Abstract
The relative significance of gene regulation and protein isovariant differences remains unexplored for most gene families, particularly those participating in multicellular development. Arabidopsis thaliana encodes three vegetative actins, ACT2, ACT7, and ACT8, in two ancient and highly divergent subclasses. Mutations in any of these differentially expressed actins revealed only mild phenotypes. However, double mutants were extremely dwarfed, with altered cell and organ morphology and an aberrant F-actin cytoskeleton (e.g., act2-1 act7-4 and act8-2 act7-4) or totally root-hairless (e.g., act2-1 act8-2). Our studies suggest that the three vegetative actin genes and protein isovariants play distinct subclass-specific roles during plant morphogenesis. For example, during root development, ACT7 was involved in root growth, epidermal cell specification, cell division, and root architecture, and ACT2 and ACT8 were essential for root hair tip growth. Also, genetic complementation revealed that the ACT2 and ACT8 isovariants, but not ACT7, fully rescued the root hair growth defects of single and double mutants. Moreover, we synthesized fully normal plants overexpressing the ACT8 isovariant from multiple actin regulatory sequences as the only vegetative actin in the act2-1 act7-4 background. In summary, it is evident that differences in vegetative actin gene regulation and the diversity in actin isovariant sequences are essential for normal plant development.
INTRODUCTION
Actin is a multifunctional protein encoded by a large gene family in plants. The most conspicuous cytoplasmic actin cytoskeleton in plant cells is essential for a wide range of cellular processes, including establishing and maintaining cell shape and polarity, tip growth, cytoplasmic streaming, organelle movement and repositioning, cell division, and responses to external signals (reviewed in Wasteneys and Galway, 2003; Smith and Oppenheimer, 2005; Hussey et al., 2006; Staiger and Blanchoin, 2006). In addition, as in vertebrates (Jockusch et al., 2006; McDonald et al., 2006; Vartiainen, 2008), actin has also been identified in the nucleus of plant cells (Skubatz et al., 2000; Cruz and Moreno Díaz de la Espina, 2009). The nuclear actin along with actin-related proteins and a host of other nuclear proteins is implicated in diverse nuclear processes, such as chromatin remodeling, regulation of gene expression, transcription, RNA processing, and nuclear export (Blessing et al., 2004; Miralles and Visa, 2006; Meagher et al., 2007; Vartiainen et al., 2007). We are exploring the contingent link between the actin gene family evolution and multicellular development in plants (Meagher et al., 1999b, 2008) and are particularly interested in establishing if the different actin genes and their encoded protein isovariants are specialized to perform a subset of the many essential actin functions in different organs and tissues (Meagher et al., 1999a).
The model dicot Arabidopsis thaliana contains eight actin genes that are grouped based on phylogeny and expression patterns into two ancient classes, vegetative and reproductive, and into five subclasses (Figure 1). Several lines of evidence strongly suggest that the two major classes of plant actin genes and protein isovariants are functionally distinct. First, plant cells show regulated class- and subclass-specific expression of actins during differentiation and organismal development (Meagher et al., 1999b). For example, during microsporogenesis, actin isovariant expression switches from the vegetative to the reproductive class (Kandasamy et al., 1999). In microspore mother cells and microspores, an abundance of vegetative actins is produced, yet when the microspores differentiate into mature pollen, they express mostly reproductive actins. Second, ectopic expression of a reproductive actin, ACT1, but not overexpression of a vegetative actin, ACT2, in vegetative tissues severely affects actin filament organization and plant morphology (Kandasamy et al., 2002a). Third, the ectopic expression phenotypes are suppressed when the reproductive ACT1 is coexpressed with reproductive, but not vegetative, actin binding proteins (ABPs; Kandasamy et al., 2007). These data suggest that there are preferential, class-specific interactions among actins and ABPs in vivo, further supporting the existence of functional specificity between the two classes of plant actins (Meagher et al., 2008). Experiments by Fyrberg et al. (1998) and Roper et al. (2005) suggested the existence of similar functional specialization among different classes of Drosophila melanogaster actin isovariants. Null mutation in the 88F gene encoding an adult muscle actin reveals a flightless phenotype, which can be rescued by a wild-type copy of 88F or the other flight muscle actin 79B, but not by the two larval muscle actins or any of the cytoplasmic actins. Of the two fly cytoplasmic actins (ACT5C and ACT42A), which differ by two amino acids, only the regulated expression of ACT5C was essential for fly development (Wagner et al., 2002). Vertebrate actins also exhibit isovariant-specific interactions with ABPs and specialized functions (e.g., ezrin interacts with β- but not α-actin; Shuster and Herman, 1995). Moreover, at least two major families of ABPs in plants, profilins and actin depolymerizing factors (ADFs), exhibit distinct class-specific differences in biochemical properties. For instance, the vegetative profilins of maize (Zea mays) have a higher affinity for poly-l-proline, sequester more monomeric actin, and disrupt the actin cytoplasmic architecture in live cells more rapidly than pollen-specific profilins (Kovar et al., 2000). Also, in Arabidopsis, the reproductive (e.g., PRF4) and vegetative (e.g., PRF2) profilins exhibit differential binding properties with plant and vertebrate actins (C.J. Staiger, unpublished data). Thus, families of actins and ABPs in multicellular eukaryotes have functional diversity manifested through differences in gene regulation, amino acid sequence, and protein–protein interaction, which direct normal multicellular development. The depth and significance of this diversity remains unexplored for most multigene families.
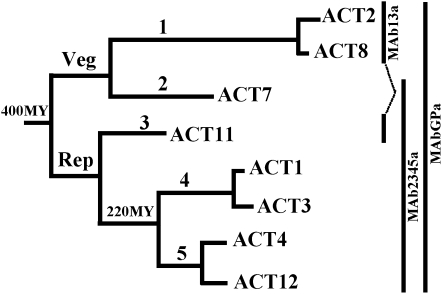
Figure 1.
Arabidopsis Actin Family.
The actin tree of Arabidopsis consists of two major classes, vegetative (Veg) and reproductive (Rep), and five subclasses (1 to 5) of protein isovariants. The specificities of the monoclonal antibodies used in this study are indicated to the right with vertical solid lines. MAb13a reacts with the vegetative Subclass 1 actins ACT2 and ACT8 and the reproductive Subclass 3 actin ACT11, whereas MAb2345a reacts with all reproductive actins (ACT11, ACT1, ACT3, ACT4, and ACT12 of Subclasses 3, 4, and 5) and the vegetative Subclass 2 actin, ACT7. MAbGPa is a general antibody reacting with all Arabidopsis actins. The tree is adapted from Kandasamy et al. (2007).
In this work, we examine if divergent actins within the vegetative class are functionally distinct in Arabidopsis. The vegetative class has three differentially but ubiquitously expressed genes that encode two distinct subclasses of actin isovariants (An et al., 1996; McDowell et al., 1996a). ACT2 and ACT8 (Subclass 1) encode actins that vary only by a single amino acid, but the two genes have been saturated with silent nucleotide substitutions since they shared a common ancestor more than 30 million years ago. However, the ACT7 (Subclass 2) encoded protein differs from ACT2 and ACT8 by 7% and from the closest reproductive actin, ACT11, by 4.25%, levels of amino acid divergence similar to that found between vertebrate cytoplasmic and muscle actins (McDowell et al., 1996b). ACT7 has not shared a common ancestral sequence with ACT2 or ACT8 for at least 200 million years; thus, there is significant potential for divergent subclass-specific isovariant functions among the three vegetative actin proteins. The relative contribution of the three vegetative actins to various cellular processes during plant development has not yet been established. Earlier genetic analyses of single mutants in the ACT2 and ACT7 genes revealed mild or selective defects affecting only certain cell types; for example, lack of the most abundant ACT2 shows stunted root hairs (Gilliland et al., 2002; Ringli et al., 2002), whereas the other strongly expressed actin, ACT7, affects seed germination and root growth (Kandasamy et al., 2001; Gilliland et al., 2003). Herein we have quantified the root phenotypes of single mutants in ACT2 and ACT7, characterized a mutant allele in ACT8, and created double mutants among the three pairs of vegetative actin genes, to dissect further the role of individual actins during plant growth. Also, to examine whether the two ancient vegetative actin subclasses have advantageous expression patterns and/or essential amino acid differences required for distinct interactions with ABPs, we performed suppression studies and synthesized various plant genotypes expressing only a single vegetative actin. Our data suggest that the two subclasses of vegetative actins exhibit unique functional properties. The synthesis of healthy plants overexpressing a single vegetative actin isovariant from multiple actin regulatory sequences demonstrates that differential actin gene regulation is also important for normal plant development.
RESULTS
Analysis of Single Mutants in Vegetative Actin Genes Reveal Subclass-Specific and Redundant Actin Functions
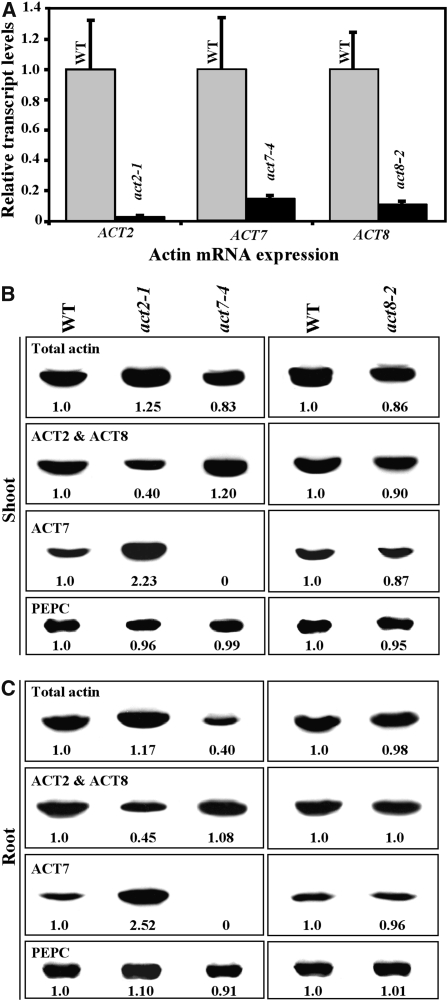
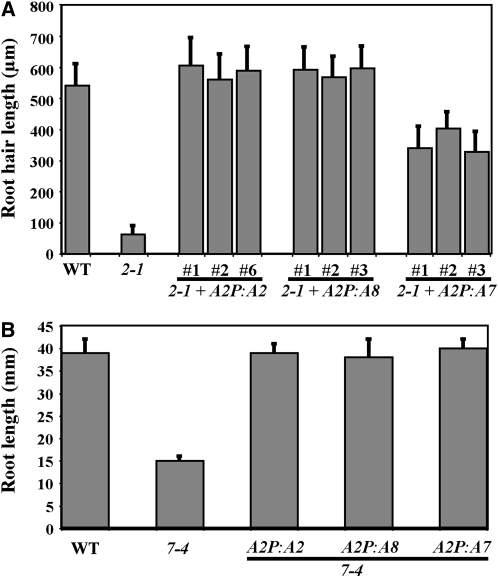
To decipher the role of individual actin genes in the vegetative class, we isolated a mutant allele of the ACT8 gene (act8-2) and further characterized the morphological and molecular phenotypes of single mutants in the other two genes (ACT2 and ACT7). The act2-1 mutant allele, which has a T-DNA insertion five nucleotides upstream of the ATG start codon and a deletion of an intron-exon splice junction in the strongly expressed ACT2 gene (Gilliland et al., 2002), is a severe knockdown with <4% of wild-type levels of ACT2 transcripts (Figure 2A). These plants contain levels of total Subclass 1 actin proteins (ACT2 + ACT8) that are reduced by 55 to 60% compared with the wild type in shoots and roots of 20-d-old seedlings (Figures 2B and 2C, second row, left column). Because none of the antibodies available so far can distinguish the nearly identical ACT2 isovariant from ACT8, we used MAb13a, an antibody that detects both Subclass 1 actins in vegetative tissue (Figure 1). However, protein gel blot analysis with a general actin antibody MAbGPa (Figure 1) revealed that the act2-1 plants have the same or even slightly higher levels of total actin in shoot and root tissues compared with the wild type (Figures 2B and 2C, first row, left column). Probing of identical blots with MAB2345a, which detects only ACT7 in the vegetative tissue, suggested that the total actin in the ACT2-deficient act2-1 mutant plants is increased due to an upregulation in the expression of the ACT7 isovariant (Figures 2B and 2C, third row, left column). Even though the act2-1 mutant plants are essentially null for ACT2, they were indistinguishable in size and aboveground morphology from the wild type at adult stages (Table 1), probably because they had similar levels of total actin as the wild type. However, these mutant plants had roots with severely stunted, bulbous, and often branched root hairs at all developmental stages (Figures 3A and 3D, Table 1). To demonstrate that the root hair growth defects are the result of mutation in the ACT2 gene, we tested wild-type ACT2 cDNA under the regulation of the homologous promoter and 5′ and 3′ regulatory sequences (A2P:A2; see Supplemental Figure 1B online) for its ability to rescue the root hair phenotypes of act2-1 mutant plants. We found that the A2P:A2 transgene fully complemented the act2-1 mutant phenotype, confirming that the root hair growth defects resulted only from loss of function in the ACT2 gene and not from other genotypic differences in the mutant (Figure 4A).
