Abstract
Glucosinolates are defensive secondary compounds that display large structural diversity in Arabidopsis thaliana and related plants. Much attention has been paid to variation in the biosynthesis of Met-derived aliphatic glucosinolates and its ecological consequences, but little is known about the genes that cause qualitative and quantitative differences in Trp-derived indole glucosinolates. We use a combination of quantitative trait locus (QTL) fine-mapping and microarray-based transcript profiling to identify CYP81F2 (At5g57220), encoding a cytochrome P450 monooxygenase, as the gene underlying Indole Glucosinolate Modifier1 (IGM1), a metabolic QTL for the accumulation of two modified indole glucosinolates, 4-hydroxy-indole-3-yl-methyl and 4-methoxy-indole-3-yl-methyl glucosinolate. We verify CYP81F2 function with two SALK T-DNA insertion lines and show that CYP81F2 catalyzes the conversion of indole-3-yl-methyl to 4-hydroxy-indole-3-yl-methyl glucosinolate. We further show that the IGM1 QTL is largely caused by differences in CYP81F2 expression, which results from a combination of cis- and trans-acting expression QTL different from known regulators of indole glucosinolate biosynthesis. Finally, we elucidate a potential function of CYP81F2 in plant–insect interactions and find that CYP81F2 contributes to defense against the green peach aphid (Myzus persicae) but not to resistance against herbivory by larvae from four lepidopteran species.
INTRODUCTION
Activated defenses are among the most fascinating adaptations that plants have evolved to ward off natural enemies. These defense systems consist of precursor molecules and activators, usually enzymes. The precursor molecules can be stored in the plant tissue without harmful effects to the plant but have the potential to give rise to highly toxic or deterrent effectors upon activation. The glucosinolate-myrosinase system is an activated defense system that protects plants from the Brassicaceae and related families effectively against most herbivorous insects. This system is often referred to as the mustard oil bomb to characterize its basic functional principle (Matile, 1980; Lüthy and Matile, 1984). The mustard oil bomb consists of glucosinolates, amino acid–derived β-thioglucoside-N-hydroxysulfates, and matching breakdown enzymes, myrosinases, which have β-thioglucoside glucohydrolase activity. In intact plant tissue, glucosinolates and myrosinases are stored in separate cell types (Koroleva et al., 2000; Husebye et al., 2002; Thangstad et al., 2004). However, upon tissue disruption, myrosinases gain access to glucosinolates and hydrolyze their β-thioglucoside ester bond. The resulting aglycone is instable and rearranges to form a variety of breakdown products, such as isothiocyanates, thiocyanates, nitriles, epithionitriles, and others, depending on reaction conditions and the presence (or absence) of modifying proteins (Lambrix et al., 2001; Bones and Rossiter, 2006; Zhang et al., 2006; Burow et al., 2007). These hydrolysis products have diverse ecological functions. They may serve as oviposition and feeding stimulants for insects specialized in glucosinolate-containing plants but usually act as toxins or deterrents toward other insect herbivores (Raybould and Moyes, 2001; Kliebenstein et al., 2005).
More than 120 different glucosinolate structures have been identified in Capparales (Daxenbichler et al., 1991; Fahey et al., 2001). They share a chemical core, consisting of a sulfonated oxime and a β-thioglucose moiety but differ in their side chain structures. Depending on the precursor amino acid, glucosinolates are grouped into different classes. Aliphatic glucosinolates originate from Ala, Met, Leu, Ile, or Val, aromatic glucosinolates from Phe or Tyr, and indole glucosinolates are derived from Trp. In Arabidopsis thaliana, ∼40 different glucosinolates have been identified, derived from Met, Phe, or Trp (Kliebenstein et al., 2001a; Reichelt et al., 2002). This enormous structural variety is thought to have evolved in response to challenges imposed by a large and diverse community of herbivores and other enemies (Kliebenstein et al., 2001a; Kroymann et al., 2003; Benderoth et al., 2006, 2008).
Natural genetic variation among Arabidopsis accessions has been exploited to map glucosinolate quantitative trait loci (QTL) in various recombinant inbred line (RIL) populations (Campos de Quiros et al., 2000; Kliebenstein et al., 2001b, 2002; Keurentjes et al., 2006; Pfalz et al., 2007). Efforts to clone metabolic QTL have mainly focused on aliphatic glucosinolates, the largest and most diverse glucosinolate class in Arabidopsis. As a result, several genes underlying major aliphatic glucosinolate biosynthesis QTL have been identified, and ecological consequences of natural variation in these genes have been investigated (Kliebenstein et al., 2001c; Kroymann et al., 2001, 2003; Kroymann and Mitchell-Olds, 2005).
Less attention has been paid to variation in indole glucosinolate structures. Here, research has mainly focused on the biosynthesis of the glucosinolate core from Trp because an early intermediate, indole-3-acetaldoxime, also serves as a precursor for auxin (Bak and Feyereisen, 2001; Bak et al., 2001; Zhao et al., 2002; Ljung et al., 2005) and camalexin biosynthesis (Glawischnig et al., 2004), such that alterations in the indole glucosinolate core pathway often feedback on plant growth and development (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995; Lehman et al., 1996; Delarue et al., 1998; Barlier et al., 2000; Bak et al., 2001; Smolen and Bender, 2002). Nonetheless, recent studies demonstrate that indole glucosinolates and their breakdown products influence ecological interactions, contributing to defense against aphids and influencing oviposition by the European cabbage butterfly (Pieris rapae) (Kim and Jander, 2007; De Vos et al., 2008; Kim et al., 2008). Arabidopsis accessions contain typically four different indole glucosinolates (Kliebenstein et al., 2001a; Brown et al., 2003), indole-3-yl-methyl (I3M), 1-methoxy-indole-3-yl-methyl (1MO-I3M), 4-hydroxy-indole-3-yl-methyl (4OH-I3M), and 4-methoxy-indole-3-yl-methyl (4MO-I3M) glucosinolate. I3M is the most abundant indole glucosinolate in Arabidopsis. 1MO-I3M, 4OH-I3M, and 4MO-I3M bear various modifications of the indole ring and are present in lower quantities than I3M. However, the genes that are responsible for indole glucosinolates structures are not yet known.
In a recent study, we mapped QTL for indole glucosinolates in an RIL population derived from a cross between the Arabidopsis accessions Da(1)-12 and Ei-2 and found a complex genetic architecture underlying variation in indole glucosinolates (Pfalz et al., 2007). To start dissecting this architecture, in this work, we focused on the bottom of chromosome 5, where a 4OH-I3M QTL colocalized with a 4MO-I3M QTL, termed Indole Glucosinolate Modifier1 (IGM1) (Figure 1). With a combination of QTL fine-mapping in near isogenic lines (NILs) and transcript profiling with whole-genome Arabidopsis microarrays, we identified candidate genes for IGM1 on chromosome 5 and used T-DNA insertion lines to verify that a single gene, CYP81F2 (At5g57220), underlies this QTL. The product of this gene was a cytochrome P450 monooxygenase, and we showed that CYP81F2 catalyzed the conversion of I3M to 4OH-I3M. We further showed that metabolic variation in modified indole glucosinolates is largely attributable to variation in gene expression, with cis- and trans-acting factors controlling CYP81F2 transcript abundance. Finally, we investigated effects of CYP81F2 on Arabidopsis–insect interactions and found that the gene did not affect herbivory by several generalist and specialist lepidopterans but, instead, contributed to resistance against the green peach aphid (Myzus persicae), a generalist phloem-feeding herbivore.
Figure 1.
Metabolic and Expression QTL for Modified Indole Glucosinolates.
Shown are chromosomes (1 and 5) with significant QTL for 4OH-I3M (top), 4MO-I3M (middle), and eQTL for CYP81F2 expression (bottom). Data for 4OH-I3M and 4MO-I3M were taken from Pfalz et al. (2007). The vertical axes quantify statistical significance. Vertical arrows indicate QTL effect directions; upwards arrows correspond to higher glucosinolate accumulation (or ΔCt values) with the Da(1)-12 allele. Note that higher ΔCt values indicate lower expression.
RESULTS
Fine-Mapping of QTL for Modified Indole Glucosinolates in DE089 × Ei-2 NILs
DE089 is an RIL from the Da(1)-12 × Ei-2 population (Pfalz et al., 2007). Most of its genome originates from the Ei-2 parental accession except for a region corresponding to ∼7 Mbp in Columbia-0 (Col-0) at the bottom of chromosome 5, where DE089 has a Da(1)-12 genotype. Therefore, DE089 was ideally suited to generate NILs for simultaneous fine-mapping of the 4OH-I3M and 4MO-I3M QTL near the bottom of chromosome 5.
We fine-mapped the QTL in F2 and F3 progeny of DE089 × Ei-2 and were able to place the 4MO-I3M QTL between markers 5FMJ01 and 5seq05 (Figure 2). This interval corresponds to 283 kb in Col-0 and contains nearly 70 genes. Low 4OH-I3M quantity precluded a comparably exact delimitation of the 4OH-I3M QTL. Nonetheless, results from single marker analyses suggested that the 4OH-I3M QTL was located near marker 5FM39, close to the 4MO-I3M QTL (see Supplemental Results online).
Figure 2.
Identification of Candidate Genes for the IGM1 QTL.
