Abstract
A general strategy is described for improving the binding properties of small-molecule ligands to protein targets. A bifunctional molecule is created by chemically linking a ligand of interest to another small molecule that binds tightly to a second protein. When the ligand of interest is presented to the target protein by the second protein, additional protein–protein interactions outside of the ligand-binding sites serve either to increase or decrease the affinity of the binding event. We have applied this approach to an intractable target, the SH2 domain, and demonstrate a 3-fold enhancement over the natural peptide. This approach provides a way to modulate the potency and specificity of biologically active compounds.
In nature, certain small-molecule ligands use a remarkable mechanism to enhance the affinity for their targets. These ligands bind to an endogenous protein, forming a new composite surface, which can bind then to a target protein. Presentation of the ligand by the larger endogenous protein vastly enlarges the surface area available for interactions with the target, facilitating additional protein–protein interactions and improved affinity.
For example, the peptide ligands of the T cell receptor use this strategy of surface enlargement to promote a high-affinity-binding event. By themselves, peptides usually have only a very low affinity for the polymorphic T cell receptor (TCR). For a high-affinity-binding event to occur, a peptide must be presented by the major histocompatibility complex (MHC). As the crystal structures of the trimeric TCR–peptide–MHC complex have shown, the TCR makes contacts not only with the peptide but also with the MHC (1, 2). The TCR-MHC contacts are important because mutations of these sites abolish the immune response and much of the remarkable specificity of the immune response is a result of this simple trimeric complex (3–5).
Certain microorganisms also make use of endogenous proteins to present and enhance the activity of their toxins. For example, the immunosuppressive drugs cyclosporin and FK506 are presented by cyclophilin and human FK506-binding protein 12 (FKBP), respectively, to inhibit the activity of a common protein target, calcineurin (6). By themselves, FK506 and cyclosporin have no measurable affinity for calcineurin. However, the FK506–FKBP and cyclosporin–cyclophilin complexes bind to calcineurin with high affinity (7). The cocrystal structure of calcineurin–FK506–FKBP reveals that the protein surfaces of calcineurin and FKBP make extensive contacts that promote the high affinity of the binding event (8, 9).
However, extensive protein–protein interactions between a presenting protein and the drug target are not always required to enhance the affinity of a ligand for its target protein. Like FK506, the macrolide drug rapamycin has no measurable affinity for its target, the cell cycle control protein FRAP. Instead, rapamycin forms a complex with FKBP to create a composite surface that binds to FRAP with high affinity. But, unlike the FK506–FKBP complex, the rapamycin–FKBP complex establishes only few contacts with its target (10).
Inspired by these natural examples, we envisioned that the specificity and affinity of a ligand–protein interaction or a drug–protein interaction could be modulated deliberately by borrowing additional surface contacts from an endogenous protein. Thus, we set out to chemically link a ligand for an abundant presenting protein to a weak binder for a target protein. By using chemical linkers of different lengths, tilt, and rotation, one may expect the resulting protein–protein interactions to be favorable or unfavorable so that binding occurs with enhanced or decreased affinity, respectively. As an extension of traditional medicinal chemistry, such an approach may be useful to modulate the potency and specificity of biologically active compounds.
As a first attempt to explore the feasibility of this approach, we selected two members of the FK506-binding protein family, FKBP12 and FKBP52, as presenter proteins. We chose the SH2 domain of the Fyn tyrosine kinase as the target protein because of its well established structure and the availability of ligands for SH2 domains (11). SH2 domains bind to peptides and proteins that contain phosphotyrosine residues, and they are commonly found in signaling proteins that regulate cell growth and differentiation. Ligands that bind to SH2 domains have been explored as possible therapeutics for cancer, osteoporosis, and inflammation and as immunosuppressive agents (12, 13). However, the development of ligands that bind to SH2 domains with high affinity and selectivity has met with little success, and SH2 domains generally are considered to be good examples of an intractable drug target.
MATERIALS AND METHODS
Synthetic Chemistry.