Figure 2.
Molecular Phenotypes of Vegetative Actin Single Mutants.
(A) qRT-PCR analysis of transcript levels in different actin mutant alleles. The data represent average values of two technical replicates, and the bars correspond to sd.
(B) and (C) Protein gel blot analysis of the levels of actin isovariants in shoot (B) and root (C) samples of different 20-d-old actin mutant seedlings. Total actin (first rows) is detected with MAbGPa, which reacts with all eight actin isovariants encoded in Arabidopsis. ACT2 and ACT8 (second rows) are detected with MAb13a that reacts with Subclass 1 (vegetative) and Subclass 3 (reproductive) isovariants. In shoot and root samples of seedlings, MAb13a detects only ACT2 and ACT8. ACT7 (third rows) is detected with MAb2345a that reacts with Subclass 2 (vegetative), 3, 4, and 5 (reproductive) actin isovariants and hence in seedling samples detects only ACT7. Blots shown in the last rows were probed with an anti-PEPC polyclonal antibody to reveal equal loading. The numbers at the bottom of each blot indicate the relative levels of different actin isovariants in the mutants and wild type.
Table 1.
Phenotypic Analysis of Arabidopsis Vegetative Actin Mutants
| Line | Plant Height (cm ± sd)a | Root Length (mm ± sd)b | Root Hair Length (μm ± sd)b | Leaf Length (mm ± sd)c | Silique Length (mm ± sd)d | Cotyledon Width (mm ± sd)b |
|---|---|---|---|---|---|---|
| Wild type | 29 ± 3 | 39 ± 3 | 496 ± 40 | 36 ± 0.9 | 14 ± 0.6 | 2.30 ± 0.15 |
| act2-1 | 28 ± 2 | 38 ± 3 | 62 ± 16 | 35 ± 0.7 | 14 ± 0.5 | 1.95 ± 0.04 |
| act7-4 | 25 ± 2 | 15 ± 1 | 458 ± 104 | 34 ± 1.4 | 14 ± 0.7 | 1.61 ± 0.07 |
| act8-2 | 29 ± 3 | 38 ± 5 | 258 ± 56 | 35 ± 0.8 | 14 ± 0.5 | 1.83 ± 0.10 |
Seven-week-old plant (n = 20).
Ten-day-old seedling (n = 50).
Two largest leaves from just bolted plants (n = 25).
Mature silique (n = 20).
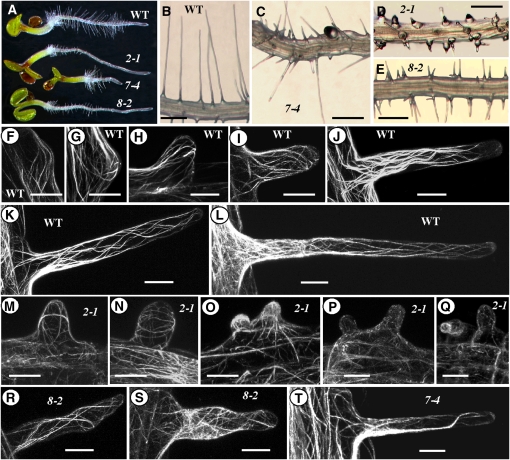
Figure 3.
Morphological and Cellular Phenotypes of Arabidopsis Vegetative Actin Single Mutants.
(A) to (E) Mophological phenotypes of root hair or root growth defects.
(A) Four-day-old wild-type and mutant (act2-1, act8-2, and act7-4) seedlings.
(B) to (E) Portions of primary roots depicting defects in root hair development. Bars = 200 μm.
(F) to (T) Cellular phenotypes of F-actin cytoskeletal organization. Confocal microscopy maximum projection images of developing root hairs of wild-type ([F] to [L]) and act2-1 ([M] to [Q]), act8-2 ([R] and [S]), and act7-4 (T) mutants. All plants express the GFP-fABD2 fusion protein construct. Bars = 20 μm.
Figure 4.
Complementation/Suppression of Root Hair and Root Growth Phenotypes of Actin Mutants.
(A) Differential rescue of the act2-1 root hair growth phenotype by different vegetative actin transgenes. The numbers indicate different transgenic T2 lines analyzed.
(B) Rescue of root growth defects in the act7-4 mutant. Note that all three vegetative actin genes restore root length to wild-type lengths. Because different lines showed same degrees of rescue, data for one independent line of each transgene are shown.
The bars in (A) and (B) represent sd (n = 50).
The act8-2 mutant allele, which contains a T-DNA insertion at the ATG deleting this codon along with 19 bases upstream of it (see Supplemental Figure 1A online), showed >90% reduction from wild-type transcript levels (Figure 2A). As ACT8 is the most poorly expressed actin gene in wild-type plants (Meagher et al., 1999b), there was only a marginal reduction (∼10%) in the levels of total Subclass 1 actin isovariants (Figure 2B, second row, right column) and total actin (Figure 2B, first row, right column) in the shoot tissue of act8-2 mutants. However, no difference in the expression of either the total actin or total Subclass 1 isovariants was observed in the root tissue between the wild-type and mutant plants because of a marginal upregulation of ACT2 in the root. The defects in the expression of ACT8 in act8-2 mutants did not alter aerial plant morphology but caused an ∼50% reduction in root hair length (Figures 3A and 3E, Table 1). The root hair growth defects of the act8-2 mutant plants were fully complemented with the A8P:A8 transgene (see Supplemental Figure 1B online; Table 2), suggesting that the reduced root hair growth is the result of the act8-2 mutation.
Table 2.
Rescue of act2-1, act8-2, and act7-4 Mutant Phenotypes with Different Transgenes
| Complementation/Suppression of Phenotype
|
|||
|---|---|---|---|
| Transgene | act2-1/Root Hair Growth | act8-2/Root Hair Growth | act7-4/Root Growth |
| A2P:A2 | Full (5) | Full (5) | Full (5) |
| A8P:A8 | Full (5) | Full (5) | Full (5) |
| A7P:A7 | Partial (4) | Partial (4) | Full (5) |
| A2P:A7 | Partial (4) | Partial (4) | Full (4) |
Full: Almost 100% rescue of root hair and root growth to wild-type level. Partial: 80% or less compared with wild-type growth level. The numbers within parentheses indicate independent T2 lines examined.
Moreover, we analyzed the role of ACT7 using the act7-4 mutant allele, which has a T-DNA insertion and deletion in the leader intron (Gilliland et al., 2003) and ∼85% reduction in ACT7 transcript levels compared with the wild type (Figure 2A). Using a highly specific monoclonal antibody, MAb2345a (Kandasamy et al., 2001), the act7-4 mutant plants were found to have no detectable expression of ACT7 protein in the shoot and root tissues (Figures 2B and 2C, third row, left column); thus, act7-4 appears to be a null allele. Protein gel blot analysis with the general actin antibody MAbGPa suggested that the act7-4 mutation caused only a marginal (∼17%) reduction in total actin in the shoot tissue, but in the root tissue, there was a significant reduction (∼60%; Figures 2B and 2C, first row, left column). Thus, loss of the ACT7 isovariant severely retarded root development by affecting root elongation and root hair density (∼20% less than that of the wild type). The roots of act7-4 plants also showed frequent radial bulging of epidermal cells not seen in the wild type (Figure 3C). However, the act7-4 mutation had very little effect on root hair growth (Figure 3A, Table 1). We previously showed that act7-4 mutant plants are defective in cell proliferation (Kandasamy et al., 2001). Although the mutant seedlings were smaller, with cotyledons that were considerably reduced in size, the adult plants grew almost to the same height as wild-type plants, and the shoot morphology in most stages was unaltered (Table 1). Complementation of the act7-4 mutant with ACT7 cDNA under the homologous ACT7 or heterologous ACT2 promoters (A7P:A7 or A2P:A7; see Supplemental Figure 1B online) restored normal growth and architecture to the roots (Figure 4B, Table 2), revealing that the root growth aberrations are the result of the act7-4 mutation.
Confocal microscopy analysis of developing root hairs of wild-type (Figures 3F to 3L) and act2-1 plants (Figures 3M to 3Q) expressing a green fluorescent protein:fimbrin Actin Binding Domain2 (GFP:fABD2) F-actin reporter revealed that the defective root hairs of act2-1 plants often contained transversely oriented actin filaments (Figures 3M and 3N) instead of longitudinal arrays as seen in the wild type (Figures 3I to 3L). At the early stages of root hair development, near the tip of roots, the bulge region of the mutant and wild-type trichoblast cells were similar, except that the mutant cells started to form transverse filaments (Figure 3M). However, after slight elongation, the stunted mutant root hairs showed predominantly transverse filaments (Figure 3N). In fully differentiated root regions, the arrested root hairs contained mostly fragmented actin filaments (Figures 3P and 3Q), whereas the fully expanded wild-type root hairs at similar regions contained intact, longitudinally oriented actin filaments and bundles (Figure 3L). By contrast, the less stunted act8-2 root hairs revealed only a mildly altered organization of actin filaments compared with act2-1 (Figures 3R and 3S). The act7-4 mutant showed no obvious defect in actin cytoskeletal organization in root hairs, except for having fewer actin filaments (Figure 3T) than the wild type (Figures 3K and 3L).