(A) NILs used for fine-mapping of IGM1. Markers are indicated on top, with distances in megabase pairs corresponding to the marker position in Col-0. NIL families have fixed genotypes [either D = Da(1)-12 or E = Ei-2] on one side but segregate (seg) at the other side. NIL labels indicate the approximate location of recombination breakpoints. As an example, family 5C7 is Ei-2 at markers 5FM39 to 5FM09 and segregates at markers MNC6 and 5FM38.
(B) Statistical support for the 4MO-I3M QTL, given as F ratios for the comparison between Da(1)-12 and Ei-2 genotypes (FD, E) and when heterozygotes are included (FD, H, E).
(C) Differences in transcript abundance between DE089 and Ei-2 in the QTL candidate region under two different experimental conditions, as determined by transcript profiling. Values are given as average fold changes for Ei-2 versus DE089 from three biological replicates each for nonmanipulated plants (light gray bars) and herbivory-treated plants (dark gray).
Identification of Candidate Genes with Transcript Profiling
QTL can be caused by structural variation in the underlying genes (i.e., by differences in the amino acid sequence of the gene products) or by regulatory variation causing differences in gene expression. Transcript profiling with microarrays is a convenient means to monitor differences in transcript levels of a large number of genes simultaneously. We used Agilent Arabidopsis 3 arrays to compare DE089 and Ei-2 transcriptomes under two different experimental conditions, each with three biological replicates per plant line. In the first set of hybridizations, we compared transcripts of unmanipulated DE089 versus unmanipulated Ei-2. In the second set of hybridizations, we compared transcript levels between DE089 and Ei-2 after 9-h herbivory with one 3-d-old larva of the Cabbage white butterfly (Pieris brassicae) per plant.
After RNA isolation, hybridization, and data processing, we focused on the genes in the QTL candidate interval between markers 5FMJ01 and 5seq05. Most genes had similar transcript levels in Ei-2 and DE089, but there were two notable exceptions (Figure 2C). In both control and herbivory experiments, At5g57123 and At5g57220 had much higher transcript levels in Ei-2 than in DE089. At5g57123 encodes an unknown protein with a putative DNA binding domain, and At5g57220 is annotated as a gene for a cytochrome P450 monooxygenase of the CYP81F subfamily, CYP81F2.
T-DNA Insertions in CYP81F2 Cause Reduced Levels of Modified Indole Glucosinolates
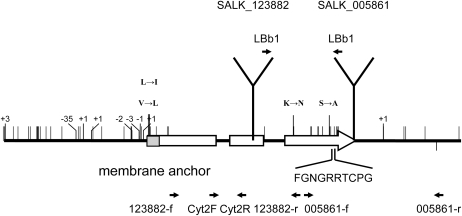
Because cytochromes P450 are often involved in the biosynthesis of plant secondary metabolites and typically catalyze oxygenation reactions (Werck-Reichhart and Feyereisen, 2000; Guengerich, 2001; Werck-Reichhart et al., 2002), CYP81F2 appeared to be a promising candidate gene for the QTL. We obtained two SALK lines with T-DNA insertions in CYP81F2 (Alonso et al., 2003) from the Arabidopsis stock centers, SALK_123882 and SALK_005861. In SALK_123882, the T-DNA is located in the second exon of CYP81F2, and in SALK_005861, the T-DNA is inserted in the third exon very close to the stop codon (Figure 3). Because we could not determine the exact position of the insertion in SALK_005861 from the sequence deposited in the T-DNA insertion database (http://signal.salk.edu/cgi-bin/tdnaexpress), we resequenced the T-DNA insertion site and found the insertion located 10 nucleotides upstream of the CYP81F2 stop codon. As a result, the gene product encoded by SALK_005861 had an altered composition of the four original C-terminal amino acids and an extension of 26 amino acids.
Figure 3.
CYP81F2 in Different Arabidopsis Accessions.
Small black vertical lines show nucleotide and insertion/deletion polymorphisms, and gray vertical lines show amino acid polymorphisms between Ei-2 and Col-0 (top) and between Da(1)-12 and Col-0 (bottom). Positive numbers correspond to an insertion and negative numbers to a deletion relative to the Col-0 reference. Positions of N-terminal membrane anchor (gray box) and Cys heme-iron ligand structure are indicated, as well as location and direction of T-DNA insertions in SALK lines. Positions of selected primers for RT-PCR are also shown.
To identify plants with homozygous mutant CYP81F2 alleles, we planted seeds from each line and screened for the presence/absence of the T-DNA insertion. We obtained homozygous mutant (CYP81F2Δ/Δ) and wild-type progeny (CYP81F2Col/Col) from both lines. We examined the number of T-DNA insertions with DNA gel blots. Hybridization experiments were compatible with the presence of a single T-DNA insertion in SALK_123882, but SALK_005861 had multiple insertions.
We grew mutant and wild-type progeny from both SALK lines together with Col-0 wild-type and conducted an analysis of variance (ANOVA) of leaf glucosinolate profiles of 3-week-old plants. We accounted for flat-to-flat variation and for position effects and nested genotype within plant line. All SALK_123882 and SALK_005861 genotypes had a typical Col-0 glucosinolate profile (Kliebenstein et al., 2001a), with two notable exceptions. Compared with CYP81F2Col/Col, CYP81F2Δ/Δ plants from both SALK lines accumulated significantly less 4MO-I3M (n = 150, df = 2, F = 769.2, P < 0.0001) and 4OH-I3M (n = 150, df = 2, F = 78.7, P < 0.0001) than control plants (Figure 4A). For all other glucosinolates, we did not detect any significant difference between CYP81F2Col/Col and CYP81F2Δ/Δ genotypes. In particular, the other indole glucosinolates, I3M and 1MO-I3M, did not show significant differences between mutant and wild-type genotypes. Hence, in comparison to the CYP81F2 wild type, T-DNA insertion mutants showed a substantial reduction in both 4OH-I3M and 4MO-I3M, indicating that a single gene encoding CYP81F2 controlled both the 4OH-I3M and the 4MO-I3M QTL.
Figure 4.
Modified Indole Glucosinolate Content and CYP81F2 Transcript Levels in SALK T-DNA Insertion Lines.
(A) 4MO-I3M (light gray) and 4OH-I3M (dark gray) concentration (±1 se) in leaves from Col-0, SALK_005861, and SALK_123882 contingent on the CYP81F2 genotype (M = mutant). Vertical lines indicate se, and N indicates sample sizes. Lowercase letters (a to d) designate statistically different means. Note that the y axis has a logarithmic scale.
(B) CYP81F2 transcript levels (±1 se) are significantly reduced in mutant genotypes from SALK_005861 and SALK_123882. SALK_005861 wild type was set as 100%. N indicates sample sizes, and lowercase letters (a to c) designate statistically different means.
SALK_123882 Mutants Lack CYP81F2 Transcripts
We investigated CYP81F2 transcript levels in SALK_123882 and SALK_005861 progeny with RT-PCR. Two control transcripts, coding for RPS18B and actin7, respectively, amplified equally well in all progeny, independent of the CYP81F2 genotype. We found CYP81F2 transcripts in mutant progeny from SALK_005861 but at a lower level than in wild types. However, in SALK_123882 progeny, we detected a clearly visible PCR product only when plants had wild-type CYP81F2 alleles. We repeated this experiment and used quantitative RT-PCR to quantify the results (Figure 4B). Again, both mutants had significantly reduced levels of PCR product, with CYP81F2Δ/Δ genotypes from SALK_123882 abolishing CYP81F2 mRNA almost completely. Nonetheless, 4OH-I3M and 4MO-I3M were detectable in both mutants, albeit at significantly reduced levels, suggesting that other genes contribute to the generation of modified indole glucosinolates (Figure 4).
Intact CYP81F2 Complements the Glucosinolate Phenotype in SALK Mutants Quantitatively
We crossed DE089 and Ei-2 with both mutant lines to test whether intact CYP81F2 can complement the glucosinolate phenotype in CYP81F2Δ/Δ genotypes. The 4MO-I3M levels in DE089 and mutant lines were indistinguishable from one another (Figure 5). Therefore, a complementation of the defective allele was invisible in CYP81F2Da(1)-12/Δ genotypes. By contrast, F1 from crosses between Ei-2, which accumulates more modified indole glucosinolates than DE089, and CYP81F2 mutants had significantly increased levels of 4MO-I3M compared with homozygous mutants but lower levels than Ei-2 wild types. Thus, intact CYP81F2 complements the defective allele in SALK_005861 and SALK_123882 in a quantitative manner, confirming that variation in CYP81F2 causes the IGM1 QTL.
Figure 5.
Quantitative Complementation of the CYP81F2ΔΔ Mutant Phenotype.
4MO-I3M concentration (±1 se) in leaves from SALK_005861 and SALK_123882 mutants (M), DE089, Ei-2 and Col-0, and the F1 from crosses between DE089 or Ei-2 and the mutant lines. N indicates sample sizes, and lowercase letters (a to e) designate statistically different means. Note that the F1s from crosses between Ei-2 and SALK mutants are intermediate between Ei-2 and mutants.
