragment VII (∼47 kb upstream of the TSS) and the rest of the b1 locus (C).
To further investigate the conformation of the b1 locus, experiments were performed using other fixed fragments. When fragment X was used as a fixed fragment, elevated interaction frequencies were measured with fragment I in B-I and B' husk, but not in B-I and B' inner tissue (Figure 2B). Moreover, the X–I interaction frequency was significantly higher in B-I than in B' husk (Student's t test, CI = 90%). In addition, in high expressing B-I husk tissue, fragment X showed high cross-linking frequencies with fragment VII (Figure 2B), confirming that sequences within fragment VII participate in the interaction between the TSS region and the hepta-repeat. This interaction was virtually absent from B' husk tissue and was not observed for inner tissues (Figure 2B). The results obtained using fixed fragment X confirm the formation of tissue-specific as well as expression level–specific interactions at the b1 locus.
To examine the role of fragment VII (∼47 kb upstream of TSS) in the expression level–specific interactions, it was used as a fixed fragment in the qPCR analysis. High interaction frequencies were detected exclusively in B-I husk tissue between fragment VII and fragments X, XII, IV, and I (Figure 2C). Fragment IV, ∼15 kb upstream of the TSS region, was previously not noticed to take part in the physical interactions at the b1 locus. The data shown in Figure 2A, however, indicate an interaction between fragments I and IV, and this interaction is significantly higher in B-I than in B' husk (Student's t test, CI = 90%). Together, these findings show that fragments XII, VII, and IV interact with the TSS and hepta-repeat region in an expression level–dependent manner. These interactions result in a multiloop conformation, which, we hypothesize, mediates high levels of b1 expression.
Multiple Repeats Required for Frequent Interaction between Hepta-Repeat and TSS
Multiple 853-bp repeats are required for paramutation and transcriptional enhancement of b1 (Stam et al., 2002a, 2002b). Fragment X, containing seven repeats, physically interacts with the TSS region (Figure 2). What drives this interaction? Is it sequence-specific, does the repetitiveness play a role, or both? To address this question, we set out to perform 3C analysis on a b1 allele containing one copy of the 853-bp sequence. To allow comparison of the data with those obtained for B-I and B', the same primer and probe sets had to be used. Therefore, the sequence of the chosen allele had to be very similar to that of B-I and B'. The B-615 allele was a good candidate fulfilling this condition; PCR amplification, cloning, and sequencing of multiple, pooled clones showed that there is only a single 853-bp sequence present. BLAST analysis revealed a 96% homology with that of the consensus B-I, B' repeat sequence (see Supplemental Figure 1 online). Moreover, the sequence just 5′ of the coding region is identical to B-I and B', and the sequence up to ∼49 kb upstream is very similar, if not identical (Stam et al., 2002b). This overall high sequence similarity between B-615 and B-I and B' suggested that the entire 3C primer set would bind. For verification, the sequence surrounding each BglII site analyzed in 3C was amplified by PCR. This resulted in similar sized fragments as those obtained for B-I and B'. In addition, when cut with BglII, fragments of the expected sized were obtained.
3C analysis was performed on husk and inner stem tissue from B-615 plants. Expression of b1 is not detectable in B-615 inner tissue and is low in husk tissue (Figure 1B; Stam et al., 2002b). The fragment containing the TSS region was used as fixed fragment (I). In inner tissue, a very low interaction frequency was detected between fragments I and X (X contains a single copy 853-bp sequence; Figure 3A), whereas in B-615 husk tissue, somewhat elevated cross-linking frequencies were detected. Overall, the interaction frequencies between fragments I and X are, however, significantly lower in B-615 than in B' and B-I tissue (Figure 3A; Student's t test, CI = 99% for B-I and B'; for comparison, the interaction frequencies for B' are also shown). The reciprocal experiment using fragment X as fixed fragment could not be performed. For efficient amplification of 3C ligation products, the primer and Taqman probe should both anneal within 50 bp of the restriction site (Hagege et al., 2007). Minor sequence differences prevented annealing of the fragment X-TaqMan probe to the B-615 allele. That probe was specifically designed for B' and B-I. The use of a different Taqman probe for the B-615 allele would have precluded a fair comparison between the data sets.
Figure 3.
Multiple Repeats Are Required for a Frequent, Tissue-Specific Interaction between the Repeats and the TSS Region.
Relative cross-linking frequencies are shown between fragment I (TSS region) and the rest of the B-615 allele (A) and between fragment VII and the rest of the B-615 allele (B). The 1 on the y axes in (A) and (B) is the same value 1 as in Figures 2A and 2B, respectively. The dark-blue line represents B-615 husk tissue, and the light blue line represents B-615 inner tissue. The single 853-bp B-615 sequence is indicated with a black arrowhead. For comparison, the relative cross-linking frequencies observed for the B' allele are shown. See Figure 2 for further details.
