Summary
Rpn1 (109 kDa) and Rpn2 (104 kDa) are components of the 19S regulatory complex of the proteasome. The central portions of both proteins are predicted to have toroidal α-solenoid folds composed of 9–11 PC (proteasome/cyclosome) repeats, each ~ 40 residues long and containing two α-helices and turns (A.V. Kajava, J. Biol. Chem. 277, 49791–8, 2002). To evaluate this prediction, we examined the full-length yeast proteins and truncated (PC) versions thereof consisting only of the repeat-containing regions by gel filtration, circular dichroism spectroscopy, and negative staining electron microscopy. All four proteins are monomeric in solution and highly α-helical – particularly, the truncated ones. The EM data were analyzed by image classification and averaging techniques. The preponderant projections, in each case, show near-annular molecules, 6–7 nm in diameter. Comparison of the full-length with the truncated proteins showed molecules similar in size and shape, indicating that their terminal regions are flexible and thus smeared to invisibility in the averaged images. We tested the toroidal model further by calculating resolution-limited projections and comparing them with the EM images. The results support the α-solenoid model, except that they indicate that the repeats are organized not as symmetrical circular toroids but in less regular horseshoe-like structures.
Keywords: negative staining electron microscopy, image classification, circular dichroism, alpha-solenoids
Energy-dependent proteolysis plays a key role in prokaryotic and eukaryotic cells by restricting the availability of certain short-lived regulatory proteins, ensuring the proper stoichiometry for multi-protein complexes, and eliminating misfolded, mislocalized or damaged proteins 1; 2. In eukaryotes, the proteasome is responsible for the degradation of ubiquitin-tagged substrates. The fully assembled 26S particle comprises two subcomplexes: the 20S proteolytic chamber formed of four heptameric rings; and the end-mounted 19S complex that regulates substrate processing. The 19S complex may be subdivided into a base and a lid 3; 4. The distally positioned lid is a complex of eight subunits implicated in deubiquitination and the initial steps of substrate processing 5; 6. The base stacks on to the apical surface of the 20S barrel and consists of a ring of six AAA+ ATPase subunits, Rpt1–6, and four other proteins - Rpn1, Rpn2, Rpn10 and Rpn13 7. Rpn10 and Rpn13 have ubiquitin-binding domains 8; 9 and Rpn13 can also bind the de-ubiquitinating enzyme, Uch37 10. The remaining base components, Rpn1 and Rpn2, are the largest proteasomal subunits and have been reported to interact with several other subunits 8; 9; 11; 12; 13 and auxiliary factors 14; 15 and with each other 16.
Whereas the structure of the 20S core particle is known to high resolution 17, structural information on the 19S regulatory particle remains limited. 3D reconstructions from electron microscopy 18; 19 and tomography 20 have provided an overall frame of reference, but there has been little information on the placement of components other than the AAA+ ring or on the detailed structures of individual subunits, apart from Rpn13 8; 21.
Based on bioinformatic analysis, it was deduced that Rpn1 and Rpn2 both have central regions containing 9–11 tandem pseudo-repeats of an α-helical motif 22. Repeats of this kinds are quite widespread and several subfamilies have been identified, including the so-called HEAT and ARM repeats 23. The Rpn1 and Rpn2 sequence repeats are of a kind associated with proteins of the Proteasome/Cyclosome family 24 and thus are called PC repeats. In tertiary structures, these repeats stack, giving folds that are generically called α-solenoids. Several crystal structures have been determined and the solenoids found to vary in shape from highly curved to nearly straight 25. Based on the periodic occurrence of residues with small side chains, it was anticipated that the α-solenoids of Rpn1 and Rpn2 would be highly curved, and toroidal models were proposed for both 22. In the present study, we aimed to test these predictions by circular dichroism spectroscopy and negative staining electron microscopy as applied to both the full-length proteins and truncated forms, restricted to the PC-containing regions.
