Abstract
ROCKII kinase activity is known to be regulated by Rho GTPase binding; however, the context-specific regulation of ROCKII is not clearly understood. We pursued the C-terminal PH domain as a candidate domain for regulating ROCKII function. A proteomics-based screen identified potential ROCKII signaling partners, a large number of which were associated with membrane dynamics. We used subcellular fractionation to demonstrate that ROCKII is localized to both the plasma membrane and internal endosomal membrane fractions, and then used microscopy to show that the C-terminal PH domain can localize to internal or peripheral membrane compartments, depending on the cellular context. Co-immunoprecipitation demonstrated that Dynamin1 is a novel ROCKII binding partner. Furthermore, blocking Dynamin function with a dominant negative mutant mimicked the effect of inhibiting ROCK activity on the actin cytoskeleton. Our data suggest that ROCKII is regulated by localization to specific membrane compartments and its novel binding partner, Dynamin1.
Keywords: ROCK, PH domain, Dynamin, Membrane, Subcellular localization
INTRODUCTION
ROCK is a ser/thr kinase that regulates cell adhesion, migration, invasion, cytokinesis, apoptosis, and oncogenic transformation. ROCK induces these effects primarily by phosphorylation of different substrates; however, the mechanisms that determine which downstream signaling pathways are activated in specific biological contexts are not presently understood[1, 2]. Based on the range of biological functions for ROCK, it is reasonable to expect that specification of downstream signaling events is regulated by cell type, subcellular localization, and extracellular stimuli.
While mechanisms that regulate activation of ROCK kinase activity are well-described, determining which substrates will be phosphorylated, and what signaling pathways will be activated, remains unclear. Generally, regulation of kinase signaling is not restricted to activation of catalytic activity but also on subcellular localization and incorporation of the kinase into multi-protein complexes. The ROCK PH domain has potential to regulate ROCK localization and interaction with binding partners since PH domains can bind to phospholipids and also act as protein-protein interaction domains [2–5].
We hypothesize that the ROCKII PH domain regulates the formation of signaling complexes and that the composition and localization of this signaling complex determines which downstream signaling pathways are activated by ROCK. Thus, we designed an experimental strategy to isolate ROCKII binding partners using a proteomics-based approach. We screened the proteome of rat brain lysate by affinity purification to isolate proteins that bind to the ROCKII C-terminal PH domain, and then identified these proteins by mass spectrometry. Unexpectedly, we found that ROCKII associated with multiple membrane regulatory proteins. Follow up experiments with subcellular fractionation and microscopy demonstrate that ROCKII can localize to both internal and peripheral membrane fractions, via its C-terminal PH domain. Furthermore, co-immunoprecipitation experiments demonstrated that ROCKII binds to the membrane regulator, Dynamin1. Significantly, blocking Dynamin function with a dominant negative mutant mimicks the effect of inhibiting ROCK activity on the actin cytoskeleton. Our results indicate that ROCKII regulation of the actin cytoskeleton is modulated by localization to membrane compartments and the membrane regulator, Dynamin1.
MATERIALS AND METHODS
Tissues and Cell culture
Rat brains were purchased from Pel-Freez® Biologicals (Rogers, Arkansas). PC12 cells were purchased from ATCC (Manassas, Virginia) and cultured in DMEM 10% FBS, 5% HS, NEAA.
Antibodies
CRMP2 (ab36201), Rab5 (ab13253), and sodium-potassium ATPase (ab7671) antibodies were purchased from Abcam Inc. (Cambridge, Massachusetts). ROCK (BL964) antibodies were purchased from Bethyl Laboratories, Inc (Montgomery, Texas). The RabGDI antibody (130001) was purchased from Synaptic Systems (Gottingen, Germany). The mouse monoclonal antibody to Dynamin 1 (05–319) and ROCKII antibodies were purchased from Upstate (Lake Placid, New York).