To examine the role of fragment VII in expression level–specific interactions in more depth, fragment VII was used as a fixed fragment in the qPCR analysis. In B-615 inner tissue, very low interaction frequencies were detected with fragment VII, whereas in husk tissue the cross-linking frequencies were only slightly higher. For both inner and husk tissue, the interaction frequencies were not significantly different from those detected for B', except for fragment V (Figure 3B; Student's t test, CI = 90%). Overall, these data indicate that the interaction between the TSS region and fragment X is sequence-specific, while the interaction frequency appears to be dependent on the repetitiveness of the 853-bp sequence. Moreover, the B-615 results fit the B' and B-I data that indicate a correlation between the b1 expression level and the interaction frequencies involving fragment VII.
FAIRE Uncovers Nucleosome-Depleted Regions Specific for High b1 Expression
The 3C data showed that regions ∼107, ∼47, and ∼15 kb upstream of the TSS participate in the formation of a chromatin hub with the TSS and the hepta-repeat, suggesting that these sites play a role in the regulation of b1 expression. To test whether they contain potential regulatory sequences, we performed FAIRE (Hogan et al., 2006; Giresi et al., 2007). FAIRE isolates nucleosome-depleted DNA regions, regions that are strongly enriched for active regulatory elements. The identification of nucleosome-depleted regions by FAIRE is based on the tendency of such sequences to end up in the aqueous phase after phenol extraction of formaldehyde cross-linked, sonicated chromatin.
We focused our FAIRE analysis on regions that revealed high 3C interaction frequencies in husk tissue. The DNA samples obtained by FAIRE were evaluated by qPCR, and enrichment at b1 was calculated by normalization against input samples and the unrelated Sam locus (see Figure 4A for primer sets used; the primer sequences are listed in Supplemental Table 1 online). Analysis of various sequence regions indicated that the FAIRE technique was working properly for maize tissue. In husk tissue, the B-I hepta-repeat is more nuclease sensitive than the B' hepta-repeat (see Supplemental Figure 2 online; Chandler et al., 2000; Stam et al., 2002a). In agreement, the B-I hepta-repeat showed significantly more FAIRE enrichment than the B' hepta-repeat (Figure 4B; amplicons b and c). Fragments IX and V showed similar, low interaction frequencies in B-I and B'. In accordance, amplicons d and i within fragment IX and V showed no significant difference in FAIRE enrichment between B-I and B' (Figure 4B). To assess which FAIRE signals can be considered low, a primer set (p) for the coding region was used. Coding regions are generally nucleosome-rich (Yuan et al., 2005) and not enriched by FAIRE (Hogan et al., 2006). In line with this, primer set p, amplifying part of exon 3, resulted in a very low FAIRE signal in both B-I and B' (Figure 4B). Based on these observations, we conclude that FAIRE analysis works well for plant tissue.
Figure 4.
FAIRE Enrichment at ∼110 kb Chromatin Domain in B-I over B' Husk Tissue.
(A) Schematic representation of the b1 and Sam locus, indicating the primer sets used for FAIRE with letters. See legend of Figure 1 for further details.
(B) Quantitative FAIRE analysis on B-I and B' husk tissue. FAIRE values obtained for b1 were normalized against those measured for Sam using amplicon q. Error bars indicate the se of six samples. Ex3, exon 3 of the b1 coding region.
(C) ChIP-qPCR analysis on B-I and B' husk tissue using an antibody against histone H3. The b1 ChIP data were normalized against the ChIP data obtained for the Sam q amplicon. Error bars indicate the se of three samples. The signals levels for the no-antibody immunoprecipitation control were negligible for all amplicons and not shown.
In (B) and (C), purple bars represent B-I husk tissue, and green bars represent B' husk tissue. Values that differ significantly between B-I and B' in a two-tailed Student's t test are indicated with one, two, or three asterisks, specifying a 90, 95, and 99% CI, respectively.
FAIRE enrichment in B-I over B' husk tissue was observed for amplicons in 3C fragments XII (Figure 4B; amplicon a), VII (e and f), and VI (g and h). This supports the idea that these fragments contain regulatory DNA elements important for high expression of the b1 gene. Besides the above-mentioned amplicons, amplicon m, 5′ of the TSS (in 3C fragment I), is significantly enriched in B-I over B' (Figure 4B), indicating that the TSS region contains more regulatory activity in the high expressed B-I husk than in the low expressed B' husk. This is in line with published genome-wide FAIRE experiments for yeast and human showing that FAIRE enrichment of promoters positively correlates with the activity level of the promoter sequences (Hogan et al., 2006; Giresi et al., 2007). Fragment IV (amplicons j-l; ∼15 kb upstream of the TSS), also involved in the physical interactions associated with high b1 expression, did not show significant differences in FAIRE signals between B-I and B' (Figure 4B). Yet, amplicon l belongs to the fragments most enriched in FAIRE, with enrichments comparable to that observed for amplicon h in B-I, for example. Thus, region IV contains candidate regulatory elements in both the B-I and B' allele. However, it is currently unclear why region IV only appears to form chromatin loops in B-I and not in B'. Finally, we note that no significant differences in FAIRE enrichment between B-I and B' were found at the transcription start or the 5′ untranslated region (amplicons n and o, respectively), indicating no differences in nucleosome organization at these regions.