Peptides were synthesized by using conventional solid-phase peptide synthesis methods. The FK506-derived mixed carbonate was synthesized as described by Spencer et al. (14). It was dissolved in dimethylformamide with triethylamine and a 2-fold excess of phosphotyrosyl-glutamyl-glutamyl-isoleucine (pYEEI). The coupled product was treated with hydrogen fluoride in acetonitrile to remove the two silyl ether protecting groups, and the desired product was purified by using reverse-phase HPLC. Synthetic ligand for FKBP (SLF) was synthesized according to the procedures of Holt et al. (15). SLF was coupled to the N terminus of the resin-bound protected pYEEI peptide by using PyBOP. The bifunctional SLFpYEEI was simultaneously deprotected and cleaved from the Novasyn TGT resin (Calbiochem) by using 25% trifluoroacetic acid and 2.5% triisopropylsilane in methylene chloride, and the desired product was isolated by using reverse-phase HPLC.
Peptide Coupling to Beads.
The peptide pYEEI (1 mg) was dissolved in 1 ml of dimethyl sulfoxide and incubated with 1 ml of Affi-Gel 10 beads (Bio-Rad) for 6 hr at room temperature. The reaction was stopped by incubating the beads in 5 ml of ethanolamine (1 M, pH 8.0) for 1 hr. The beads were washed and resuspended 1:2 in 20 mM Tris, pH 7.2/150 mM NaCl.
Protein Expression.
The human Fyn SH2 domain (residues 102–205, SIQA-LVVP) and human FKBP12 were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli. The cDNAs were cloned into the pGEX2TK expression vector (Pharmacia). When required, the recombinant proteins were labeled with [γ-32P]ATP on glutathione beads (Pharmacia) using protein kinase A (PKA) and the pGEX2TK-derived PKA site at the N terminus of FKBP12 and the Fyn SH2 domain. FKBP12 and the Fyn SH2 domain were cleaved from GST by using thrombin (Sigma) and the thrombin cleavage site between the PKA site and GST. Human FKBP52 was expressed in pET28c (Novagen) with a His tag at the N terminus. The recombinant protein was purified with Ni2+ nitrilotriacetic acid agarose beads (Qiagen).
Isothermal Titration Calorimetry.
The binding constants of FKBP12, FKBP52, and the Fyn SH2 domain for SLF, SLFpYEEI, and FKpYEEI were determined by using an Omega Isothermal Titration Calorimeter (Microcal, Northampton, MA). Protein in aqueous buffer (20–60 μM: 150 mM NaCl/20 mM Tris, pH 7.2) was equilibrated in the microcalorimeter cell at 25°C for 1–2 hr after being degassed for 10 min. Ligand (170–600 μM) in identical buffer as protein was taken into a 250-μl syringe after being degassed for 10 min. The syringe was loaded into the cell and spun at 400 rpm, and the system was allowed to equilibrate for 1–2 hr. Ligand was then injected into the cell (25 × 10 μl, 6-min intervals), and the heat evolved was quantitated. Binding constants were calculated from a numerical fit to the experimental data as described in ref. 16.
RESULTS
Molecular Design.
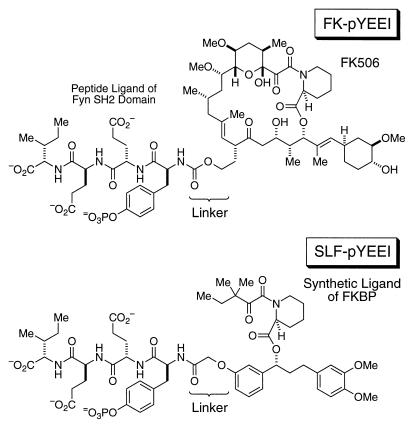
Beginning with a tetrapeptide that binds to the Fyn SH2 domain, pYEEI (17), we synthesized two bifunctional molecules. Both molecules are capable of simultaneously binding to FKBP and the Fyn SH2 domain. The pYEEI peptide was linked covalently to two FKBP ligands, FK506 and SLF (15), to provide the desired bifunctional molecules, FKpYEEI and SLFpYEEI (Fig. 1). SLF is smaller than FK506 and does not project as far from the FKBP protein surface. By using the three-dimensional structures of FKBP12 (15, 18), FKBP52 (19), and the Fyn SH2 domain (20), the linkers between the two halves of the bifunctional molecules were designed to bring the FKBP surface into close proximity to the SH2 domain surface. The affinities of FKpYEEI and SLFpYEEI for recombinant FKBP12, FKBP52, and the Fyn SH2 domain were measured by using isothermal titration calorimetry (ITC, Table 1).