Rpn1 and Rpn2, truncated to their PC regions, are alpha-helical monomers
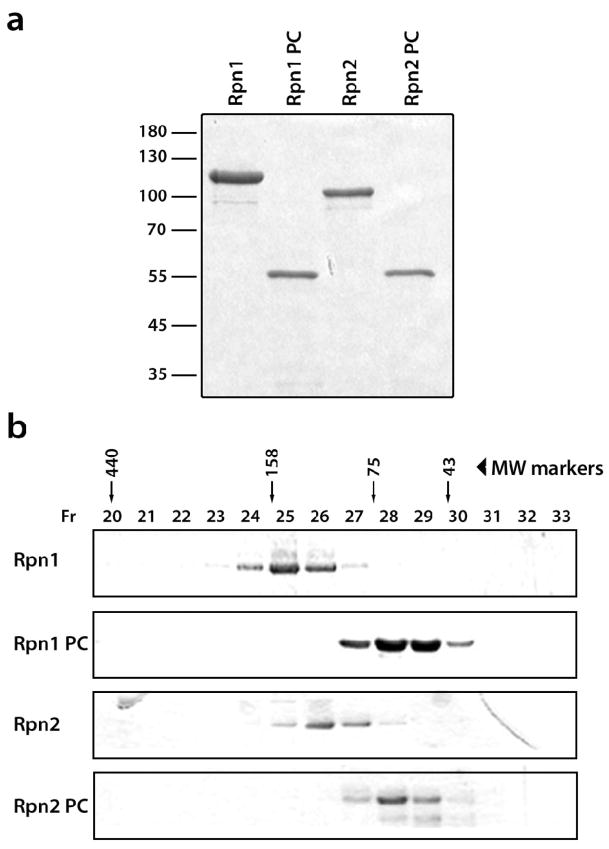
The full-length and truncated forms of Rpn1 and Rpn2 were expressed in E. coli and purified (see Fig. 1 legend). In gel filtration, all four proteins elute at positions corresponding to their monomeric molecular weights (Fig. 1). These data are consistent with prior evidence from analytic ultracentrifugation that the full-length proteins are monomeric in solution 16, and establish that the truncated proteins share this property.
Fig. 1.
Expression and purification of Rpn1 and Rpn2 and truncated versions thereof. (a) SDS-PAGE (10% gel) of purified recombinant proteins stained with Coomassie Blue: Rpn1 (lane 1), Rpn1PC (lane 2), Rpn2 (lane 3), and Rpn2PC (lane 4). (b) Gel-filtration chromatography of Rpn1 and Rpn2. The purified proteins were loaded separately on a Superdex200 column. Molecular mass standards eluted in fractions 20, 24, 27 and 29. The elution profiles are shown in Supplemental Figure 4. i) Using a calibration chart, the peak of Rpn1 was calculated at a mass of 114.7 kDa (calculated monomer mass, 110 kDa); Rpn1PC eluted at 53.5 kDa (calculated monomer mass, 50 kDa); Rpn2 eluted at 100.1 kDa (calculated monomer mass, 104 kDa); and Rpn2PC eluted at 54.0 kDa (calculated monomer mass, 50 kDa). Expression. Vectors were prepared for Rpn1, Rpn1PC (residues 438–876), Rpn2 and Rpn2PC (residues 350–730). The full-length coding sequences of RPN1 and RPN2 or base pairs 1314 to 2628 of RPN1 and 1050–2190 of RPN2 were amplified by PCR from genomic DNA (S. cerevisiae wt strain Sub62), using Pfu DNA Polymerase (Promega). The 5′ and 3′ amplification primers were designed to add BamH1 and Sac1 restriction sites to the ends to facilitate subsequent cloning in the pQE30 vector (Qiagen) for expression with a His6 tag. Clones were verified by DNA sequencing and then subcloned into E. coli M15 for expression. Purification. Transformants were grown at 37°C in liquid LB media supplemented with 1 mM ampicillin to A600 values of 0.6–0.8, followed by 30 min heat shock at 42°C. Next, 0.1 mM isopropyl-1-thio-β-D-galactopyranoside was added for induction and cultures were grown overnight at 16°C, then harvested. Cells were lysed and lysates were clarified at 16,000 rpm for 20 min at 4°C, and the supernatant loaded onto a Ni2+-nitrilotriacetic acid column (Qiagen) previously