Thus, knocking out or severely knocking down the expression of any of the vegetative actin genes resulted only in selective phenotypes affecting certain cell types in roots. All the vegetative actin genes are expressed constitutively in all vegetative tissues and organs, suggesting that there is functional redundancy among these genes in the unaffected cell types. Moreover, there is some upregulation in the expression of other actin genes when one becomes nonfunctional. The mutations in vegetative actin genes, however, show subclass-specific functional differences among the actin isovariants with regard to root hair development.
Suppression Studies Illustrate Functional Differences between Subclass 1 and 2 Actins with Regard to Root Hair Growth but Not Root Growth
To determine if the two subclasses of vegetative actin genes are functionally different with regard to root hair growth, we performed complementation and suppression studies with the act2-1 mutant, which is severely defective in root hair development. First, we complemented the mutant plants with ACT2 cDNA or suppressed them with ACT8 cDNA under the regulation of the ACT2 promoter and regulatory sequences (see Supplemental Figure 1B online). We found that the A2P:A2 transgene fully complemented the act2-1 mutant phenotype and similarly the A2P:A8 construct completely suppressed the act2-1 root hair growth defects (Figure 4A), indicating that the ACT2 and ACT8 protein isovariants are mutually interchangeable with regard to the regulation of root hair tip growth and development. Then, we examined whether the Subclass 2 actin isovariant ACT7 can substitute for the Subclass 1 isovariant ACT2 and hence suppress the root hair growth defects of act2-1 mutant plants. We introduced the ACT7 cDNA under the regulation of the heterologous ACT2 promoter, which is highly active in root hairs, into the strongly defective act2-1 mutant plants. The various transgenic mutant lines analyzed revealed partial suppression of root hair growth, as the A2P:A7 transgene only restored 65 to 70% of wild-type root hair lengths (Figure 4A). Similar results were observed when the homologous ACT7 regulatory sequences were used to direct the expression of ACT7 cDNA (Table 2). Consequently, the ACT7 isovariant appears functionally different from ACT2 or ACT8, especially with reference to root hair tip growth.
When we tested the complementation of the act7-4 root growth defect, the wild-type ACT7 cDNA driven by homologous ACT7 or heterologous ACT2 regulatory sequences (A7P:A7 or A2P:A7) fully restored root growth to the act7-4 mutant allele. Surprisingly, ACT2 and ACT8 cDNAs also completely rescued the root growth defects of act7-4 mutants when expressed under the regulation of ACT2 regulatory sequences (Figure 4B). Thus, any one of the three vegetative actins, when expressed in adequate levels, supported normal growth and architecture of roots.
Subclass 1 Isovariants ACT2 and ACT8 Are Essential for Tip Growth and Maturation Phases of Root Hair Development
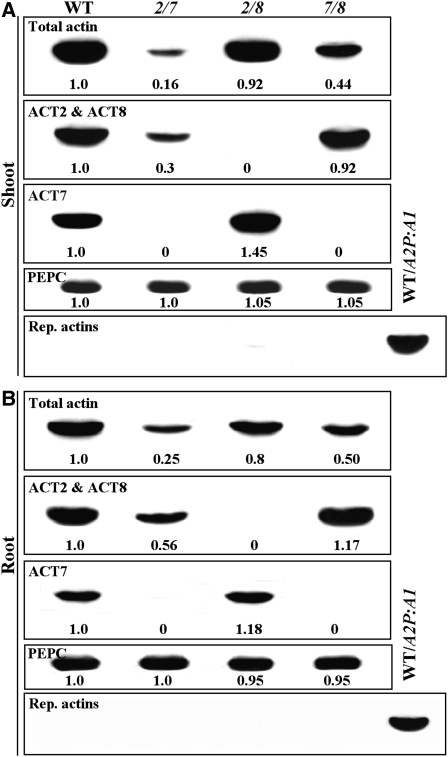
Because analysis of single mutants indicated that both ACT2 and ACT8 isovariants have prominent roles in root hair tip growth, we wanted to further examine their function in root hair development by generating act2-1 act8-2 double mutants. By knocking out the expression of both these genes, we should be able to drastically affect root hair development and dissect their exact role in regulating different phases of this complex morphogenetic process. Protein gel blot analysis showed that the act2-1 act8-2 plants had no detectable levels of ACT2 and ACT8 isovariants in shoot and root tissues, as revealed by MAb13a, which detects only these two actin proteins in the vegetative tissue (Figures 5A and 5B, second row). This confirms that both act2-1 and act8-2 are null alleles. However, analysis with the general actin antibody MAbGPa suggested that the act2-1 act8-2 plants show only a marginal (10 to 20%) reduction in total actin (Figures 5A and 5B, top row). A full analysis of vegetative actin protein expression suggests that the ACT7 isovariant is upregulated in the act2-1 act8-2 double mutant plants, especially at the young stages of development (see third rows in Figures 5A and 5B). No reproductive actins were induced in the shoot or root tissue due to the knocking out of both ACT2 and ACT8 expression (Figures 5A and 5B, bottom rows).
Figure 5.
Protein Gel Blot Analysis of Actin Protein Expression in Different Vegetative Actin Double Mutants.
(A) Expression of total actin or different actin isovariants in shoot tissue.
(B) Expression of total actin or different actin isovariants in root tissue.
Top rows in (A) and (B) show total actin (first panels) detected with MAbGPa. The second rows reveal total ACT2 and ACT8 isovariants detected with MAb13a. The third row shows ACT7 expression as detected with MAb2345a. Blots shown in the forth rows were probed with an anti-PEPC polyclonal antibody to reveal uniform loading of proteins in different lanes. The bottom rows probed with MAb45a, which detects reproductive actin Subclasses 4 and 5, show no induction of reproductive actin in the double mutants. The numbers at the bottom of each blot indicate the relative levels of different actins and are the average of two different experiments. 2/7, act2-1 act7-4; 2/8, act2-1 act8-2; 7/8, act7-4 act8-2. WT/A2P:A1, protein sample from a wild-type plant ectopically expressing reproductive ACT1 in vegetative tissue (positive control for the bottom row in [A] and [B]).
Interestingly, the act2-1 act8-2 double mutant completely lacked root hair development at all stages of plant growth (Figures 6B and 6F). Otherwise, root architecture and length were normal, with a similar number of branches and regular files of cells at the root expansion zone as in the wild type (Figures 6D, 7O, and 7P; see Supplemental Figure 2A online). The act2-1 act8-2 double mutant lines, which express simply ACT7, were slightly (∼15%) smaller than the wild type at the seedling stage (Figure 6C), but at the adult stage, they did not reveal any obvious difference in plant height (Figure 6E), leaf size (Figure 7J), or flower morphology. The leaf epidermal cells (Figures 7A to 7D and 7I) and trichomes (Figures 7K and 7L) closely resembled those of the wild type in size and shape, with a similar number of lobes and branches, respectively, suggesting that the lack of ACT2 and ACT8 has no effect on the expansion of the diffusely growing cell types of the epidermis or trichomes.
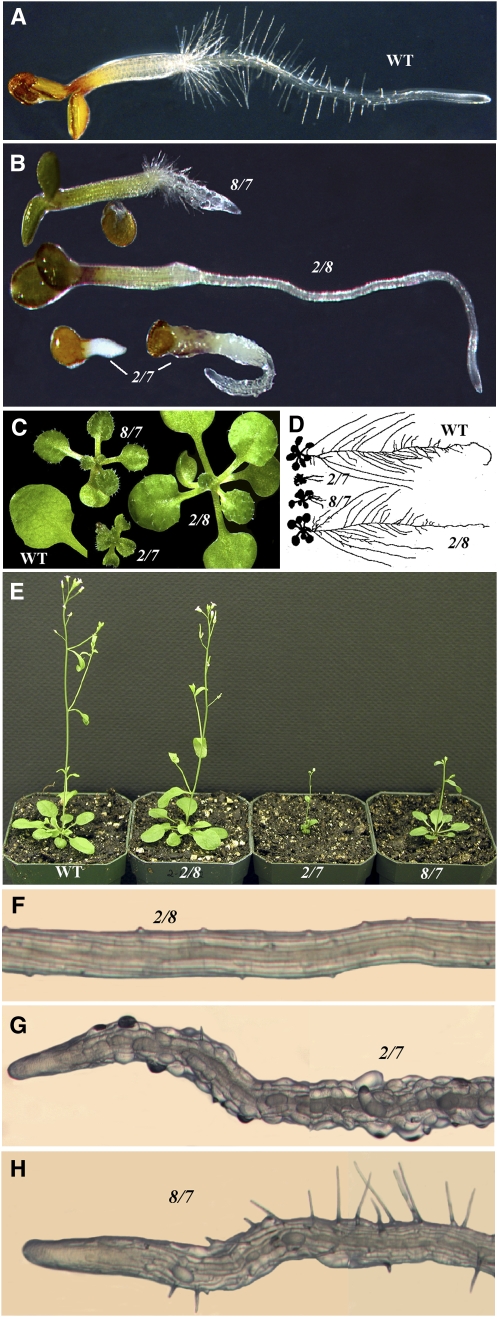
Figure 6.
Phenotypic Analysis of Arabidopsis Vegetative Actin Double Mutants.
(A) and (B) Four-day-old wild-type and actin double mutant (act2-1 act7-4 [2/7], act8-2 act7-4 [8/7], act2-1 act 8-2 [2/8]) seedlings.
(C) Shoots of 20-d-old double mutant seedlings. For wild-type control, a single leaf from a seedling of the same age is shown for comparison.
(D) Scanned images of ∼18-d-old wild-type (Wassilewskija) and double mutant seedlings showing differential defects in root development.
(E) Five-week-old plants.
(F) to (H) Portions of the primary roots of double mutants depicting different defects in root hair development.
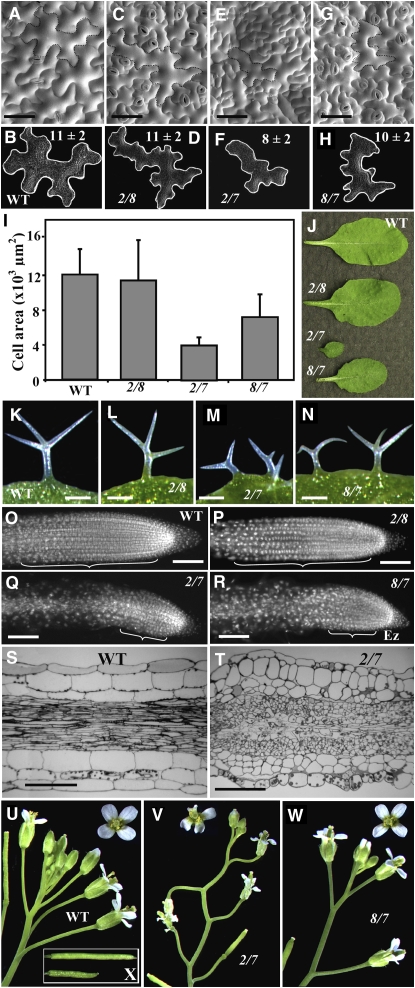
Figure 7.