Figure 1.
Structures of FKpYEEI and SLFpYEEI. The C21 allyl group of FK506, which is required for calcineurin binding, was used to link the pYEEI peptide, effectively eliminating any immunosuppressive activity of the FKpYEEI molecule.
Table 1.
Dissociation constants (Kd) for protein–ligand combination
| FKpYEEI, nM | SLFpYEEI, nM | FK506, nM | SLF, nM | |
|---|---|---|---|---|
| FKBP12 | 45 | 60 | 0.4 (refs. 28 and 29) | 20 |
| FKBP52 | 150 | 5,000 | 50 (ref. 30 and 31) | 3,000 |
| Fyn SH2 domain | 340 | 180 | ND | ND |
ND, not determined.
Binding Assay.
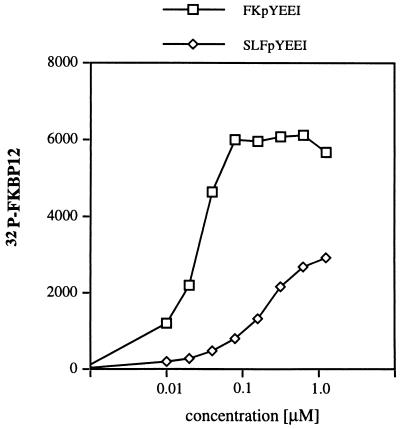
To determine whether the bifunctional molecules allow the formation of a trimeric complex between Fyn and FKBP, GST-Fyn SH2 domain fusion proteins bound to glutathione beads were incubated with radioactively labeled FKBP12 and increasing concentrations of FKpYEEI or SLFpYEEI (Fig. 2). After equilibrium was established, the beads were sedimented and the radioactively labeled FKBP12 associated with the beads was quantified. Both FKpYEEI and SLFpYEEI support the formation of a trimeric complex between the Fyn SH2 domain and FKBP12. However, FKpYEEI forms the trimeric complex more efficiently. FKpYEEI and SLFpYEEI can also form a trimeric complex between immobilized FKBP52 and the radioactively labeled Fyn SH2 domain (data not shown).
Figure 2.
FKpYEEI and SLFpYEEI form trimeric complexes with the Fyn SH2 domain and FKBP12. Recombinant GST-Fyn SH2 domain was expressed, bound to glutathione beads (7.5 μl beads, 0.55 nmol protein), and was incubated with 100 nM 32P-labeled FKBP12 (10,000 cpm) in 100 μl of binding buffer (20 mM Tris, pH 7.2/150 mM NaCl). Increasing concentrations of FKpYEEI or SLFpYEEI were added and the reactions were rotated for 2 hr. The binding reactions were centrifuged in a pierced PCR tube to separate the beads from the supernatant. The radioactive protein associated with the beads was resuspended in 100 μl of PBS, which was added to EcoLite scintillation fluid (ICN) and counted in an LS5000CE liquid scintillation counter (Beckman).
