Abstract
Nucleoside degradation and salvage are important metabolic pathways but hardly understood in plants. Recent work on human pathogenic protozoans like Leishmania and Trypanosoma substantiates an essential function of nucleosidase activity. Plant nucleosidases are related to those from protozoans and connect the pathways of nucleoside degradation and salvage. Here, we describe the cloning of such an enzyme from Arabidopsis thaliana, Uridine-Ribohydrolase 1 (URH1) and the characterization by complementation of a yeast mutant. Furthermore, URH1 was synthesized as a recombinant protein in Escherichia coli. The pure recombinant protein exhibited highest hydrolase activity for uridine, followed by inosine and adenosine, the corresponding Km values were 0.8, 1.4, and 0.7 mM, respectively. In addition, URH1 was able to cleave the cytokinin derivative isopentenyladenine-riboside. Promoter β-glucuronidase fusion studies revealed that URH1 is mainly transcribed in the vascular cells of roots and in root tips, guard cells, and pollen. Mutants expressing the Arabidopsis enzyme or the homolog from rice (Oryza sativa) exhibit resistance toward toxic fluorouridine, fluorouracil, and fluoroorotic acid, providing clear evidence for a pivotal function of URH1 as regulative in pyrimidine degradation. Moreover, mutants with increased and decreased nucleosidase activity are delayed in germination, indicating that this enzyme activity must be well balanced in the early phase of plant development.
INTRODUCTION
Nucleotides are uniquely important since they represent building blocks of genetic information (DNA and RNA), represent major energy carriers, and are also core elements of cofactors such as NAD, FAD, S-adenosylmethionine, or CoA, which serve in essential biochemical reactions, such as the synthesis of phospholipids and polysaccharides. Additionally, nucleotides are components of secondary metabolites like caffeine, cAMP, cGMP, and cytokinins (Boldt and Zrenner, 2003; Stasolla et al., 2003; Schmidt et al., 2004; Geigenberger et al., 2005). By reviewing the recently published work on nucleotide metabolism, it becomes obvious that many facets of this important biochemical aspect of plant metabolism are still poorly understood. One possible reason for this is the complexity of a multitude of biochemical reactions that facilitate de novo synthesis, degradation, and interconversion (partial degradation and recycling) of nucleotides, nucleosides, and nucleobases. In contrast with nucleotides, nucleosides do not possess phosphate groups, and nucleobases lack the ribose moeity. The recycling of nucleosides and nucleobases is also known as salvage.
In the salvage pathway, nucleobases and nucleosides are converted to nucleoside monophosphates by action of phosphoribosyl-pyrophosphatases and nucleoside kinases, respectively. Both phosphoribosyl-pyrophosphatases and nucleoside kinases have been identified in plants on the biochemical and molecular level (Moffatt et al., 2002; Zrenner et al., 2006; Islam et al., 2007). A great benefit of the salvage pathway is the lower energy cost compared with de novo synthesis. For example, purine de novo synthesis requires five ATP molecules, whereas the salvage of nucleosides only requires one ATP. Another difference is the high degree of conservation in the de novo synthesis compared with the salvage pathway. The de novo synthesis pathway is highly similar in prokaryotic and eukaryotic organisms, whereas nucleotide salvage pathways exhibit more pronounced variations (Katahira and Ashihara, 2002; Kafer et al., 2004).
Most of our understanding about the salvage pathway and the importance of this process originates from work on protists like Trypanosoma brucei, Leishmania major, and Crithidia fasciculata (Gopaul et al., 1996; Mäser et al., 1999; Shi et al., 1999; Stein et al., 2003). This group of organisms, which includes some human pathogens, does not possess the enzymatic equipment for nucleotide de novo synthesis and therefore completely relies on the import and salvage of nucleotide derivatives (Gopaul et al., 1996). Thus, much of our knowledge on nucleotide salvage in plants is based on comparisons to the situation in protists and awaits further experimental confirmation.
Besides other salvage pathway enzymes, nucleosidases have been identified and characterized from protists. These enzymes, also known as nucleoside hydrolases, catalyze the splitting of nucleosides into a ribose and a free base (Magni et al., 1975). Such enzyme activities have also been found in Escherichia coli and in Saccharomyces cerevisiae. By contrast, mammals do not contain nucleosidase activity (Parkin et al., 1991) but do contain nucleoside phosphorylase activity, resulting in the generation of a nucleobase and ribose-phosphate.
Such alterations in the biochemistry of enzyme-catalyzed reactions between different organisms are often beneficial for the development of new pharmaceutical drugs acting as enzyme inhibitors (Mitterbauer et al., 2002). In cancer research, the complete unraveling of nucleotide metabolism is of high importance because nucleobase analogs such as 5-fluorouracil are used to inhibit cell division. Moreover, understanding of nucleotide metabolism is key for understanding disease; for instance, a drastic example of the impact of an unbalanced ratio between nucleotide de novo synthesis and salvage pathway activity is given by the Lesch-Nyhan syndrome. Here, a genetically based loss of hypoxanthine-guanosine phosphoribosyltransferase activity, a salvage pathway enzyme, leads to a strong increase in de novo synthesis and in succession to increased levels of uric acid. Patients suffering from Lesch-Nyhan are characterized by neurological dysfunction and cognitive and behavioral disturbances (Sculley et al., 1992).
In plants, two examples are known that illustrate the importance of a functioning salvage pathway: mutants deficient in adenine phosphoribosyltransferase are characterized by nonfertile pollen (Moffatt and Somerville, 1988), and mutants lacking adenosine kinase activity show reduced fertility, transmethylation, and abnormal cell walls (Moffatt et al., 2002).
Such examples have led to a deeper investigation of salvage pathway enzymes in different organisms. Nucleosidases, which are in the focus of this work, are best characterized in protists and yeast, and crystal structures of the homologs from C. fasciculata and Trypanosoma vivax have been resolved (Degano et al., 1996; Versées et al., 2001). In higher plants, only few reports exist that describe nucleosidase activity, e.g., in white lupin (Lupinus luteus; Guranowski 1982), pea (Pisum sativum; Christensen and Jochimsen, 1983), soybean (Glycine max; Christensen and Jochimsen, 1983), and coffee (Coffea arabica; Campos et al., 2005). However, to our knowledge, nucleoside cleavage is the only way to degrade nucleotides and thus to liberate NH3 trapped in the cellular pool of RNA, DNA, and nucleotides.
Therefore, a main issue of this work was to identify such proteins in Arabidopsis thaliana. We aimed to identify homologs to the well-characterized nucleosidase of C. fasciculata in Arabidopsis, generate recombinant proteins, and analyze their enzymatic capacity. In addition, mutant plants with increased or decreased activities of nucleosidase were generated to shed light on the physiological function of this enzyme.
RESULTS
Identification of cDNAs Encoding Putative Nucleosidase Isoforms in Arabidopsis
As a first step in identifying putative nucleosidases in Arabidopsis, we looked for the biochemical activity by analyzing nucleosidase activity in crude, desalted extracts from Arabidopsis rosette leaves. We observed that [14C]-labeled uridine, provided at a concentration of 200 μM, was converted to [14C]-uracil by Arabidopsis leaf extracts in a time-dependent manner, with an activity of 68.7 ± 3.0 nmol mg−1 total protein h−1.
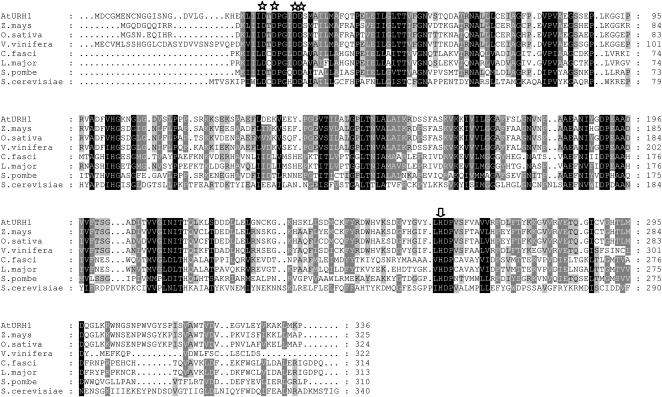
To identify the nucleosidase at the molecular level, we looked for Arabidopsis sequences homologous to the well-studied proteins from protists. An Arabidopsis cDNA encoding a protein with high similarity to URH1 from S. cerevisiae was identified. During the biochemical analysis, it turned out that uridine was the main substrate of this enzyme; therefore, it was named Arabidopsis URH1 in analogy to the Uridine-Ribohydrolase 1 (URH1) from bakers' yeast (Kurtz et al., 2002). Figure 1 displays a protein alignment between the protist, yeast, and Arabidopsis nucleosidase isoforms. At URH1 shares higher sequence similarity to the protist nucleosidases from L. major and C. fasciculata, both 28%, when compared with that from bakers' yeast (S. cerevisiae; 20%). The highest sequence identity outside the plant kingdom exists with the protein from Schizosaccharomyces pombe (34%; Figure 1). Among plants, many further sequences homologous to At URH1 can be identified. Only the closest relatives from Zea mays, Oryza sativa, and Vitis vinifera are shown to illustrate this finding (Figure 1).
Figure 1.
Multiple Alignment of the Deduced Amino Acid Sequences of Nucleosidases from Protozoans, Yeast, and Higher Plants.
Stars indicate conserved amino acid residues of the nucleosidase typical N-terminal Asp cluster. The arrow marks a conserved His residue responsible for release of the liberated nucleobase in IU-NH–type nucleosidases.
Arabidopsis URH1 contains the common sequence motive for inosine/uridine preferring nucleosidase isoforms that contains four Asp residues in a conserved domain at the N terminus of the proteins (Figure 1, stars) (Degano et al., 1996) and a His residue responsible for protonation of the departing nucleobase that is also conserved in all sequences shown (Figure 1, arrow). By comparing the deduced amino acid sequence from At URH1 with Arabidopsis open reading frames by a translated BLAST search, At URH2 was identified, sharing 49% identical amino acids. The sequence identity of At URH1 to Os URH1, the closest homolog found in rice is 69% (83% similarity; Table 1).