Defects in Cell Division and Expansion in the Vegetative Actin Double Mutants.
(A) to (H) Scanning electron micrographs of leaf epidermis (adaxial) of the wild type ([A] and [B]) and act2-1 act8-2 ([C] and [D]), act2-1 act7-4 ([E] and [F]), and act8-2 act7-4 ([G] and [H]) double mutants. Pavement cells indicated with dotted lines in the top row ([A], [C], [E], and [G]) are marked along their edges and shown in the second row ([B], [D], [F], and [H]). The numbers on the top of images in the second row indicate the average number of lobes ± sd, calculated from 30 representative cells for each sample.
(I) Cell surface area. Vertical bars represent sd (n = 30).
(J) Rosette leaves.
(K) to (N) Leaf trichomes.
(O) to (R) Organization of cells in 4',6-diamidino-2-phenylindole–stained root apices of 10-d-old seedlings. The brackets indicate the zone of elongation.
(S) and (T) Light micrographs of longitudinal sections of wild-type (S) and act2-1 act7-4 double mutant (T) hypocotyls.
(U) to (W) Inflorescences. Single flower insets are shown at the top of each panel.
(X) Mature siliques of the wild type (top) and the act2-1 act7-4 double mutant (bottom).
Bars = 50 μm in (A), (C), (E), (G), and (O) to (R) and 100 μm in (K) to (N), (S), and (T).
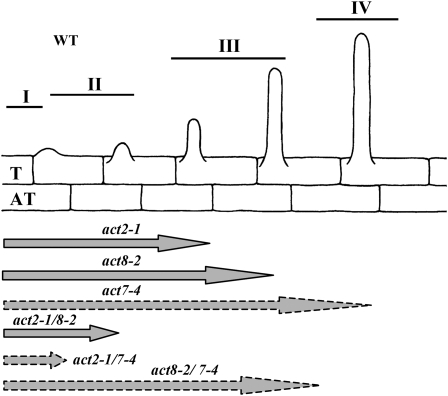
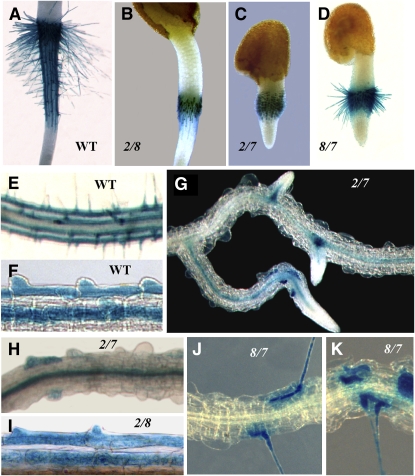
In Arabidopsis, root hair development comprises four distinct stages: cell specification, root hair initiation/bulge formation, tip growth, and maturation (Dolan et al., 1994; Schiefelbein, 2000; Figure 8). To determine the exact stage at which root hair development is arrested in the act2-1 act8-2 double mutant, we transformed this plant with a ADF8P:β-glucuronidase (GUS) reporter gene consisting of the ADF8 promoter and the 5′ flanking regulatory sequence fused to a GUS reporter gene and NOS terminator. The ADF8P:GUS reporter is strongly active early in developing root trichoblast cells and then in root hairs (Ruzicka et al., 2007). Analysis of GUS staining revealed that the act2-1 act8-2 double mutants had normal root hair cell specification and root hair initiation because we observed separate rows of trichoblast and atrichoblast cells. Also, the stained trichoblast cells included distinct bulges at the proximal end of the cells, as did the trichoblast cells of the wild type (Figures 9F and 9I). Furthermore, at the junction between the hypocotyl and radicle, in germinating seedlings, a group of dormant epidermal cells stained positive for ADF8P:GUS gene expression, and in the wild type, all of these cells developed into root hairs (Figures 9A and 9B). Hence, in act2-1 act8-2 double mutants, root hair development never proceeds beyond the initiation/bulge formation stage at both the hypocotyle-radicle junction and in differentiated roots. In the single act2-1 or act8-2 mutant, however, root hair development is arrested at various times during the tip growth phase (Figure 8).
Figure 8.
A Schematic Representation of Root Hair Growth in the Wild Type and Different Vegetative Actin Mutants.
In the wild type, there are four major phases during root hair development: (I) root epidermal cell specification into trichoblast (T) and atrichoblast cells (AT); (II) root hair initiation/bulge formation in the trichoblast cells; (III) tip growth; and (IV) maturation. Root hair development is differentially affected in various single and double actin mutants. The gray arrows below the diagram indicate the stages at which root hair development is arrested. For the act7-4 single mutant and the act2-1 act7-4 and act8-2 act7-4 double mutants, the arrows are shown in dashed lines to suggest partial differentiation of root epidermal cells into trichoblast and nontrichoblast cells. This defect in cell specification reduces the number of root hairs formed compared with the wild type (act7-4 and act8-2 act7-4) or development of no root hairs (act2-1 act7-4).
Figure 9.
Spatial Expression of the ADF8P:GUS Reporter Gene in Different Actin Double Mutants.
(A) to (D) Hypocotyl-radicle junction of 36-h-old seedlings of the wild type (A) and of act2-1 act8-2 (B), act2-1 act7-4 (C), and act8-2 act7-4 (D) double mutants.
(E) to (K) Portions of the root of the wild type ([E] and [F]) and of various actin double mutants ([G] to [K]). ADF8 promoter activity is generally restricted to the trichoblast cells and root hairs in the wild type (Ruzicka et al., 2007). 2/7, act2-1 act7-4; 2/8, act2-1 act8-2; and 7/8, act7-4 act8-2.
Confocal microscopy analysis of act2-1 act8-2 plants expressing GFP:fABD2 suggested that the bulges at the time of initiation (near the root tip) revealed an F-actin cytoskeletal architecture almost indistinguishable from that of the wild type (Figures 10A, 10B,10G, and 10H). However, the arrested bulges of the trichoblast cells within older and more differentiated root regions contained no or only fragmented actin filaments (Figures 10I and 10J). By contrast, the non-root hair–bearing atrichoblast and cortical cells contained normal levels of cytoskeletal filaments (Figure 10I). Thus, based on both single and double mutant analyses, it is clear that both of the Subclass 1 actin genes are essential for actin dynamics that regulate the tip growth and maturation phases of root hair development.
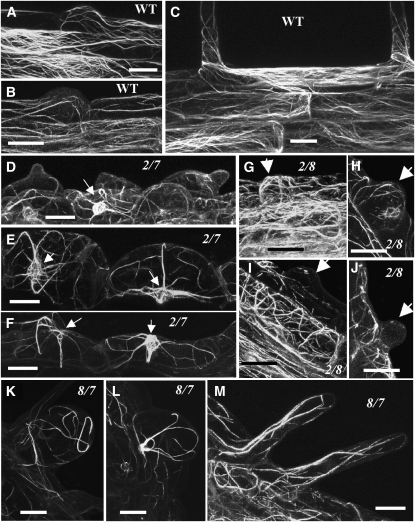
Figure 10.
Organization of the Actin Cytoskeleton in Different Actin Double Mutants.
Confocal microscopy images of root hairs or root epidermal cells showing actin filament organization in the wild type ([A] to [C]) and in act2-1 act7-4 ([D] to [F]), act2-1 act8-2 ([G] to [J]), and act8-2 act7-4 ([K] to [M]) double mutants. All plants express the GFP-fABD2 fusion protein construct. The small arrows in (D) to (F) point to the nuclear basket of actin filaments surrounding the nucleus, and the large arrows in (G) to (J) point to the root hair bulges. Bars = 20 μm.
Subclass 2 Actin ACT7 Plays a Central Role in Cell Morphogenesis and Plant Development
Considering that ACT7 is predominantly expressed in all young tissues (Meagher et al., 1999b) and strongly influenced by hormones (Kandasamy et al., 2001), and that the act7-4 null mutant plants were mainly defective in root elongation (Figure 3A; Gilliland et al., 2003), we wanted to explore the role of ACT7 in whole plant development by producing double mutants lacking both ACT7 and one of the two Subclass 1 actins. Knocking out the expression of ACT7 and ACT2 resulted in act2-1 act7-4 double mutant plants that were extremely dwarfed from seedlings to the adult stage of development, with adult mutant plants growing only up to 15% of the height of wild-type plants (Figures 6A to 6E). Protein gel blot analysis revealed that these double mutant plants contained only ∼15% of wild-type levels of total actin in the shoot and ∼25% in the root (Figures 5A and 5B, top row). At 18 d, the roots of these plants were only ∼10% the length of wild-type roots (Figure 6D; see Supplemental Figure 2A online). The severely stunted double mutant roots contained excessively swollen root epidermal cells with occasional bulges and no root hairs (Figure 6G). To determine the stage at which the root hair development was blocked in the act2-1 act7-4 double mutant, we examined plants expressing the ADF8P:GUS reporter. In germinating seedlings at the junction between the hypocotyl and radicle, all the cells stained blue for GUS gene expression, suggesting root hair cell specification occurred, but there was no root hair growth (Figure 9C). However, in the roots of young seedlings, only a few epidermal cells were stained blue (Figures 9G and 9H), signifying that there is a defect in the specification of trichoblast and atrichoblast cells. Thus, the lack of ACT2 and ACT7 isovariants in the act2-1 act7-4 roots severely disrupted epidermal cell specification and bulge formation and fully arrested tip growth, as summarized in Figure 8. When we examined the actin cytoskeletal organization of act2-1 act7-4 plants expressing GFP:fABD2, we found that the swollen root epidermal cells contained fewer, thick, and transversely oriented actin cables or rod-like structures that were often radiating from the nuclear basket (Figures 10D to 10F). By contrast, the wild-type root hairs and epidermal cells contained longitudinal arrays of dense, fine actin filaments (Figures 10A to 10C).
In addition, the act2-1 act7-4 double mutants were defective in cell division and expansion, as revealed by their small leaves (Figure 7J), containing smaller epidermal cells with significantly reduced surface area (Figures 7E and 7I) and ∼25% fewer lobes (Figure 7F; see Supplemental Figure 2E online) than those of the wild type, small leaf trichomes with a reduced number of branches (one or two branches instead of three; Figure 7M), and stunted roots having about a 60% shorter elongation zone compared with the wild type (Figures 7O and 7Q). The aberrant roots contained irregular files of cells as a result of defects in cytokinesis and formation of oblique transverse cell walls (Figure 7Q; see Supplemental Figure 2C online). Additionally, the defects in cell division and elongation were quite obvious in hypocotyls (Figure 7T) where the epidermal cells were much shorter than those of the wild type and the cortical and central vascular cylinder cells were smaller and irregularly arranged compared with the long, regularly arranged cells in all regions of wild-type hypocotyls (Figure 7S). The act2-1 act7-4 plants also had defects in flower morphology, inflorescence architecture, silique development, and fertility. The flower petals were curled backward, and the flower pedicel and inflorescence stem were highly twisted compared with that of the wild type (Figures 7U and 7V). Moreover, the act2-1 act7-4 inflorescences contained only a few flowers/buds compared with the wild type (Figures 7U and 7V). The siliques of the double mutants were only 50% or less the length of the wild type (Figure 7X) and had very few seeds (3 to 10 instead 40 to 60 seeds per silique).