ROCKII PH domain affinity column
His- ROCK II C-terminus (amino acids 1135–1381) was generated in Rosetta 2(DE3) (EMD Chemicals, San Diego, California), purified with a HisTRAP column (GE Healthcare, Pittsburgh, Philadelphia) and size exclusion chromatography. Recombinant protein was rebound to HisTRAP beads to form an affinity column and used in a pull-down experiment: Rat brain was homogenized in 50 mM Tris-HCl, pH 8 protease inhibitors (Roche, Indianapolis, Indiana) by sonication. Cleared homogenate was incubated with the affinity column (16 hrs, 4°C), washed 3 times with binding buffer (50 mM Tris-HCl, pH 8), and the bound proteins were eluted with a Immobiline-pH-gradient (IPG)- strip rehydration buffer (1.5 M NaCl, 20% glycerol, 2 M Thiourea, 7 M urea, 4% (w/v) CHAPS, 20 mM Tris pH 8.5, 0.5% (v/v) ampholines/biolytes (pH 3–10), 200 mM Imidazole and 0.005% (w/v) bromophenol blue).
2D-SDS-PAGE
Eluted proteins from the experimental column were subjected to SDS-PAGE and stained with Coomassie Blue. 2D SDS-PAGE 1st dimension separation was performed with 18 cm, 3-10NL IPG strips on an Ettan IPGphor II Isoelectric focusing system (GE Healthcare; Pittsburgh, Philadelphia) followed by SDS-PAGE.
Mass Spectrometry and Identification of Proteins
Protein spots were subjected to Maldi-TOF mass spectrometry with partial sequence determination. The generated peptide masses and partial sequences from MS/MS were used to identify binding partners by querying the NCBInr DATABASE (Mammalia) using the MASCOT search algorithm (oxidation and methionine modifications considered). With a significance threshold of p<0.05).
Subcellular Fractionation
To isolate early endosomes, we used the flotation gradient described by [6]. 4.8 mg of were typically used for subcellular fractionation to purify and enrich for early endosomes. Plasma membranes were isolated using a method described by [7]. 20 mg of total protein was typically used as starting material for plasma membrane isolation.
Immunofluorescence
Cells were plated onto collagen coated coverslips, then transfected with dominant negative HA-dynamin1 (K44A mutation), Constitutively active myc-ROCKII, or treated with the ROCK inhibitor, Y-27632 (Calbiochem, San Diego, CA). Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% saponin, blocked with 5% normal goat serum, and stained with Alexa Fluor594 Phalloidin and FITC-conjugated HA antibody or FITC-conjugated myc antibody. Actin structures of transfected cells were analyzed on an inverted Olympus IX81 fluorescent microscope and images collected on a Hamamatsu CCD camera.
RESULTS & DISCUSSION
Identification & analysis of proteins interacting with the ROCKII PH domain
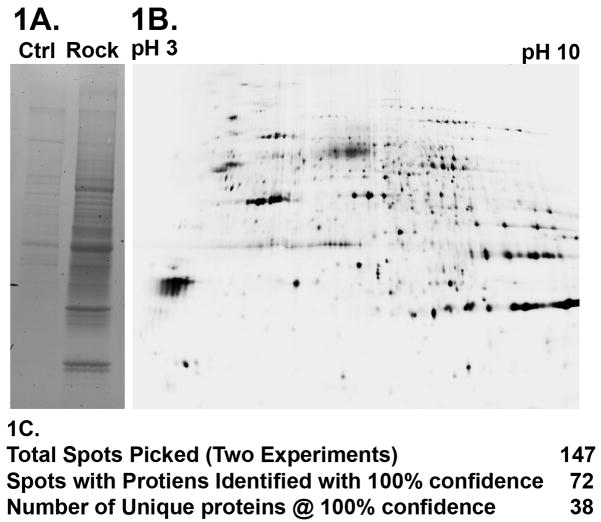
To investigate whether the PH domain of ROCK functions as a protein-protein interaction domain; we used a proteomics-based screen to identify potential signaling partners. Highly purified recombinant ROCKII PH domain and optimized wash and elution conditions reduced non-specific interactions, in the control column (Fig. 1A). 2D SDS-PAGE resolved the different proteins into individual spots followed by Sypro Ruby staining for total protein (Fig. 1B). These were subsequently processed for analysis by MALDI-TOF MS MS mass spectrometry. We identified 38 unique proteins, seven were identified in duplicate experiments and eleven were present in multiple spots (Fig. 1C). Several of the proteins have been previously identified as ROCKII binding partners and/or substrates; endogenous ROCKII [2, 8], Pfn2 [9, 10], Crmp2 [11], and Gfap [12]. Thus, our approach to isolate ROCKII PH domain signaling partners is confirmed by our successful identification of previously documented ROCKII signaling partners. A full list of the positively identified proteins, along with experimental statistics is provided in supplementary data table 1.