Table 1.
Comparison between At URH1 and 2 and the Closest Homolog from Rice
| Protein | Code | Size (aa) | Identity to At URH1 (%) | Similarity to At URH1 (%) |
|---|---|---|---|---|
| At URH1 | At2g36310 | 336 | – | – |
| At URH2 | At1g05620 | 323 | 49 | 67 |
| Os URH1 | Os08g05579 | 324 | 69 | 83 |
aa, amino acids.
Biochemical Characteristics of URH1 Expressed in Yeast
We next characterized the activity of URH1 using the Arabidopsis cDNA to complement a yeast mutant in pyrimidine synthesis and salvage. After expression of Arabidopsis URH1 in the yeast mutant YRZM18 (Mitterbauer et al., 2002) under control of a constitutive promoter, growth of the corresponding cells on selective media was analyzed. Due to multiple gene deletions, YRZM18 represents a mutant unable to perform both pyrimidine de novo synthesis and pyrimidine nucleoside salvage. Consequently, this mutant is unable grow on uridine as sole source for pyrimidines, whereas control cells are able to grow (see Supplemental Figure 1A online). YRZM18-expressing URH1 grows as efficiently as control cells on uridine, indicating that this nucleoside is metabolized and that Arabidopsis URH1 complements the yeast mutation.
For a more detailed analysis of the properties of the heterologously expressed nucleosidase, two different analysis methods were applied, and for both, yeast extracts were desalted and incubated with potential nucleosidase substrates. First, these substrates were [14C]-radiolabeled and the reaction products were subsequently subjected to thin layer chromatography and analyzed with a phosphor imager. When analyzing the time dependence of uridine hydrolysis in extracts from URH1-expressing yeast cells, most of the label shifted from the upper band in the thin layer chromatography comigrating with the uridine standard toward the lower band, matching the uracil standard, during the course of 20 min of incubation (see Supplemental Figure 1B online). With help of a phosphor imager, the hydrolysis of uridine (200 μM) was quantified, and an almost linear time dependence for the first 5 min was obtained (see Supplemental Figure 1C online). No uridine degradation was observed upon the addition of a boiled yeast extract. The observed enzyme activity after the first 5 min was 0.3 μmol uridine mg−1 protein min−1. In a subsequent experiment, the uridine hydrolase activity of URH1 was determined by separation and quantification of the reaction products by HPLC. With both methods, essentially similar results were obtained. The hydrolysis of uridine in the presence of extract from yeast cells expressing URH1 followed Michaelis-Menten kinetics. From a Hanes plot analysis, an apparent Km value of 2.1 mM was determined (see Supplemental Figure 1D online). The maximal activity was 30.2 μmol uridine mg−1 protein h−1.
Purification of Recombinant URH1 Expressed in E. coli
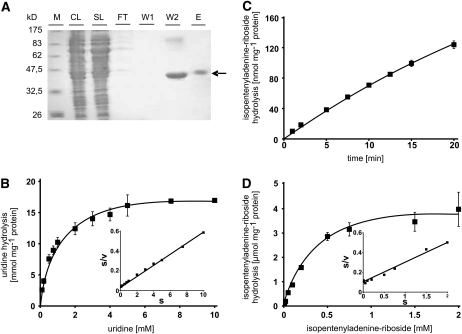
The yeast mutant used for complementation with Arabidopsis URH1 is deficient in the sole uridine nucleosidase but contains activities to cleave other nucleosides. From inhibitor experiments, indications arose that URH1 exhibits the capacity to use further substrates in addition to uridine (see Supplemental Figure 1 E online). Therefore, it was decided to purify the recombinant Arabidopsis enzyme after heterologous expression to allow a detailed biochemical characterization. For this, URH1 was expressed in E. coli as a hexa-his–tagged fusion protein and subsequently purified on nickel nitrilotriacetic acid (Ni-NTA) agarose. In the crude lysates derived by centrifugation, the recombinant protein could be visualized in the Coomassie blue–stained SDS-PAGE (Figure 2A) and was also detected by a his-tag–specific antibody. A high amount of recombinant URH1 protein is released from the Ni-NTA column in the second washing step. However URH1 in the eluate fraction is purified to a higher degree and is almost free of contaminating proteins (Figure 2A). This purified protein was desalted and used for further, more detailed, characterization of URH1 enzyme activity.
Figure 2.
Purification of Recombinant URH1 Synthesized in E. coli and Biochemical Properties of the Protein.
(A) SDS-PAGE, showing results of immobilization affinity chromatography. M, marker bands showing protein size in kilodaltons; CL, crude lysate; SL, solubilized lysate, FT, flow-through, W1 and W2, wash steps 1 and 2; E, eluate. Recombinant At URH1 is marked by an arrow.
(B) Substrate dependency of uridine hydrolysis by recombinant, purified URH1. Inset shows Hanes plot.
(C) Time dependency of IPR hydrolysis by recombinant, purified URH1.
(D) Substrate dependency of IPR hydrolysis by recombinant, purified URH1. Inset shows Hanes plot.
Data given in (B) to (D) represent means of three individual experiments, each with three to four replicate samples, ± se.
Biochemical Characteristics of Purified URH1
The substrate-dependent nucleoside cleavage revealed that uridine is the preferred substrate of URH1, showing a maximal catalytic activity of 18.2 mmol mg−1 protein h−1 and an apparent affinity constant of 0.8 mM (Figure 2B, Table 2). Inosine is hydrolyzed at a much lower maximal activity of 0.87 mmol mg−1 protein h−1 and an apparent Km value of 1.4 mM (Table 2). A further but less effective substrate that could be identified is adenosine. With this substrate, the maximal activity of URH1 is further reduced compared with uridine and inosine and accounted only for 0.03 mmol mg−1 protein h−1. The apparent Km value for adenosine was determined to be 0.7 mM (Table 2). Cytidine is not a substrate of URH1 (data not shown). Enzyme activity was linear for at least 10 min for all substrates tested.
Table 2.
Comparison of Apparent Affinities and Maximal Catalytic Activities of Arabidopsis URH1 Expressed in Yeast or E. coli
| Substrate/Expression System | Km (mM) | Vmax (mmol h−1 mg−1 Protein) |
|---|---|---|
| Uridine yeast | 2.1 | 0.030 |
| Uridine E. coli | 0.8 | 18.281 |
| Inosine E. coli | 1.4 | 0.869 |
| Adenosine E. coli | 0.7 | 0.028 |
| IPR E. coli | 0.4 | 0.005 |
Isopentenyladenine-riboside (IPR) inhibited uridine hydrolysis in a competition study and was thus suspected to function also as substrate for URH1 (see Supplemental Figure 1E online). When IPR was used as substrate in corresponding experiments, hydrolysis to isopentenyl-adenine was clearly demonstrated. IPR was cleaved in a time- and substrate-dependent manner, and a constant activity was observed for at least 10 min (Figure 2C). The calculated apparent Km value was 0.44 mM (Figure 2D, Table 2). A boiled enzyme preparation was used as control and showed no cleavage of IPR.
Expression Pattern and Subcellular Localization of URH1
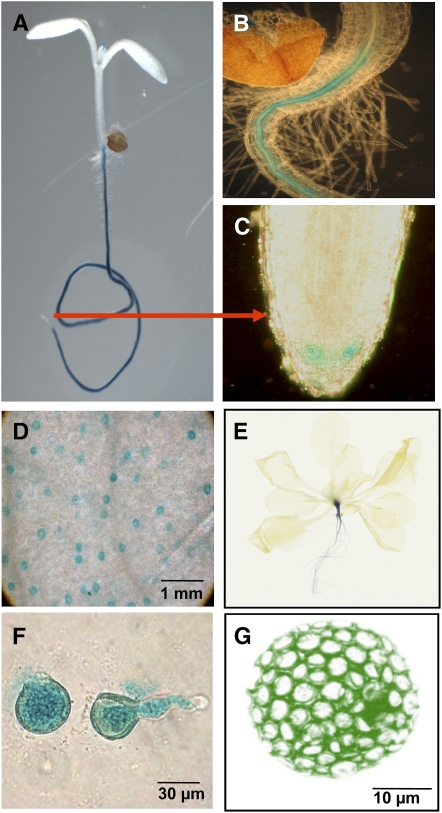
Real-time PCR data showed expression of URH1 in leaves, flowers, stems, and most prominently in roots (see Supplemental Figure 2 online). These results are in line with transcript measurements provided by the Genevestigator database (Zimmermann et al., 2004). To analyze the expression pattern in more detail, promoter β-glucuronidase (GUS) studies were performed. A 1-kb promoter fragment upstream of the start codon was fused to the GUS gene, and Arabidopsis plants were transformed. Analysis of 10 independent lines showed a similar pattern of GUS staining. Histochemical analysis of GUS activity showed URH1 expression in roots of young seedlings, and this expression remains intense in older plants (Figures 3A to 3C and 3E). Within the root, staining was restricted to the vasculature and the root tip meristem (Figures 3B and 3C). Furthermore, GUS staining was observed in guard cells of young leaves and in mature pollen cells (Figures 3D and 3F).
Figure 3.
Histochemical Localization of URH1 Gene Expression and Subcellular Localization of a URH1-GFP Fusion Protein.
(A) to (C) GUS stained 8-d-old seedling (A) and close-up view of roots (B) and root tip (C).
(D) Leaf area showing stained guard cells.
(E) Twenty-one-days old plant.
(F) Germinating pollen cells.
(G) GFP fluorescence from transiently transformed tobacco protoplast.