A competition binding assay was used to determine whether the pYEEI peptide bound more tightly to the Fyn SH2 domain when presented by FKBP (Fig. 3A). The pYEEI tetrapeptide was covalently attached to beads. The beads were incubated with radioactively labeled Fyn SH2 domain, and the bound protein was quantified by centrifuging the beads and counting the associated radioactivity. When FKpYEEI or SLFpYEEI was added to the binding reaction, the pYEEI peptide of the bifunctional molecules competed with the peptide beads for binding to the 32P-labeled Fyn SH2 domain. As a consequence of this competition, less 32P-labeled Fyn SH2 domain bound to the beads. Hence, low levels of radioactivity bound to the beads reflect high occupancy of the Fyn SH2 domain by a competing ligand in the solution (Fig. 3B).
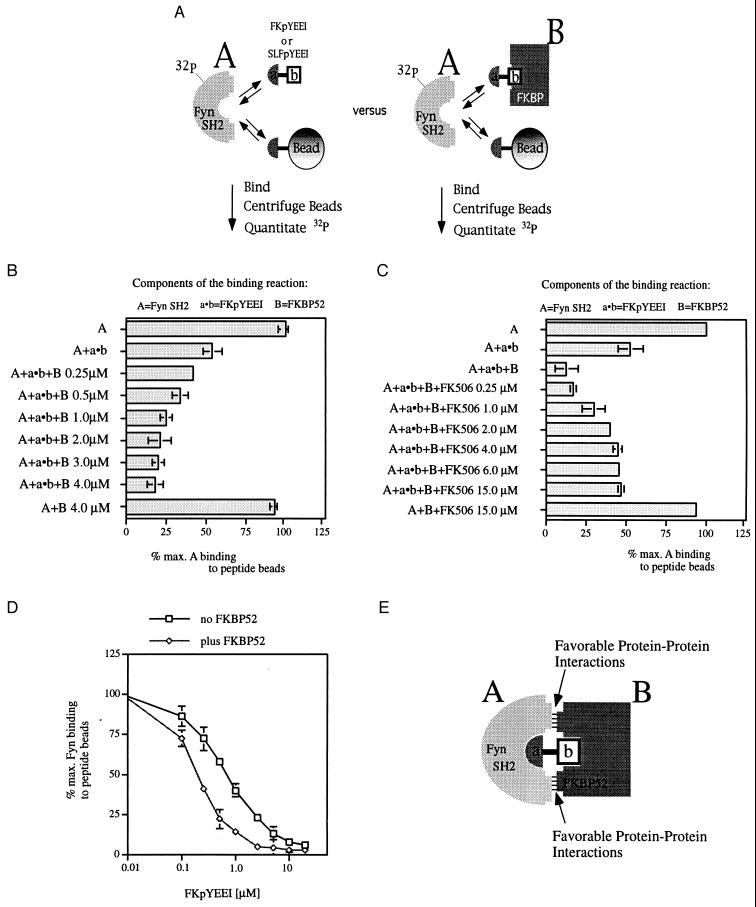
Figure 3.
Fyn SH2 domain binding to FKpYEEI alone or in a complex with FKBP52. (A) Model of the competition binding assay. (B–D) In 100 μl of binding buffer, peptide beads (7.5 μl) were incubated with the 32P-labeled Fyn SH2 domain (200 nM, 14,000 cpm) and the molecules stated below. After a 2-hr incubation time, the beads were centrifuged and the radioactive Fyn SH2 domain bound was quantitated. (B) Addition of FKpYEEI (1.0 μM) and increasing concentrations of FKBP52 (0.25–4.0 μM). (C) Addition of FKpYEEI (1.0 μM), FKBP52 (2.0 μM), and increasing concentrations of FK506 (0.25–15 μM). The maximal radioactivity associated with the beads (=100%) was 9,139 cpm for b and 10,467 cpm for C. (D) Competition binding curves: 32P-labeled Fyn SH2 domain (200 nM, 16,000 cpm) was incubated without or with FKBP52 (4.5 μM) in 100 μl of binding buffer. Increasing concentrations of FKpYEEI (0.1–20.0 μM) were added to the binding reactions. The maximal radioactivity associated with the beads was 12,569 cpm (=100%) for the 32P-labeled Fyn SH2 domain alone and 10,250 cpm (=100%) for the 32P-labeled Fyn SH2 domain in the presence of FKBP52. All data points were taken in triplicate, and the average is plotted. (Bars = SE.) (E) Model for favorable, affinity-enhancing protein–protein interactions.