equilibrated with 50 mM Tris, pH 7.4, 500 mM NaCl, and 10 mM Imidazole. Washes were performed with the same buffer and bound proteins were eluted using 50 mM Tris, pH 7.4, 500 mM NaCl, and 250 mM Imidazole. Samples were then concentrated via a 30 ml Centricon (Vivascience), and further purified by gel filtration on a 24 ml Superdex200 column) eluted with 50 mM Hepes, 150mM NaCl, pH 7.4. The resulting proteins were stored at −80°C. Purity was assessed by Coomassie blue staining and immunoblotting. Protein concentrations were determined using an ND-1000 Spectrophotometer (NanoDrop). Gel Filtration. Gel filtration was performed with a Superdex200 column HR 10/30 (Pharmacia), eluted with 50mM HEPES buffer, pH 7.4, containing 150 mM NaCl; the flow rate was 0.3ml/min. The column was calibrated with the following markers: ovalbomin (43 kDa), conalbumin (75 kDa), aldolase (158kDa), and apoferritin (440 kDa).
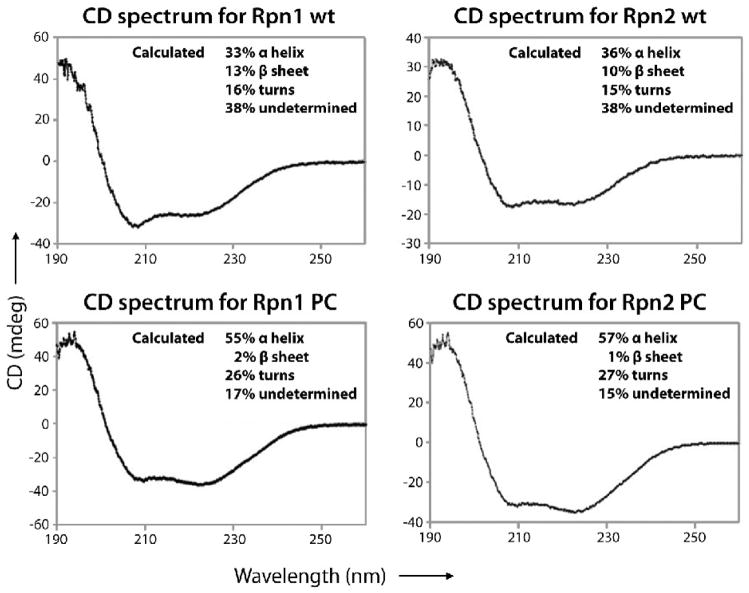
We investigated their secondary structures by circular dichroism spectroscopy. The resulting spectra (Fig. 2), which are characteristic of α-helical proteins 26, were used to estimate their secondary structure contents. In both cases, the truncated proteins have a higher helical content than the full-length proteins (~ 55% vs ~ 35%) and a negligible content of β-sheets. When allowance is made for the presence of inter-helical turns and a reasonable margin of experimental error, these data are consistent with the PC proteins being almost entirely α-solenoidal. This analysis also assigns a substantial amount of non-regular secondary structure to the full-length proteins.
Fig. 2.
Circular dichroism spectra in the far-UV range (190–260 nm) of recombinant Rpn1, Rpn1PC, Rpn2, and Rpn2PC. Prior to CD analysis, each protein was dialyzed (three times, 100 vol. of sample) against 25mM Borate buffer (pH 7.4). Data were recorded using a Jasco J-810 spectropolarimeter set to the amide band (190–260 nm), a protein concentration of 30 μg/ml, a 0.05 cm cell path length, a scan speed of 30 nm/min, a 0.1nm bandwidth, and a 4ms response. All spectra were measured 5 times, averaged and baseline-corrected by subtraction of blank buffer spectrum. The spectra were analyzed on the DichroWeb website 34, using the CDSSTR 35; 36 and SELCON3 37; 38 algorithms and a reference set containing the spectra of 43 proteins with solved structures 39.