To analyze the subcellular localization of URH1, the protein sequence was explored by standard prediction software. However, no indications for organelle targeting information within the N-terminal part of the URH1 protein were obtained. For a deeper analysis, a green fluorescent protein (GFP) fusion construct was used for transient tobacco (Nicotiana tabacum) protoplast transformation. Green fluorescence in these protoplasts was evenly distributed in the cytosol, as shown in Figure 3G. Circular areas showing no fluorescence signals were where cytoplasm was excluded by chloroplasts (Figure 3G).
Analysis of Mutants with Altered Uridine Hydrolase Activity
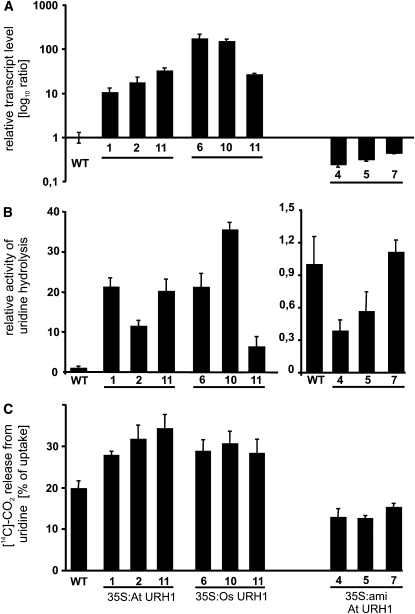
To deepen our physiological understanding of uridine nucleosidase activity, three types of mutants were generated. First, mutants expressing URH1 under control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter were generated. Second, as we could not exclude cosuppression effects from such mutant lines, we choose to express the closest homolog from rice, Os URH1 (Table 1). Third, activity of uridine hydrolase was reduced in mutants expressing an artificial microRNA (amiRNA) construct. Expression of Os URH1 and At URH1-microRNAs were also controlled by the CaMV 35S promoter. For each construct, three lines were chosen for further investigations.
The relative transcript levels of all types of mutants were determined by real-time PCR. All selected overexpressor lines exhibit at least 10-fold elevated transcript levels; in the case of Os URH1 overexpression lines #6 and #10, transcript increased up to 100-fold (Figure 4A). By contrast, microRNA lines showed 3- to 7-fold reduced transcript levels in lines #7, #5, and #4, respectively (Figure 4A). These alterations in transcript levels were accompanied by similar alterations in enzyme activity. At URH1 overexpressor lines showed 10- to 20-fold higher degradation of uridine compared with wild-type activities (Figure 4B). In the mutants expressing the rice homolog, the strongest line (#10) even showed a 30-fold higher uridine hydrolysis activity (Figure 4B). In the microRNA lines, the differences in uridine hydrolase activity were less pronounced. However, lines #4 and #5 exhibited significantly reduced activities accounting for 30 and 57% of wild-type level, respectively (Figure 4B).
Figure 4.
Biochemical Analysis of Mutants with Altered Nucleosidase Activity.
(A) Relative transcript level of At URH1 (in 35S:At URH1 and 35S:amiRNA lines) and of Os URH1 (in 35S:Os URH1 lines).
(B) Activity of uridine hydrolysis in plant extracts.
(C) Release of [14C]-CO2 from imported uridine in wild-type and mutant seedlings. 35S:At URH1, plants expressing At URH1 under control of CaMV 35S promoter; 35S:Os URH1, mutants expressing Os URH1 under control of CaMV 35S promoter; 35S:amiRNA, mutants expressing a microRNA construct directed against At URH1 under control of CaMV 35S promoter.
All data given represent means of three individual experiments, each with three to four replicate samples, ± se. Data are given relative to the wild-type levels for each graph.
Subsequent to the observation of altered enzyme activities in extracts of mutants, the ability to degrade imported uridine in vivo was analyzed using intact, 7-d-old seedlings. In accordance to the results obtained before, all overexpressor mutants released significantly more [14C]-CO2 from labeled, imported uridine compared with wild-type seedlings. At URH1–overexpressing lines #1, #2, and #11 released 28, 32, and 34% CO2, respectively, whereas wild-type seedlings only gave rise to 20% CO2 of the previously imported uridine (Figure 4C). For Os URH1–expressing mutants #6, #10, and #11, the corresponding liberation was 29, 31, and 28%, respectively (Figure 4C). All three analyzed microRNA mutants exhibited CO2 release in the range of 13% of the imported uridine (Figure 4C).
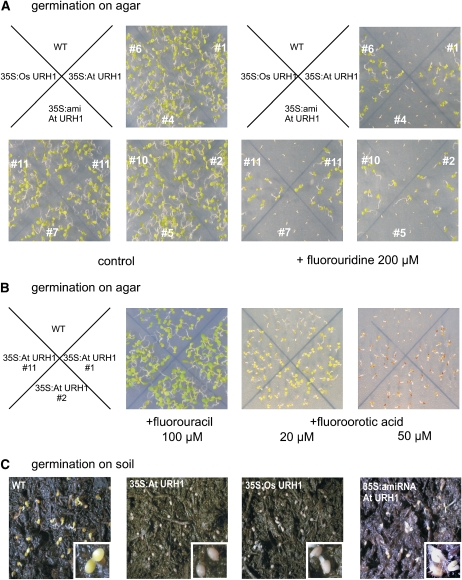
Nucleotide analogs like 5-fluorouracil (5-FU) and 5-fluorouridine (5-FD) are used in cancer therapy because after incorporation into DNA and RNA they are toxic to cells. Furthermore, they have proven useful in the exploration of enzyme functions in many facets of nucleotide metabolism. When wild-type seeds were germinated on 5-FD, growth was arrested shortly after onset of germination. Interestingly, growth of both At URH1 and Os URH1 overexpressor mutants was almost unaffected by 5-FD (Figure 5A). By contrast, microRNA lines performed like wild-type seeds and did not grow (Figure 5A). Obviously, 5-FD is degraded to a much higher extent in mutants with increased uridine hydrolase activity. In addition to the substrate of URH, its product uracil was provided to seedlings in form of the toxic fluorinated analog 5-FU. Similarly, increased expression of URH enhanced resistance of the corresponding seedlings toward 5-FU (100 μM; Figure 5B). Moreover, 35S:At URH1 clearly showed higher resistance toward 5-fluoroorotic acid (5-FO) when provided at 20 and 50 μM (Figure 5B). This substrate analog to orotic acid is an intermediate of pyrimidine de novo synthesis. Similar results were obtained with mutants expressing Os URH1.
Figure 5.
Growth Analysis of Mutants with Altered Nucleosidase Activity.
(A) Wild-type and nucleosidase mutants grown on agar medium for 7 d containing toxic fluorouridine. The # symbol indicates individual mutant lines.
(B) Growth of 35S:At URH1 mutants in presence of toxic fluorouracil.
(C) Growth of 35S:At URH1 mutants in presence of toxic 5-FO.
(D) Growth of seeds on standard soil. 35S:At URH1, plants expressing At URH1 under control of CaMV 35S promoter; 35S:Os URH1, mutants expressing Os URH1 under control of CaMV 35S promoter; 35S:amiRNA, mutants expressing a microRNA construct directed against At URH1 under control of CaMV 35S promoter. All experiments were repeated at least three times showing similar results.
These results point to altered overall pyrimidine nucleotide metabolism in mutants. Therefore, it was of interest to analyze the germination efficiency of corresponding seeds, as these fast-growing tissues show substantial demand for nucleotides to serve DNA, RNA, and nucleotide sugar synthesis on the one hand and for amino acids to serve protein synthesis on the other hand. Exact balancing of the respective synthesis and degradation pathways is therefore essential in the early phase of germination. Thus, germination on standard soil was monitored for all mutants. Seeds imbibed for 24 h were transferred to a growth chamber with a 10-h/14-h day/night regime. All seeds used for this experiment were of the same age. It was obvious that wild-type seeds germinated much faster. Compared with mutants, the radicle was visible in wild-type plants 38 h after transfer to the growth chamber. Sixty hours after transfer to the growth chamber, wild-type plants showed green, expanded cotyledons, whereas At URH1 overexpressor mutants still showed no germination (Figure 5C). In Os URH1 overexpressor mutants, the radicle emerged, and in microRNA mutants, hypocotyls and radicle started growth 60 h after transfer to the growth chamber (Figure 5C).
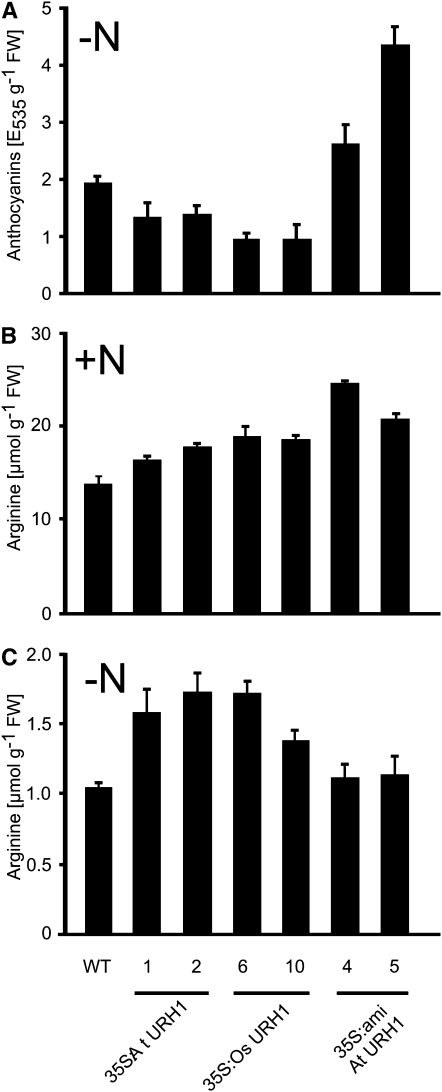
In previous experiments, we observed increased nucleoside degradation in liquid-grown seedlings under conditions of nitrogen starvation (M. Flörchinger, S. Hach, B. Jung, M. Traub, R. Wartenberg, C. Salzig, P. Lang, and T. Möhlmann, unpublished results). Therefore, we took advantage of the seedling liquid culture (Scheible et al., 2004) to analyze the performance of our mutants. As a marker for nitrogen starvation stress response, the accumulation of anthocyanins was chosen. Wild-type seedlings accumulated anthocyanins after 3 d of starvation up to 1.9 absorbance units g−1 fresh weight (FW), whereas the corresponding level for all overexpressor mutants was significantly lower (Figure 6A). In detail, the anthocyanin levels for At URH1 overexpressors were 1.3 absorbance units g−1FW and 0.9 absorbance units g−1FW for Os URH1, respectively. By contrast, microRNA lines accumulated anthocyanins to a much higher extent. The anthocyanin levels are in the range of 2.6 to 4.5 absorbance units g−1FW, respectively.