Using a Borrowed Protein Surface to Enhance Affinity.
By using this assay, FKBP12 or FKBP52 was added at various concentrations to a binding reaction to form a complex with FKpYEEI or SLFpYEEI. When presented by FKBP12, FKpYEEI bound to the Fyn SH2 domain as well as free FKpYEEI (data not shown). However, the FKpYEEI–FKBP52 complex competed more effectively for binding to the Fyn SH2 domain than did FKpYEEI alone (Fig. 3B). To confirm that the observed effect depends on binding of FKpYEEI to the FK506-binding pocket of FKBP52, FK506 was added to the binding reaction. FK506 binds more tightly to FKBP12 and FKBP52 than either bifunctional molecule (Table 1). In the binding assay, as FK506 reaches an equimolar concentration with respect to FKBP52, most of the FKBP52 is bound to FK506. Under these conditions, the affinity-enhancing effect of FKBP52 was abolished (Fig. 3C). These results demonstrate that the enhanced affinity of FKpYEEI for the Fyn SH2 domain depends on FKpYEEI binding to the FK506-binding pocket of FKBP52.
To quantify the increase in affinity of the Fyn SH2 domain for pYEEI presented by FKBP52, we measured the binding of the 32P-labeled Fyn SH2 domain to peptide beads as a function of the concentration of FKpYEEI (Fig. 3D). FKBP52 enhances the affinity by a factor of 3, which is reflected in a shift of the IC50 from 750 nM in the absence of FKBP52 to an IC50 of 250 nM in the presence of FKBP52. ITC measurements confirmed the approximately 3-fold affinity enhancement (FKpYEEI plus Fyn SH2 domain: Kd = 340 nM versus the FKpYEEI–FKBP52 complex plus Fyn SH2 domain: Kd = 120 nM).
We considered the possibility that FKBP12 and FKBP52 present the pYEEI peptide in different orientations. However, the structures of the FK506-binding domains from both proteins are very similar (19), and it is unlikely that preorganization of the relatively flexible pYEEI peptide is responsible for the observed binding enhancement. Thus, we conclude that when the Fyn SH2 domain binds to the FKpYEEI-FKBP52 complex, the SH2 domain makes additional favorable protein–protein interactions with FKBP52 that enhance the overall stability of the trimeric complex (Fig. 3E).
Using a Borrowed Protein Surface to Reduce Affinity.
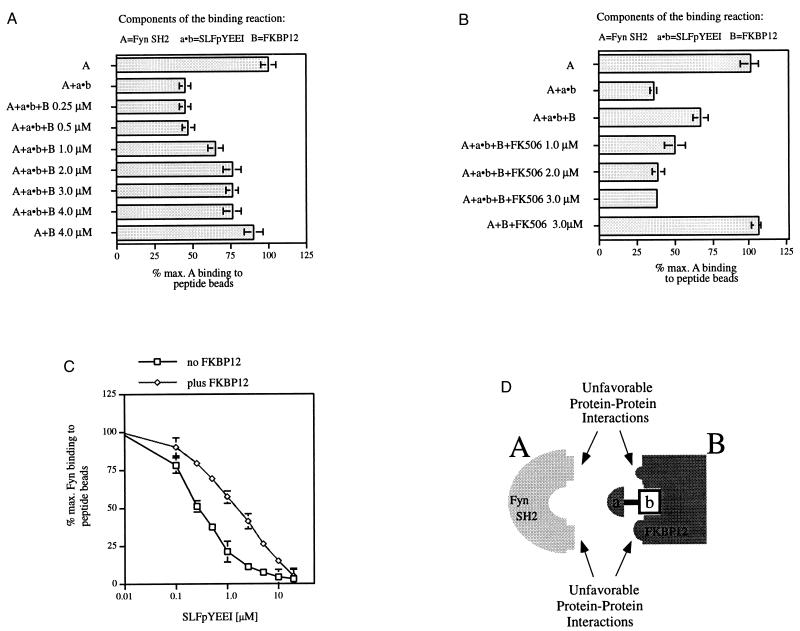
In contrast to the FKpYEEI–FKBP52 complex, the SLFpYEEI–FKBP12 complex displays decreased affinity for the Fyn SH2 domain. In the competition binding assay, greater amounts of the Fyn SH2 domain bind to the peptide beads in response to increasing concentrations of FKBP12 (Fig. 4A). This result indicates that the FKBP12–SLFpYEEI complex is a less effective competitor than SLFpYEEI alone. The effect is reversed by the addition of FK506, which indicates that SLFpYEEI binds to the FK506-binding pocket of FKBP12 (Fig. 4B). The IC50 of SLFpYEEI for the Fyn SH2 domain when presented by FKBP12 was increased 