Visualized by negative staining, Rpn1 and Rpn2 have central stain-accumulating regions
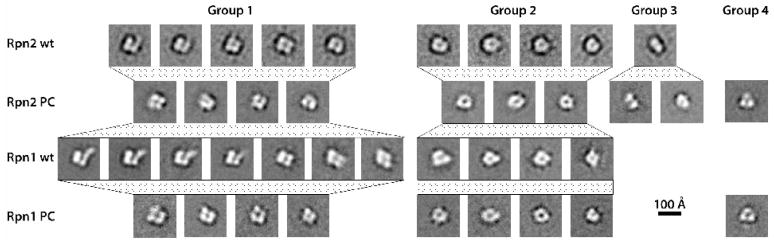
The four proteins were examined by negative staining EM. In all cases, the predominant species observed was small globular particles ( Supp. Fig. 1a–d), many with a stain accumulation at their center. The micrographs were digitized and the resulting data analyzed to distinguish the various classes of particles, and then averaged within each class to improve the signal-to-noise ratio and thus the interpretability of the images. The 10 classes distinguished for full-length Rpn2 are shown in the top row of Figure 3, ordered according to shape similarity. We assign the classes to four groups. Five of the classes (group 1), accounting for ~ 58% of the data, present tetrads of stain-excluding densities, 6 to 7 nm in total diameter, surrounding a central stain accumulation, ~ 1.5 to 2 nm across. The group 1 classes differ in relatively subtle features, such as how clearly densities are resolved from their neighbors or the extent to which their distribution departs from a square. In the leftmost class, the top right density appears to extend out through the surrounding stain layer. The four group 2 classes (~ 30% of the data) show a generally similar molecule, i.e. of about the same size and also with a stain-accumulating center, but with the densities less clearly resolved. The single group 3 class depicts a pair of densities whose length matches the diameter of the other classes. It is plausibly explained as a side-view, in which case its central transverse stripe indicates that the stain-accumulating features at the centers of the other classes represent a hole passing through the molecule.
Fig. 3.
Negative stain electron microscopy. 2D class averages are shown for Rpn2 (row 1), Rpn2PC (row 2), Rpn 1 (row 3) and Rpn1PC (row 4). Microscopy. The purified proteins were brought to final concentrations of 7.5 to 15 μg/mL in 50 mM HEPES and 150 mM NaCl. Grids were prepared by applying 3.5 μl drops to carbon film substrates; after 1 min, blotting away excess sample; then soaking the grid on two successive drops of stain; blotting and allowing to dry. Freshly prepared 1% uranyl formate was used as stain except for Rpn2 (1% uranyl acetate). Micrographs were recorded Kodak SO-163 film, using a Philips CM120 electron microscope operating at 120kV and 45000 to 60,000 magnification. Image analysis. Drift-free micrographs were scanned on a Zeiss scanner at 14 μm step size except for Rpn2 images which were scanned on a Nikon CoolScan 9000 at 6.35 μm. Pixel sizes, after binning the Nikon-scanned data, were between 2.11 and 3.11 Å/pixel. All image preprocessing, including particle boxing and extraction, CTF parameter determination, and correction by phase flipping, was done with the Bsoft package 40. 1025, 1084, 1951, and 961 particles were picked for Rpn2, Rpn2PC, Rpn1, and Rpn1PC. Computation of reference-free class averages was done as described previously 41. Initial sets of 2D class averages were generated with the EMAN package with the Refine2D.py script 42. Class averages were then refined in SPIDER 43 by several rounds of multi-reference alignment, dimension reduction by correspondence analysis, hierarchical ascendant clustering, and averaging until the set of classes obtained was stable from one cycle to the next. The resolution of the class averages varied between 2.1 and 2.9 nm, according to the Fourier ring correlation 44 at 0.5 cutoff.