To further dissect the role of ACT7 in plant development, we created the act8-2 act7-4 double mutant combination, where knocking out the expression of the least abundant Subclass 1 actin ACT8 and the Subclass 2 ACT7 caused about a 55 and 50% reduction in total actin levels in the shoot and root tissues, respectively (Figures 5A and 5B, top row). This drastic reduction in total actin affected both shoot and root development and resulted in a significant decrease in plant size (Figures 6A to 6E). The leaves of act7-4 act8-2 double mutants were ∼35% smaller than those of the wild type (Figures 6C and 7J) and were composed of 30 to 40% smaller epidermal cells (Figures 7G and 7I) with a small reduction (10%) in the number of lobes compared with the wild-type pavement cells (Figure 7H; see Supplemental Figure 2E online). The leaf trichomes of act8-2 act7-4 plants contained two or three branches (Figure 7N), whereas two-branched trichomes were seldom seen in the wild type. There was only a marginal effect on reproductive development and fertility. The act8-2 act7-4 flowers were quite distinct, having narrow petals compared with the wild type (Figures 7U and 7W). Even though the aerial parts of these plants exhibited slightly milder phenotypes than act2-1 act7-4 plants, the root growth and architecture were severely affected (Figures 6B and 6D; see Supplemental Figure 2A online). The root epidermal cells were extremely swollen, although a few cells produced almost normally shaped, but short root hairs (Figure 6H). The root tips contained markedly smaller elongation zones and irregular files of cells (Figure 7R; see Supplemental Figure 2D online). Observation of GUS reporter staining in germinating act8-2 act7-4+ADF8P:GUS seedlings suggested that, in the hypocotyl-radicle junction, all epidermal cells stained blue and developed root hairs (Figure 9D). However, in the differentiated roots there were discontinuous files or only small sectors of root hair–bearing trichoblast cells that stained positively for ADF8P:GUS expression (Figures 9J and 9K). Consequently, in the act7-4 act8-2 plants lacking ACT7 and ACT8 isovariants, there was partial root epidermal cell specification and poor root hair tip growth (Figure 8). The actin cytoskeleton in the swollen epidermal cells of act7-4 act8-2 roots was aberrant and closely resembled that in the act2-1 act7-4 double mutant (Figures 10E and 10L). The enlarged cells also showed thick, irregularly oriented, and coiled actin filaments (Figure 10K). Furthermore, the remaining root hairs contained a few, longitudinally organized thick actin filaments (Figure 10M). Thus, knocking out ACT7 along with ACT2 or ACT8 impaired root epidermal cell differentiation, root hair initiation, and root organization, in addition to having significant effects on shoot architecture.
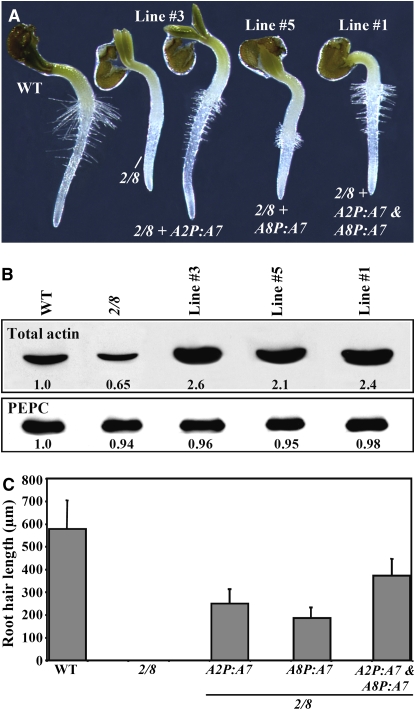
Overexpression of ACT7 Partially Restores Root Hair Growth to the Root Hairless act2-1 act8-2 Double Mutant
Our above results indicate that ACT7 primarily plays a role in cell specification during root hair development because only the act7-4 single mutant and double mutants involving the act7-4 allele have a significant loss of root hair cells. Moreover, the suppression studies of the aberrant root hair phenotype of the act2-1mutant suggest that ACT7 is less efficient than ACT2 and ACT8 in carrying out root hair elongation. To further demonstrate that the ACT7 isovariant is less capable of supporting root hair tip growth and hence is functionally different from the other two vegetative actin proteins, we overexpressed ACT7 in act2-1 act8-2 plants. Because these double mutant roots normally lack any root hairs, they are ideal to assess the effect of the ACT7 isovariant on root hair growth. We expressed ACT7 cDNA under the regulation of ACT2 and ACT8 regulatory flanking sequences, which are very active in root hairs, as shown from a previous promoter-GUS reporter analysis (An et al., 1996; Meagher et al., 1999b). When ACT7 was overexpressed from either the A2P:A7 or A8P:A7 transgene, the transformed plants produced more than twice as much total actin than the wild type (Figure 11B, top row). However, the double mutant plants with overexpressed ACT7 still showed only poor restoration of root hair tip growth and elongation (30 to 40% of the wild type) at the hypocotyl-radicle junction or in the root proper (Figures 11A and 11C, lines 3 and 5). When transformed with both A2P:A7 and A8P:A7 transgenes, the transgenic act2-1 act8-2 plants contained approximately the same level of total actin as plants expressing either transgene (Figure 11B, line 1), but showed only a slight improvement in root hair growth (Figure 11A, line 1). Thus, the root hairs of act2-1 act8-2 plants overexpressing ACT7 from both constructs were still 30 to 40% shorter than those of the wild type (Figure 11C). This demonstrates that the ACT7 isovariant is functionally not equivalent to ACT2 or ACT8, as it has only a limited role in root hair tip growth.
Figure 11.
Overexpression of ACT7 Partially Suppresses the Root Hairless Phenotype of the act2-1 act8-2 Double Mutant.
(A) Germinating seedlings (36 h old) of wild-type, act2-1 act8-2 double mutant (2/8), and act2-1 act8-2 plants transformed with different transgenes.
(B) Protein gel blot analysis of total actin (top row, probed with MAbGPa) in leaf samples of the plant lines shown in (A). The bottom row, probed with anti-PEPC antibody, reveals equal loading of total protein. The numbers at the bottom of the blots indicate the relative levels of the respective proteins.
(C) A bar graph showing the root hair length of the wild type, the act2-1 act8-2 double mutant, and different transgenic lines. The bars represent sd (n = 50).
Synthesis of a Healthy Arabidopsis Plant Expressing ACT8 as the Only Vegetative Actin Isovariant
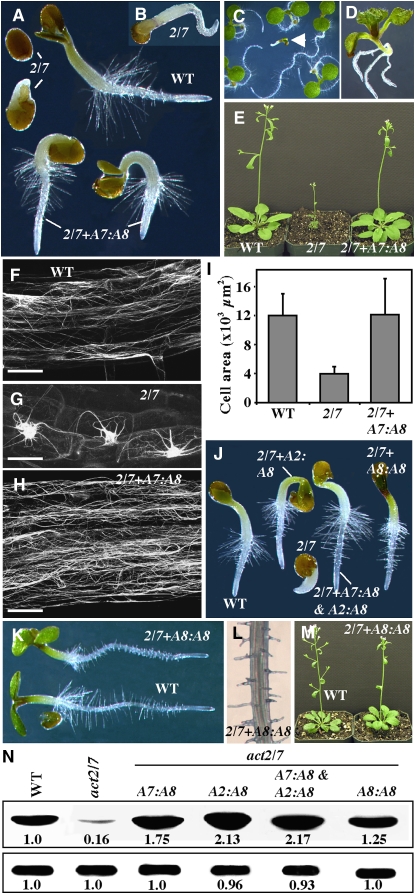
It is clear from our characterization of act7-4 single and act2-1 act7-4 and act8-2 act7-4 double mutants that ACT7 is involved in most developmental pathways (except root hair tip growth). However, the overexpression of ACT2 and ACT8 isovariants, which play essential roles in root hair development, could fully rescue the phenotypes of an act7-4 mutant null for the expression of ACT7. So, if the amino acid sequence differences among the vegetative isovariants are not that important for the majority of actin functions, except the root hair tip growth, and any one of the Subclass 1 actins can compensate for the function of the other vegetative actins, we should be able to synthesize a normal plant complete in all developmental pathways by sufficiently expressing ACT8 or ACT2 as the only actin in all vegetative organs and cell types. In other words, we wanted to determine if we could suppress the dwarf plant phenotype of the act2-1 act7-4 double mutant by overexpressing its only functional vegetative actin, ACT8, under the regulation of a hormonally responsive ACT7 promoter. Because the double mutants were extremely dwarfed (Figure 6E) and highly sterile, we first introduced the A7P:A8 transgene into single mutants. Next, we identified single A7P:A8 insertion lines in both single mutants (act2-1 A7P:A8 and act7-4 A7P:A8) that expressed significantly higher levels of ACT8 protein than the wild type and crossed them. If high-level expression of ACT8 (from native ACT8 and transgenic ACT7 promoters) could suppress all the double mutant phenotypes, we should be able to generate normal act2-1 act7-4 homozygous plants expressing the A7P:A8 transgene in the F2 generation. After screening several hundred F2 plants by PCR, we identified four plants that were homozygous for act2-1 and act7-4 mutant alleles that contained at least one copy of the A7P:A8 transgene and that looked morphologically very similar to the wild type, as shown in Figure 12E. Protein gel blot analysis revealed that the rescued plants contained severalfold higher levels of total actin compared with the act2-1 act7-4 double mutant and almost 75% more actin than the wild type (Figure 12N, A7:A8). Upon germination, the complemented act2-1 act7-4 seedlings produced root hairs very similar to those of the wild type (Figure 12A), whereas the uncomplemented act2-1 act7-4 line exhibited a delay in germination and a lack of root hair formation (Figures 12A and 12B). Moreover, the rescued adult plants grew to normal heights (see Supplemental Figure 3A online) and formed leaves with epidermal cells indistinguishable in size, surface area, and architecture from those of the wild type (Figure 12I). Also, the leaf pavement cells and trichomes of the rescued plants contained ∼11 lobes and three branches, respectively, which were similar to the wild type (Figures 7B and 7K).
Figure 12.
Suppression of act2-1 act7-4 Double Mutant Phenotypes by Overexpression of ACT8.
(A) Germinating seedlings (36 h old) of wild-type, act2-1 act7-4 (2/7), and act2-1 act7-4 double mutants complemented with A7P:A8 (2/7+A7:A8).
(B) Six-day-old act2-1 act7-4 seedling.
(C) Segregation of F3 act2-1 act7-4 plants complemented with the A7P:A8 transgene. Arrowhead points to a seedling that lost the transgene.
(D) Sixteen-day-old seedling indicated in (C).
(E) Thirty-day-old plants. Normal growth is seen in the act2-1 act7-4 mutant overexpressing ACT8 from ACT7 regulatory sequences.
(F) to (H) Actin filament organization in the wild type, act2-1 act7-4 double mutant, and the rescued plant (2/7+A7:A8), all expressing GFP-fABD2. Bars = 50 μm.
(I) Restoration of leaf epidermal cell size to wild-type sizes in act2-1 act7-4 plants complemented with A7P:A8 (n = 30). The error bars represent sd.