CYP81F2 Catalyzes the Conversion of I3M to 4OH-I3M
The current model of indole glucosinolate biosynthesis suggests that 4MO-I3M is generated from I3M via 4OH-I3M as an intermediate. Variation in a single gene, CYP81F2, causes quantitative difference in the accumulation of both 4OH-I3M and 4MO-I3M among Arabidopsis accessions and in T-DNA insertion lines. This observation led us to postulate that CYP81F2 catalyzes the conversion of I3M to 4OH-I3M. To test this hypothesis, we expressed CYP81F2 in insect cells and conducted enzyme assays with the intact I3M, isolated from seeds of Dyer's woad (Isatis tinctoria), as a substrate. Because of an N-terminal membrane anchor in the mature protein, we used the microsome fraction of the insect cells for our assays. We included two negative controls, microsomes isolated from Sf9 cells expressing a gene from the European cabbage butterfly and a buffer control. In all samples, the peak corresponding to I3M was clearly visible with HPLC (Figure 6). However, we obtained a second major peak in the samples with heterologously expressed CYP81F2 but not in the controls. We converted the respective compound to its desulfo form and subjected it to liquid chromatography–mass spectrometry. The mass spectrum of this reaction product showed a base peak of mass-to-charge (m/z) 385 [M+H]+ and a typical fragment of m/z 223 [M-Glucose+H]+. Hence, the molecular weight of this reaction product was +16 compared with the substrate, I3M (desulfo form m/z 369 [M+H]+), corresponding to the addition of a hydroxy group. Furthermore, we compared the retention time of the reaction product with known glucosinolate profiles from different Arabidopsis accessions (Kliebenstein et al., 2001a; Pfalz et al., 2007) and found it to elute at exactly the same retention time as 4OH-I3M. We also tested desulfo-I3M, but CYP81F2 had no activity with this substrate. Thus, CYP81F2 was indeed capable of catalyzing the suspected reaction, the conversion of I3M to 4OH-I3M.
Figure 6.
CYP81F2 Catalyzes the Conversion of I3M to 4OH-I3M.
Compared with buffer controls (A) or heterologously expressed major allergen (B), HPLC runs of I3M incubated with heterologously expressed CYP81F2 (C) have an additional peak, corresponding to hydroxylated I3M and having the same retention time as 4OH-I3M in an extract of leaf glucosinolates from Col-0 (D), as shown with a mix of separate extracts of I3M incubated with CYP81F2 and of Col-0 leaf glucosinolates (E). All HPLC runs were done with desulfo-glucosinolates. The figure shows representative examples from several independent experimental replicates. Note that Col-0 leaves contain much more 4MO-I3M than 4OH-I3M. mAU = milliabsorption units at 226 nm wavelength.
CYP81F2 Sequence Variation between Col-0, Da(1)-12, and Ei-2
At5g57220 is annotated as a member of the CYP81F subfamily of cytochrome P450s (http://www.Arabidopsis.org). The gene has three exons and two introns (Figure 3) and encodes a protein of 491 amino acids. TargetP predicts an N-terminal signal peptide of 27 amino acids in the translated sequence, serving as a membrane anchor (Emanuelsson et al., 2000; Nielsen et al., 1997). ScanProsite identified a cytochrome P450 Cys heme-iron ligand signature, comprising amino acids 422 to 431 (www.expasy.ch/tools/scanprosite/). Furthermore, we detected a Pro-rich membrane hinge next to the signal peptide, an I-helix involved in oxygen binding and activation (amino acids 295 to 299), and an E-R-R triad (amino acids 352 to 355) with a PERF consensus (amino acids 407 to 410) in the primary protein sequence. These features are typical of conserved structures in cytochrome P450s (Paquette et al., 2000; Werck-Reichhart et al., 2002).
We sequenced an ∼3.9-kb fragment containing CYP81F2 from Da(1)-12, DE089, and Ei-2. This fragment included ∼1200 bp of sequence upstream of the start and 900 bp downstream of the stop codon. The Da(1)-12 and DE089 sequences were nearly identical to the Col-0 sequence, except for one single nucleotide polymorphism in the intergenic region between At5g57220 and At5g57230 (Figure 3). By contrast, we identified numerous sequence differences between Ei-2 and Col-0 or Da(1)-12. However, only four nucleotide substitutions altered the meaning of codons in the open reading frame, and all caused conservative amino acid exchanges. For example, two nonsynonymous substitutions were found in the N-terminal membrane anchor, and in both cases a hydrophobic amino acid was replaced by another hydrophobic amino acid (Figure 3). In the promoter region, we found more substantial differences. This region contained at least 25 single nucleotide polymorphisms and several insertion/deletion polymorphisms, the largest consisting of a stretch of 35 nucleotides absent from the Ei-2 sequence. These data suggest that the indole glucosinolate QTL near the bottom of chromosome 5 is controlled by variation in the CYP81F2 promoter, although we cannot entirely exclude the possibility that structural variation in the amino acid sequence causes differences in the biochemical properties of CYP81F2 variants.
Two Expression QTL Control CYP81F2 Transcript Levels
Microarray hybridization experiments enabled us to identify CYP81F2 as a candidate gene underlying the 4OH-I3M and 4MO-I3M QTL because DE089 and Ei-2 displayed strong differences in CYP81F2 transcript accumulation under two different experimental conditions. In both experiments, Ei-2 accumulated substantially more transcript than DE089 (Figure 2C). This difference in expression correlated with glucosinolate phenotypes (Figure 4). Likewise, Da(1)-12 × Ei-2 RILs with an CYP81F2 Ei-2 genotype produced on average more 4OH-I3M and 4MO-I3M than Da(1)-12 genotypes (Figure 1; Pfalz et al., 2007).
To confirm our microarray data, we analyzed CYP81F2 transcript levels in Da(1)-12, Ei-2, and DE089 with quantitative RT-PCR. We included three biological replicates per line and compared ΔCt values between lines (see Supplemental Figure 1 online). As expected, we found low CYP81F2 expression in DE089, corresponding to ∼31% of the Ei-2 transcript level. Surprisingly, the other parental line, Da(1)-12, had CYP81F2 transcript levels comparable to Ei-2. This led us to suspect that additional factors, encoded outside of the chromosomal segment segregating in DE089 × Ei-2 progeny, contributed to CYP81F2 expression control.
To test this hypothesis, we analyzed CYP81F2 transcript levels in Da(1)-12 × Ei-2 RILs with quantitative RT-PCR to map expression QTL (eQTL). We grew plants randomized in 96-celled flats, with one replicate per RIL and performed RT-PCR in 96-well PCR plates, with control (RPS18B) and test (CYP81F2) gene assays on the same plate. Altogether, we analyzed 181 RILs; thus, each marker/genotype combination was replicated ∼90 times. For each RIL, we calculated ΔCt(test gene – control gene). To control for plate-to-plate variation in RT-PCR efficiency, we normalized data such that all plates had identical sums of ΔCt values. These data were then used as the input for QTL cartographer (http://statgen.ncsu.edu.qtlcart/WQTLCart.htm) and R/qtl (Broman et al., 2003).
As expected, we found a cis-acting eQTL near CYP81F2, but we also detected an eQTL in trans near the bottom of chromosome 1 (Figure 1). This eQTL colocalized with another metabolic QTL that affected 4MO-I3M but not 4OH-I3M accumulation (Pfalz et al., 2007). To test for a potential epistatic interaction between both eQTL, we used markers MSAT1.1, closely linked to the eQTL on chromosome 1, and MMN10, near CYP81F2, with the following statistical model:
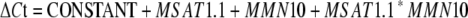 |
Both markers, MSAT1.1 (n = 174, df = 1, F = 22.80, P < 0.0001) and MMN10 (n = 174, df = 1, F = 27.36, P < 0.0001), were significantly associated with CYP81F2 expression. The interaction term MSAT1.1 * MMN10 was not significant (n = 174, df = 1, F = 0.28, P = 0.60), indicating independence of the two eQTL. Furthermore, eQTL effects had opposite directions. For MSAT1.1, we obtained ΔΔCt(Ei-2 – Da(1)-12) = 0.74, indicating higher expression in RILs with a Da(1)-12 genotype. For MMN10, a ΔΔCt(Ei-2 – Da(1)-12) value of −0.80 indicated higher expression in the Ei-2 genotype. Hence, the eQTL were additive and eQTL effects compensated each other in Da(1)-12 and Ei-2, resulting in nearly equal CYP81F2 expression in these accessions.
Finally, we tested for an association between CYP81F2 transcript quantity and indole glucosinolate accumulation in Da(1)-12 × Ei-2 RILs. Indole glucosinolate data were taken from a previously published analysis (Pfalz et al., 2007). ΔCt values were negatively correlated with 4OH-I3M (rG = −0.22, t = −2.93, P = 0.0038) and 4MO-I3M (rG = −0.39, t = −5.62, P < 0.0001) but not with I3M and 1MO-I3M (both: P > 0.35; nonsignificant). Hence, because large ΔCt values indicate low and small values high transcript levels, the expression of CYP81F2 is positively correlated with the production of both 4OH-I3M and 4MO-I3M.
CYP81F2 Contributes to Resistance against Aphids but Not Lepidopteran Herbivores
To determine a potential ecological function of CYP81F2, we tested the performance of several insects on Col-0 wild type and on SALK_005861 and SALK_123882 progeny with CYP81F2Col/Col and CYP81F2Δ/Δ genotypes. We used larvae from four lepidopteran species, the crucifer specialists diamondback moth (Plutella xylostella) and P. brassicae and the generalist herbivores cabbage looper (Trichoplusia ni) and beet armyworm (Spodoptera exigua). We conducted herbivory assays to examine whether larvae fed differentially on mutant and wild-type plants but found that CYP81F2 genotype did not significantly influence any of the lepidopterans (Table 1).