6-fold from 0.25 to 1.5 μM (Fig. 4C). This increase was confirmed by ITC (SLFpYEEI plus Fyn SH2 domain: Kd = 180 nM versus the SLFpYEEI–FKBP12 complex plus Fyn SH2 domain: Kd = 1.0 μM; 5.5-fold increase). We interpret this decrease in affinity to indicate that the FKBP12 surface establishes unfavorable interactions with the Fyn SH2 domain surface in the trimeric complex (Fig. 4D). The SLFpYEEI–FKBP52 complex does not affect the binding of the Fyn SH2 domain to pYEEI (data not shown). Considering the four possible complexes for presentation of the pYEEI peptide (two FKBPs and two bifunctional molecules), one complex improves binding of the pYEEI peptide to the Fyn SH2 domain, one complex diminishes binding, and two complexes have no measurable effect.
Figure 4.
Fyn SH2 domain binding to SLFpYEEI in a complex with FKBP12. (A–C) In 100 μl of binding buffer, peptide beads (7.5 μl) were incubated with the 32P-labeled Fyn SH2 domain (200 nM, 18,000 cpm) and the molecules stated below. After a 2-hr incubation time, the beads were centrifuged and the radioactive Fyn SH2 domain bound was quantitated. (A) Addition of SLFpYEEI (1.0 μM) and increasing concentrations of FKBP12 (0.25–4.0 μM). (B) Addition of SLFpYEEI (1.0 μM), FKBP12 (2.0 μM), and increasing concentrations of FK506 (1.0–3.0 μM). The maximal radioactivity associated with the beads (=100%) was 12,583 cpm for A and 12,187 cpm for B. (C) Competition binding curves: 32P-labeled Fyn SH2 domain (200 nM, 13,000 cpm) was incubated without or with FKBP12 (20 μM) in 100 μl of binding buffer. Increasing concentrations of FKpYEEI (0.1–20.0 μM) were added to the binding reactions. The maximal radioactivity associated with the beads was 9,284 cpm (=100%) for the 32P-labeled Fyn SH2 domain alone and 10,492 cpm (=100%) for the 32P-labeled Fyn SH2 domain in the presence of FKBP12. All data points were taken in triplicate, and the average was plotted. (Bars = SE.) (D) Model for unfavorable, destabilizing protein–protein interactions.
DISCUSSION
Borrowing Endogenous Proteins to Enhance the Characteristics of Small-Molecule Ligands.
Our findings demonstrate that a phosphopeptide ligand for the Fyn SH2 domain can be engineered to bind more tightly to its protein target by inducing the formation of a trimeric complex. In the case of FKBP52, FKpYEEI, and the Fyn SH2 domain, the trimeric complex is more stable than would be expected based on the stabilities of individual bimolecular complexes (Table 1). Although our data do not definitively demonstrate new protein–protein interactions resulting from the covalent linkage of FK506 to the pYEEI peptide, the observation that the change in affinity is related to both the structure of the ligand (SLF vs. FK506) and the borrowed endogenous protein (FKBP12 vs. FKBP52) points to the surface between the SH2 domain and the FKBP as the origin of the altered affinity.