Figure 6.
Contents of Anthocyanin and Arg in Mutants Grown in Liquid Culture.
(A) Anthocyanin accumulation in nitrogen-starved seedlings grown in liquid culture. Nitrogen was omitted from the medium for 3 d.
(B) Arg contents in seedlings grown under full nutrition.
(C) Arg contents in nitrogen-starved seedlings as given in (A). 35S:At URH1, plants expressing At URH1 under control of CaMV 35S promoter; 35S:Os URH1, mutants expressing Os URH1 under control of CaMV 35S promoter; 35S:amiRNA, mutants expressing a microRNA construct directed against At URH1 under control of CaMV 35S promoter. All experiments were repeated at least three times showing similar results.
Quantitation of amino acids in extracts of liquid-grown seedlings revealed most prominent changes in the contents of Arg. Arg is of special interest in this context because it shares the precursor carbamoyl phosphate with pyrimidine de novo synthesis. Under conditions of full nitrogen supply (for details, see Methods) 35S:At URH and 35S:Os URH plants contained 19 to 38% higher Arg levels and amiRNA mutants even 51 to 78% higher levels compared with wild-type seedlings, the latter with a content of 13.7 μmol g−1FW (Figure 6B). Under conditions of nitrogen starvation, only 35S:At URH and 35S:Os URH accumulated higher Arg levels, accounting for 32 to 65% of the wild-type content (Figure 6C). Here, control seedlings contained much lower contents compared with growth conditions under full nitrogen supply, accounting for 1.05 μmol g−1FW (Figure 6C). The overall contents of amino acids in nitrogen supplied and nitrogen starved seedlings were not significantly altered between the wild type and mutants (data not shown).
To obtain a deeper understanding of the integration of URH function in nucleotide metabolism, the response of de novo synthesis and salvage pathway in addition to degradation in our mutants was analyzed. When the transcript levels of key enzymes of pyrimidine de novo synthesis (CPSase, ATCase, DHODH, and UMPSase; see Figure 7) were quantified by real-time PCR, no significant differences between wild-type plants, overexpressors, and amiRNA mutants became obvious (see Supplemental Figure 3 online). To analyze salvage of pyrimidines, the activities of uridine kinase (UK) and uracil phosphoribosyltransferase (UPRT) were determined on 20-d-old soil-grown seedlings. Again, no significant differences in the activities between the wild type and mutants appeared (see Supplemental Figure 2 online). The wild-type activity for UK was 15.9 nmol mg protein−1 h−1 and 30.6 nmol mg−1 protein h−1 for UPRT (see Supplemental Figure 4 online). Similar results were obtained with liquid-grown seedlings.
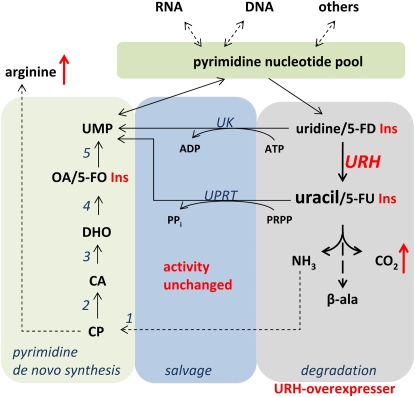
Figure 7.
Schematic Overview of Pyrimidine de Novo Synthesis, Salvage, and Degradation.
Nucleotides synthesized de novo enter the nucleotide pool and become further converted to nucleic acids and other metabolic intermediates. Breakdown of nucleotides to uridine and uracil may lead to complete degradation or the resynthesis of UMP in the salvage pathway. Increasing the activity of URH in overexpressor mutants leads to marked stimulation of pyrimidine degradation but not of salvage pathway activity or de novo synthesis. This is illustrated by the insensitivity of mutants with enhanced URH activity toward 5-FO, 5-FD, and 5-FU. 1, Carbamoyl phosphate synthase (CPSase); 2, Asp transcarbamoylase (ATCase); 3, dihydroorotase (DHOase); 4, dihydroorotate dehydrogenase (DHODH); 5, UMP-synthase (UMPSase); CP, carbamoyl phosphate; CA, carbamoyl aspartate; DHO, dihydro-orotate; OA, orotic acid.
DISCUSSION
Importance of Nucleoside Salvage
Salvage of purines and pyrimidines is an essential biochemical process operating in all living organisms (Zrenner et al., 2006). The importance of this pathway is illustrated by the Lesch-Nyhan syndrome of humans, wherein victims suffer from a deficiency in the salvage enzyme hypoxanthine-guanosine-phosphoribosyltransferase (Sculley et al., 1992). This deficiency leads to increased degradation of purine nucleotides and in succession to the accumulation of high levels of uric acid in the blood leading to mental retardation, choreoathetosis, and compulsive self-mutilation. Nucleotide salvage is also important in protists such as L. major and C. fasciculata, or T. brucei, which rely on purine uptake from the host since they lack nucleotide de novo synthesis. Nucleosidases are key enzymes in purine salvage in these organisms, cleaving imported nucleosides to liberate the base, which then is recycled to the nucleoside monophosphate. Thus, nucleosidases from pathogenic protozoans are potential targets for drugs or antibiotics because homologs do not exist in mammals. These enzymes are well characterized in protists (Degano et al., 1996). Here, we report on the characterization of URH1, an Arabidopsis protein with high homology to nucleoside hydrolases from protists and yeast. Purified, recombinant URH1 catalyzes highest degradation rates for uridine and the resistance of mutants with increased nucleosidase activity toward toxic 5-FD indicates that this enzyme exerts high control over pyrimidine degradation in plants in vivo.
URH1 Is a Member of the Inosine-Uridine Nucleosidase Protein Family of Prokaryotes and Eukaryotes, Excluding Mammals
In plants, several sequences with high similarity to Arabidopsis URH1 can be found (Figure 1). Among eukaryotes, but outside the plant kingdom, At URH1 exhibits highest similarity to the inosine/uridine nucleosidase from the fission yeast S. pombe. The well-studied inosine/uridine nucleosidases from the protozoan protists L. major and C. fasciculata both share 28% identical amino acids with At URH1 (Figure 1).
The presence of the sequence motif DXDXXXDD groups At URH1 into a cluster of nucleosidases from prokaryotes and eukaryotes and is called a hallmark of nucleosidase activity (Versées and Steyaert, 2003). The crystal structures of the nucleosidases from C. fasciculata and L. major have been resolved (Degano et al., 1996; Shi et al., 2004) and revealed the function of conserved amino acids in the active site of the enzymes, including a His residue in addition to the Asp cluster, important for the release of the nucleobase. This residue is also conserved in At URH1, further substantiating that At URH1 belongs to the type of inosine/uridine nucleosidases found in prokaryotes and eukaryotes.
In Arabidopsis, a homolog to URH1, named URH2, exists, sharing a relatively high degree of similarity with URH1. However, the function of URH2 is unclear, and we were unable to detect nucleosidase activity in URH2 expressing yeast or E. coli cells.
Uridine Is the Preferred Substrate of URH1, while Inosine, Adenosine, and Cytokinin Ribosides Are Accepted
After expression in yeast strain YRZM18, URH1 catalyzes the hydrolysis of uridine, as revealed by complementation analysis and by direct measurement of enzyme activity in a radioactive assay with subsequent analysis by thin layer chromatography. The results obtained by quantifying radiolabeled hydrolysis products could be confirmed by separation and quantification of unlabeled reaction products by HPLC.
URH1 expressed as recombinant protein in E. coli could be purified to near homogeneity by affinity chromatography (Figure 2A). In addition, a high enzyme activity was recovered in the purified enzyme. This is illustrated by a 600 times higher activity of the enzyme purified from E. coli compared with the crude yeast extract (Table 2). These changes in total activity might also explain the differences in affinity between both enzymes. It has to be anticipated that further cellular components in the yeast extract interfere with URH1 activity and thus lead to the observed lower affinity of URH1 toward uridine when expressed in yeast (Table 2).
The high specific activity of purified, recombinant URH1 enabled us to determine that URH1 could act on additional substrates such as inosine and adenosine. Thus, the substrate spectrum of URH1, converting purine and pyrimidine nucleosides, is generally similar to that of the homologs from C. fasciculata and L. major. In addition, the fact that At URH1 is unable to use cytidine as a substrate concurs with the finding that cytosine cannot be metabolized by plants (Moffatt et al., 2002; Boldt and Zrenner, 2003; Stasolla et al., 2003). Instead, plants convert cytidine to uridine by cytidine deaminase, which is encoded by a multigene family (Stasolla et al., 2003; Kafer et al., 2004).