To verify whether FAIRE-enriched regions are indeed nucleosome depleted in plant chromatin, as has been reported for yeast and mammals (Hogan et al., 2006; Giresi et al., 2007), we performed chromatin immunoprecipitation (ChIP) experiments using an antibody against an invariant domain of histone H3 (Haring et al., 2007). The precipitated DNA was analyzed by qPCR, focusing on the FAIRE-enriched regions and using the FAIRE primer sets. The data were normalized against the data for amplicon q of the Sam locus. Our results show that also for plant chromatin a strong negative correlation was observed between FAIRE enrichment and nucleosome occupancy (cf. Figures 4B and 4C). The sequences strongly enriched by FAIRE (B-I amplicons a, b, and f and B' amplicon a) exhibited a low histone H3 signal, while sequence regions with a low FAIRE signal in general displayed a high histone H3 ChIP signal (B-I amplicon p and B' amplicons b, c, e, g, h, and p). Furthermore, consistent with B-I sequences being FAIRE enriched over B' sequences, B-I sequences resulted in lower H3 signal levels than B' sequences.
In conclusion, FAIRE analysis indicated that BglII fragments XII, VII, and VI contain nucleosome-depleted regions specifically in B-I husk. This supports our hypothesis that the regions ∼107 and ∼47 kb upstream, picked up by 3C, are important for the regulation of b1 expression and contain B-I–specific regulatory sequences. Fragment IV may contain b1 regulatory sequences as well.
DISCUSSION
Gene regulation in higher eukaryotes often involves physical interactions between genomic sequence elements that are tens of kilobases apart on the same chromosome (Carter et al., 2002; Tolhuis et al., 2002; Spilianakis and Flavell, 2004; Lanzuolo et al., 2007; Vernimmen et al., 2007). At the maize b1 locus, multiple repeats ∼100 kb upstream of the TSS are required for high b1 expression, indicating the involvement of long-range interactions in the regulation of b1 expression. Using the 3C technique (Dekker et al., 2002; Tolhuis et al., 2002), we show the occurrence of several tissue-, epiallele-, and expression level–specific long-distance interactions at the maize b1 locus. These interactions play a role in controlling b1 expression. Using the FAIRE technology (Hogan et al., 2006; Giresi et al., 2007), we obtained evidence suggesting that multiple interacting regions, previously not characterized as regulatory, are involved in the control of b1 expression.
3C analysis of the maize b1 locus indicated that the TSS region interacts with the hepta-repeat in a tissue-specific manner. The interaction frequency is high in B-I and B' husk tissue and relatively low in inner tissue. The frequency depends on the epiallele; it is significantly higher in B-I husk than in B' husk. Interestingly, three other regions of the b1 locus, ∼107, ∼47, ∼15 kb upstream of the TSS, interact with the TSS region in an epiallele- and expression level–specific manner. The interactions are observed exclusively in high expressing B-I husk tissue, indicating that, besides the hepta-repeat, the region upstream of the b1 transcription start contains several other regulatory sequences that are involved in inducing high expression of B-I. The FAIRE data strengthens this hypothesis. Recombination experiments had previously already hinted at the involvement of additional DNA regions, between 8.5 and 49 kb upstream of the TSS, in the regulation of b1 expression, but these regions had not been fine-mapped (Stam et al., 2002b). The region ∼107 kb upstream contains nuclease hypersensitive sites, matches a Unigene cluster, and encodes a hypothetical protein (Stam et al., 2002b; Unigene ZM.5756; Figures 1A and 4A). The function of this sequence is unknown. Together, our results indicate that the formation of a multiloop structure, involving interactions between the hepta-repeat, the TSS region, and regions ∼107, ∼47, and ∼15 kb upstream, is required to achieve the high B-I expression level. The formation of a single, less stable loop between the TSS region and the hepta-repeat in B' husk tissue seems not sufficient for high b1 expression. It is important to realize that the formation of chromatin loops is a dynamic process. The 3C data reflect an average structure in a large number of cells (de Laat et al., 2008), and the exact chromosomal structures are likely to vary between the different cells at a specific point in time. Furthermore, although we anticipate the interactions between different sequence elements to be in cis, we cannot exclude that these interactions occur in trans.