One explanation for the increase in affinity is that additional protein–protein interactions between FKBP52 and the SH2 domain surface make a significant, direct energetic contribution to the stability of the complex. Alternatively, the additional distal interactions may indirectly enhance the free energy of binding of the peptide ligand to the SH2 domain. This possibility is based on the analysis of the energetic contributions of single amino acid side chains to protein–protein interactions. The area of contact between two proteins is often large and flat (21, 22), but, interestingly, a major part of the free energy of binding of two interacting protein surfaces can be contributed by a limited number of clustered amino acids. These clusters have been designated as “hot spots” (23), and the surrounding contacts may serve to insulate the critical amino acids from bulk solvent. Thus, the SH2 domain-FKBP52 contacts may limit the accessibility of water to pYEEI and the binding pocket so that their energetic interactions are increased. Ultimately, structural studies will be helpful for confirming the role of FKBP-SH2 domain contacts in the affinity modification demonstrated here.
Plasticity of Binding Surfaces.
The establishment of favorable or, at least, nondetrimental contacts will depend on the juxtaposition of the presenter protein surface to the surface of the target protein. Recent studies of human growth hormone and the TCR suggest that the plasticity of protein surfaces can act favorably in the attempt to establish beneficial protein–protein contacts between the presenter protein and the target. Mutations in growth hormone that compensate for a mutation in the core region of its receptor have been shown to lead to major rearrangements at the protein–protein interface (24). The crystal structure of the mutant proteins reveals that amino acid side chains reorganize to establish new interactions or to avoid unfavorable interactions. The plasticity of protein surfaces is also apparent in the binding of the TCR to peptide–MHC, which can lead to large conformational changes in the complementarity determining regions of the TCR to gain new contacts (25).
We also have demonstrated that borrowing the surface of an endogenous protein can reduce the affinity of interactions of a phosphopeptide with its SH2 domain. Steric hindrance or electrostatic repulsion are probably the basis for the decreased stability of the SH2 domain–SLFpYEEI–FKBP12 complex. In general, the creation of unfavorable contacts should be easier to achieve than favorable contacts, and it can be exploited to enhance the specificity of a molecule of interest. If, for example, a ligand binds to one desired and several undesired targets, a bifunctional molecule that causes unfavorable protein–protein interactions with the undesired targets may be selected. If the obtained bifunctional molecule shows favorable or at least neutral interactions with the desired molecule, specificity for the desired molecule will be created.
General Strategy to Modulate Ligand-Binding Affinities.
The introduction of secondary binding interactions through synthetic modifications to small-molecule ligands has been examined in some detail (26, 27). Our approach differs by borrowing surface area from presenting proteins. Features of our strategy include the ability to (i) recruit different endogenous presenting proteins by changing one-half of the bifunctional molecule, (ii) vary the length and rigidity of the linker that joins the two halves of the bifunctional molecule, and (iii) vary the affinity of the small-molecule ligand that binds to the presenting protein. Changes to any of these three variables have the potential to directly affect the overall stability of the trimeric complex. Ultimately, this general strategy may prove useful to improve the affinity and/or specificity of small-molecule drugs for their targets. It can also provide a useful tool for the, so far, intractable problem of developing agonists or antagonists of protein–protein interactions.
Acknowledgments
We thank Lewis Cantley for providing the Fyn SH2 domain expression construct; David Smith for providing the FKBP52 expression construct; Jay Boniface, Dan Lyons, and Steve Biggar for helpful discussion; Bob Flowers for help designing and Lincoln Bickford for help executing the ITC measurements; and Kurt Vogel for assistance in the HPLC purification of compounds.