Only few reports exist on nucleosidases purified from plant tissues. For example, uridine nucleosidase has been purified from mung bean (Phaseolus radiatus) (Achar and Vaidyanathan, 1967), and an adenosine-specific nucleosidase was purified from young leaves of coffee (Campos et al., 2005). The latter enzyme exhibits a Km value for adenosine of 6.3 μM, which is substantially lower than the observed Km of 0.7 mM of URH1. A further purine-specific nucleosidase was purified from yellow lupine seeds; this enzyme also exhibits a quite high apparent affinity for adenosine of 65 μM (Guranowski, 1982). The enzymes characterized from coffee and lupine were purified from leaves or seeds, respectively. By contrast, URH1 reported here shows an almost exclusive localization within the roots (Figure 4). The different localization might be an indication for the presence of further nucleosidase isoforms exhibiting different biochemical properties. However, the lower catalytic activity for adenosine and inosine may be sufficient to allow for purine degradation, as we found out in previous experiments, that exogenously supplied adenosine is degraded to only 5% in Arabidopsis, whereas exogenously provided uridine is degraded to 30% (M. Flörchinger, S. Hach, B. Jung, M. Traub, R. Wartenberg, C. Salzig, P. Lang, and T. Möhlmann, unpublished results). Adenylates are catabolized via inosine or xanthosine (Zrenner et al., 2006), whereas the physiological role of adenosine to adenine conversion in plants is not yet understood. To the best of our knowledge, the only enzyme taking adenine as substrate is adenine phosphoribosyltransferase. Remarkably, a lack of this enzyme in Arabidopsis leads to male-sterile plants (Moffatt and Somerville, 1988). Recently, a cell wall–bound adenosine nucleosidase was identified in Solanum tuberosum involved in salvage of extracellular ATP, which acts as a signal molecule (Riewe et al., 2008). Because of the cytosolic localization, it is improbable that URH1 fulfils such a function. However, besides URH1 and 2, Arabidopsis harbors three further putative URH isoforms, albeit they differ significantly in molecular mass and share only limited sequence identity to URH1 and 2 (below 23%). One of these (At5g18860) seems to represent a tandem protein composed of two complete URH open reading frames both sharing sequence motives with an adenosine-specific nucleosidase from T. vivax being functional as a homodimer (Versées et al., 2001). Furthermore, the At5g18860 translation product was identified in cell walls in two proteome studies (Borderies et al., 2003; Kwon et al., 2005) and may thus be a good candidate for the cell wall–bound adenosine nucleosidase.
Apart from a high nucleosidase activity, URH1 is characterized by the ability to cleave the cytokinin-riboside IPR (Figures 2C and 2D). This finding of a dual function of URH1 in nucleoside and cytokinin metabolism is surprising at a first glance. However, such a dual function has also been reported for other proteins of the salvage pathway, namely, adenosine kinase, adenine phosphoribosyl-transferase, and rice Equilibrative Nucleoside Transporter2 (ENT2). The latter protein transports cytokinin-ribosides in addition to nucleosides (von Schwartzenberg et al., 1998; Moffatt et al., 1991, 2000; Hirose et al., 2005).
It is well accepted that the main site of cytokinin biosynthesis in plants is the root (Kieber, 2002). Subsequent to synthesis, cytokinins can be distributed within the plant through the vascular system (Sakakibara, 2006; Sakakibara et al., 2006), and it is supposed that cytokinin-ribosides represent the transport form of this plant hormone (Mok and Mok, 2001; Kieber, 2002). Activities of enzymes that hydrolyze cytokinin-ribosides have been detected in plants, but the identification of corresponding genes was not successful so far. The tissue-specific expression of URH1 within the root vasculature fits very well to a supposed function of activating cytokinins produced in the roots. However, at first sight, the relatively low affinity of URH1 toward IPR is amazing because measured cytokinin levels in plants are in the low nanomolar range (Beck, 1996). Nevertheless, it is quite possible that local concentrations restricted to few cell layers may reach much higher concentrations. This assumption is supported by a relatively low affinity of rice ENT2 toward its substrates IPR and trans zeatin riboside. The corresponding apparent Km values are 32 μM for IPR and 660 μM for trans zeatin riboside (Hirose et al., 2005). However, so far we have no indication for a function of URH1 in cytokinin metabolism in vivo after analysis of the available mutants.
Recently, a novel cytokinin activating enzyme has been discovered in rice, converting cytokinin monophosphates to the active base form. This protein, named lonely guy (LOG), is essentially important for the correct development of the shoot meristem and is specifically expressed in the meristem tip (Kurakawa et al., 2007). Possibly, two independent pathways of cytokinin activation exist in plants.
Altered Nucleosidase Activity Effects Uridine Degradation, Nitrogen Starvation, Stress Response, and Germination
Mutants generated in the course of this work clearly differed in nucleosidase transcript levels as well as in their in vitro nucleosidase activities from the corresponding wild-type plants (Figures 4A and 4B). Plants expressing Os URH1 also show the functionality of the rice homolog. The alterations in transcript level and enzyme activity were accompanied by increased uridine degradation in vivo in At URH1 and Os URH1 overexpressor lines (Figure 4C). This finding in itself is a clear indication for a shift from uridine salvage toward uridine degradation as response to increased uridine nucleosidase activity (Figure 7). The latter result is clearly confirmed by increased resistance of uridine nucleosidase overexpressor lines toward 5-FD and 5-FU. Following uptake, 5-FD and 5-FU can become converted to fluoro-desoxyuridine-monophosphate acting as inhibitor of thymidylate synthetase, known to be essential for dTTP synthesis and thus for cell division. Furthermore, 5-FD and 5-FU can be metabolized to fluorouridine-triphosphate and then incorporated into RNA leading to the synthesis of abnormal proteins (Hakala and Rustrum, 1979). When 5-FD is degraded to 5-FU and further catabolized, nontoxic fluoro-β-alanine appears, which may further become defluorinated to β-aminoacrylate and fluorid (Porter et al., 1995).
In principle, both uridine and uracil, the product of nucleosidase activity, can be salvaged to UMP by UK or UPRT (Moffat and Ashihara, 2002; Boldt and Zrenner, 2003; Zrenner et al., 2006). Consequently, mutants lacking major UK and UPRT activities are resistant toward 5-FD and 5-FU (Islam et al., 2007).
The salvage pathway operates in the cytosol, and accordingly URH1 is located in this compartment as observed by GFP-based localization studies (Figure 4G). The same is true for so far identified UK and UPRT isoforms (Islam et al., 2007). The measured maximal activities for UK and UPRT were unchanged in the mutants generated in the course of this work (see Supplemental Figure 2 online). It has to be mentioned that the activities for both enzymes are severalfold in excess compared with uptake rates of uridine (Wormit et al., 2004; Traub et al., 2007). However, it cannot be excluded that the cosubstrate of UPRT phosphoribosyl pyrophosphate limits uracil salvage as mutants in phosphoribosyl pyrophosphate synthesis showed that this metabolite may colimit growth rates in Arabidopsis (Koslowsky et al., 2008). Anyhow, plants with increased activities of URH exhibit resistance toward 5-FD and 5-FU, and it can be supposed that this is achieved by increasing the uracil-to-uridine ratio and subsequent further breakdown of uracil. Thus, URH is clearly an enzyme involved in pyrimidine breakdown rather than in pyrimidine salvage (Figure 7). This is further strengthened by the observation of a lower sensitivity of 35S:URH1 mutants toward 5-FO, which enters the pyrimidine nucleotide pool after conversion to 5-fluoro UMP. As part of the nucleoside pool, 5-fluoro UMP is then degraded under control of URH to an extent that causes the observed lower sensitivity of mutants. From this we have to conclude that (1) there is cycling among nucleotides nucleosides and nucleobases, and (2) that under conditions of increased URH activity, the capacity of the salvage pathway is not sufficient to recycle additional uracil, thus leading to increased degradation and resistance toward the fluorinated intermediates. One may speculate that such cycling functions in balancing nitrogen or removing rare nucleotide species present in t-RNA, rRNA, or even from damaged DNA.
Attenuation of nitrogen starvation stress response in form of reduced anthocyanin accumulation in nucleosidase overexpressors further indicates that reprogramming from uridine salvage to degradation occurs in corresponding liquid grown seedlings (Figure 6A). By contrast, microRNA lines showed increased anthocyanin levels, supporting this view. By increasing nucleoside degradation, nitrogen bound in the large pool of nucleotides and RNA may become available for amino acid synthesis. Liquid-grown seedlings with higher URH activity are characterized by increased contents of Arg, and, interestingly, both Arg synthesis and pyrimidine de novo synthesis depend on the shared precursor carbamoyl phosphate (Figure 7). As the transcript levels of de novo synthesis genes were unchanged in mutants, this result may be indicative for a reduced rate of pyrimidine de novo synthesis in URH overexpressor lines, based on allosteric regulation of key enzymes. For example, the reaction catalyzed by ATCase in pyrimidine de novo synthesis (Figure 7) is known to be strictly controlled by pyrimidine levels (Chen and Slocum, 2008). From mutants with reduced ATCase activity, it is known that limitation of pyrimidine de novo synthesis leads to an increase in UPRT transcript accompanied by downregulation of pyrimidine breakdown. Moreover, supply of these mutants with uracil could reverse growth retardations in the mutants (Chen and Slocum, 2008). A similar crosstalk between de novo synthesis and salvage was described in potato plants with reduced UMPsase activity. Latter plants could overcompensate for the loss in de novo synthesis by activating salvage pathway activity, leading to even increased uridine nucleotide levels and enhanced carbohydrate synthesis (Geigenberger et al., 2005). By contrast, increasing pyrimidine degradation does not evoke such measurable crosstalk effects (Figure 7), representing a marked difference to the situation described above.