Table 1.
Performance of Lepidopteran Larvae Contingent on CYP81F2 Genotype
| SALK_005861
|
SALK_123882
|
|||||
|---|---|---|---|---|---|---|
| Insect Species | N | Fdf=1 | P | N | Fdf=1 | P |
| P. xylostella | 685 | 0.880 | 0.349 | 479 | 0.300 | 0.584 |
| P. brassicae | 88 | 1.129 | 0.292 | 392 | 0.121 | 0.728 |
| T. ni | 516 | 3.502 | 0.062 | 207 | 0.481 | 0.489 |
| S. exigua | 246 | 0.911 | 0.341 | 413 | 0.220 | 0.639 |
For all tested lepidopterans, leaf area consumed by larvae from mutant plants (either SALK_005861 or SALK_123882) did not significantly differ from wild types. Sample sizes (N), F ratios, and P values are indicated.
We also investigated the performance of the green peach aphid, a generalist phloem-feeding insect (Pollard, 1972) and natural pest on Arabidopsis and other crucifers (Blackman and Eastop, 2006). We measured aphid proliferation contingent on plant genotype. Here, we reared aphids for several generations either on Col-0 or on CYP81F2Δ/Δ genotypes before we transferred the aphids to test plants and started our experiments. We included this factor origin in our statistical models. Plant genotype had a significant effect on aphid proliferation (Figure 7). We found fewer aphids on CYP81F2Col/Col than on CYP81F2Δ/Δ genotypes (n = 82, df = 1, F = 9.19, P = 0.0033) irrespective of whether aphids had been previously reared on plants with wild-type or with mutant CYP81F2 alleles. Furthermore, the preexperimental rearing procedure strongly influenced aphid performance. Aphids that originated from wild-type plants proliferated less well than aphids that came from mutant plants (n = 82, df = 1, F = 13.04, P = 0.0006). However, we found no interaction between both variables (n = 82, df = 1, F = 0.89, P = 0.37, nonsignificant), indicating that plant genotype effects during the experiment were independent of genotype effects in the preexperimental treatment. This showed that aphids did not adapt to the genotype on which they had been reared.
Figure 7.
Aphid Proliferation Contingent on CYP81F2 Genotype.
Shown are aphid numbers (±1 se) with different combinations of preexperimental rearing (Origin, either on Col-0 or on homozygous SALK_123882 mutants) and experimental test plants (Test Plants, either with CYP81F2 wild-type alleles or with mutant alleles [M]). Sample sizes (N) are indicated. We conducted these assays three times with the same results. Letters a to c indicate statistically different means in t tests.
DISCUSSION
The Role of CYP81F2 in the Biosynthesis of Modified Indole Glucosinolates
Most reaction steps in the biosynthesis of I3M from Trp are known, and the underlying genes have been cloned (Figure 8). Trp is synthesized in the plastids (Li et al., 1995; Zhao and Last, 1995; Bak et al., 1998), and CYP79B2 and CYP79B3, which convert Trp to indole-3-acetaldoxime, contain plastid transit peptides (Hull et al., 2000; Mikkelsen et al., 2000). All other enzymes of the indole glucosinolate core pathway have no discernable plastid targeting sequences, indicating that subsequent reaction steps take place in other cell compartments. CYP83B1 N-hydroxylates indole-3-acetaldoxime to the corresponding aci-nitro compound, 1-aci-nitro-2-indolyl-ethane (Bak and Feyereisen, 2001; Bak et al., 2001; Hansen et al., 2001; Naur et al., 2003). This compound is conjugated with an S-donor, most likely Cys (Wetter and Chisholm, 1968). The respective genes and enzymes are not yet known (Halkier and Gershenzon, 2006). Next, a C-S-lyase converts S-indolyl-thiohydroximate to the corresponding thiohydroximate (Mikkelsen et al., 2004), followed by a glycosylation that is catalyzed by an S-glycosyltransferase to form desulfo-I3M (Grubb et al., 2004). The final reaction in the core structure pathway is performed by a family of three desulfoglucosinolate sulfotransferases, with At ST5a preferably acting on desulfo-I3M (Piotrowski et al., 2004).
Figure 8.
Biosynthesis of Indole Glucosinolates from Trp.
Shown are intermediates and known biosynthesis enzymes. Glucosinolates are boxed. 1OH-I3M, 1-hydroxy-indol-3-yl-methyl; GT, S-glucosyltransferase; ST, sulfotransferase. Note that we postulate a two-step synthesis of 1MO-I3M from I3M with 1OH-I3M (gray) as an intermediate, in contrast with previous models that propose direct conversion of I3M to 1MO-I3M.
This model of indole glucosinolate biosynthesis suggests that 4OH-I3M and 4MO-I3M originate from I3M by a common pathway (Figure 8). Detached leaves of Arabidopsis cyp79B2 cyp79B3 double mutants (Zhao et al., 2002), blocked early in the indole glucosinolate pathway, can convert artificially supplied I3M to 4MO-I3M but only when they are infested with aphids (Kim and Jander, 2007). We show that CYP81F2 catalyzes the first reaction in this pathway, the hydroxylation at position 4 of the indole ring, which results in the formation of 4OH-I3M. Generation of 4MO-I3M, however, requires the subsequent methylation of the hydroxy group, which is probably catalyzed by an as yet unidentified methyltransferase. The respective gene appears to be not variable in Da(1)-12 × Ei-2 and therefore remained undetected during QTL mapping. Nonetheless, this reaction sequence explains why natural variation in a single gene, CYP81F2, causes both a 4OH-I3M and a 4MO-I3M QTL and why mutations in this gene affect both compounds simultaneously; reduction of the metabolic flux from I3M to the 4OH intermediate reduces the quantity of the substrate available for the methylation reaction and, hence, the accumulation of the end product, 4MO-I3M. Levels of 1MO-I3M, the third modified indole glucosinolate in Arabidopsis, remained unaffected by mutations in CYP81F2, corroborating our previous finding that different QTL control natural variation in 1MO-I3M versus 4OH-I3M and 4MO-I3M accumulation in Da(1)-12 × Ei-2 RILs (Pfalz et al., 2007). This is consistent with 1MO-I3M originating from I3M by another pathway than 4OH-I3M and 4MO-I3M. However, in contrast with the indole glucosinolate biosynthesis model, we surmise that I3M is not directly converted to 1MO-I3M but rather via a 1OH intermediate, in analogy to the reaction scheme for the generation of 4MO-I3M (Figure 8). Such an intermediate, however, would be highly instable because of the hydroxy group attached to the nitrogen of the indole ring. This may explain why this compound has not yet been isolated from Arabidopsis or other crucifers (Fahey et al., 2001).
Natural Variation in the Quantitative Genetic Architecture of Modified Indole Glucosinolate Biosynthesis
T-DNA insertions in CYP81F2 strongly impair 4OH-I3M and 4MO-I3M production. RT-PCRs failed to amplify CYP81F2 cDNA in SALK_123882 CYP81F2Δ/Δ genotypes, while control genes had wild-type levels of PCR product. Hence, the T-DNA insertion in SALK_123882 causes a complete knockout of CYP81F2 function. Nonetheless, mutant plants produced low levels of 4OH-I3M and 4MO-I3M. Likewise, DE089 leaves accumulated ∼60% of the Ei-2 level in modified indole glucosinolates but had only ∼30% of CYP81F2 transcript levels (see Supplemental Figure 1 online). This suggests that other gene products are also capable of catalyzing the oxygenation of I3M to 4OH-I3M. Candidate genes are the three other members of the Arabidopsis CYP81F subfamily, At4g37400, At4g37410, and At4g37430, which form a small gene cluster on Arabidopsis chromosome 4. We are currently testing this hypothesis with appropriate insertion lines and expression constructs.
To identify the molecular basis for the IGM1 QTL, we investigated differences between the Col-0, Da(1)-12, and Ei-2 CYP81F2 sequences. While the sequenced region was almost identical in Col-0 and Da(1)-12, Ei-2 deviated at numerous positions, both in the promoter and in the coding region. However, nonsynonymous substitutions were rare and all involved conservative amino acid replacements, suggesting that the 4OH-I3M and 4MO-I3M QTL are most likely caused by quantitative variation in CYP81F2 transcript levels. This matches the observation of Wentzell et al. (2007) who found that many metabolic QTL in glucosinolate biosynthesis are caused by differences in transcript abundance of the underlying genes and, hence, colocalize with corresponding eQTL. Indeed, CYP81F2 transcript levels correlated well with 4OH-I3M and 4MO-I3M quantity in Da(1)-12 × Ei-2 RILs, and mapping revealed the presence of two eQTL, one in cis and another one in trans on chromosome 1. These QTL compensated each other in the parental lines, Da(1)-12 and Ei-2, which had nearly equal CYP81F2 steady state transcript levels. Transcript levels were highest in RILs with a combination of Ei-2 alleles at the cis and Da(1)-12 alleles at the trans eQTL and lowest in the reciprocal combination, as exemplified by the DE089 RIL. This suggests that the Ei-2 CYP81F2 promoter contains an activating element that is absent from Da(1)-12 [or, vice versa, lacks a repressing element present in Da(1)-12]. The corresponding transcription factor remains to be identified. However, it is not likely that this factor is encoded by the gene that underlies the trans eQTL in Da(1)-12 × Ei-2 because cis and trans eQTL act additively and are, hence, independent of each other. Likewise, the gene causing the trans eQTL represents a novel regulator specifically acting on CYP81F2 expression. Most other regulators known to be involved in the formation of indole glucosinolates, ALTERED TRYPTOPHAN REGULATION1/MYB34 (Bender and Fink, 1998; Celenza et al., 2005), IQ-DOMAIN1 (IQD1) (Levy et al., 2005), and HIGH INDOLE GLUCOSINOLATE1/MYB51 (Gigolashvili et al., 2007), appear to affect all indole glucosinolates equally and are encoded by genes whose position does not match the QTL location. A notable exception is At Dof1.1 (= OBF BINDING PROTEIN2), whose overexpression causes an increase in I3M but a decrease in 4MO-I3M (Skirycz et al., 2006), but again, the location of this gene does not correspond to the eQTL position.
