Abstract
The mammalian Sonic hedgehog (Shh) signalling pathway is essential for embryonic development and the patterning of multiple organs. Disruption or activation of Shh signalling leads to multiple birth defects, including holoprosencephaly, neural tube defects and polydactyly, and in adults results in tumours of the skin or central nervous system. Genetic approaches with model organisms continue to identify novel components of the pathway, including key molecules that function as positive or negative regulators of Shh signalling. Data presented here define Tulp3 as a novel negative regulator of the Shh pathway. We have identified a new mouse mutant that is a strongly hypomorphic allele of Tulp3 and which exhibits expansion of ventral markers in the caudal spinal cord, as well as neural tube defects and preaxial polydactyly, consistent with increased Shh signalling. We demonstrate that Tulp3 acts genetically downstream of Shh and Smoothened (Smo) in neural tube patterning and exhibits a genetic interaction with Gli3 in limb development. We show that Tulp3 does not appear to alter expression or processing of Gli3, and we demonstrate that transcriptional regulation of other negative regulators (Rab23, Fkbp8, Thm1, Sufu and PKA) is not affected. We discuss the possible mechanism of action of Tulp3 in Shh-mediated signalling in light of these new data.
INTRODUCTION
Birth defects remain the greatest cause of infant mortality in the Western world. These developmental anomalies can affect any organ in the body and are often severely disabling or fatal. In the UK, over 90 000 pregnancies per year (∼2.3%) are affected by a major congenital malformation, and amongst the most common anomalies are defects of the nervous system and limbs, which are observed in 10 and 16% of affected fetuses, respectively (1). Reducing the clinical burden from birth defects requires a more complete understanding of the causative factors, including knowledge of the genetic, cellular and molecular mechanisms involved, in order to permit the development of appropriate preventative therapies. Studies with model organisms are essential in helping to unravel the complex network of processes involved in normal development, and the identification of mutant animals provides key insights into the pathways that are involved in these disorders. Mutagenesis of mice with ethylnitrosourea (ENU) has proven to be a powerful method for the generation of new mutations that provide good models of human developmental disorders (2–4). Investigation of these mutants promises to shed new light on the causes of these defects as well as identifying novel components of known signalling pathways.
Development of the mammalian nervous system involves the initial formation of the neural tube, the precursor of the brain and spinal cord, and the subsequent differentiation of the neural tissue into spatially appropriate neuronal cell types. The rolling up and closure of the neural plate to form the neural tube constitutes the process of neurulation, and this occurs with a characteristic sequence of events and varying neural fold morphology in different regions of the embryo (5–7). Initiation of neural tube closure occurs at the level of the cervical/hindbrain boundary (Closure 1) at E8.5 in mouse, and continues from here both cranially into the hindbrain and caudally along the developing spine. Two further sites of closure initiation are observed in the cranial region, at the forebrain–midbrain boundary (Closure 2) and at the most rostral extent of the forebrain (Closure 3). Neurulation in the head is completed with closure of the intervening regions, termed the anterior neuropore and hindbrain neuropore, by E9.5. Neural tube closure continues until E10.5 in a cranio-caudal direction from the site of Closure 1 along the developing spine. The region immediately caudal to the closed neural tube, in which the neural folds are elevating and about to close, is termed the posterior neuropore (PNP). The morphology of the neural folds within the PNP varies along the spinal axis, and this has led to the characterization of three modes of spinal neurulation (6). In mode 1 (E8.5–E9), the neural plate exhibits pronounced bending at the ventral midline, the median hinge point (MHP), but straight lateral folds, whereas in mode 2 (E9–E9.75) the neural folds bend both at the MHP and also at paired dorso-lateral hinge points (DLHPs). Closure of the upper sacral level involves mode 3 neurulation (E9.75–E10.5), in which the neural plate bends solely at the DLHPs.