During germination of Arabidopsis seeds, storage proteins are degraded to amino acids to allow synthesis of new proteins, nucleotide de novo synthesis, and synthesis of other nitrogen-containing metabolites. It's a matter of debate whether nucleotide de novo synthesis or salvage pathway activity prevails during germination. From germination studies with different plant systems, it was concluded that the salvage pathway is very active within the first hour after imbibition and de novo synthesis is delayed (Stasolla et al., 2003). However, recently, Brittle1 from Arabidopsis was identified as the plastidial nucleotide uniport carrier protein strictly required for export of newly synthesized adenylates into the cytosol. Mutants lacking this protein were nearly unable to germinate, indicating that purine de novo synthesis is required early during seed germination (Kirchberger et al., 2008). Moreover, the importance of de novo synthesis during the first 5 d of seedling growth is underlined by an increasing expression of ATCase during this time and an arrest of growth when this enzyme is specifically inhibited by PALA (Bassett et al., 2003). The delay in germination of mutants with increased and decreased uridine nucleosidase activity shows that balancing uridine degradation versus salvage is important at this developmental stage to allow liberation of nitrogen, only from excessive pyrimidine compounds.
In young seedlings, the expression pattern of URH is highly similar to that of ATCase (Chen and Slocum, 2008; pyrB), further indicating that de novo synthesis and degradation occur simultaneously. In older plants, the predominant expression of URH1 in roots may function in redistribution of nucleosides from aboveground parts to the roots and thus provide uracil delivered as uridine through the phloem to allow salvage of this nucleobase. In addition, nucleobases could be further degraded to provide reduced nitrogen to the heterotrophic root tissue.
Another possible function may be the processing of uridine after it has been imported from the soil. Interestingly, import of uridine via the roots is catalyzed by ENT3, an equilibrative nucleoside transporter, which is also expressed in the root vasculature (Traub et al., 2007; M. Flörchinger, S. Hach, B. Jung, M. Traub, R. Wartenberg, C. Salzig, P. Lang, and T. Möhlmann, unpublished results). ENT3-deficient plants show resistance toward fluorouridine as observed for URH1 overexpressor mutants (Traub et al., 2007). Possibly both enzymes are functionally integrated.
In sum, we conclude that uridine nucleosidase, URH1, plays a pivotal role in the regulation of nucleoside, mainly uridine, degradation.
METHODS
Cultivation of Plants
Wild-type and transgenic Arabidopsis thaliana plants (ecotype Columbia) were used throughout. Prior to germination, seeds were incubated for 1 d in the dark at 4°C for imbibition in standardized ED73 soil (Weigel and Glazebrook, 2002). Plant growth was performed at 22°C and 120 μmol quanta m−2 s−1 in a 10-h-light/14-h-dark regimen. For growth experiments with 5-FU, 5-FD, and 5-FO, surface-sterilized seeds were sown on Murashige and Skoog plates as described earlier (Reiser et al., 2004). Growth of seedlings in hydroponic culture was performed as described by Scheible et al. (2004). Therefore, 50 surface-sterilized Arabidopsis seeds grew for 7 d in 250-mL Erlenmeyer glass flasks with full nutrition medium on an orbital shaker (80 rpm). The full nutrition medium contained 2 mM KNO3, 1 mM NH4NO3, 1 mM Gln, 3 mM KH2PO4, 1 mM CaCl2, 1 mM MgSO4, 2 mM K2SO4, 0.5% sucrose, 50 μM KCl, 50 μM H3BO4, 10 μM MnSO4, 2 μM ZnSO4, 1.5 μM CuSO4, 0.075 μM (NH4)6Mo7O24, 72 μM Fe-EDTA, and 5 mM MES-KOH, pH 5.8. After 7 d, medium was changed against medium without nitrogen. The N medium included 3 mM KCl, but no KNO3, NH4NO3, and Gln. To prevent nutrient limitation, all medium was changed daily from day 8 onwards. Seedlings were harvested at an age of 10 d and stored in liquid nitrogen.
Quantification of Anthocyanin and Amino Acid Content
Frozen plant tissue was ground in liquid nitrogen using mortar and pestle and stored at −80°C.
For determination of the total anthocyanin content, 100 mg frozen tissue was boiled for 3 min in 1 mL H2O:HCl:1-propanol (81:18:1) and stored for 1 d at room temperature in the dark. After centrifugation (14,000g, 15 min) the anthocyanin content was determined as absorbance measured at 535 nm. This value was corrected according to the Rayleigh's formula (Acorrected = A535 − 2.2A650) (Lange et al., 1971).
To determine free amino acids, 50 mg of frozen plant material was extracted with 80% ethanol at 80°C and centrifuged. After evaporating the supernatant to dryness, the amino acids and standards were derivatized as described previously (Rolletschek et al., 2002) using AQC (Watrex). The AQC amino acids were measured using a Dionex P680-HPLC system with an UV170U detector (Dionex) and the column system consisting of CC8/4 ND 100- 5 C18-ec and Nucleodur 100-5 C18-ec (Macherey-Nagel). For separation of amino acids, a gradient consisting of 100 mM sodium acetate and acetonitrile (0 to 15%) was used. The AQC amino acids were detected by fluorescence with excitation at 250 nm and emission at 395 nm.
Cloning of At URH1 amiRNA
A suitable target site for the microRNA was identified by following the instructions on http://wmd2.weigelworld.org. Cloning of the 35S:amiRNA At URH1 construct was done as previously described (Schwab et al., 2006) with minor changes. To use the benefits of Gateway Cloning Technology, a CACC site was added to 5′ of primer A. Primer sequences are listed in Supplemental Table 1 online. The product of PCR amplification including the specific amiRNA was subsequently cloned into pENTR/ D-TOPO vector to obtain an entry clone. pGWB2 (Nakagawa et al., 2007) was used in the L/R reaction to generate a destination clone. Transgenic plants were selected based on kanamycin resistance conferred by the nptII gene contained in pGWB2.
Generation of 35S:At URH1 and 35S:Os URH1 Mutant Plants
To obtain overexpression plants, we amplified the entire gene of At URH1 from Arabidopsis cDNA and Os URH1 from rice (Oryza sativa) cDNA by PCR using the Pfu-DNA polymerase (Stratagene). The primers used are listed in Supplemental Table 1 online. The obtained PCR products were cleaved with HindIII/XhoI (At URH1) and XbaI/XhoI (Os URH1), respectively, and inserted in the gene expression cartridge of the pHannibal (Wesley et al., 2001) vector downstream of the CaMV 35S promoter. The entire expression cartridge was removed from pHannibal as a NotI fragment and introduced directly into the binary vector pArt27 (Gleave, 1992) and used for Agrobacterium tumefaciens–mediated plant transformation (Clough and Bent, 1998).
Quantitation of Respiratory Use of [14C]-Uridine
For analysis of the [14C]-nucleoside-driven respiratory activity, we used 7-d-old Arabidopsis seedlings grown in hydroponic culture (Scheible et al., 2004). These were incubated in 2-mL reaction tubes with 800 μL of 5 μM of the indicated [14C]-labeled substrate in growth media. On the inside at the top of this tube, a small 0.5-mL reaction vessel containing 100 μL of 1 M KOH was fixed with grease to allow [14C]-CO2 absorption. The seedlings were allowed to float on the solution and incubation continued for 24 h in the dark. The reaction was stopped by adding 100 μL of 2 M HCl with a syringe through the closed lid. The hole in the lid was sealed with grease and after subsequent incubation for 12 h the released radioactively labeled [14C]-CO2 was quantified in a scintillation counter.
Construction of the Sequence Alignment
Multiple alignments of protein sequences were performed with the program ClustalW (Thompson et al., 1994).
Complementation of Yeast Cells
For heterologous expression of URH1 in yeast cells, the corresponding cDNA was cloned into the yeast expression vector pA1NE181 (Riesmeier et al., 1992) using the primer URH1yeastfor and URH1yeastrev (see Supplemental Table 1 online). Plasmids were propagated in Escherichia coli cells (XL1Blue, Stratagene; DH5α, Clontech) grown in YT medium (0.8% peptone, 0.5% yeast extract, and 0.25% NaCl) with or without ampicillin (50 mg L−1) and tetracycline (2.5 mg L−1). YRZM18 yeast cells (YYM8; Mat α; ura3-1; his3-Δ200; leu2-Δ1; trp1∷P3xERE−URA3; Δpdr5∷hisG; Δsnq2; hisG; Δurk1∷TRP1; Δurh1∷KanMX, [2 μm; LEU2; PADH1]) (Mitterbauer et al., 2002) were transformed with pA1NE (Riesmeier et al., 1992) harboring URH1 applying the method of Ito et al. (1983). Cells were grown on minimal medium containing a 0.67% yeast nitrogen base (Remel, distributed by Ceratogene Biosciences), 2% glucose, and supplements as required to maintain auxotrophic selection. Cells were harvested at an A600 of 10 by centrifugation (10 min, 1500g, 4°C) and collected in medium consisting of 50 mM Tris-HCl, pH 7.5, and 1 mM Pefabloc. Disruption of yeast cells was performed using a Bead Beater (Biospec Products) at 4600 rpm for 90 s. After centrifugation (1 min, 13,000g, 4°C), the crude extract was transferred to Nap5 column (GE Healthcare) and used for nucleosidase measurement.
Heterologous Expression of At URH1 in E. coli
For heterologous expression of URH1 in E. coli, the entire URH1 cDNA was amplified from Arabidopsis cDNA with Pfu-DNA polymerase. The primers URH1colifor with NdeI restriction site and URH1colirev (see Supplemental Table 1 online) with XhoI restriction site were used. The obtained PCR product for URH1 was cloned into the EcoRV site of the cloning vector pBSK (Stratagene). To construct E. coli plasmids expressing URH1 with a C-terminal His tag, the gene was cloned into the NdeI/XhoI restriction site of the bacterial expression vector pET21a (Novagen) and transformed in Rosetta 2 (pLysS) cells (Novagen). Cells were grown at 37°C in TB medium (2.31 g L−1 KH2PO4, 12.54 g L−1 K2HPO4, 1.2% peptone, 2.4% yeast extract, and 0.4% glycerol, pH 7.0) with ampicillin (50 mg L−1) and chloramphenicol (25 mg L−1). At an A600 of 0.5 induction of T7 RNA polymerase was initiated by addition of 0.2% isopropyl-β-D thiogalactoside. One hour after induction, cells were harvested by centrifugation for 10 min at 5000g (22°C). The sediments were resuspended in 25 mL breaking buffer (15% glycerol, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.5) and stored at −20°C.