Multiple 853-bp repeats, ∼100 kb upstream, are required for high b1 expression (Stam et al., 2002a). The presence of repeated sequences in enhancers and other regulatory elements appears to influence transcription in other systems as well (Chandler et al., 2002; Greene et al., 2007; Shadley et al., 2007; Romney et al., 2008; Espley et al., 2009). Intriguingly, two of the newly identified b1 regions that seem to have regulatory activity contain repeated sequences as well (Figure 4), in a direct (∼43 kb upstream) and an inverted orientation (∼15 kb upstream). If a particular sequence contains binding sites for regulatory proteins, repetition of such a sequence allows binding of more proteins. The promoter of the apple (Malus domestica) MYB10 gene, for example, contains a 23-bp sequence capable of binding a regulatory protein (Espley et al., 2009). The more 23-bp sequences in tandem, the higher the expression driven by this promoter. Similarly, it may be that a local increase in binding sites for proteins mediating chromatin looping results in a higher frequency of physical interactions. In agreement with this idea, multiple copies of the 853-bp repeat appear necessary for a frequent interaction between the TSS and fragment X ∼100 kb upstream (Figures 2 and 3).
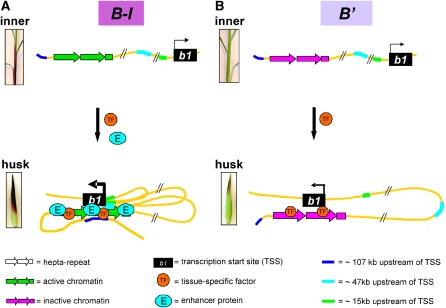
Chromatin looping is tissue-specifically regulated at the B-I and B' epialleles. High interaction frequencies are only observed in husk (Figure 2), the tissue in which the b1 gene is transcriptionally activated (Figure 1B). This tissue-specific chromatin looping is to some extent independent of the hepta-repeat chromatin structure. The B-I and B' hepta-repeat differ in chromatin structure (Figure 4; Stam et al., 2002a), but the TSS and hepta-repeat physically interact in both B-I and B' husk. We propose that the tissue-specific interaction between the TSS and hepta-repeat region (Figure 5) is mediated by sequence-specific transcription factors that are strongly upregulated in husk. We postulate that the binding efficiency and/or functionality of these transcription factors is affected by the chromatin structure at the hepta-repeat, resulting in the observed differences in interaction frequencies.
Figure 5.
Model of Tissue- and Expression Level–Specific Chromatin Looping at the b1 Locus.
In the cartoons a blow-up of part of the hepta-repeat is shown. The hepta-repeat tissue specifically interacts with the TSS region, and this interaction is mediated by a transcription factor (TF).
(A) Tissue-specific, multi-loop formation at the B-I epiallele. The enhancer function of the B-I hepta-repeat is tissue-specifically activated by binding of a transcription factor and proteins mediating enhancer activity (E). The activated enhancer interacts with the TSS region. Regulatory sequences ∼107, ∼47, and ∼15 kb upstream of the TSS interact with the TSS and hepta-repeat as well, resulting in the formation of a multiloop structure that mediates high b1 expression levels in B-I husk.
(B) Tissue-specific, single-loop formation at the B' epiallele. The chromatin state at the B' hepta-repeat is inactive. In husk tissue, binding of the TF occurs and mediates the interaction between the hepta-repeat and the TSS region. However, the inactive B' chromatin state prevents the formation of a multiloop structure and results in a single-loop structure. This single-loop structure is associated with a low expression level in B' husk.
Apart from the tissue-specifically regulated physical interactions, we identified interactions that are expression level specific and only occur at the B-I epiallele. These are the interactions with the newly identified b1 regions. We propose that the latter interactions depend nearly entirely on the chromatin structure of the B-I and B' epialleles in husk and that the B-I chromatin structure, but not that of B', allows formation of the multiloop structure associated with high expression. The TSS and hepta-repeat regions of B-I are more nuclease sensitive than those of B' (see Supplemental Figure 2 online; Chandler et al., 2000; Stam et al., 2002a), suggesting a more open chromatin structure at the B-I epiallele. The FAIRE and ChIP data indicate that the b1 regions that interact in an expression level–specific manner are more nucleosome depleted in high expressing B-I husk than in low expressing B' husk (Figure 4). Nucleosome depletion indicates the binding of proteins associated with gene regulation (Mellor, 2005; Yuan et al., 2005; Workman, 2006). For B-I, we propose the binding of proteins involved in the formation of a multiloop structure mediating enhanced b1 expression (Figure 5A). The less active chromatin state at the B' locus is likely to prevent the binding of these proteins (Bird, 2002; Martin and Zhang, 2005).