Figure 1. Proteomics-Based Screen for ROCKII Interactors.
1A. 1D SDS PAGE and Coomassie staining of eluted proteins from a control column and ROCKII affinity column shows strong enrichment for protein binding to the affinity column over the control column. 1B. 2D SDS-PAGE of eluted proteins from both control and affinity columns facilitated spot-picking prior to mass spectrometry analysis and protein identification. 1C. Listing of numbers of proteins isolated and identified via MALDI TOF analysis.
The database search identified a number of proteins associated with cytoskeletal dynamics, which was expected. However, a significant number of identified proteins were associated with membrane trafficking (see highlighted proteins in supplementary data table 1). This was unexpected and suggested that apart from activation by RhoA, ROCK might also be regulated by membrane trafficking. Based on these observations, we predicted that ROCKII is localized to membrane compartments within the cell, as a mechanism for regulating its signaling.
Localization of ROCKII to membrane compartments
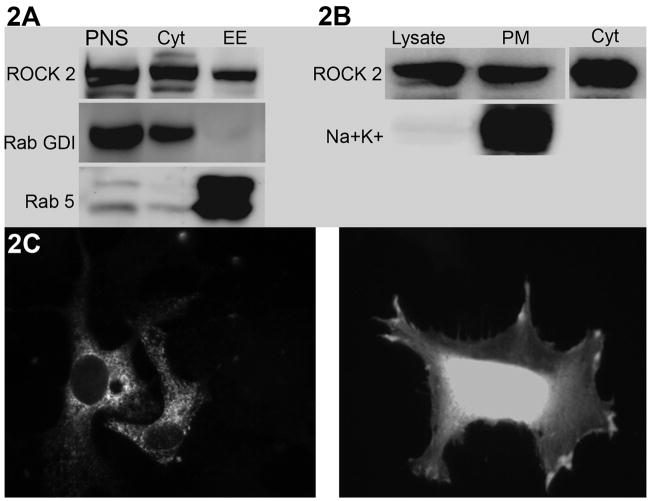
Immunolocalization experiments have previously shown that ROCKII can be localized to multiple sites, including the cytosol, nucleus [13], myosin fibrils [14] and plasma membrane [15]. However, the results of our proteomics screen suggest a significant and unexplored association between ROCKII and multiple membrane compartments. There are several issues that contribute to the interpretation of immunolocalization experiments, including the influence of fixation and permeabilization techniques, and limitations due to suitable antibodies. Thus, we chose to analyze ROCKII localization with a biochemical approach, to build on the findings from the existing immunolocalization studies. To test our hypothesis that ROCKII is localized to membrane compartments we used subcellular fractionation to purify specific membrane domains. First, we purified a Rab5 positive endosomal compartment because many of the ROCKII binding partners identified in our screen are associated with vesicle trafficking. Figure 2A demonstrates that ROCKII is present in this fraction, a compartment of the cell previously unrecognized as containing ROCKII. In a separate experiment, we isolated plasma membrane and found ROCKII in this membrane compartment, as well (figure 2B.) Because we had used the C-terminal PH domain of ROCKII in our proteomics screen, we proposed that the PH domain may be responsible for localizing ROCKII to the different membrane compartments. PH domains are generally recognized as phospholipid binding domains, and can influence the localization of proteins [16]. We expressed GFP-tagged PH domain from ROCKII to determine its localization. We chose two different cell types for analysis, based on the idea that localization of ROCKII is likely to depend on the specific cellular context. First, we analyzed HeLa cells after 2 hours of spreading on Collagen1 (Fig. 2C, right). In these cells, the GFP-PH domain localized to peripheral membranes in areas of cell spreading. Second, we analyzed Cos7 cells cultured for 48 hours prior to analysis (Fig. 2C, left). These cells showed a distinctly different distribution of GFP-PH domain, with localization in an internal punctate distribution, resembling vesicle membranes. Thus, the PH domain is likely to physically link ROCKII to membrane domains, but the molecular determinants that specify the particular compartment is outside of this domain. These patterns of localization are consistent with both the finding of vesicle trafficking proteins in the proteomics screen, as well as the subcellular fractionation data. Taken together, our data demonstrates that ROCKII resides in not only in peripheral membranes, but also in internal membrane compartments. Furthermore, it suggests that the function of ROCKII may be regulated by membrane trafficking.