(J) Seedlings (36 h old) showing full or partial rescue of the act2-1 act7-4 double mutant phenotype by overexpression of ACT8 from ACT2 (A2:A8), ACT8 (A8:A8), and ACT7 and ACT2 (A7:A8 and A2:A8) regulatory sequences.
(K) to (M) Phenotypes of act2-1 act7-4 plants complemented with A8P:A8.
(K) Four-day-old seedlings.
(L) Root.
(M) Five-week-old plants.
(N) Protein gel blot analysis of total actin expression in the wild type, act2-1 act7-4 double mutant, and different rescued plants (top row). PEPC levels are shown in the bottom row. The numbers at the bottom of the blots indicate the relative levels of respective proteins.
To examine the actin cytoskeletal organization in the complemented double mutant plants, we transformed these plants with the GFP:fABD2 F-actin reporter (Wang et al., 2004). The double homozygous (act2-1 act7-4) mutant plants harboring A7P:A8 and GFP:fABD2 transgenes revealed full restoration of wild-type F-actin organization, as shown by longitudinal arrays of fine actin filaments, instead of the fewer and thick transverse actin cables seen in the double mutant root cells (Figures 12F to 12H). Moreover, to demonstrate that the suppression of double mutant phenotypes is only due to the overexpression of ACT8, we segregated out the A7P:A8 transgene in the F3 generation, and the plants lacking the transgene all reverted to the double mutant phenotype (Figures 12C and 12D). In addition, we performed quantitative RT-PCR (qRT-PCR) with native gene- and transgene-specific primers to show that the rescued double mutant plants contained ACT8 as the only expressed vegetative actin. The qRT-PCR analysis revealed that there was hardly any detectable ACT2 transcript in the rescued plants, and they contained <5% of ACT7 transcripts as in the act2-1 act7-4 double mutant plants (see Supplemental Figure 3B online). In the act2-1 act7-4 mutant background, plants had no detectable levels ACT7 protein (Figures 5A and 5B). Also, these rescued plants contained ∼17 times higher levels of ACT8 (native + transgenic ACT8) transcripts than the wild type (see Supplemental Table 1 online). Unexpectedly, knocking out the expression of ACT2 and ACT7 in act2-1 act7-4 double mutant plants caused a twofold upregulation in the level of native ACT8 transcripts. However, the rescued plants (act2-1/7-4+A7P:A8) overexpressing ACT8 from the transgene showed only a slight increase in native ACT8 transcripts (see Supplemental Figure 3B online).
In addition, we wanted to explore whether overexpression of ACT8 under the regulation of the strongly constitutive ACT2 regulatory sequences could also restore normal stature to act2-1 act7-4 double mutants. We generated the act2-1 act7- 4 double mutant plants harboring A2P:A8 or both A2P:A8 and A7P:A8 transgenes as described above. The seeds of the act2-1 act7- 4 plants overexpressing ACT8 under the regulation of ACT2 or ACT2 and ACT7 regulatory sequences germinated normally, and the seedlings produced root hairs very similar to those of the wild type (Figure 12J). The adult plants from these seedlings grew to the same height as the wild type (see Supplemental Figure 3A online). qRT-PCR analysis demonstrated a 33-fold increase in total ACT8 transcripts when overexpressed from the ACT2 promoter and a 38-fold increase when transgenic expression was from both ACT7 and ACT2 promoters (see Supplemental Table 1 online). Protein gel blot analysis revealed that there was slightly more than a twofold increase in total actin levels over wild-type levels in these plants (Figure 12N). Thus, overexpression of ACT8 from ACT2 or ACT2 and ACT7 regulatory sequences also fully restored growth to the act2-1 act7-4 double mutant.
However, when ACT8 was overexpressed from its own promoter (A8P:A8), there was a fourfold increase in ACT8 transcript levels (see Supplemental Table 1 online), and the plants produced ACT8 protein levels that were ∼25% higher than the total actin in the wild type (Figure 12N). However, the complementation was incomplete. The act2-1 act7-4 seedlings overexpressing ACT8 from its own promoter produced ∼50% shorter root hairs at the junction between the hypocotyl and radicle (Figures 12J and 12K) and ∼65% shorter root hairs in the root compared with the wild type (Figures 12K and 12L). Otherwise, the plants grew almost to the same height (∼95%) as the wild type (Figure 12M; see Supplemental Figure 3A online). The overexpression of ACT8 from its own promoter, which is weak compared with that of ACT2 or ACT7 (Meagher et al., 1999b), might not have produced sufficient levels of actin in all root cells and hence could not restore complete growth to root hairs. Full rescue of all the act2-1 act7-4 double mutant phenotypes may be possible only when ACT8 gene is regulated from the native ACT8 promoter and at least one other vegetative actin promoter (ACT2 or ACT7) and ACT8 protein is expressed in high levels in all shoot and root tissues. Thus, we were able to synthesize stable, healthy Arabidopsis plant lines overexpressing ACT8 as a single vegetative actin isovariant only when the ACT8 gene was driven by multiple vegetative actin regulatory sequences. These results demonstrate the importance of actin gene regulation, along with the appropriate actin isovariants, in directing multicellular development.
DISCUSSION
Angiospserms, like vertebrates and flies, have multigene families encoding actin. In plants, the actin family members exhibit distinct temporal and spatial expression patterns and phylogenetic grouping into vegetative and reproductive classes (McDowell et al., 1996b; Meagher et al., 1999b; Kandasamy et al., 2007). ABPs, such as profilins and ADFs, which are encoded by multigene families and are essential for the regulation of actin dynamics, also display regulational diversity and differential expression in vegetative and reproductive tissues (Staiger et al., 1993; Christensen et al., 1996; Kandasamy et al., 2002b; Maciver and Hussey, 2002; Ruzicka et al., 2007). Furthermore, these two major classes of ABPs show differential biochemical and binding properties with actin (Kovar et al., 2000; Kandasamy et al., 2007). Thus, the ABPs preferentially interact with the diverse classes of actin isovariants and regulate actin dynamics to coordinate different aspects of plant growth and development. In previous ectopic expression and suppression studies, we have shown that the ancient classes of vegetative and reproductive plant actins and ABPs have unique functional properties (Kandasamy et al., 2002a, 2007). However, each actin class also includes ancient subclasses of genes encoding protein isovariants with significant differences in their amino acid sequences, and they are generally coexpressed in various cell types. Prior to this work, the question of whether the different subclasses of actin isovariants are also specialized to perform subsets of the many functions required of actin in plants remained unanswered. Here, we have demonstrated that the two subclasses of vegetative actin isovariants of Arabidopsis are functionally distinct. By knocking out the expression of any two of the three vegetative actins and concomitantly inducing strong cellular and morphological phenotypes, we have characterized the essential and differential functions of actin isovariants in the regulation of multicellular plant development. Also, we have demonstrated the additional importance of gene regulation by synthesizing novel and stable plant lines overexpressing a single vegetative actin isovariant under the regulation of multiple actin regulatory sequences.
Vegetative Actins Play Isovariant-Specific Roles during Root Hair Morphogenesis
Our results on the characterization of single and double actin mutants and their complementation/suppression show that the two ancient subclasses of vegetative actins play specific and universal roles during plant development. The best example for the subclass-specific roles of vegetative actins is found during the four phases of root hair morphogenesis (Figure 8). Genetic analysis revealed that the Subclass 1 actin genes ACT2 and ACT8 participate in root hair tip growth and maturation, whereas the Subclass 2 actin ACT7 has roles in root hair cell specification and root hair initiation/bulge formation. Knocking out the expression of either ACT2 or ACT8 severely affected root hair elongation, resulting in stunted and sometimes highly aberrant and excessively bulged or branched root hairs. Moreover, effectively knocking out the expression of both ACT2 and ACT8 in the act2-1 act8-2 double mutants resulted in totally root hairless plants, but the roots still showed clear differentiation of root epidermal cells into trichoblast and nontrichoblast cells and bulge formation at the proximal ends of trichoblast cells (Figures 6F, 8, and 9I). Thus, ACT2 and ACT8 are essential for the later phases of root hair development, in particular root hair tip growth, a specialized form of polar cell expansion occurring in rapidly growing cell types, such as root hairs and pollen tubes (Hepler et al., 2001; Campanoni and Blatt, 2007). Our complementation/suppression studies revealed that ACT2 fully suppressed the root hair phenotypes of act8-2 and similarly ACT8 rescued the root hair defects of act2-1 mutants, but the Subclass 2 actin ACT7 only partially complemented both mutants, even when expressed from the ACT2 regulatory sequences that are strongly active in root hairs (Table 2, Figure 4A). These data suggest that the highly similar and coexpressed ACT2 and ACT8 isovariants are redundant and can mutually substitute for the function of each other but are functionally divergent from ACT7. Also, the overexpression of ACT7 under the regulation of ACT2 or ACT8 or both regulatory sequences could only partially restore root hair growth to the bald act2-1 act8-2 double mutant roots (Figures 11A to 11C), further signifying that ACT7 is not functionally equivalent to ACT2 or ACT8, especially with regard to root hair growth. It appears that the ACT7 protein isovariant has been subfunctionalized during plant evolution to such an extreme that it can no longer fully substitute for all the functions of its vegetative actin relatives. However, the reproductive ACT1, which is expressed mainly in mature pollen and pollen tubes, could have a function similar to that of ACT2 or ACT8 and suppress the root hair growth defects of the act2-1 mutant (Gilliland et al., 2002). This may not seem very surprising because both root hairs and pollen tubes show polar tip growth, which requires active actin cytoskeletal remodeling, and ACT2 and ACT1 isovariants may have some identical properties that promote this type of growth. Still, the result is unexpected because the ACT2 and ACT1 isovariants have not shared a common ancestral actin sequence for over 300 million years (McDowell et al., 1996b).
Furthermore, characterization of the act7-4 single mutant or double mutants defective in the expression of ACT7 and one of the Subclass 1 actins suggests that ACT7 has a very limited role in root hair tip growth. However, ACT7 appears to participate in the earlier stages of root hair formation because plants lacking in the expression of ACT7 revealed severe defects in the specification of trichoblast and atrichoblast cells and formation of root hairs (Figures 8, 9H, and 9J). Such defects in root epidermal specification were not observed in plants deficient in ACT2 or ACT8 or both. Thus, ACT7 may have some direct or indirect, but specific, role in root hair cell specification. The essential contribution of actin to the regulation of polar growth in root hairs and pollen tubes is well recognized (Hepler et al., 2001; Smith and Oppenheimer, 2005); however, here, we show that the two subclasses of vegetative actin genes differentially regulate different phases of root hair development, with the ACT2 and ACT8 isovariants mainly participating in tip growth and ACT7 involved in cell specification. Because the act2-1 act8-2 double mutants revealed an additive phenotype, completely lacking root hair growth (Figure 6F), which differed from the stunted root hair phenotype in either single mutant (Figures 3D and 3E), ACT2 and ACT8 may act as quantitative loci and operate at the same level in the root hair developmental pathway.