Consequences of Variation in Indole Glucosinolates for Plant–Enemy Interactions
Aphids largely avoid damaging the cells surrounding the phloem when they insert their stylet into the plant tissue to access the phloem sap (Tjallingii and Hogen Esch, 1993), thereby preventing a contact with the content of cells that contain defensive compounds. However, the phloem sap itself contains glucosinolates (Brudenell et al., 1999; Chen et al., 2001). Several studies have indicated that glucosinolates can play an important role in Arabidopsis defense against aphids. High IQD1 expression that increases total glucosinolate levels made Arabidopsis less attractive to M. persicae in choice assays (Levy et al., 2005). Likewise, relative performance of two aphid species, M. persicae and the cabbage aphid (Brevicoryne brassicae), was negatively correlated with total glucosinolate content in Arabidopsis genotypes with mutations in different phytohormone signaling pathways (Mewis et al., 2005). Subsequently, this inhibitory effect on aphid proliferation was more specifically attributed to indole glucosinolates and their breakdown products (Kim and Jander, 2007; Kim et al., 2008), although a contribution of aliphatic glucosinolates to aphid resistance cannot yet be excluded. Aphid-induced changes in Arabidopsis leaf glucosinolate profiles are largely negligible, with some variation in short-chain (Mewis et al., 2005, 2006) or long-chain (Kim and Jander, 2007) aliphatic glucosinolates, but with a high and statistically significant increase in 4MO-I3M levels (Kim and Jander, 2007).
In assays with artificial diet supplemented with I3M, 4MO-I3M, 1MO-I3M, or the aliphatic glucosinolate sinigrin 4MO-I3M turned out as the strongest inhibitor of aphid proliferation. Addition of myrosinase, the plant glucosinolate breakdown enzyme, increased the inhibitory effect of I3M and 1MO-I3M to a level equivalent to artificial diet supplemented with 4MO-I3M alone (Kim and Jander, 2007). Thus, 4MO-I3M has a particularly strong impact on M. persicae proliferation in vitro. Our assays, comparing aphid proliferation on CYP81F2 wild-type versus mutant plants, show that this also holds true in planta and demonstrate that CYP81F2 is an important player in Arabidopsis defense against aphids.
Only recently it was discovered that 4MO-I3M also plays a major role in Arabidopsis innate immunity (Bednarek et al., 2009; Clay et al., 2009). This defense response against microbial pathogens relies on a novel metabolic pathway that involves glucosinolate hydrolysis via an atypical myrosinase, PENETRATION2 (PEN2). Mutants in CYP81F2 or PEN2 have impaired innate immunity. While classical glucosinolate hydrolysis, directed against herbivores, largely depends on passive diffusion of glucosinolates and myrosinases upon tissue maceration, glucosinolate-dependent innate immunity involves active transport of PEN2 to the infection site. 4MO-I3M–derived metabolites may either have direct antimicrobial activity or prevent the entry of pathogens by triggering callose deposition at the infection site. In this context, Clay et al. (2009) have proposed that CYP81F2 may produce 4MO-I3M via 4-methoxylation of I3M. However, CYP81F2 rather catalyzes 4-hydroxylation of I3M, with an unknown methyltransferase subsequently acting to convert 4OH-I3M to 4MO-I3M.
Thus, 4MO-I3M exerts its effects in at least three different ways: (1) by myrosinase-catalyzed hydrolysis in combination with modifying proteins to influence oviposition preferences of P. rapae (De Vos et al., 2008), (2) through postingestive breakdown in M. persicae independent of classical myrosinases (Kim et al., 2008), and (3) via PEN2-dependent 4MO-I3M hydrolysis in plant–microbe interactions (Bednarek et al., 2009; Clay et al., 2009). Neither classical myrosinase-catalyzed nor PEN2-mediated 4MO-I3M hydrolysis appears to influence tissue damage caused by herbivorous lepidopteran larvae. A potential participation of the PEN2-dependent innate immunity response in plant defense against aphids and other phloem-feeding insects remains to be tested.
Perspectives for Targeted Manipulation of Glucosinolate Profiles
Most transcription factor genes known to be involved in the regulation of the indole glucosinolate pathway affect all indole glucosinolates similarly. Likewise, mutations in functional genes typically disturb this entire class of glucosinolates. For example, cyp79B2 cyp79B3 double knockouts are almost completely devoid of indole glucosinolates (Zhao et al., 2002), cyp83B1 was found to have reduced levels of all indole glucosinolates (Naur et al., 2003), and impaired C-S lyase function blocked glucosinolate synthesis completely (Mikkelsen et al., 2004). Furthermore, alterations in the indole glucosinolate core pathway often have side effects including aberrant plant growth because pathways for the synthesis of the plant hormone auxin and for indole glucosinolate biosynthesis partially overlap. Thus, genetic alteration of known transcription factors or of structural genes involved in the indole glucosinolate core pathway does not yet allow a targeted manipulation of specific glucosinolates and is rarely likely to yield the desired results, a healthy plant with increased herbivore resistance. The identification of CYP81F2 controlling the IGM1 QTL may offer a new perspective for metabolic engineering of glucosinolates against aphids because mutations in CYP81F2 specifically act on 4OH-I3M and 4MO-I3M, while the levels of all other glucosinolates do not change detectably and plants appear to be perfectly normal. It remains to be seen, however, whether this is also the case when CYP81F2 expression is artificially increased above wild-type levels.
METHODS
Plant Material and Growth Conditions
The Da(1)-12 × Ei-2 RIL population was developed at the Max Planck Institute for Chemical Ecology, Jena, and has been described previously (Pfalz et al., 2007). This RIL population is available from the Nottingham Arabidopsis Stock Center (NASC; stock number N722034). All other plant material was obtained from NASC. Seeds were sown into damp potting medium and covered with clear plastic grow domes, and seeds were stratified for 3 to 4 d at 6°C in the dark. Grow domes were removed 5 d after transfer to the light. Seedlings were transferred to 1:3 vermiculate/potting soil mix with 20 mL time release fertilizer (Osmocote) per flat. Plants were grown in 11.5 h day/12.5 h night cycles at 22°C and 60% relative humidity (day) and 16°C and 80% relative humidity (night) in an environment-controlled growth room. Light was supplied by NH 360 FLX Sunlux ACE bulbs with an intensity of 200 μmol s−1 m−2. Assays were, in general, performed with 3-week-old plants.
Glucosinolate Extraction and Analysis
For glucosinolate analyses from DE089 × Ei-2 progeny, 100 mg of fresh leaf material was harvested and immediately frozen in liquid nitrogen, lyophilized to dryness, and ground to a fine powder with seven 2.3-mm ball bearings in a paint shaker. Glucosinolates were extracted in a 96-well format as described by Kliebenstein et al. (2001a). HPLC separation and identification and quantification of extracted desulfo-glucosinolates were performed according to Kroymann et al. (2001). Molar glucosinolate concentration was calculated using response factors from Brown et al. (2003).
Genotyping and DNA Gel Blot Analyses
For mapping purposes, DNA was extracted from one to two freeze-dried leaves as described by Kroymann et al. (2001). Genotyping was performed with PCR-based markers (see Supplemental Table 1 online). PCR reactions contained, in general, ∼30 ng DNA, 2.3 μL 10× PCR buffer (Qiagen), 4 nmol of each deoxynucleotide triphosphate, 1.25 pmol of each of both primers, 70 nmol MgCl2, and 0.15 units Taq DNA polymerase (Qiagen) in a 23 μL volume. Cycling conditions were 94°C for 2 min, followed by 38 cycles of 94°C for 15 s, 50°C or 55°C for 15 s, and 72°C for 30 s, with a final extension of 72°C for 2 min on an Applied Biosystems 9700 thermocycler. PCR products obtained with primers 5seq01f/r, 5seq02f/r, 5seq03f/r, and 5seq05f/r harbor single nucleotide polymorphisms (accession numbers AM412009 to AM412016). These products were gel purified with QiaQuick columns (Qiagen) and sequenced directly. The other PCR products were separated on 4% MetaPhor (BMA) agarose gels.
Progeny from SALK T-DNA insertion lines SALK_123882 and SALK_005882 (Alonso et al., 2003) was tested for the presence of T-DNA insertions in CYP81F2 (At5g57220). Primers 123882-f and 123882-r produced a PCR product when SALK_123882 progeny carried a CYP81F2 wild-type allele and primers LBb1 and SALK_123882-r when plants carried a mutant allele. Similarly, in SALK_005861 progeny, primers 005861-f and 005861-r generated a PCR product from the wild type and primers LBb1 and 005861-f from the mutant allele (Figure 3).