Figure 2. Subcellular Localization of ROCK to Membrane Compartments.
2A. ROCKII was found in Rab5+ vesicles isolated by subcellular fractionation. RabGDI was used as a cytosolic marker. 2B. ROCKII was found in plasma membrane fraction, marked by presence of the Sodium-Potassium pump (Na+ K+). 50μg of total protein in the post-nuclear supernatant (PNS) or cell lysate (Lysate) were loaded as a reference to compare with the fractions. 2C. GFP-ROCKII PH domain was expressed by transient transfection in either Cos7 cultured for 48 hours (left) or HeLa plated for 2 hours on Collagen1 (right). ROCKII PH domain is localized to both peripheral and internal membrane compartments.
Interestingly, Sturge et al show that disruption of endocytosis by manipulation of Endo180 affects activation of RhoA-ROCK myosin phosphorylation [17]. Significantly, the location of Endo180-containing vesicles and ROCKII is essential for retraction of the trailing edge of migrating cells. In addition, Norman et al show that inhibition of the Rab4-dependent “short loop” of endocytic recycling promotes activation of ROCK-dependent phosphorylation of Cofilin [18]. Thus, it is now clear that it is important to understand how ROCKII subcellular localization is regulated as a critical mechanism governing its function.
Interaction between ROCKII and the membrane regulator Dynamin1
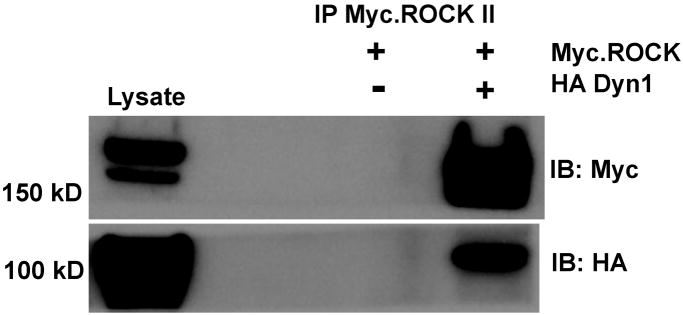
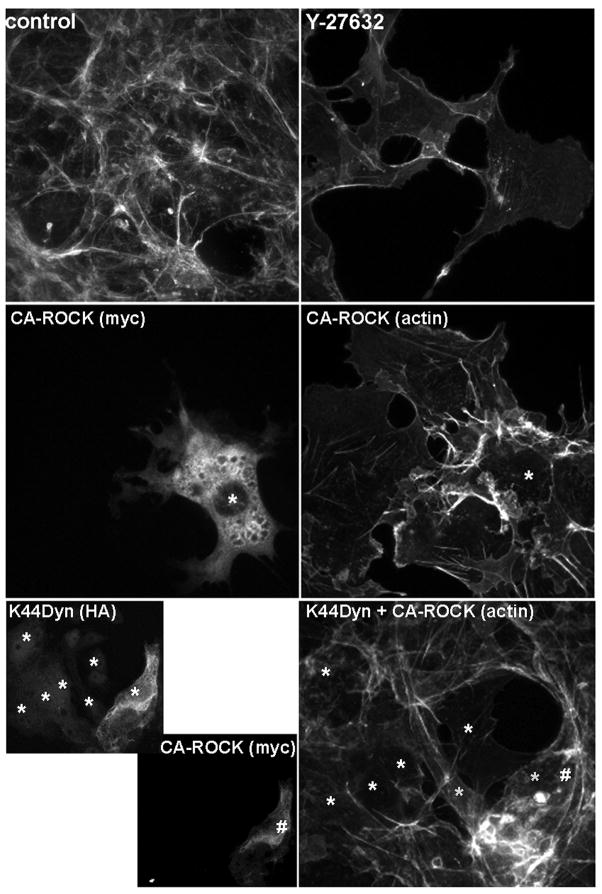
We selected Dynamin1 for further analysis of its interaction with ROCKII, because it is a novel ROCKII binding partner and is an important regulator of membrane trafficking [19, 20]. In addition, Dynamin1 was identified in two independent experiments, was present in multiple spots on the 2D gel, and was identified with high statistical significance (protein score 335 – see supplemental data.) For biochemical confirmation, we expressed HA-Dynamin1 with or without myc-ROCKII and analyzed myc immunocomplexes for the presence of HA-Dynamin1. We found that Dynamin1 specifically co-immunoprecipitated with myc-ROCKII, confirming the results of our proteomic screen (figure 3.) These experiments demonstrate a previously undescribed relationship and signaling interaction between ROCKII and Dynamin1. To test the functional significance between Dynamin1 and ROCKII, we used microscopic