ACT7 Plays a Role in Diffuse Growth and Is Essential for Most Aspects of Plant Development
Among the three vegetative actin genes, the Subclass 2 ACT7, which includes several hormone responsive elements in its regulatory sequences and responds to various environmental signals (McDowell et al., 1996a; Kandasamy et al., 2001), is the most active in all young tissues (Meagher et al., 1999b). Thus, our working hypothesis has been that ACT7 plays a greater role in the regulation of early aspects of plant growth and development. This hypothesis was supported by our observation that, as the only remaining functional actin gene, ACT7 could sufficiently support most cellular and developmental processes in act2-1 act8-2 double mutants, which resulted in normal aerial plant morphology, almost identical to that of the wild type. Except for defects in root hair development, the morphogenesis of all other cell types (e.g., leaf trichomes and epidermal pavement cells) was unaffected in the act2-1 act8-2 plants. Moreover, knocking out the expression of ACT7 along with either one of the Subclass 1 actins (act2-1 act7-4 and act8-2 act7-4) severely retarded the growth of the plants by interfering with cell division, expansion, and morphogenesis of most cell types. For example, the roots of the dwarf double mutant plants lacking ACT7 were extremely stunted with uneven and excessive lateral expansion of epidermal cells, irregular orientation of root apical cells due to oblique cell wall formation, and unusual aggregation of F-actin into thick transverse rod-like structures. The leaf epidermis was also defective in having smaller pavement cells with reduced numbers of lobes and smaller trichomes with fewer branches. Moreover, in act2-1 act7-4 double mutants, the shape of cells in the hypocotyl epidermis was also significantly affected.
In plants, the different cell types can be grouped into two broad categories based on the area over which growth is spread during cell morphogenesis: tip growing cells and diffusely growing cells (Mathur, 2004). From our studies, it is clear that ACT2 and ACT8 have essential roles in polarized cell expansion, such as in the tip growth of root hairs, whereas ACT7 has roles in cell expansion by diffuse growth in various cell types, such as hypocotyl epidermal and cortical cells, leaf trichomes, and pavement cells. The aberrant cellular morphologies and actin cytoskeletal organization in different cell types in the ACT7-deficient plants, in addition to poor cell proliferation and hormone-induced callus formation (Kandasamy et al., 2001), are indicative of the specific participation of ACT7 during various early stages of cell morphogenesis and its significant contribution to overall plant development. Our data on the effect of ACT7 on root cell specification and epidermal cell morphogenesis are consistent with previous suggestions that the ACT7 gene is essential for determining cell polarity (Gilliland et al., 2003). While earlier studies (Bannigan and Baskin, 2005; Smith and Oppenheimer, 2005) suggested that actin promotes growth of cells that expand by diffuse growth, this study clearly defines the specific involvement of the ACT7 isovariant in this common form of growth in most cell types. In summary, in the act2-1 act7-4 double mutants, the lack of ACT7 disrupts cell division and diffuse growth of cells, and the lack of ACT2 inhibits polar tip growth, resulting in an aberrant plant with multiple defects in root and shoot development. Loss of function of these two predominantly expressed isovariants caused a severe disruption in the organization and dynamics of the actin cytoskeleton, thereby affecting several actin-dependent cellular activities that are essential for normal plant development. By contrast, because actins appear to play a role in gene regulation as a component of different chromatin remodeling complexes in the nucleus (Blessing et al., 2004; Chen and Shen, 2007; Meagher et al., 2007), the loss of these two major isovariants might also affect the expression of different genes and produce pleiotropic effects. Considering the major defects we observed in F-actin organization in the act2-1 act7-4 plants, the former seems more likely.
Overexpression of ACT8 from Multiple Actin Regulatory Sequences Is Sufficient for Normal Development of Arabidopsis
Our complementation and suppression analysis of single actin mutants suggested that the Subclass 1 isovariants ACT2 and ACT8 are functionally equivalent, as they can fully complement for the loss of one another. Moreover, either one of them can also substitute for the function of Subclass 2 isovariant ACT7, although ACT7 can only partially rescue the root hair defects of ACT2 or ACT8 deficient plants. Therefore, sufficient levels of ACT2 or ACT8 are expected to support all actin functions and normal plant development. To substantiate this, we synthesized plants overexpressing ACT8 alone as the only vegetative actin isovariant using the act2-1 act7-4 double mutant. It was surprising that we were able to synthesize a complete, healthy, and fully fertile plant from the extremely dwarfed and highly sterile act2-1 act7-4 double mutant plant just by overexpressing the usually poorly expressed ACT8 protein. However, we could regenerate fully normal plants only when we overexpressed ACT8 from more than one (native ACT8 + transgenic ACT2 or ACT7 or both ACT7 and ACT2) actin regulatory sequences. When we overexpressed ACT8 from its own promoter, which is relatively a weaker promoter compared with that of ACT2 or ACT7, there was only an incomplete rescue of plant growth because the complemented plants were still defective in root hair development. The partially rescued plants contained ACT8 protein at levels similar to or slightly (∼25%) higher than the levels of total actin in the wild type. However, the fully rescued plants expressing ACT8 from multiple promoters (native ACT8P and at least one other promoter, ACT7P or ACT2P or both) had twice as much ACT8 as the total amount of actin in the wild type. It is possible that overexpression of ACT8 from more than one promoter might have sufficiently increased the levels of ACT8 in all cell types. Thus, the regulation of ACT8 from multiple actin regulatory sequences and its overexpression to significant levels is essential to achieve complete plant development. In addition, those plants complemented with the single vegetative actin (ACT8) revealed fully restored cytoskeletal organization. Therefore, our results suggest that plants could survive normally with a single vegetative actin isovariant as long as the isovariant is ubiquitously regulated and expressed in all cell types in sufficient quantities. Thus, our synthesis of healthy Arabidopsis plants expressing ACT8 as the single vegetative actin isovariant from multiple regulatory sequences demonstrates that, besides amino acid differences, gene regulation among the vegetative class of actins is very important for the functions of these actins in the regulation of whole plant development.
In parallel studies, we showed that, because of the subfunctionalization of the differentially expressed vegetative and reproductive plant actin sequences, the reproductive actin ACT1 isovariant does not function correctly when expressed in vegetative tissues and produces extreme, dominant cytoskeletal, cellular, and developmental phenotypes (Kandasamy et al., 2002a, 2007). However, the aberrant phenotypes were suppressed when ACT1 was overexpressed along with reproductive, but not vegetative, ABPs, such as profilins and ADFs. This is, at a minimum, due to coevolved specific interactions between ACT1 and reproductive ABP isovariants. The actin and ABP isovariants must have evolved class-specific interactions during the ∼400 million years following their divergence from common ancestral sequences that were coexpressed in ancient reproductive structures (Meagher et al., 1999b, 2008). Testing and interpreting the extent to which the three vegetative actin genes and isovariants were subfunctionalized into two ancient subclasses, as performed herein, was a more subtle and complex undertaking because ACT2, ACT7, and ACT8 expression patterns overlap dramatically (Meagher et al., 1999b). Considering that the Subclass 1 and 2 vegetative actin genes have been preserved from their common ancestry for over 200 million years, we proposed that they also had evolved functional differences. We performed a detailed dissection of vegetative actin mutants and suppression studies with various combinations of vegetative actin gene regulatory sequences and protein isovariants to prove their subfunctionalization. Mutant suppression studies illustrated that ACT7, even when overexpressed at high levels, did not fully restore the growth phase of root hair development. The ACT7 isovariant has become subfunctionalized to the point that it can no longer carry out all the activities required of vegetative actins, while the ACT2 and ACT8 isovariants appear to have retained full functional activity. In addition, the full rescue of double mutants by overexpression of the third vegetative actin isovariant from multiple regulatory sequences suggests that no single vegetative actin regulatory sequence is sufficient to carry out all the vegetative phases of multicellular development, particularly root hair development. Thus, the necessary regulatory elements are also subfunctionalized among the multiple gene sequences. We found that ACT8 overexpressed from a combination of, at a minimum, ACT8 with ACT7 or ACT8 with ACT2 regulatory sequences was crucial to the restoration of normal plant and cell morphology in the act2-1 act7-4 double mutant background. We were surprised that regulatory sequences from the most highly and constitutively expressed ACT2 and the hormonally responsive ACT7, which is strongly active in young tissue, were mutually interchangeable and that plants could survive without the existence of one of them. However, neither regulatory sequence could be replaced with the ubiquitous, but weakly expressed, ACT8 regulatory sequence.
In summary, we conclude that the two ancient subclasses of vegetative actins play specific and universal roles during plant development. For instance, during root hair morphogenesis, the Subclass 1 actins participate in root hair growth and the subclass 2 actin is involved root hair cell specification and root hair initiation/bulge formation. While ACT2 and ACT8 have essential roles in tip growing root hairs, ACT7 contributes more significantly to cell shape development in diffusely growing cell types. Although the ACT7 isovariant was important for root growth, it was inadequate to carry out normal root hair development. Also, this study demonstrates that any one of the two Subclass 1 actin isovariants, ACT2 or ACT8, is sufficient to regulate whole plant development, provided that it is overexpressed from more than one actin regulatory sequence. These data suggest that the regulated expression of vegetative actins is the key to the regulation of normal plant development and that during its subfunctionalization the ACT7 encoded isovariant lost some essential activities. Synthetic plants expressing a single vegetative actin should be an enormous aid in the isolation of individual functional actin isovariants and the biochemical characterization of their interaction with numerous actin binding proteins.
METHODS
Plant Material and Generation of Double Mutants and Transgenic Plants
Arabidopsis thaliana wild-type ecotype Wassilewskija or Columbia and act2-1 (Gilliland et al., 2002), act7-4 (Gilliland et al., 2003), and act8-2 (GABi-Kat line 480C07; Rosso et al., 2003) mutant alleles were cultivated in growth chambers at 22°C with 16-h-light/8-h-dark periods. By crossing homozygotes of the appropriate single actin mutant alleles, various double mutant lines were generated. The F1 plants heterozygous for both alleles were selfed, and the homozygous double mutants were then identified by PCR screening of the F2 population (see primers in Supplemental Table 2 online). For generation of act2-1 act7-4 double mutant plants expressing various constructs (e.g., A7P:A8, A2P:A8, A8P:A8, GFP-fABD2, and ADF8P:GUS; made as described in Kandasamy et al. [2007] and Ruzicka et al. [2007]), the transgenes were first introduced into single mutants by vacuum infiltration, and then the transformants carrying single insertions were identified by selecting T2 plants on Murashige and Skoog medium containing suitable antibiotics. The act2-1 and act 7-4 single mutants containing a single copy of a transgene were crossed and the double mutant plants carrying various constructs were then identified from the F2 population by PCR. Because act2-1 act8-2 double mutants were morphologically normal except for lack of root hairs, they were directly transformed with different transgenes to study suppression. Transgenic single and double mutant lines expressing AFD8P-GUS and GFP-fABD2 transgenes were confirmed by staining with X-Gluc and examining with a fluorescence microscope, respectively.
Plasmid Construction
Nine different actin overexpression constructs were made for the complementation/suppression of different single and double mutants. Each construct contained a 1.1-kb full-length actin cDNA inserted between either homologous or heterologous actin promoter and terminator sequences, as shown in Supplemental Figure 1B online. The various steps involved in cloning were identical to those described previously for A2P:A2 in Kandasamy et al. (2002a). The expression plasmids were mobilized into the Agrobacterium tumefaciens strain C58C1 and transformed into various mutant plants by vacuum infiltration of inflorescences and flower buds.