The physical interactions between the different b1 sequence regions are anticipated to be in cis. It is tempting to speculate that the hepta-repeat, which is required for paramutation, also physically interacts in trans with the hepta-repeat on the homologous chromosome. RNA is implicated in paramutation; however, transcription of the repeats is not sufficient for paramutation to occur (Alleman et al., 2006; Chandler, 2007). We speculate that in addition to RNAs, trans-interactions play a crucial role in paramutation (Chandler, 2004; Louwers et al., 2005; Stam and Mittelsten Scheid, 2005). To study in trans interactions with 3C technology, the sequence of the DNA fragments of interest needs to be sufficiently different to allow the design of allele-specific primers. None of the b1 alleles known to participate in paramutation show sufficient sequence divergence. Fluorescence in situ hybridization is a potential alternative to establish whether in trans interactions play a role in paramutation.
Formation of a multiloop structure, such as observed for the b1 locus (Figure 5A) and also for other loci (Tolhuis et al., 2002; Spilianakis and Flavell, 2004; Liu and Garrard, 2005; Lanzuolo et al., 2007; Vernimmen et al., 2007), adds another level of complexity to the regulation of gene expression. To fully understand the regulation at the b1 locus, the identification of proteins mediating the physical interactions at the locus is essential and a challenging task for future research. Transgenic experiments analyzing the contribution of each region to b1 expression will provide insight into the function of each of the interactions at the locus and will shed light on the role of physical interactions in general. Importantly, the establishment of the 3C technique for plant tissue opens the exciting possibility to explore the role of long-distance in cis and in trans interactions in gene regulation in plants.
METHODS
Plant Stocks and Tissues
The plant stocks containing the b1 alleles examined (B-I, B', and B-615) were obtained from V.L. Chandler (University of Arizona, Tucson, AZ) and were grown in a greenhouse. The B-I and B', but not B-615, plants had dominant functional alleles for the anthocyanin biosynthetic genes required in vegetative plant tissues. For inner stem tissue, we used the young sheaths and leaves surrounding the shoot meristem inside the maize (Zea mays) stem. This tissue was harvested from 5 to 6-week-old plants. For husk tissue, we used the leaves surrounding the maize corncob, whereby the tough, outer leaves were discarded. Depending on the amount of daylight, husks were harvested when the plants were between 2 and 3 months old, before silks appeared; the actual maize cobs were between 3 and 6 cm long.
Cloning and Sequence Analysis of B-615
The B-615 single copy 853 bp and its flanking sequences were amplified using primers M14 and M80 (see Supplemental Table 2 online). The 1257-bp PCR fragment was cloned into pGEMT-easy (Promega), and multiple, pooled clones were sequenced.
RNA Gel Blot Analysis
Total RNA was isolated from liquid N2–ground plant tissue using the TRIzol method (Invitrogen). To this end, material was collected from the same tissues used in the 3C, FAIRE, and ChIP analyses. For RNA gel blot analysis, 10 μg of RNA was separated on a formaldehyde agarose gel in 1× MOPS (0.2M 3-CN-morpholinol propane sulfonic acid, 50 mM natrium acetate, and 10 mM EDTA, pH 7). Next, the RNA was transferred to Hybond-N+ membrane (Amersham Biosciences) by overnight capillary blotting using 10 mM NaOH. The following day, the RNA was fixed to the membrane by UV cross-linking (0.120 J; Stratalinker). The RNA gel blots were hybridized essentially as described by Stam et al. (1997) using probes recognizing the coding regions of the b1 or Sam gene. The probes were generated by PCR (see Supplemental Table 2 online for the primer sequences) using plasmid DNA as template. For the b1 gene, a B-Peru cDNA clone was used (Radicella et al., 1991). Three b1 probes together recognized exons 3 to 9. For Sam, the insert of pML17 was PCR amplified. pML17 was generated by PCR amplification of part of the Sam coding region with primers M854 and M855 (see Supplemental Table 2 online), followed by cloning of the fragment into pGEMT-easy. The RNA gel blot results were visualized and the relative band intensities quantified using a phosphor-imager and ImageQuant software (Storm; GE Healthcare).