To start autolysis of E. coli harboring URH1 by endogenous lysozyme, cells were thawed at 37°C and 1 mM PMSF, 1 mg mL−1 DNase, and 1 mg mL−1 RNase were added. Additional cell disruption was performed by sonification. After centrifugation (30 min, 100,000g, 4°C), the crude extract was used for URH1 purification with immobilized-metal affinity chromatography according to a modified protocol form the supplier (Qiagen). For purification of URH1, a crude extract from E. coli cells was incubated for 1 h with Ni-NTA under constant stirring and transferred to an Econo-Pac chromotography column (Bio-Rad). Nonspecific proteins were removed by washing the column with binding buffer consisting of 10 mM imidazole, 10 mM NaCl, and 50 mM Na2PO4, pH 8.0, and washing buffer containing 60 mM imidazole, 10 mM NaCl, and 50 mM Na2PO4, pH 8.0. The synthesized At URH1 protein was eluted with buffer consisting of 500 mM imidazole, 10 mM NaCl, and 50 mM Na2PO4, pH 8.0. The elution buffer was exchanged with 50 mM Tris-HCl, pH 7.5, using a Nap5 column (GE Healthcare) and used for nucleosidase measurement and SDS-PAGE. SDS-PAGE was performed as described by Laemmli (1970) and stained with Coomassie Brillant Blue R 250.
Staining for GUS Activity
For the histochemical localization of promoter activity, a 1-kb upstream fragment of URH1 was inserted to pGPTV (Becker et al., 1992) using the primers URH1gusfor and URH1gusrev (see Supplemental Table 1 online). The resulting construct was transformed in Agrobacterium strain GV3101. Transformation of Arabidopsis was conducted according to the floral dip method (Clough and Bent, 1998). Tissue from transgenic plants was collected in glass vials, filled with ice-cold 90% acetone, and incubated for 20 min at room temperature. Subsequently, the samples were stained according to standard protocols (Weigel and Glazebrook, 2002).
Transient Expression of a URH1-GFP Fusion Construct
To construct the URH1-GFP fusion proteins, we first amplified the entire At URH1 cDNA by PCR using the Pfu-DNA polymerase (Stratagene). The primers used are listed in Supplemental Table 1 online. The obtained DNA fragments were cleaved with XbaI and XhoI, respectively, and fused translationally with the GFP coding region of the vector pGFP2 (Kost et al., 1998), leading to the final URH1-GFP construct under the control of a 35S promoter. Protoplasts isolated from sterile-grown tobacco (Nicotiana tabacum cv W38) were transformed with column-purified plasmid DNA (30 mg/1.5 106 cells) as given (Wendt et al., 2000). After 1 d of incubation at 24°C in the dark, protoplasts were checked for the presence of green fluorescence by use of an Axiovert 200 Apo-tome fluorescence microscope (Carl Zeiss) equipped with a Plan Neofluar ×40/1.3 oil objective using Zeiss filter set 10.
Quantitative RT-PCR
Quantitative RT-PCR was performed as given by Leroch et al. (2005). Total RNA was prepared from Arabidopsis leaf tissues using the RNeasy plant mini kit (Qiagen). To remove any contaminating DNA, the samples were treated with DNase (RNase-free DNase kit; Qiagen). Quantitative PCR was performed using MyIQ-Cycler (Bio-Rad) and IQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions with the following cycler conditions: 20 min at 50°C, 15 min at 95°C, and 55 cycles of 15 s at 95°C, 25 s at 58°C, and 40 s at 72°C. The gene-specific oligonucleotides used for real-time RT-PCR are listed in Supplemental Table 1 online. The gene At1g07930 encoding elongation factor 1α (EF1α) was used for quantitative normalization (Curie et al., 1991). The specificity of all RT-PCR products obtained was controlled on 2% agarose gels.
Determination of Nucleosidase Activity
Nucleosidase measurement of radioactive-labeled uridine was performed according to the assay described by Kurtz et al. (2002). Four microliters of a desalted, crude yeast extract were added to 4 μL of [14C]-uridine in 50 mM Tris-HCl, pH 7.5, and incubated at 30°C. The reaction was terminated by heating the protein to 98°C. PEI-Cellulose F (Merck) thin layer chromatography was used to identify the amounts of produced uracil. Separation was performed for 60 min with water and detected by Cyclone Storage phosphorscreen (Perkin-Elmer Life Sciences).
For measuring hydrolysis of nucleosides by HPLC, 30 μL of yeast crude extract or purified URH1 were incubated with 30 μL of corresponding substances at 30°C. The reaction was terminated as described above. A Dionex P680-HPLC system with an UV170U detector (Dionex) and the column system CC8/4 ND 100-5 C18-ec and Nucleodur 100-5 C18-ec (Macherey-Nagel) were used. For separating the nucleosides from nucleobases, an eluent consisting of 50 mM ammoniumacetat and 1% methanol, pH 5.2, was used, whereas for uridine, inosine, adenosine, and isoplentenyladenine-riboside, 0, 2.5, 7.5, or 12.5% acetonitrile were added, respectively.
Determination of Pyrimidine Salvage Activities
For the determination of enzyme activities, 20-d-old soil-grown seedlings or liquid-grown seedlings were homogenized in extraction medium (50 mM HEPES-KOH, pH 7.4, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 50 μg/mL PMSF, 10% [v/v] glycerol, and 0.1% [v/v] Triton X-100) using a Retsch ball mill MM 301. After centrifugation (10 min, 20,000g, 4°C) the supernatant was used for the determination of enzyme activity. Measurement of salvage pathway enzymes was performed as given by Moffatt et al. (2000) with [5-3H]-uridine as substrate for UK and [2-14C]- uracil as substrate for determination of UPRT activity according to the method of Lee and Moffatt (1993).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At URH1 (At2g36310; Q9SJM7); Schizosaccharomyces pombe (Q9P6J4); Leishmania major (P83851); Crithidia fasciculata (Q27546); Saccharomyces cerevisiae (Q04179); Zea mays (ACF80359; Oryza sativa (Os08g05579; XP483754); and Vitis vinifera (CAO15947).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of a Yeast Mutant and Biochemical Properties of URH1 Synthesized in Yeast.
Supplemental Figure 2. Tissue-Specific Expression of URH1 Determined by Real-Time PCR
Supplemental Figure 3. Expression Level of Pyrimidine de Novo Synthesis Genes in URH Mutants.
Supplemental Figure 4. Activities of Salvage Pathway Enzymes.
Supplemental Table 1. Oligonucleotides Used in This Work.
Supplementary Material
Acknowledgments
This work was financially supported by the Deutsche Forschungsgemeinschaft (Grant MO 1032/3-1). We thank R. Mitterbauer (University of Natural Resources and Applied Life Sciences, Vienna) for providing the yeast strain YRZM18 and G. Amoroso (University of Kaiserslautern) for critical reading of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Torsten Möhlmann (moehlmann@biologie.uni-kl.de).
Online version contains Web-only data.
References
- Achar, B.S., and Vaidyanathan, C.S. (1967). Purification and properties of uridine hydrolase from mung-bean (Phaseolus radiatus) seedlings. Arch. Biochem. Biophys. 119 356–362. [DOI] [PubMed] [Google Scholar]
- Bassett, E.V., Bouchet, B.Y., Carr, J.M., Williamson, C.L., and Slocum, R.D. (2003). PALA mediated pyrimidine starvation increases expression of aspartate transcarbamoylase (pyrB) in Arabidopsis seedlings. Plant Physiol. Biochem. 41 695–703. [Google Scholar]
- Beck, H.E. (1996). Regulation of shoot/root ratio from roots in Urtica dioica: Opinion. Plant Soil 185 3–12. [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Boldt, R., and Zrenner, R. (2003). Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant. 117 297–304. [DOI] [PubMed] [Google Scholar]
- Borderies, G., Jamet, E., Lafitte, C., Rossignol, M., Jauneau, A., Boudart, G., Monsarrat, B., Esquerre-Tugaye, M.T., Boudet, A., and Pont-Lezica, R. (2003). Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: A critical analysis. Electrophoresis 24 3421–3432. [DOI] [PubMed] [Google Scholar]
- Campos, A., Rijo-Johansen, M.J., Carneiro, M.F., and Fevereiro, P. (2005). Purification and characterisation of adenosine nucleosidase from Coffea arabica young leaves. Phytochemistry 66 147–151. [DOI] [PubMed] [Google Scholar]
- Chen, C.T., and Slocum, R.D. (2008). Expression and functional analysis of aspartate transcarbamoylase and role of de novo pyrimidine synthesis in regulation of growth and development in Arabidopsis. Plant Physiol. Biochem. 46 150–159. [DOI] [PubMed] [Google Scholar]
- Christensen, T.M., and Jochimsen, B.U. (1983). Enzymes of ureide synthesis in pea and soybean. Plant Physiol. 72 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Curie, C., Liboz, T., Bardet, C., Gander, E., Medale, C., Axelos, M., and Lescure, B. (1991). Cis and trans-acting elements involved in the activation of Arabidopsis thaliana A1 gene encoding the translation elongation factor EF-1 alpha. Nucleic Acids Res. 19 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degano, M., Gopaul, D.N., Scapin, G., Schramm, V.L., and Sacchettini, J.C. (1996). Three-dimensional structure of the inosine-uridine nucleoside N-ribohydrolase from Crithidia fasciculata. Biochemistry 35 5971–5981. [DOI] [PubMed] [Google Scholar]
- Geigenberger, P., Regierer, B., Nunes-Nesi, A., Leisse, A., Urbanczyk-Wochniak, E., Springer, F., van Dongen, J.T., Kossmann, J., and Fernie, A.R. (2005). Inhibition of de novo pyrimidine synthesis in growing potato tubers leads to a compensatory stimulation of the pyrimidine salvage pathway and a subsequent increase in biosynthetic performance. Plant Cell 17 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gopaul, D.N., Meyer, S.L., Degano, M., Sacchettini, J.C., and Schramm, V.L. (1996). Inosine-uridine nucleoside hydrolase from Crithidia fasciculata. Genetic characterization, crystallization, and identification of histidine 241 as a catalytic site residue. Biochemistry 35 5963–5970. [DOI] [PubMed] [Google Scholar]
- Guranowski, A. (1982). Purine catabolism in plants: Purification and some properties of inosine nucleosidase from yellow lupin (Lupinus luteus L.). seeds. Plant Physiol 70 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakala, M.T., and Rustrum, Y.M. (1979). The potential of in vitro screening. In Cancer Drug Development, Part A, V.T. DeVita and H. Busch, eds (New York: Academic Press), pp. 271–290.