Protein Gel Blot Analysis
Protein samples were prepared from frozen root and shoot tissues of 20-d-old seedlings or leaf tissues of adult plants, and SDS-PAGE and protein gel blotting were performed as described previously (Kandasamy et al., 1999, 2007). We detected actin proteins on protein gel blots using MAbGPa, a general anti-actin monoclonal antibody reacting with all actin isovariants, MAb13a, an actin subclass-specific antibody reacting only with Subclass 1 (ACT2 and ACT8) and Subclass 3 (ACT11) actins, or MAb2345a, a monoclonal antibody reacting with all actins except ACT2 and ACT8 (Figure 1). In vegetative tissue samples, MAb13a detects ACT2 and ACT8, and MAb2345a detects only the ACT7 protein isovariant. Equal loading of proteins was monitored by Coomassie Brilliant Blue staining of polyaccrylamide gels and probing of high molecular mass (>80 kD) strips of blots or duplicate blots with an anti-PEP carboxylase polyclonal antibody (Rockland). The protein bands were detected using appropriate horseradish peroxidase–conjugated secondary antibodies (Amersham) and the ECL kit. Quantification of actin bands was done using films with short exposure times and the NIH Sci Image program. Care was taken to make sure the bands used for quantification were not oversaturated and were in the linear range of exposure by comparing them with blots containing known quantities of actin samples. Experiments were repeated at least twice, and the average readings were included in different figures showing protein gel blots. Separate protein extractions from duplicate shoot and root samples of the single and double mutant seedlings grown independently were prepared, and each extract was subjected to immunoblotting.
qRT-PCR Analysis
Ribonucleic acid was isolated from the various plant tissues of Arabidopsis using the RNeasy plant mini kit (Qiagen). The RNA was treated with RQ1 RNase-free DNase (Promega) before RT. Three micrograms of treated RNA was added to RT reactions using the Invitrogen SuperscriptIII first-strand synthesis kit (Invitrogen) with random hexamer primers to make cDNA. Quantitative real-time PCR was used to analyze cDNA populations using ribosomal protein RP16A transcript as the endogenous normalization control. qRT-PCR was performed on an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry (Applied Biosystems). The 2−(dCT)*10,000 method of relative quantification was used in all experiments as described by Ruzicka et al. (2007). Primer sequences for each real-time reaction are listed in Supplemental Table 2 online.
Microscopy
To study cellular organization, roots were fixed for 1 h with 4% paraformaldehyde containing traces of Tween 20, stained with 4',6-diamidino-2-phenylindole for 10 min (Sigma-Aldrich; 0.1 μg mL−1), and observed under a Zeiss fluorescence microscope. Root and hypocotyl samples were fixed in 3% glutaraldehyde for 2 to 3 h, processed, and embedded in Spurr's resin (Kandasamy et al., 2005), and semithin sections cut with a Diatome diamond knife were stained with Toluidine blue and observed with a light microscope.
Scanning electron microscopy observations of adaxial surfaces of cryopreserved leaf samples were made using a Leo field emission scanning electron microscope (LEO Electron Microscopy; Kandasamy et al., 2005). The surface area of 30 epidermal pavement cells for each sample was measured from the scanning electron microscopy images using the ImageJ program (NIH), and the average values were calculated and plotted using Microsoft Excel. The same cells that were used for surface area measurement were selected and copied to an Adobe Photoshop document and opened again in the ImageJ program to mark the edges, and the resulting cells as shown for example in Figure 7B were then used to count the number of lobes. For root hair length measurements, 50 fully grown root hairs from different mutants or transgenic plants were measured using low magnification microscopy images of differentiated root segments of 4- or 10-d-old seedlings captured with a Leica stereomicroscope. During complementation or suppression analyses of the root phenotype, the length of the root hairs or roots was measured from four or five independent lines for each transgenic construct.
To examine actin cytoskeletal organization, the single and double mutant plants were either transformed with the GFP-fABD2 construct (Wang et al., 2004) or crossed with the wild type expressing a GFP-fABD2 transgene (Sheahan et al., 2004). Roots from vertically grown seedlings expressing GFP-fABD2 were examined live with a Leica TCS-SP2 confocal laser scanning microscope (Leica Microsystems).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT2, U41998; ACT8, U42007; ACT7, U27811; and RP16A, NM_126785.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ACT8 Gene and Different Transgenic Constructs Used in Complementation/Suppression.
Supplemental Figure 2. Phenotypic Analysis of Vegetative Actin Single and Double Mutants.
Supplemental Figure 3. Overexpression of ACT8 Restores Normal Plant Growth to act2-1 act7-4 Double Mutants.
Supplemental Table 1. Relative Transcript Levels in the act2-1 act7-4 Double Mutant Plants Overexpressing ACT8 under the Control of Various Actin Promoters.
Supplemental Table 2. Primers Used for Detection of Mutant Alleles and qRT-PCR.
Supplementary Material
Acknowledgments
We thank Suganthi Kandasamy for her help with PCR screening to identify the rescued, single actin overexpressing act2-1 act7-4 double mutant plants, D. Ruzicka and L. King-Reid for critically reading the manuscript, D.W. McCurdy for providing a GFP-fABD2 expressing line, and E.B. Blancaflor for the GFP-fABD2 construct. Scanning electron microscopy and confocal microscopy observations were made at UGA's Center for Ultrastructural Research facility. This work was supported by a grant from the National Institutes of Health (GM36397).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard B. Meagher (meagher@uga.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. [DOI] [PubMed] [Google Scholar]
- Bannigan, A., and Baskin, T.I. (2005). Directional cell expansion–Turning toward actin. Curr. Opin. Plant Biol. 8 619–624. [DOI] [PubMed] [Google Scholar]
- Blessing, C.A., Ugrinova, G.T., and Goodson, H.V. (2004). Actin and ARPs: Action in the nucleus. Trends Cell Biol. 14 435–442. [DOI] [PubMed] [Google Scholar]
- Campanoni, P., and Blatt, M.R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58 65–74. [DOI] [PubMed] [Google Scholar]
- Chen, M., and Shen, X. (2007). Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 19 326–330. [DOI] [PubMed] [Google Scholar]
- Christensen, H.E.M., Ramachandran, S., Tan, C.-T., Surana, U., Dong, C.-H., and Chua, N.-H. (1996). Arabidopsis profilins are functionally similar to yeast profilins: Identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 10 269–279. [DOI] [PubMed] [Google Scholar]
- Cruz, J.R., and Moreno Díaz de la Espina, S. (2009). Subnuclear compartmentalization and function of actin and nuclear Myosin I in plants. Chromosoma 118 193–207. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120 2465–2474. [Google Scholar]
- Fyrberg, E.A., Fyrberg, C.C., Biggs, J.R., Saville, D., Beall, C.J., and Ketchum, A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36 271–287. [DOI] [PubMed] [Google Scholar]
- Gilliland, L.U., Kandasamy, M.K., Pawloski, L.C., and Meagher, R.B. (2002). Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 130 2199–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., Pawloski, L.C., Kandasamy, M.K., and Meagher, R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33 319–328. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Ketelaar, T., and Deeks, M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57 109–125. [DOI] [PubMed] [Google Scholar]
- Jockusch, B.M., Schoenenberger, C.A., Stetefeld, J., and Aebi, U. (2006). Tracking down the different forms of nuclear actin. Trends Cell Biol. 16 391–396. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., Burgos-Rivera, B., McKinney, E.C., Ruzicka, D.R., and Meagher, R.B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19 3111–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., Gilliland, L.U., McKinney, E.C., and Meagher, R.B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., Deal, R.B., and Meagher, R.B. (2005). Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol. 138 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (1999). The late pollen-specific actins in angiosperms. Plant J. 18 681–691. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002. a). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002. b). Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil. Cytoskeleton 52 22–32. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R., Drobak, B.K., and Staiger, C.J. (2000). Maize profilin isoforms are functionally distinct. Plant Cell 12 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver, S.K., and Hussey, P.J. (2002). The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 3 reviews3007.1–3007.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J. (2004). Cell shape development in plants. Trends Plant Sci. 9 583–590. [DOI] [PubMed] [Google Scholar]
- McDonald, D., Carrero, G., Andrin, C., de Vries, G., and Hendzel, M.J. (2006). Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 172 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., An, Y.Q., Huang, S., McKinney, E.C., and Meagher, R.B. (1996. a). The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 111 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Huang, S., McKinney, E.C., An, Y.Q., and Meagher, R.B. (1996. b). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142 587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., Kandasamy, M.K., Deal, R.B., and McKinney, E.C. (2007). Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 17 325–332. [DOI] [PubMed] [Google Scholar]
- Meagher, R.B., Kandasamy, M.K., and McKinney, E.C. (2008). Multicellular development and protein-protein interactions. Plant Signal. Behav. 3 333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasamy, M.K. (1999. a). Isovariant dynamics expands and buffers the responses of complex systems: The diverse plant actin gene family. Plant Cell 11 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Vitale, A.V. (1999. b). The evolution of new structures: Clues from plant cytoskeletal genes. Trends Genet. 15 278–284. [DOI] [PubMed] [Google Scholar]
- Miralles, F., and Visa, N. (2006). Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 18 261–266. [DOI] [PubMed] [Google Scholar]
- Ringli, C., Baumberger, N., Diet, A., Frey, B., and Keller, B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, K., Mao, Y., and Brown, N.H. (2005). Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J. Cell Sci. 118 3937–3948. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Ruzicka, D.R., Kandasamy, M.K., McKinney, E.C., Burgos-Rivera, B., and Meagher, R.B. (2007). The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 52 460–472. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J.W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan, M.B., Staiger, C.J., Rose, R.J., and McCurdy, D.W. (2004). A green fluorescent protein fusion to actin-binding domain of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster, C.B., and Herman, I.M. (1995). Indirect association of ezrin with F-actin: Isoform specificity and calcium sensitivity. J. Cell Biol. 128 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubatz, H., Orellana, M.V., and Yablonka-Reuveni, Z. (2000). Cytochemical evidence for the presence of actin in the nucleus of the voodoo lily appendix. Histochem. J. 32 467–474. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., and Oppenheimer, D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21 271–295. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., and Blanchoin, L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9 554–562. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Goodbody, K.C., Hussey, P.J., Valenta, R., Drobak, B.K., and Lloyd, C.W. (1993). The profilin multigene family of maize: Differential expression of three isoforms. Plant J. 4 631–641. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M.K. (2008). Nuclear actin dynamics - From form to function. FEBS Lett. 582 2033–2040. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M.K., Guettler, S., Larijani, B., and Treisman, R. (2007). Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316 1749–1752. [DOI] [PubMed] [Google Scholar]
- Wagner, C.R., Mahowald, A.P., and Miller, K.G. (2002). One of the two cytoplasmic actin isoforms in Drosophila is essential. Proc. Natl. Acad. Sci. USA 99 8037–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.-S., Motes, C.M., Mohamalawari, D.R., and Blancaflor, E.B. (2004). Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil. Cytoskeleton 59 79–93. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54 691–722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.