3C Analysis
3C analysis was basically performed according to the method described for mammals by Hagege et al. (2007) but with plant-specific adjustments (M. Louwers, E. Splinter, R. van Driel, W. de Laat, and M. Stam, unpublished data). In short, maize tissue was cross-linked using formaldehyde, and nuclei were isolated. Overnight BglII digestion was followed by ligation in a large volume and overnight de-cross-linking. Next, the DNA was phenol-chloroform extracted and precipitated. Real-time PCR quantification of ligation products was performed on an Applied Biosystems 7500 real-time PCR system using Platinum Taq (Invitrogen) and double-dye oligonucleotides (5′ 6-FAM and 3′ TAMRA) TaqMan probes (Applied Biosystems). In case of AT-rich sequences, MGB-TaqMan probes were used (5′ 6-FAM and 3′ MGB; Applied Biosystems). Primer and probe sequences are listed in Supplemental Table 3 online. The following PCR program was used: 10 min at 95°C, 45 cycles of 15 s at 95°C, and 90 s at 60°C. To account for differences in quality and quantity of the DNA templates, data were normalized to interaction frequencies measured at the Sam locus using the following formula: enrichment = 2 − ((Ct b1 − Ct Sam) 3C sample − (Ct b1 − Ct Sam) BAC mix). For each genotype and tissue, 3C-qPCR was performed on four to eight different plants. For each normalized data point, the mean and standard error were calculated. For statistical analysis, a two-sample, two-tailed student t test, with a 99, 95, or 90% CI, was used to determine whether the two samples are likely to come from distributions with equal population means. The two-sample t test assumes that the two data sets came from distributions with the same variances. A two-tailed t test is required for a nondirectional hypothesis. Such a hypothesis merely holds that the means of the two populations are different, but does not predict in which direction.
3C Control Templates
To correct for differences in quality and quantity of the template, an internal control was required. To this end, the ligation product of an unrelated locus was used in the normalization of the qPCR data. Sam was identified to fulfill the required criteria (Dekker, 2006). It showed a similar expression level in all tissues examined (Figure 1B), indicating it adopts the same spatial configuration in these tissues. Sequences derived from database searches suggested that sufficient (five) BglII sites were present in the available Sam sequence. To verify the presence of BglII sites at the Sam locus in the genetic background we were using, and to accurately design 3C primers and a TaqMan probe, 300 bp surrounding each BglII site were cloned and sequenced. In our search for a suitable internal control, a variety of bioinformatics tools and websites were used: electronic RNA expression data were combined with data from Highly Expressed Gene Finder (http://www.maizearray.org) and data from Fernandes et al. (2002). To obtain sufficient sequence information, GSS BLAST (http://www.maizegdb.org/blast.php), MAGI (http://magi.plantgenomics.iastate.edu), and other sequence tools (http://www.maizecdna.org, http://www.genome.arizona.edu/cgi-bin/gbrowse/gbrowse, and http://www.maizeseq.org) were applied.
To control for the differences in primer set efficiency during PCR amplification, a control template was required that contains all possible ligation products of the loci of interest (b1 and Sam) in equimolar amounts. The b1 template was obtained as follows: 5 μg pBACB'1 DNA (Stam et al., 2002b) was digested overnight with BglII, extracted with phenol-chloroform and precipitated. The pellet was washed with 70% ethanol, the DNA taken up in ligation mixture (5 μL ligation buffer [Roche], 5 μL PEG-4000, 5 μL ligase [Roche; 1 unit/ μL], and 35 μL water), and incubated for 1 h at room temperature, followed by 4 h at 16°C. The ligated DNA was purified by phenol-chloroform extraction, precipitated, washed, and dissolved in water. The Sam control template was amplified from one of our 3C samples using primers M932 and M934 (see Supplemental Table 3 online for primer sequences). The amplified fragment was purified from gel, followed by a second round of PCR. The resulting fragment was once more purified from gel; the DNA concentration was determined and mixed in an equimolar amount with the b1 PCR efficiency template. Serial dilutions were made to obtain standard curves that covered the same range of qPCR signals as obtained with the concentrations of the ligation products in the 3C samples.
FAIRE Analysis
FAIRE analysis was basically performed as described for yeast cells by Hogan et al. (2006), but with plant-specific adjustments. Maize tissue was cross-linked, and chromatin was isolated and sonicated (Haring et al., 2007). Approximately 1/20th of the total sample was taken as an input sample and de-cross-linked overnight at 65°C in the presence of 2.5 μL proteinase K (10 μg/μL), followed by phenol-chloroform extraction and ethanol precipitation. To the rest of the sample an equal volume of phenol-chloroform-isoamyl alcohol 25:24:1 (saturated with 10 mM Tris at pH 8.0, 1 mM EDTA) was added, and the tube was vortexed and centrifuged at 13,000 rpm (10 min, room temperature). The aqueous phase was transferred to a new tube and the DNA precipitated by addition of 2 M NaOAc, pH 5.6, up to 0.3 M, glycogen up to 20 μg/mL, and two volumes of 95% ethanol, followed by incubation at −80°C till frozen (1 h). The tube was spun at 13,000 rpm for 45 min at 4°C, and the pellet was washed with 70% ethanol and air-dried. The pellet was resuspended in 10 mM Tris, pH 8.0, and allowed to dissolve completely by incubating a few hours at room temperature, followed by overnight incubation at 4°C. The isolated DNA was analyzed by qPCR using the Applied Biosystems 7500 real-time PCR system (primers are listed in Supplemental Table 1 online). For each reaction, 1 μL of DNA template was amplified using the Platinum SYBR Green qPCR supermix-UDG (Invitrogen) in a 25-μL reaction according to the manufacturer's protocol. In all experiments, input samples were taken along for every primer set used. First, data were normalized to input levels to correct for template quantity. Second, to correct for template quality, the data were normalized to FAIRE values measured for the Sam locus (amplicon q) using the following formula: enrichment = 2 − ((Ct b1 − Ct Sam,q) aqueous phase− (Ct b1 − Ct Sam,q) input sample ). Each FAIRE experiment was performed on six different B-I and six different B' husk tissues. For each normalized data point, the mean and standard error were calculated. For statistical analysis a two-sample, two-tailed Student's t test, with a 99, 95, or 90% CI, was used (see text for further details).