Analysis of full-length Rpn1 (Fig. 3, row 3) had a similar outcome. Here, the large majority of the data were assigned to 11 classes and again, tetrad-presenting classes predominated (seven of ten classes, with ~ 71% of the data, are in group 1). Compared with Rpn2, more of these images show extensions of the top right density, to varying lengths, in classes 1 to 4 of group 1.
The PC repeat regions form the central cores of Rpn1 and Rpn2
The class averages obtained for the truncated proteins are similar in shape and size to those of the full-length proteins, despite missing nearly half their masses (Fig. 3, rows 2 and 4). Moreover, they can be matched fairly consistently with the group 1 and 2 images defined for the full-length proteins. The truncated proteins also having an abundance of tetrad-presenting particles (40% and 50% of the total data for Rpn2PC and Rpn1PC). Both truncated proteins also have a single triad-presenting class (group 4, ~ 8% of both data sets). However, no class average of either truncated protein showed an “extending arm” domain, as seen for both of the full-length proteins (Rpn2; group 1, class 1 and Rpn1; group1, classes 1 – 4). These observations clearly assign the tetrads of density to the central PC repeat-containing regions of Rpn1 and Rpn2. It is noteworthy that neither truncated protein shows the “extending arm” seen in class averages of the full-length proteins. This observation leads us to infer that the arm is contributed by a terminal region. On the other hand, the variability of this feature implies that the terminal regions of the full-length proteins are relatively flexible.
Modeling the PC repeat regions of Rpn1 and Rpn2 as curved α-solenoids
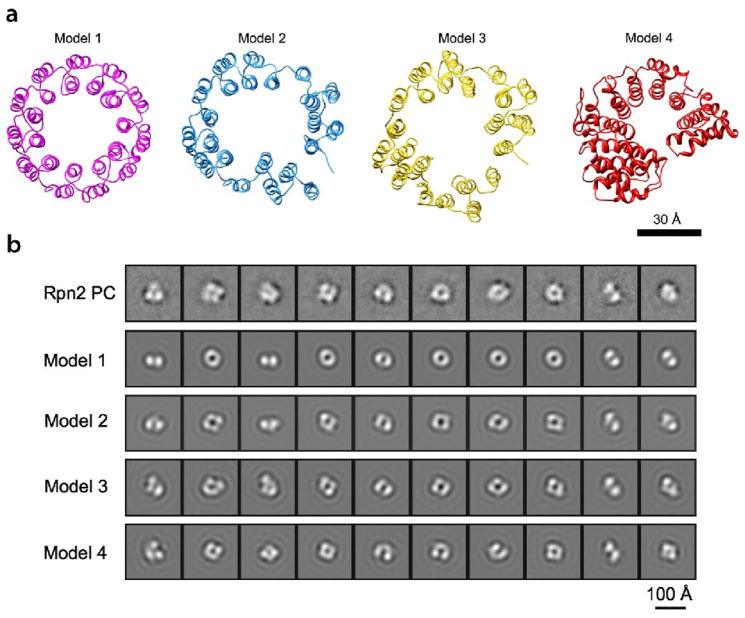
The PC repeat regions of both proteins have been predicted to form toroidal α-solenoids 22. The size of the molecules that we observe (Fig. 3) and their hollow centers are consistent with this prediction. To test it further, we calculated a set of projections corresponding to different views of this model and of other related models (Fig. 4a), and band-limited them to 2.5 nm resolution (Fig. 4b, row 2–5) for comparison with the EM data (Fig. 4b, row 1). The diameters of the EM class averages are slightly more consistent with a 11-repeat model (6.0 nm vs. 5.5 nm for the 9-repeat model, Supp. Fig. 2), although they do not rule out the latter model when the possibility of molecules spreading laterally when dried in negative stain and the limited resolution are borne in mind.
Fig. 4.