The Sonic hedgehog (Shh) signalling pathway is essential for the normal growth and patterning of many organs including the nervous system and limbs (8,9). Disruption of Shh signalling results in a range of congenital defects, including holoprosencephaly, craniofacial abnormalities and skeletal malformations (8,9). Conversely, inappropriate activation of the pathway leads to neural tube defects, polydactyly, basal cell carcinomas and tumours of the central nervous system (7,10,11). Shh plays a key role in regulating closure of the neural tube since Shh expression in the notochord is necessary and sufficient to inhibit formation of DLHPs, through a mechanism involving antagonism of Bmp signalling (12,13). Shh signalling from the notochord and, subsequently, the floor plate is also important for correct dorso-ventral (DV) patterning of the nervous system (14–18). The spatially restricted specification of neuronal subtypes is essential for the subsequent assembly of functional neuronal circuits. High concentrations of Shh induce floor plate markers, whereas lower concentrations specify increasingly more dorsal interneuron subtypes within the ventral neural tube. Absence of Shh signalling results in loss of ventral markers and complete dorsalization of the spinal cord, whereas ectopic Shh signalling leads to expansion of ventral markers at the expense of more dorsal marker domains. Patterning along the anterior–posterior (AP) axis of the limb is also dependent on the Shh pathway (19–21). Shh is expressed in the zone of polarizing activity (ZPA) in the posterior domain of the limb buds, and the spatial and temporal gradients of Shh activity across the limb determine digit number and identity. Increased or ectopic Shh activity leads to polydactyly, whereas decreased Shh activity results in narrowed limbs and reduced number of digits.
The vertebrate Shh pathway shares fundamental similarities with that of Drosophila (22–24). Hh ligand binds to Patched (Ptch) receptor, releasing the repressive activity of Ptch on Smoothened (Smo) and, thereby, allowing Smo to initiate intracellular signalling. Downstream of Smo the signal culminates with the activation of the Ci/Gli transcriptional activators and removal of the Ci/Gli repressors. Vertebrates express three Gli family members, Gli1–3; Gli1 functions as a transcriptional activator, whereas Gli2 and Gli3 can act both as transcriptional activators and, following proteolytic removal of the C-terminal region, as transcriptional repressors. The relative importance of the Gli activator and repressor forms differs in the patterning of the neural tube and limb. Neural tube patterning is mostly dependent on ventral Gli1 and Gli2 activators, with less important roles of Gli3 repressor dorsally (15,25–29). In contrast, limb patterning is generated predominantly by Gli3, with opposing gradients of Gli3 activator and Gli3 repressor (30–32). Although vertebate and Drosophila hedgehog pathways are grossly similar, the intermediate steps between the release of Smoothened and the activation or processing of Ci/Gli are distinct between species and, indeed, are not yet fully understood (9,22,33,34).
Forward genetic screens in mouse have successfully identified additional components of the vertebrate Shh pathway, with the advantage of no prior bias on the type of molecule that may be involved (35,36). A number of positive regulators of Shh signalling have recently been discovered, based on loss of ventral cell types in the neural tube, including proteins involved in the formation/maintenance of the primary cilia or intraflagellar transport (37). Several Shh pathway components localize within these subcellular organelles (38–42), and the general consensus is that some aspects of vertebrate Smo-mediated Gli processing occur within cilia. Specific proteins have been revealed as negative regulators of the pathway, since disruption of their function leads to activation of Shh signalling, as observed by the expansion of ventral marker domains within the neural tube. The known negative regulators include Ptch1, Sufu, protein kinase A (PKA), Rab23, Fkbp8 and Thm1 (42–51). PKA phosphorylates Gli3 prior to processing by the ubiquitin-proteasome pathway (52), whereas Sufu inhibits the transcriptional activity of Gli activators in the nucleus and may also sequester Gli in the cytoplasm (49,50,53,54). Thm1 is involved in intraflagellar transport (42). The functions of Rab23 and Fkbp8 are not clear, although they may have roles in vesicular trafficking (55,56). Clearly, the vertebrate Shh pathway is complex, and our incomplete understanding of the processing and regulation of intermediate steps suggests that additional components remain to be discovered.
Here, we report a new ENU-induced mouse mutant that we have named hitchhiker (hhkr), which exhibits a range of developmental abnormalities including neural tube defects and polydactyly. We reveal that hitchhiker carries a strongly hypomorphic mutation in Tulp3, and we show that mutants have ventralization of the caudal spinal cord. We demonstrate that Tulp3 functions genetically downstream of Shh and Smo, to give activation of Shh pathway targets in the absence of Shh and Smo; complementary results are reported in an accompanying paper (Norman et al., submitted for publication). In addition, we show that the hitchhiker mutant exhibits a genetic interaction with Gli3 in limb development, but does not substantially affect Gli3 processing or expression. We demonstrate that transcriptional regulation of other negative regulators (Rab23, Fkbp8, Thm1, Sufu and PKA) is not affected in hitchhiker mutants. We propose that Tulp3 functions as a novel negative regulator of the Shh pathway and we discuss the possible mechanism of action of Tulp3 in Shh-mediated signalling in light of these new data.