- Hirose, N., Makita, N., Yamaya, T., and Sakakibara, H. (2005). Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinine transport. Plant Physiol. 138 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.R., Kim, H., Kang, S.W., Kim, J.S., Jeong, Y.M., Hwang, H.J., Lee, S.Y., Woo, J.C., and Kim, S.G. (2007). Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Mol. Biol. 63 465–477. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, J., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer, C., Zhou, L., Santoso, D., Guirgis, A., Weers, B., Park, S., and Thornburg, R. (2004). Regulation of pyrimidine metabolism in plants. Front. Biosci. 9 1611–1625. [DOI] [PubMed] [Google Scholar]
- Katahira, R., and Ashihara, H. (2002). Profiles of pyrimidine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta 215 821–828. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J. (2002). Cytokinins. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Physiologists), doi/11.1199/tab.0018, http://aspb.org/publications/arabidopsis/.
- Kirchberger, S., Tjaden, J., and Neuhaus, H.E. (2008). Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 56 51–63. [DOI] [PubMed] [Google Scholar]
- Koslowsky, S., Riegler, H., Bergmuller, E., and Zrenner, R. (2008). Higher biomass accumulation by increasing phosphoribosylpyrophosphate synthetase activity in Arabidopsis thaliana and Nicotiana tabacum. Plant Biotechnol. J. 6 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16 393–401. [DOI] [PubMed] [Google Scholar]
- Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., Sakakibara, H., and Kyozuka, J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655. [DOI] [PubMed] [Google Scholar]
- Kurtz, J.E., Exinger, F., Erbs, P., and Jund, R. (2002). The URH1 uridine ribohydrolase of Saccharomyces cerevisiae. Curr. Genet. 41 132–141. [DOI] [PubMed] [Google Scholar]
- Kwon, H.K., Yokoyama, R., and Nishitani, K. (2005). A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 46 843–857. [DOI] [PubMed] [Google Scholar]
- Laemmli, K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lange, H., Shropshire, W., and Mohr, H. (1971). An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol. 47 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D., and Moffatt, B.A. (1993). Purification and characterization of adenine Phosphoribosyltransferase from Arabidopsis thaliana. Physiol. Plant. 87 483–492. [Google Scholar]
- Leroch, M., Kirchberger, S., Haferkamp, I., Wahl, M., Neuhaus, H.E., and Tjaden, J. (2005). Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J. Biol. Chem. 280 17992–18000. [DOI] [PubMed] [Google Scholar]
- Magni, G., Fioretti, E., Ipata, P.L., and Natalini, P. (1975). Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties. J. Biol. Chem. 250 9–13. [PubMed] [Google Scholar]
- Mäser, P., Sütterlin, Ch., Kralli, A., and Kaminsky, R. (1999). A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285 242–244. [DOI] [PubMed] [Google Scholar]
- Mitterbauer, R., Karl, T., and Adam, G. (2002). Saccharomyces cerevisiae URH1 (encoding uridine-cytidine N-ribohydrolase): Functional complementation by a nucleoside hydrolase from a protozoan parasite and by a mammalian uridine phosphorylase. Appl. Environ. Microbiol. 68 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B., and Somerville, C. (1988). Positive selection for male-sterile mutants of Arabidopsis lacking adenine phophoribosyl transferase activity. Plant Physiol. 86 1150–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, B.A., and Ashihara, H. (2002). Purine and pyrimidine nucleotide synthesis and metabolism. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Physiologists), doi/11.1199/tab.0018, http://aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Moffatt, B., Pethe, C., and Laloue, M. (1991). Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol. 95 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B.A., Stevens, Y.Y., Allen, M.S., Snider, J.D., Pereira, L.A., Todorova, M.I., Summers, P.S., Weretilnyk, E.A., Martin-McCaffrey, L., and Wagner, C. (2002). Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 128 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B.A., Wang, L., Allen, M.S., Stevens, Y.Y., Qin, W., Snider, J., and von Schwartzenberg, K. (2000). Adenosine kinase of Arabidopsis. Kinetics properties and gene expression. Plant Physiol. 124 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, D.W.S., and Mok, M.C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 89–118. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104 34–41. [DOI] [PubMed] [Google Scholar]
- Parkin, D.W., Horenstein, B.A., Abdulah, D.R., Estupinan, B., and Schramm, V.L. (1991). Nucleoside hydrolase from Crithidia fasciculata. Metabolic role, purification, specificity, and kinetic mechanism. J. Biol. Chem. 266 20658–20665. [PubMed] [Google Scholar]
- Porter, D.J., Harrington, J.A., Almond, M.R., Chestnut, W.G., Tanoury, G., and Spector, T. (1995). Enzymatic elimination of fluoride from alpha-fluoro-beta-alanine. Biochem. Pharmacol. 50 1475–1484. [DOI] [PubMed] [Google Scholar]
- Reiser, J., Linka, N., Lemke, L., Jeblick, W., and Neuhaus, H.E. (2004). Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 136 3524–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Willmitzer, L., and Frommer, W.B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe, D., Grosman, L., Fernie, A.R., Zauber, H., Wucke, C., and Geigenberger, P. (2008). A cell wall-bound adenosine nucleosidase is involved in the salvage of extracellular ATP in Solanum tuberosum. Plant Cell Physiol. 49 1572–1579. [DOI] [PubMed] [Google Scholar]
- Rolletschek, H., Hajirezaei, M.R., Wobus, U., and Weber, H. (2002). Antisense-inhibition of ADP-glucose pyrophosphorylase in Vicia narbonensis seeds increases soluble sugars and leads to higher water and nitrogen uptake. Planta 214 954–964. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Takei, K., and Hirose, N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11 440–448. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., Schindelasch, D., Thimm, O., Udvardi, M.K., and Stitt, M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., Su, Y.H., Kunze, R., Warner, S., Hewitt, M., Slocum, R.D., Ludewig, U., Frommer, W.B., and Desimone, M. (2004). UPS1 and UPS2 from Arabidopsis mediate high affinity transport of uracil and 5-fluorouracil. J. Biol. Chem. 279 44817–44824. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sculley, D.G., Dawson, P.A., Emmerson, B.T., and Gordon, R.B. (1992). A review of the molecular basis of hypoxanthine-guanine phosphoribosyltransferase (HPRT) deficiency. Hum. Genet. 90 195–207. [DOI] [PubMed] [Google Scholar]
- Shi, W., Schramm, V.L., and Almo, S.C. (1999). Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure. J. Biol. Chem. 274 21114–21120. [DOI] [PubMed] [Google Scholar]
- Shi, W., Ting, L.M., Kicska, G.A., Lewandowicz, A., Tyler, P.C., Evans, G.B., Furneaux, R.H., Kim, K., Almo, S.C., and Schramm, V.L. (2004). Plasmodium falciparum purine nucleoside phosphorylase: Crystal structures, immucillin inhibitors, and dual catalytic function. J. Biol. Chem. 279 18103–18106. [DOI] [PubMed] [Google Scholar]
- Stasolla, C., Katahira, R., Thorpe, T.A., and Ashihara, H. (2003). Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 160 1271–1295. [DOI] [PubMed] [Google Scholar]
- Stein, A., Vaseduvan, G., Carter, N.S., Ullman, B., Landfear, S.M., and Kavanaugh, M.P. (2003). Equilibrative nucleoside transporter family members from Leishmania donovani are electrogenic proton symporters. J. Biol. Chem. 278 35127–35134. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, D.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub, M., Flörchinger, M., Piecuch, J., Kunz, H.-H., Weise-Steinmetz, A., Deitmer, J.W., Neuhaus, H.E., and Möhlmann, T. (2007). The furl (fluorouridine insensitive 1) mutant is defective in equilibrative nucleoside transporter 3 (ENT3), thus representing an important pyrimidine nucleoside uptake system in Arabidopsis thaliana. Plant J. 49 855–864. [DOI] [PubMed] [Google Scholar]
- Versées, W., Decanniere, K., Pelle, R., Depoorter, J., Brosens, E., Parkin, D.W., and Steyaert, J. (2001). Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax. J. Mol. Biol. 307 1363–1379. [DOI] [PubMed] [Google Scholar]
- Versées, W., and Steyaert, J. (2003). Catalysis by nucleoside hydrolases. Curr. Opin. Struct. Biol. 13 731–738. [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg, K., Kruse, S., Reski, R., Moffatt, B., and Laloue, M. (1998). Cloning and characterization of an adenine kinase from Physcomitrella involved in cytokinin metabolism. Plant J. 13 249–257. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wendt, U.K., Wenderoth, I., Tegeler, A., and Von Schaewen, A. (2000). Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L). Plant J. 23 723–733. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Wormit, A., Traub, M., Flörchinger, M., Neuhaus, H.E., and Möhlmann, T. (2004). Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem. J. 383 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Henning, L., and Gruissem, W. (2004). Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner, R., Stitt, M., Sonnewald, U., and Boldt, R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57 805–836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.