ChIP-qPCR Experiments
ChIP was performed as described previously (Haring et al., 2007) using an antibody recognizing histone H3 (Abcam ab6002). DNA was isolated using a spin column purification kit (Qiagen) and analyzed by qPCR (see Figure 4C and Supplemental Table 1 online for the primers used). For each PCR reaction, 5 μL of DNA template was amplified according to the same procedure as described for the FAIRE analysis. In every experiment, input samples and no-antibody controls were taken along for every primer set used. The samples were quantified using a calibration line made of DNA isolated from cross-linked, sonicated B' chromatin. The quantified b1 data were normalized to the quantified Sam data for amplicon q using the following formula: ChIP sample quantified/Sam,q quantified = ChIP sample normalized. For both B' and B-I, the ChIP experiments were repeated three times with chromatin from different plants. For each normalized data point, the standard error of the mean was calculated. For statistical analysis, a two-sample, two-tailed Student's t test was used (see text for further details).
Accession Number
Sequence data on the repeat sequence of the B-615 allele can be found in the GenBank/EMBL data libraries under accession number FJ200249.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the B-I, B' Repeat Consensus with That of the B-615 Allele.
Supplemental Figure 2. The B-I Repeats Are More DNase I Sensitive than the B' Repeats.
Supplemental Table 1. List of Primers Used in the FAIRE and ChIP Analysis.
Supplemental Table 2. List of Primers Used for Cloning the 853-bp B-615 Sequence and for Amplifying Probes for RNA and DNA Gel Blot Analysis.
Supplemental Table 3. List of Primers and TaqMan Probes Used in the 3C Analysis.
Supplemental Methods. DNase I Hypersensitivity Assay.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Jack Gardiner and Karen McGinnis for help with the electronic RNA expression data and the search for additional sequences for the internal control. We greatly appreciate the technical assistance of Michael Ignarski. We thank Erik Splinter, Damon Lisch, and Daniel Zilberman for critical reading of the manuscript, Vicki Chandler for providing seeds, and Ludek Tikovsky, Harold Lemereis, and Thijs Hendrix for taking excellent care of the maize plants. M. Stam was supported by the Royal Netherlands Academy of Arts and Sciences (KNAW).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Maike Stam (m.e.stam@uva.nl).
Online version contains Web-only data.
References
- Alleman, M., Sidorenko, L., McGinnis, K., Seshadri, V., Dorweiler, J.E., White, J., Sikkink, K., and Chandler, V.L. (2006). An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. [DOI] [PubMed] [Google Scholar]
- Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16 6–21. [DOI] [PubMed] [Google Scholar]
- Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. (2002). Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32 623–626. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L. (2004). Poetry of b1 paramutation: cis- and trans-chromatin communication. Cold Spring Harb. Symp. Quant. Biol. 69 355–361. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L. (2007). Paramutation: From maize to mice. Cell 128 641–645. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., Eggleston, W.B., and Dorweiler, J.E. (2000). Paramutation in maize. Plant Mol. Biol. 43 121–145. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., Stam, M., and Sidorenko, L.V. (2002). Long-distance cis and trans interactions mediate paramutation. Adv. Genet. 46 215–234. [DOI] [PubMed] [Google Scholar]
- Coe, E.H.J. (1959). A regular and continuing conversion-type phenomenon at b locus in maize. Maydica 24 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E.H.J. (1966). The properties, origin and mechanism of conversion-type inheritance at the b locus in maize. Genetics 53 1035–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat, W., Klous, P., Kooren, J., Noordermeer, D., Palstra, R.J., Simonis, M., Splinter, E., and Grosveld, F. (2008). Three-dimensional organization of gene expression in erythroid cells. Curr. Top. Dev. Biol. 82 117–139. [DOI] [PubMed] [Google Scholar]
- Dekker, J. (2006). The three 'C's of chromosome conformation capture: Controls, controls, controls. Nat. Methods 3 17–21. [DOI] [PubMed] [Google Scholar]
- Dekker, J., Rippe, K., Dekker, M., and Kleckner, N. (2002). Capturing chromosome conformation. Science 295 1306–1311. [DOI] [PubMed] [Google Scholar]