analysis of actin structures as a readout of ROCKII function. Figure 4 shows that inhibition of ROCK activity with Y-27632 disrupts actin stress fibers, while expression of a constitutively active ROCK mutant enhances actin bundles. When we expressed dominant negative Dynamin1, we found that the transfected cells had disrupted actin stress fibers, mimicking the ROCK inhibition phenotype. These results suggested that Dynamin1 is necessary for ROCKII regulation of the actin cytoskeleton. To definitively test this, we co-expressed dominant negative DynaminI (K44A mutant) with constitutively active ROCKII to determine if active ROCKII could rescue the K44A-DynaminI phenotype. The lower panels of figure 4 show that cells expressing only K44A-DynaminI (*) had disrupted actin structures, while co-expressing active ROCK (#) restored actin bundling. Our results show that Dynamin1 and ROCKII form a complex, and that this interaction is necessary for appropriate regulation of ROCKII function on the actin cytoskeleton. Thus, our study, in combination with previous studies, highlight the underappreciated relationship between ROCKII and membrane trafficking and point towards Dynamin1 as a potential regulator of ROCKII function via subcellular localization.
Figure 3. Confirmation of ROCKII Interaction with Dynamin 1.
Cos7 cells were transfected with HA-Dynamin1 and Myc-ROCKII, followed by immunoprecipitation of HA-Dynamin. As a negative control, cells expressing only Myc-ROCKII were also analyzed. Immunoblot for HA confirms the expression (lysate) and immunoprecipitation of HA-Dynamin. Immunoblot for Myc confirms expression (lysate) and demonstrates co-immunoprecipitation only when HA-Dynamin is co-expressed.
Figure 4. Dynamin1 is Required for ROCKII Regulation of Actin Structures.
Cos7 cells were transfected with dominant negative dynamin1 (HA-K44A-Dyn1) or constitutively active ROCKII (Myc-CA ROCKII) as indicated. Some samples were treated with the ROCK inhibitor Y-27632 as indicated. Treated cells were fixed and stained with fluorescent phalloidin and anti-HA or anti-Myc antibodies. Transfected cells are indicated with * or # to aid visualization. HA-K44A-Dyn1 disrupts actin structures, similar to that seen with Y-27632. This disruption of actin structures is rescued by co-expression of CA-ROCK.