The number of T-DNA insertions in SALK_005861 and SALK_123882 was analyzed with DNA gel blots, using 2 μg XbaI-restricted DNA per plant and a fragment of ∼300 bp amplified from the left border region of the T-DNA with primers T-DNA_L1F and T-DNA-L1R (see Supplemental Table 1 online) as a hybridization probe.
RNA Isolation for Hybridization of Agilent Arrays and for Quantitative Real-Time PCR
Per plant, three leaves were used for RNA extraction. Leaf material was ground to a fine powder in liquid N2, and total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. A DNase (Turbo DNase; Ambion) treatment followed to eliminate any contaminating DNA. A second purification step was performed with RNeasy MinElute columns (Qiagen) to remove DNase and any contaminating polysaccharides and proteins. RNA integrity was verified on an Agilent 2100 bioanalyzer using RNA nanochips. RNA quantity was determined on a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies) or with a BioPhotometer 6131 (Eppendorf).
Amplification, Labeling, and Hybridization
Total RNA was amplified using the Agilent low-input linear amplification kit according to the process outlined by the manufacturer (Agilent). One to five micrograms of amplified target cRNA was labeled with either cy5 or cy3 using the Micromax kit (Applied Biosystems). The labeled material was passed through Zymo RNA Clean-up Kit-5 columns (Zymo Research) to remove any unincorporated label and eluted in 15 to 20 μL of RNase-free water (Ambion). Concentration of labeled cRNA and label incorporation was determined by Nanodrop-1000 spectrophotometer analysis. Labeled material was hybridized onto Agilent Arabidopsis 3 Arrays overnight in a rotating oven at 60°C, and slides were processed as outlined in the Agilent processing manual. Per condition, three biological replicates were used, with a total of six arrays for this experimental group. Arrays were scanned using the Agilent G2565BA fluorescent microarray scanner. Image processing was performed with Agilent's feature extraction software (version 7.5). Further analysis was done with Rosetta Luminator (at MOgene LC) and with GeneSifter (VizXlabs) software. Arrays were normalized with the Lowess algorithm. Artificial RNA spike-in controls that anneal specifically to complementary control probes on the arrays were used to monitor linearity, sensitivity, accuracy, and dynamic range. Less than 0.001% of the data points were saturated, and none of the genes of interest were affected by saturation. After image processing, spot intensity was summarized by the mean or median pixel intensity and by a measurement of inter-pixel variability within each spot. P values were calculated for each spot within each array using the more conservative of two error models and taking signal-to-noise ratios into account. Across-array comparisons were conducted with ANOVA to obtain P values for calculation of the false discovery rate with the MULTTEST procedure of the SAS software (SAS Institute).
Quantitative Real-Time PCR
DNA-free total RNA (500 ng) was converted into single-stranded cDNA using a mix of random and oligo(dT20) primers according to the ABgene protocol (ABgene). Real-time PCR oligonucleotide primers were designed using the online Primer3 internet-based interface (http://frodo.wi.mit.edu). Gene-specific primers were designed on the basis of sequence obtained for CYP81F2 and several additional genes (At1g54270 = EIF-4A2, At1g34030 = RPS18B, At2g18110 = EF1-β, At2g29550 = Tubulin β-7 chain, and At5g09810 = Actin7) as potential housekeeping genes to serve as the endogenous control (normalizer). Primer design, quantitative RT-PCR conditions, dissociation curve analysis, tests of dynamic range, and quantification were performed as described previously (Freitak et al., 2007). All primers are listed in Supplemental Table 1 online.
The comparative quantitation method (ΔΔCt) was used to contrast the different treatments and tissues (Livak and Schmittgen, 2001). Ct values quantify the number of PCR cycles necessary to amplify a template to a chosen threshold concentration, ΔCt values quantify the difference in Ct values between a test and a control gene for a given sample, and ΔΔCt values are used for the comparison between two samples. ΔΔCt values were transformed to absolute values with 2−ΔΔCt for obtaining relative fold changes. Except for the RIL population, all assays were run in triplicate (biological replication) and duplicate (technical replication) to control for overall variability. Relative fold changes for each gene were set to 1 for the control.
Expression Constructs and Heterologous Expression in Sf9 Cells
Total RNA was extracted from Arabidopsis thaliana Col-0 with TRIzol (Invitrogen) and reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. CYP81F2 cDNA was amplified with AccuPrime Taq polymerase (Invitrogen) with primers IGM1f and IGM1r (see Supplemental Table 1 online). Primers were chosen such that the PCR product contained a Kozak translation initiation sequence in addition to the start codon. The native stop codon was omitted to include a C-terminal peptide encoded by the vector from the pIB/V5-His TOPO TA expression kit (Invitrogen) that enables detection with a V5 antibody. PCR was as follows: 94°C for 1 min, 30 cycles at 94°C for 15 s, 55°C for 30 s, and 72°C for 1.45 min, followed by a final elongation at 72°C for 5 min on a PE Applied Biosystems 9700 thermal cycler. Purified product was cloned into a pIB/V5-His TOPO vector and transformed into Escherichia coli TOP-10 cells (Invitrogen). Plasmids were isolated with the HiPure Plasmid Filter Midiprep kit (Invitrogen) and sequenced to verify correct cDNA sequence, reading frame, and cloning direction.
Sf9 cells (Invitrogen) were grown at 27°C in Sf-900 II SFM with 50 μg/mL Gentamycin (both Gibco). Per culture dish, 660 μL SFM was mixed with 66 μL Insect GeneJuice Transfection Reagent (Novagen). In parallel, 12 μg of expression construct was added to 660 μL SFM. Plasmid containing solution and transfection reagent were combined, left for 15 min at ambient temperature, and added to Sf9 cells. After 4 h of incubation at 27°C, SFM was replaced once.
After 48 h, cells were suspended in their culture medium, centrifuged with 500g at 4°C for 10 min, and washed twice with ice-cold 1× PBS. Next, cells were resuspended in 2 mL hypotonic buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail [Pierce]) and kept on ice for 20 min. Cells were homogenized and the lysate was mixed with an equal volume of sucrose buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 mM DTT, 500 mM sucrose, and protease inhibitors) and centrifuged with 1200g at 4°C for 10 min. The supernatant was stored on ice, and the homogenization step was repeated once with the pellet. Supernatants were pooled and centrifuged with 10,000g at 4°C for 15 min. The supernatant from this step was centrifuged with 100,000g at 4°C for 1 h. The resulting pellet was resuspended in 0.5 mL 100 mM phosphate buffer, pH 7.4, with 20% (v/v) glycerol and protease inhibitors. Finally, heterologous expression was confirmed with SDS-PAGE and immunoblot analysis.
Isolation of Native Indol-3-yl-Methyl Glucosinolate from Seeds of Dyer's Woad
Isatis tinctoria seeds contain high amounts of I3M (Mohn et al., 2007) and were therefore used for the isolation of intact glucosinolates. Fifty grams of seeds (Saatzucht Quedlinburg) were homogenized with 250 mL 80% (v/v) aqueous methanol using a Polytron PT3100. After centrifugation with 7000g at ambient temperature for 10 min, the supernatant was collected. The extraction step was repeated with 200 mL 80% (v/v) aqueous methanol, and the combined supernatant was loaded onto an anion-exchange column filled with 1 g DEAE-Sephadex A-25. The column was washed three times with 5 mL of a 3:2:5 (v/v/v) mixture of formic acid, isopropanol, and water, and four times with 5 mL water. Glucosinolates were eluted with 25 mL 0.5 M K2SO4/3% (v/v) aqueous isopropanol and collected in 25 mL ethanol (Thies, 1988). This first elution step preferably elutes aliphatic glucosinolates. For isolation of indole glucosinolates, an additional 75 mL of eluent was added to the column and the eluate was collected in 100 mL ethanol. This eluate was centrifuged with 3000g at ambient temperature for 10 min, and the supernatant was lyophilized with a rotary evaporator. Indole glucosinolates were dissolved in 3 × 0.5 mL water and fractionated on an Agilent HP 1100 Series system, equipped with a SUPELCOSIL LC-18-DB SEMI-PREP 250 × 10 mm, 5 μm, column (Supelco) and a fraction collector. The program for the separation of intact I3M from other glucosinolates was as follows: start with 5% solvent B (acetonitrile) and 95% A (0.02% trifluoroacetic acid), 8 min 13% B, 8.1 min 95% B, 10 min 95% B, 10.1 min 5% B, and 15 min 5% B. The fraction containing intact I3M was split into several aliquots, lyophilized with a Genevac HT-4X Series II vacuum evaporator (Ipswich), and stored at −20°C. Prior to enzyme assays, I3M was dissolved in 100 mM KPi, pH 7.4.
Enzyme Assays
CYP81F2 activity was measured in microsomes isolated from Sf9 insect cells. Microsomes isolated from Sf9 cells expressing a major allergen gene from the European cabbage butterfly (Pieris rapae), amplified with primers MA-ExF and MA-ExR (see Supplemental Table 1 online), served as a negative control (Fischer et al., 2008). A second negative control contained phosphate buffer instead of microsomes but was otherwise treated the same. Microsome protein concentration was determined with a Bio-Rad DC protein assay. Typical protein concentrations of microsomes transfected with CYP81F2 were 2.5 μg/μL. Concentration of microsomes with control plasmids was adjusted accordingly.