Modeling of Rpn2 PC and comparison of the resulting structures with the EM class averages. (a) Ribbon representations of four models. Model 1: a symmetric 11-repeat toroid; Models 2 and 3: manually distorted 11-repeat toroid; Model 4: predicted PC repeat model based on karyopherin-®2 structure (PDB code: 2h4m) (b) Row 1: 10 EM 2D class averages of Rpn2PC (see Fig. 3). Row 2–5: Each model in (a) was band-limited to (2.5 nm)−1 and used to generate a grid of 2D projections. The projection most similar to each EM class average was identified by cross-correlation, and these projections are shown in rows 2 to 5. Modeling. PDB coordinates for model 1 (Fig. 4) were provided by Dr. A.V. Kajava. Models 2 and 3 were generated manually from this model by distorting it in Chimera 45. In an alternative approach, amino acid sequences of Rpn1 and Rpn2 were probed for structural homologs 46, and segments of karyopherin β2 (PDB code: 2h4m 47) were identified as the most likely homolog. Model 4 was obtained by running the sequence alignment from the prediction server into Modeller 48. For comparison with the EM class averages, the 4 models were band-limited to 2.5 nm resolution and projected on an evenly spaced grid of viewing orientations. To simulate EM image formation, a CTF corresponding to the imaging conditions was applied to the 2D projections. They were then compared to the EM class averages by multi-reference alignment and the projection most similar to each class average was identified by cross correlation.
The most pronounced feature of the EM class averages is the resolution of the ring into four (or three) densities of approximately equal size. This feature is not reproduced in any viewing direction of a symmetrical circular toroid (Fig. 4b, row 2). However, it may be produced by distorting the toroid in any of several ways. Models 2 and 3 in Figure 4a were generated by distorting the 11-repeat circular planar toroid and slightly opening the ring at the gap where the N and C-termini of the PC repeat region meet. We also generated another 11-repeat model for Rpn2PC, using structure of the nuclear transport protein, karyopherin β2 - a known α-solenoid 27 - as template (model 4). Near-axial projections of models 2 to 4 have an overall horseshoe shape and, at lower resolution, display tetrads of density (Fig. 4b, row 3–5), in agreement with the EM class averages (Fig. 4b, row 1).
We conclude that this analysis supports the prediction that the PC repeat portions of Rpn2 and Rpn1 (Supp. Fig. 3) form monomeric α-solenoids with highly curved folds, although they appear to be less regular than in the simple symmetrical model 22. The slightly opened model that we favor over a closed one (model 3), allows the N- and C-terminal regions to extend away from the PC repeat regions.
Implications for 19S structure
In a previous study 16, isolated molecules of full-length Rpn1 and Rpn2 imaged by AFM were reported to have a thickness and annular appearance consistent with the toroidal α-solenoid model 22. AFM affords precise height measurements but tip convolution effects compromize its lateral resolution. Nevertheless, our class-averaged EM images and modeling experiments support and extend this conclusion. In particular, the close resemblance between the images of the full-length and the truncated proteins assigns the α-solenoid fold to the PC repeat-containing portions of these molecules. Moreover, the “extending arm” seen only with full-length proteins, particularly Rpn1 (Fig. 3, row 3), is likely to represent part of their terminal regions. As the C-terminal part of Rpn1 is rather short (117 residues), its “extending arm” is probably contributed by the N-terminal region (438 residues). Our EM class average images of Rpn1 and Rpn2 also show some similarities with the Blm10 protein of S. cerevisiae, an activator that binds to the 20S proteasome, and is composed almost entirely of HEAT repeats 28. In negatively stained class averages, isolated Blm10 molecules exhibit a variety of coiled shapes of a filament of about the same thickness as we observe for Rpn1 and Rpn2 29. However, its shape appears to be specifically defined when bound to the distal surface of the 20S particle 30.
As to the deployment of Rpn1 and Rpn2 in the 19S particle, two models are currently in play. In one of them, based on AFM and cross-linking 16, these proteins’ toroids stack axially through the center of the ATPase ring, with Rpn2 making contact with the apical surface of 20S and Rpn1 protruding on the distal side. Size considerations of the axial holes through AAA+ hexamers of known structure indicate that if the Rpt ring is similarly configured, it cannot accommodate a 6 nm ring; for instance, in FtsH (PDB: 2DHR 31) and P97 (PDB: 1R7R 32), whose AAA+ modules are ~ 45% sequence-similar to Rpt1, this hole is only ~ 2 nm across, widening to ~ 4 nm at the distal surface. However, it is not ruled out that Rpn2 may adjust its shape when incorporated into the 19S particle; for example β-karyopherin, another alpha-solenoid, exhibits considerable plasticity 25.