- Espley, R.V., Brendolise, C., Chagne, D., Kutty-Amma, S., Green, S., Volz, R., Putterill, J., Schouten, H.J., Gardiner, S.E., Hellens, R.P., and Allan, A.C. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, J., Brendel, V., Gai, X., Lal, S., Chandler, V.L., Elumalai, R., Galbraith, D.W., Pierson, E., and Walbot, V. (2002). Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol. 128 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi, P.G., Kim, J., McDaniell, R.M., Iyer, V.R., and Lieb, J.D. (2007). FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, E., Mahishi, L., Entezam, A., Kumari, D., and Usdin, K. (2007). Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 35 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege, H., Klous, P., Braem, C., Splinter, E., Dekker, J., Cathala, G., de Laat, W., and Forne, T. (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protocols 2 1722–1733. [DOI] [PubMed] [Google Scholar]
- Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., and Stam, M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., and Jacobsen, S.E. (2007). Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Hogan, G.J., Lee, C.K., and Lieb, J.D. (2006). Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2 e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo, C., Roure, V., Dekker, J., Bantignies, F., and Orlando, V. (2007). Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9 1167–1174. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and Garrard, W.T. (2005). Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25 3220–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers, M., Haring, M., and Stam, M. (2005). When alleles meet: Paramutation. In Plant Epigenetics, P. Meyer, ed (Oxford, UK: Blackwell Scientific Publications), pp. 134–173.
- Martin, C., and Zhang, Y. (2005). The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6 838–849. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Zhang, Y. (2007). Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 19 266–272. [DOI] [PubMed] [Google Scholar]
- Mellor, J. (2005). The dynamics of chromatin remodeling at promoters. Mol. Cell 19 147–157. [DOI] [PubMed] [Google Scholar]
- Palstra, R.J., de Laat, W., and Grosveld, F. (2008). Beta-globin regulation and long-range interactions. Adv. Genet. 61 107–142. [DOI] [PubMed] [Google Scholar]
- Patterson, G.I., Thorpe, C.J., and Chandler, V.L. (1993). Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135 881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella, J.P., Turks, D., and Chandler, V.L. (1991). Cloning and nucleotide sequence of a cDNA encoding B-Peru, a regulatory protein of the anthocyanin pathway in maize. Plant Mol. Biol. 17 127–130. [DOI] [PubMed] [Google Scholar]
- Romney, S.J., Thacker, C., and Leibold, E.A. (2008). An iron enhancer element in the FTN-1 gene directs iron-dependent expression in Caenorhabditis elegans intestine. J. Biol. Chem. 283 716–725. [DOI] [PubMed] [Google Scholar]
- Selinger, D.A., and Chandler, V.L. (1999). Major recent and independent changes in levels and patterns of expression have occurred at the b gene, a regulatory locus in maize. Proc. Natl. Acad. Sci. USA 96 15007–15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadley, J.D., Divakaran, K., Munson, K., Hines, R.N., Douglas, K., and McCarver, D.G. (2007). Identification and functional analysis of a novel human CYP2E1 far upstream enhancer. Mol. Pharmacol. 71 1630–1639. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C.G., and Flavell, R.A. (2004). Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5 1017–1027. [DOI] [PubMed] [Google Scholar]
- Splinter, E., Heath, H., Kooren, J., Palstra, R.J., Klous, P., Grosveld, F., Galjart, N., and deLaat, W. (2006). CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Belele, C., Dorweiler, J.E., and Chandler, V.L. (2002. a). Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Belele, C., Ramakrishna, W., Dorweiler, J.E., Bennetzen, J.L., and Chandler, V.L. (2002. b). The regulatory regions required for B' paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., de Bruin, R., van Blokland, R., van der Hoorn, R.A., Mol, J.N., and Kooter, J.M. (1997). Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 12 63–82. [Google Scholar]
- Stam, M., and Mittelsten Scheid, O. (2005). Paramutation: An encounter leaving a lasting impression. Trends Plant Sci. 10 283–290. [DOI] [PubMed] [Google Scholar]
- Suzuki, M.M., and Bird, A. (2008). DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 9 465–476. [DOI] [PubMed] [Google Scholar]
- Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F., and deLaat, W. (2002). Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10 1453–1465. [DOI] [PubMed] [Google Scholar]
- Vernimmen, D., De Gobbi, M., Sloane-Stanley, J.A., Wood, W.G., and Higgs, D.R. (2007). Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman, J.L. (2006). Nucleosome displacement in transcription. Genes Dev. 20 2009–2017. [DOI] [PubMed] [Google Scholar]
- Yuan, G.C., Liu, Y.J., Dion, M.F., Slack, M.D., Wu, L.F., Altschuler, S.J., and Rando, O.J. (2005). Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626–630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.