Fifty microliters of transfected microsomes were incubated overnight at ambient temperature with I3M in 100 mM KPi, pH 7.4, 2 mM NADPH, 2 mM glucose-6-phosphate, and 0.1 units of glucose-6-phosphate dehydrogenase in a total volume of 200 μL. The reaction mixture was loaded onto an anion-exchange column filled with DEAE-Sephadex A-25 and washed three times with 1 mL 67% (v/v) aqueous methanol, two times with 1 mL water, and three times with 1 mL MES buffer, pH 5.2. Fifty microliters (30 units) sulfatase (Sigma-Aldrich) were added to the column. After 6 h of incubation at ambient temperature, samples were eluted with 6 × 0.5 mL water and concentrated on a Genevac HT-4X Series II vacuum evaporator. Finally, the reaction products were dissolved in 220 μL water, and 95 μL were injected in an Agilent HP 1100 Series system with the following program: start with 1.5% solvent B (acetonitrile) and 98.5% A (water), 1 min 1.5% B, 6 min 5% B, 8 min 7% B, 18 min 21% B and 23 min 29% B, 30 min 43% B, 30.5 min 100% B, 33 min 100% B, 33.1 min 1.5% B, and 38 min 1.5% B. Identity of the reaction product was confirmed by liquid chromatography–mass spectrometry.
Herbivory Screens with Lepidopterans
Herbivory assays were essentially performed as described by Pfalz et al. (2007). Diamondback moth (Plutella xylostella) was obtained from New York State Agricultural Experiment Station, Geneva, NY, and a colony was maintained at the Max Planck Institute for Chemical Ecology, Jena, Germany. Cabbage looper (Trichoplusia ni) eggs were obtained from Benzon Research and beet armyworm (Spodoptera exigua) from Bayer Crop Sciences. All larvae were reared for 5 to 6 d on artificial diet (Shelton et al., 1991) prior to experiments. Cabbage white butterfly (Pieris brassicae) eggs were obtained from Seritech. After hatching, larvae were preexperimentally reared on Brassica napus var oleifera for 2 to 3 d.
Lepidopteran performance was analyzed separately for SALK_005861 and SALK_123882 progeny. Each experiment included several homozygous mutant and wildtype lines and Col-0 wildtype, randomized over 96-celled flats. Plant diameter was recorded prior to herbivory screens. Each plant received one larva and larvae were allowed to move freely. P. xylostella, T. ni, and S. exigua fed for 2 d, P. brassicae for 24 h. Afterwards, the leaf area removed by the insects was assessed visually, and an artificial scale was established to quantify tissue damage as described by Stotz et al. (2000). In general, the following statistical model was used:
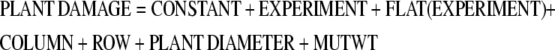 |
In this linear ANOVA model, COLUMN and ROW are variables to control for position effects that may result from larval movement during the experiments. EXPERIMENT accounts for variation between experimental replicates and FLAT(EXPERIMENT) for variation between flats within an experiment. PLANT DIAMETER controls for potential effects of plant size on insect herbivory. MUTWT estimates the genotype effect, mutant or wild type, on herbivory.
Aphid Proliferation Screens
A colony of the green peach aphid (Myzus persicae) was maintained since 2005 on Col-0 wild type at the Max Planck Institute for Chemical Ecology in Jena. Prior to experiments, aphids were either raised for several generations on 4- to 5-week old Col-0 wild-type plants or on SALK_123882 CYP81F2Δ/Δ mutant progeny. Adult aphids were collected and transferred to fresh 5-week-old plants of the same genotypes as before. Adults were allowed to proliferate for 2 d. Afterwards, adults were removed and only the aphid progeny remained on the plants. This procedure ensured that all aphids were approximately of the same age (±1 d old) at the time of the experiment.
After 7 d, aphids were transferred to 18-d-old test plants, with three aphids of the same origin per test plant. After five further days (i.e., within the linear proliferation phase), the number of aphids per plant was counted with the help of a magnifying glass. Test plants were grown randomized in 96-celled flats, with 48 plants per flat and every other cell left empty. Each experiment included three lines derived from homozygous mutant progeny of SALK_005861, two lines from homozygous wild-type progeny of SALK_005861, three lines from homozygous mutant progeny of SALK_123882, two lines derived from homozygous wild-type progeny of SALK_123882, and Col-0 wild type as test plants, with a sample size of six to nine plants per line. Here, the following ANOVA model was used:
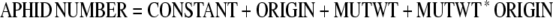 |
Because position effects were absent, it was not necessary to include COLUMN or ROW variables. Also, plant size had no detectable effect on aphid number. ORIGIN accounts for preexperimental and MUTWT for genotype effects during the experiment.
Sequencing of Genomic At5g57220 from Da(1)-12 and Ei-2
The At5g57220 region was obtained as a single fragment using primers 5-CYP and 3-CYP (see Supplemental Table 1 online). PCR products were gel purified with QiaQuick columns (Qiagen) and cloned into TOPO XL vectors (Invitrogen). Four clones each were sequenced on an automated Applied Biosystems 3730xl DNA Analyzer using BigDye terminators version 3.1 with universal primers m13u and m13f and with insert-specific primers 23204480F, 23205533R, 23206263R, 23206821R, 23207360R, Seq57720F, and Seq57720R (see Supplemental Table 1 online). The DNAstar software package (DNASTAR) was used for sequence assembly and alignment. Sequence data are deposited at EMBL (accession numbers FM208178 and FM208179).
Statistics Programs
Systat Version 10 (SPSS) was used for ANOVA. Expression QTL were mapped with Windows QTL Cartographer V2.5 (http://statgen.ncsu.edu.qtlcart/WQTLCart.htm) and with R/qtl (Broman et al., 2003). For Bayesian interval mapping, 400,000 Markov-Chain-Monte-Carlo steps were simulated, and iterations were recorded at every 400th step, with 1000 pre-burn-in and 20,000 burn-in steps. Prior for the number of QTL was Poisson, with zero initial QTL. qBase (Hellemans et al., 2007) was used for the analysis of qRT-PCR data from DE089, Da(1)-12, Ei-2, and from the SALK T-DNA insertion lines.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AM412009 to AM412016 [genetic markers 5seq01, 5seq02, and 5seq03 from Ei-2 and Da(1)-12], FM208178 (CYP81F2 from Ei-2), and FM208179 [CYP81F2 from Da(1)-12].
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Modified Indole Glucosinolate and CYP81F2 Transcript Abundance in Da(1)-12, Ei-2, and DE089.
Supplemental Table 1. Primer Sequences Used in This Work.
Supplemental Results. Details for Fine-Mapping of QTL for Modified Indole Glucosinolates in DE089 × Ei-2 NILs.
Supplementary Material
Acknowledgments
We thank Grit Kunert for sharing her M. persicae colony with us and John D'Auria, Michael Reichelt, and Jonathan Gershenzon for inspiring discussions and advice on indole glucosinolate biochemistry and for comments on the manuscript. We also thank Choon Wei (Jeffrey) Wee for his help with heterologous expression of cytochrome P450s in insect cells and Yannick Pauchet for his protocol on microsome isolation. This work was supported by the Max Planck Society, the Centre National de la Recherche Scientifique, and the Université Paris-Sud in Orsay, France.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Juergen Kroymann (juergen.kroymann@u-psud.fr).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bak, S., and Feyereisen, R. (2001). The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak, S., Nielsen, H.L., and Halkier, B.A. (1998). The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38 725–734. [DOI] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. [DOI] [PubMed] [Google Scholar]
- Bender, J., and Fink, G.R. (1998). A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 5655–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderoth, M., Pfalz, M., and Kroymann, J. (2009). Methylthioalkylmalate synthases: Genetics, ecology and evolution. Phytochem. Rev. 8 255–268. [Google Scholar]
- Benderoth, M., Textor, S., Windsor, A.J., Mitchell-Olds, T., Gershenzon, J., and Kroymann, J. (2006). Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. USA 103 9118–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.L., and Eastop, V.F. (2006). Aphids on the World's Herbaceous Plants and Shrubs. (Chichester, UK: John Wiley & Sons).