Alternatively, in their negative stain reconstruction of the human 26S proteasome, da Fonseca & Morris assigned an extended peripheral density in the base region to Rpn1 or Rpn2 19. Similar densities were observed in an earlier cryo-EM analysis of the 26S proteasome 18. This position is also in line with a proposal based on comparing averaged side-views of the 26S proteasome with those of the 20S-PAN ATPase complex, which has no Rpn1 or Rpn2 33. Part of the peripheral density of da Fonseca & Morris (their Figure 4a-ii) appears to be approximately annular and about 6 nm in diameter, raising the possibility that it might represent the PC repeat region of Rpn1 or Rpn2. However, settling this important issue will require more detailed structural information, of which cryo-EM at higher resolution and/or specific labeling experiments, appear to be likely potential sources.
Acknowledgments
We thank Dr A. V. Kajava for providing PDB coordinates for his models. This work was supported by the Intramural Research Program of NIAMS and by grants from the Israel Science Foundation and the USA-Israel Binational Science Foundation to M.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zwickl P, Baumeister W, Steven A. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–50. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–9. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 4.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–62. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrell K, Wilkinson CR, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem Sci. 2000;25:83–8. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- 8.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–8. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–61. [PubMed] [Google Scholar]
- 10.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–53. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 12.Gorbea C, Taillandier D, Rechsteiner M. Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J Biol Chem. 2000;275:875–82. doi: 10.1074/jbc.275.2.875. [DOI] [PubMed] [Google Scholar]
- 13.Davy A, Bello P, Thierry-Mieg N, Vaglio P, Hitti J, Doucette-Stamm L, Thierry-Mieg D, Reboul J, Boulton S, Walhout AJ, Coux O, Vidal M. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–8. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–30. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 15.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–22. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 16.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–80. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groll M, Ditzel L, Löwe J, Stock D, Böchtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–71. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 18.Nickell S, Beck F, Korinek A, Mihalache O, Baumeister W, Plitzko JM. Automated cryoelectron microscopy of “single particles” applied to the 26S proteasome. FEBS Lett. 2007;581:2751–6. doi: 10.1016/j.febslet.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 19.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–14. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickell S, Mihalache O, Beck F, Hegerl R, Korinek A, Baumeister W. Structural analysis of the 26S proteasome by cryoelectron tomography. Biochem Biophys Res Commun. 2007;353:115–20. doi: 10.1016/j.bbrc.2006.11.141. [DOI] [PubMed] [Google Scholar]
- 21.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–52. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajava AV. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J Biol Chem. 2002;277:49791–8. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 23.Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 24.Lupas A, Baumeister W, Hofmann K. A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome (anaphase-promoting complex) Trends Biochem Sci. 1997;22:195–6. doi: 10.1016/s0968-0004(97)01058-x. [DOI] [PubMed] [Google Scholar]
- 25.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–44. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Martin SR, Schilstra MJ. Circular dichroism and its application to the study of biomolecules. Methods Cell Biol. 2008;84:263–93. doi: 10.1016/S0091-679X(07)84010-6. [DOI] [PubMed] [Google Scholar]
- 27.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–7. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 28.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146:425–30. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 30.Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J. Structure of the Blm10–20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–59. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suno R, Niwa H, Tsuchiya D, Zhang X, Yoshida M, Morikawa K. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol Cell. 2006;22:575–85. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6 Å. J Struct Biol. 2003;144:337–48. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–2. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 35.Compton LA, Johnson WC., Jr Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem. 1986;155:155–67. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 36.Manavalan P, Johnson WC., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 38.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–80. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 40.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–7. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 43.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 44.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127:127–38. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–8. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 47.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–58. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–26. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]