ABBREVIATIONS
- FKBP
human FK506-binding protein 12
- pYEEI
tetrapeptide phosphotyrosyl-glutamyl-glutamyl-isoleucine
- FKpYEEI
FK506 covalently linked to the peptide pYEEI
- SLF
synthetic ligand for FKBP
- SLFpYEEI
SLF covalently linked to pYEEI
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- TCR
T cell receptor
Footnotes
A Commentary on this article begins on page 1826.
References
- 1.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 2.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 3.Ajitkumar P, Geier S S, Kesari K V, Borriello F, Nakagawa M, Bluestone J A, Saper M A, Wiley D C, Nathenson S G. Cell. 1988;54:47–56. doi: 10.1016/0092-8674(88)90178-x. [DOI] [PubMed] [Google Scholar]
- 4.Sun R, Shepherd S E, Geier S S, Thomson C T, Sheil J M, Nathenson S G. Immunity. 1995;3:573–582. doi: 10.1016/1074-7613(95)90128-0. [DOI] [PubMed] [Google Scholar]
- 5.Smith K D, Lutz C T. J Immunol. 1997;158:2805–2812. [PubMed] [Google Scholar]
- 6.Liu J, Farmer J D, Jr, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Albers M W, Wandless T J, Luan S, Alberg D G, Belshaw P J, Cohen P, MacKintosh C, Klee C B, Schreiber S L. Biochemistry. 1992;31:3896–3901. doi: 10.1021/bi00131a002. [DOI] [PubMed] [Google Scholar]
- 8.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 9.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, et al. Nature (London) 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 10.Choi J, Chen J, Schreiber S L, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 11.Koch C A, Anderson D, Moran M F, Ellis C, Pawson T. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 12.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 13.Bolen J B, Brugge J S. Annu Rev Immunol. 1997;15:371–404. doi: 10.1146/annurev.immunol.15.1.371. [DOI] [PubMed] [Google Scholar]
- 14.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 15.Holt D A, Luengo J I, Yamashita D S, Oh H-J, Konialian A L, Yen H-K, Rozamus L W, Brandt M, Bossard M J, Levy M A, et al. J Am Chem Soc. 1993;115:9925–9938. [Google Scholar]
- 16.Wiseman T, Williston S, Brandts J F, Lin L N. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 17.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 18.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 19.Craescu C T, Rouviere N, Popescu A, Cerpolini E, Lebeau M C, Baulieu E E, Mispelter J. Biochemistry. 1996;35:11045–11052. doi: 10.1021/bi960975p. [DOI] [PubMed] [Google Scholar]
- 20.Mulhern T D, Shaw G L, Morton C J, Day A J, Campbell I D. Structure. 1997;5:1313–1323. doi: 10.1016/s0969-2126(97)00283-9. [DOI] [PubMed] [Google Scholar]
- 21.Janin J, Chothia C. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 22.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 24.Atwell S, Ultsch M, De Vos A M, Wells J A. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 25.Garcia K C, Degano M, Pease L R, Huang M, Peterson P A, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 26.Jain A, Huang S G, Whitesides G M. J Am Chem Soc. 1994;116:5057–5062. [Google Scholar]
- 27.Jain A, Whitesides G M, Alexander R S, Christianson D W. J Med Chem. 1994;37:2100–2105. doi: 10.1021/jm00039a023. [DOI] [PubMed] [Google Scholar]
- 28.Bierer B E, Mattila P S, Standaert R F, Herzenberg L A, Burakoff S J, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siekierka J J, Hung S H, Poe M, Lin C S, Sigal N H. Nature (London) 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 30.Tai P K, Albers M W, Chang H, Faber L E, Schreiber S L. Science. 1992;256:1315–1318. doi: 10.1126/science.1376003. [DOI] [PubMed] [Google Scholar]
- 31.Yem A W, Reardon I M, Leone J W, Heinrikson R L, Deibel M R., Jr Biochemistry. 1993;32:12571–12576. doi: 10.1021/bi00210a004. [DOI] [PubMed] [Google Scholar]