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Oncklen, H.V., Montagu, M.V., and Inzé, P. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bones, A.M., and Rossiter, J.T. (2006). The enzymatic and chemically induced decomposition of glucosinolates. Phytochemistry 67 1053–1067. [DOI] [PubMed] [Google Scholar]
- Broman, K.W., Wu, H., Sen, S., and Churchill, G.A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 889–890. [DOI] [PubMed] [Google Scholar]
- Brown, P.D., Tokuhisa, J.G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481. [DOI] [PubMed] [Google Scholar]
- Brudenell, A.J.P., Griffiths, H., and Baker, D.A. (1999). The phloem mobility of glucosinolates. J. Exp. Bot. 50 745–756. [Google Scholar]
- Burow, M., Bergner, A., Gershenzon, J., and Wittstock, U. (2007). Glucosinolate hydrolysis in Lepidium sativum – Identification of the thiocyanate-forming protein. Plant Mol. Biol. 63 49–61. [DOI] [PubMed] [Google Scholar]
- Campos de Quiros, H., Magrath, R., McCallum, D., Kroymann, J., Schnabelrauch, D., Mitchell-Olds, T., and Mithen, R. (2000). α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor. Appl. Genet. 101 429–437. [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root-formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., Quiel, J.A., Smolen, G.A., Merrikh, H., Sivestro, A.R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Petersen, B.L., Olsen, C.E., Schulz, A., and Halkier, B.A. (2001). Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 127 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, N.K., Adio, A.M., Denoux, C., Jander, G., and Ausubel, F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxenbichler, M.E., Spencer, G.F., Carlson, D.G., Rose, G.B., Brinker, A.M., and Powell, R.G. (1991). Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry 30 2623–2638. [Google Scholar]
- Delarue, M., Prinsen, E., van Onckelen, H., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 603–611. [DOI] [PubMed] [Google Scholar]
- De Vos, M., Kriksunov, K.L., and Jander, G. (2008). Indole-3-acetonitrile production from indole glucosinolates deters oviposition by Pieris rapae. Plant Physiol. 146 916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fahey, J.W., Zalcmann, A.T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51. [DOI] [PubMed] [Google Scholar]
- Fischer, H.M., Wheat, C.W., Heckel, D.G., and Vogel, H. (2008). Evolutionary origins of a novel host plant detoxification gene in butterflies. Mol. Biol. Evol. 25 809–820. [DOI] [PubMed] [Google Scholar]
- Freitak, D., Wheat, C.W., Heckel, D.G., and Vogel, H. (2007). Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 5 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili, T., Berger, B., Mock, H.P., Müller, C., Weisshaar, B., and Flügge, U.I. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50 886–901. [DOI] [PubMed] [Google Scholar]
- Glawischnig, E., Hansen, B.G., Olsen, C.E., and Halkier, B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, C.D., Zipp, B.J., Ludwig-Müller, J., Masuno, M.N., Molinski, T.F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. [DOI] [PubMed] [Google Scholar]
- Guengerich, F.P. (2001). Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14 611–650. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Du, L.C., Naur, P., Olsen, C.E., Axelsen, K.B., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001). CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276 24790–24796. [DOI] [PubMed] [Google Scholar]
- Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8 R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, H., Chadchawan, S., Winge, P., Thangstad, O.P., and Bones, A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes, J.J.B., Fu, J., de Vos, C.H., Lommen, A., Hall, R.D., Bino, R.J., van der Plas, L.H.W., Jansen, R.C., Vreugdenhil, D., and Koornneef, M. (2006). The genetics of plant metabolism. Nat. Genet. 38 842–849. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., and Jander, G. (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49 1008–1019. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Lee, B.W., Schroeder, F.D., and Jander, G. (2008). Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J. 54 1015–1026. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Gershenzon, J., and Mitchell-Olds, T. (2001. b). Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001. a). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., and Mitchell-Olds, T. (2005). The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 8 264–271. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Lambrix, V.M., Reichelt, M., Gershenzon, J., and Mitchell-Olds, T. (2001. c). Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Pedersen, D., Barker, B., and Mitchell-Olds, T. (2002). Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Davies, A., Deeken, R., Thorpe, M.R., Tomos, A.D., and Hedrich, R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann, J., Donnerhacke, S., Schnabelrauch, D., and Mitchell-Olds, T. (2003). Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. USA 100 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann, J., and Mitchell-Olds, T. (2005). Epistasis and balanced polymorphism influencing complex trait variation. Nature 435 95–98. [DOI] [PubMed] [Google Scholar]
- Kroymann, J., Textor, S., Tokuhisa, J.G., Falk, K.L., Bartram, S., Gershenzon, J., and Mitchell-Olds, T. (2001). A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127 1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Lambrix, V., Reichelt, M., Mitchell-Olds, T., Kliebenstein, D.J., and Gershenzon, J. (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influence Trichoplusia ni herbivory. Plant Cell 13 2793–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194. [DOI] [PubMed] [Google Scholar]
- Levy, M., Wang, Q., Kaspi, R., Parrella, M.P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43 79–96. [DOI] [PubMed] [Google Scholar]
- Li, J., Chen, S., Zhu, L., and Last, R.L. (1995). Isolation of cDNAs encoding the tryptophan pathway enzyme indole-3-glycerol phosphate synthase from Arabidopsis thaliana. Plant Physiol. 108 877–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J.L., Yamada, M., Estelle, M., Normaly, J., and Sandberg, G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthy, B., and Matile, P.H. (1984). The mustard oil bomb - Rectified analysis of the subcellular organization of the myrosinase system. Biochem. Physiol. Pflanz. 179 5–12. [Google Scholar]
- Matile, P.H. (1980). “Die Senfölbombe”: Zur Kompartmentierung des Myrosinasesystems. Biochem. Physiol. Pflanz. 175 722–731. [Google Scholar]
- Mewis, I., Appel, H.M., Hom, A., Raina, R., and Schultz, J.C. (2005). Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 138 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis, I., Tokuhisa, J.G., Schultz, J.C., Appel, H.M., Ulrichs, C., and Gershenzon, J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67 2450–2462. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of glucosinolates and indole-3-acetic acid. J. Biol. Chem. 43 33712–33717. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Naur, P., and Halkier, B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 770–777. [DOI] [PubMed] [Google Scholar]
- Mohn, T., Cutting, B., Ernst, B., and Hamburger, M. (2007). Extraction and analysis of intact glucosinolates – A validated pressurized liquid extraction/liquid chromatography–mass spectrometry protocol for Isatis tinctoria, and qualitative analysis of other cruciferous plants. J. Chromatogr. A 1166 142–151. [DOI] [PubMed] [Google Scholar]
- Naur, P., Petersen, B.L., Mikkelsen, M.D., Bak, S., Rasmussen, H., Olsen, C.E., and Halkier, B.A. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Paquette, S., Bak, S., and Feyereisen, R. (2000). Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19 307–317. [DOI] [PubMed] [Google Scholar]
- Pfalz, M., Vogel, H., Mitchell-Olds, T., and Kroymann, J. (2007). Mapping of QTL for resistance against the crucifer specialist insect herbivore Pieris brassicae in a new Arabidopsis inbred line population, Da(1)-12 × Ei-2. PLoS One 2 e578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski, M., Schemenewitz, A., Lopukhinat, A., Müller, A., Janowitz, T., Weiler, E.W., and Oecking, C. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725. [DOI] [PubMed] [Google Scholar]
- Pollard, D.G. (1972). Plant penetration by feeding aphids (Hemiptera, Aphidoidea): A review. Bull. Entomol. Res. 62 631–714. [Google Scholar]
- Raybould, A.F., and Moyes, C.L. (2001). The ecological genetics of aliphatic glucosinolates. Heredity 87 383–391. [DOI] [PubMed] [Google Scholar]
- Reichelt, M., Brown, P.D., Schneider, B., Oldham, N.J., Stauber, E., Tokuhisa, J., Kliebenstein, D.J., Mitchell-Olds, T., and Gershenzon, J. (2002). Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59 663–671. [DOI] [PubMed] [Google Scholar]
- Shelton, A.M., Cooley, R.J., Kroening, M.K., Wilsey, W.T., and Eigenbrode, S.D. (1991). Comparative analysis of two rearing procedures for diamond-back moth, Plutella xylostella (Lepidoptera: Plutellidae). J. Entomol. Sci. 26 17–26. [Google Scholar]
- Skirycz, A., Reichelt, M., Burow, M., Birkemeyer, C., Rolcik, J., Kopka, J., Zanor, M.I., Gershenzon, J., Strnad, M., Szopa, J., Mueller-Roeber, B., and Witt, I. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47 10–24. [DOI] [PubMed] [Google Scholar]
- Smolen, G., and Bender, J. (2002). Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U., Pittendrigh, B.R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., and Mitchell-Olds, T. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangstad, O.P., Gilde, B., Chadchawan, S., Seem, M., Husebye, H., Bradley, D., and Bones, A.M. (2004). Cell specific cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum. Plant Mol. Biol. 54 597–611. [DOI] [PubMed] [Google Scholar]
- Thies, W. (1988). Isolation of sinigrin and glucotropaeolin from cruciferous seeds. Fat Sci. Technol. 90 311–314. [Google Scholar]
- Tjallingii, W.F., and Hogen Esch, T. (1993). Fine structure of aphid styet routes in plant tissues in correlation with EPG-signals. Physiol. Entomol. 18 317–328. [Google Scholar]
- Wentzell, A.M., Rowe, H.C., Hansen, B.G., Ticconi, C., Halkier, B.A., and Kliebenstein, D.J. (2007). Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 3 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart, D., Bak, S., and Paquette, S. (2002). Cytochromes P450. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/ http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Werck-Reichhart, D., and Feyereisen, R. (2000). Cytochromes P450: a success story. Genome Biol. 1 REVIEWS3003. [DOI] [PMC free article] [PubMed]
- Wetter, L.R., and Chisholm, M.D. (1968). Sources of sulfur in thioglucosides of various higher plants. Can. J. Biochem. 46 931–935. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Ober, J.A., and Kliebenstein, D.J. (2006). The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., and Last, R.L. (1995). Immunological characterization and chloroplast localization of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. J. Biol. Chem. 270 6081–6087. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.A., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYB79B2 and CYP79B3. Genes Dev. 16 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.