Acknowledgments
We thank the LSUHSC Proteomics Core Facility for technical contributions, and Dr. Andy Catling for productive scientific discussions. The project was supported by Grant Number P20RR020160 from the National Center for Research Resources (NCRR), Grant Number 0435253N from American Heart Association; and Grant Number LEQSF(2006–09)-RD-A-16 from the Louisiana Board of Regents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loirand G, Guerin P, Pacaud P. Rho Kinases in Cardiovascular Physiology and Pathophysiology. Circ Res. 2006;98:322–334. doi: 10.1161/01.RES.0000201960.04223.3c. [DOI] [PubMed] [Google Scholar]
- 2.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 3.Yoneda A, Multhaupt HA, Couchman JR. The Rho kinases I and II regulate different aspects of myosin II activity. J Cell Biol. 2005;170:443–453. doi: 10.1083/jcb.200412043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touhara K, Inglese J, Pitcher JA, Shaw G, Lefkowitz RJ. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J Biol Chem. 1994;269:10217–10220. [PubMed] [Google Scholar]
- 5.Tsukada S, Simon MI, Witte ON, Katz A. Binding of beta gamma subunits of heterotrimeric G proteins to the PH domain of Bruton tyrosine kinase. Proc Natl Acad Sci U S A. 1994;91:11256–11260. doi: 10.1073/pnas.91.23.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Zhang L, Yeung TK, Chen X. Endocytosis deficiency of epidermal growth factor (EGF) receptor-ErbB2 heterodimers in response to EGF stimulation. Mol Biol Cell. 1999;10:1621–1636. doi: 10.1091/mbc.10.5.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi H, Kasa M, Amano M, Kaibuchi K, Hakoshima T. Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil. Structure. 2006;14:589–600. doi: 10.1016/j.str.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 9.Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. Embo J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimura N, Inagaki N, Chihara K, Menager C, Nakamura N, Amano M, Iwamatsu A, Goshima Y, Kaibuchi K. Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J Biol Chem. 2000;275:23973–23980. doi: 10.1074/jbc.M001032200. [DOI] [PubMed] [Google Scholar]
- 12.Kosako H, Amano M, Yanagida M, Tanabe K, Nishi Y, Kaibuchi K, Inagaki M. Phosphorylation of glial fibrillary acidic protein at the same sites by cleavage furrow kinase and Rho-associated kinase. J Biol Chem. 1997;272:10333–10336. doi: 10.1074/jbc.272.16.10333. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Nishimura D, Wu RC, Amano M, Iso T, Kedes L, Nishida H, Kaibuchi K, Hamamori Y. Nuclear Rho kinase, ROCK2, targets p300 acetyltransferase. J Biol Chem. 2006;281:15320–15329. doi: 10.1074/jbc.M510954200. [DOI] [PubMed] [Google Scholar]
- 14.Kawabata S, Usukura J, Morone N, Ito M, Iwamatsu A, Kaibuchi K, Amano M. Interaction of Rho-kinase with myosin II at stress fibres. Genes Cells. 2004;9:653–660. doi: 10.1111/j.1356-9597.2004.00749.x. [DOI] [PubMed] [Google Scholar]
- 15.Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 16.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 17.Sturge J, Wienke D, Isacke CM. Endosomes generate localized Rho-ROCK-MLC2-based contractile signals via Endo180 to promote adhesion disassembly. J Cell Biol. 2006;175:337–347. doi: 10.1083/jcb.200602125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White DP, Caswell PT, Norman JC. alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J Cell Biol. 2007;177:515–525. doi: 10.1083/jcb.200609004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruchten AE, McNiven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119:1683–1690. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Chang S. Ever-expanding network of dynamin-interacting proteins. Mol Neurobiol. 2006;34:129–136. doi: 10.1385/MN:34:2:129. [DOI] [PubMed] [Google Scholar]