RESULTS
hitchhiker is a novel mutant with defects in neural tube and limb development
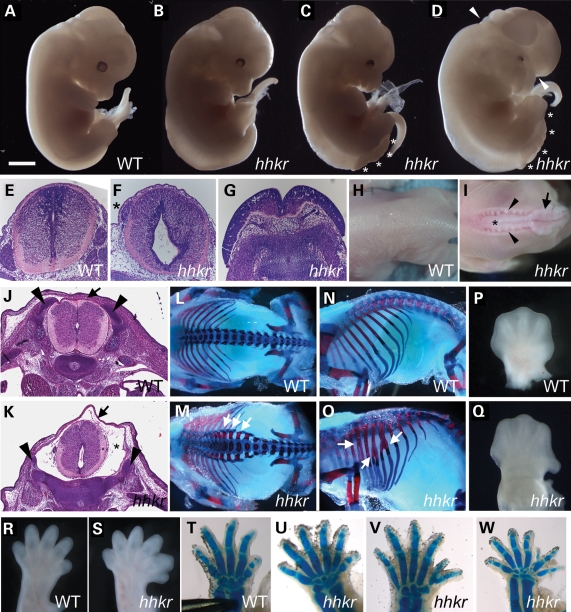
hitchhiker (hhkr) arose during an on-going screen for recessive ENU-induced mutations that affect the morphology of mid-gestation embryos. The mutant was initially identified through the presence of neural tube defects, with exencephaly and/or lumbosacral spina bifida aperta, and mutants also commonly exhibited oedema (Fig. 1A–D). Histological sections revealed abnormal morphology of the lower spinal neural tube in mutants in which the neural tube had closed, with an expanded lumen and reduced thickness of the neuroepithelium, particularly in the ventral half of the spinal cord (Fig. 1F, compare with E). Mutants also demonstrated oedema in the surrounding tissue, and small, dorsally misplaced dorsal root ganglia (Fig. 1F). Sections through other mutants demonstrated the widely splayed neural folds characteristic of spina bifida (Fig. 1G). Some hitchhiker fetuses exhibited a closed thoracolumbar neural tube but which was not enclosed within the bony vertebral column, either with or without lumbosacral spina bifida (Fig. 1I, compare with H), and this phenotype may be similar to the human defect of spina bifida occulta. Histological sections through E15.5 hitchhiker embryos with this phenotype confirmed the presence of a closed neural tube, but with a grossly enlarged spinal canal, a thin dorsal covering of surface ectoderm and flanked by the widely spaced cartilage primordia of the developing vertebrae (Fig. 1K, compare with J). Skeletal preparations of E18.5 fetuses with the exposed thoracolumbar neural tube (without spina bifida) clearly demonstrated splayed vertebrae in the thoracolumbar region (Fig. 1M, compare with L) and also revealed abnormal morphology of the ribs, commonly with bifurcations (Fig. 1O, compare with N). Since the ribs, vertebrae and dorsal dermis all derive from the somites (57), the observation of defects in these tissues might suggest possible abnormalities in somitic development in hitchhiker.
Figure 1.
hitchhiker mutant mouse embryos exhibit defects in neural tube, skeleton and limb development. (A–D) E13.5 wild-type (A) and hitchhiker embryos (B–D) with a range of phenotypes, including oedema (B), oedema with spina bifida (C) and oedema with spina bifida and exencephaly (D). Asterisks mark region with spina bifida and exencephalic region is marked with arrowheads. (E–G) Transverse sections stained with haematoxylin and eosin (H&E) through spinal neural tissue in wild-type (E), mutant with caudal oedema and dorsally positioned dorsal root ganglia (asterisk) (F) and mutant with spina bifida (G). (H and I) Dorsal views of E18.5 wild-type (H) and hitchhiker mutant (I) fetuses demonstrating splayed vertebrae (arrowheads) and visible neural tube (asterisk) adjacent to a region of open spina bifida (arrow) in mutants. (J and K) H&E-stained sections through the thoracolumbar region of E15.5 wild-type (J) and hitchhiker (K) fetuses. The mutant exhibited the splayed vertebrae phenotype, and sections demonstrate a closed but abnormally shaped neural tube within a grossly enlarged spinal canal (asterisk), covered dorsally with a thin layer of surface ectoderm (arrow) and flanked by widely spaced vertebral primordia (arrowheads), compared with wild-type. (L–O) Skeletal preparations of E18.5 wild-type (L and N) and hitchhiker mutant (M and O) fetuses stained with alcian blue for cartilage and alizarin red for bone, demonstrating splayed vertebrae (M; arrows) and rib bifurcations (O; arrows) in mutants. (P and Q) Forelimbs from wild-type (P) and hitchhiker mutants (Q) at E13.5, demonstrating malposition of digit 1 in mutants. (R and S) E15.5 wild-type (R) and hitchhiker (S) limb buds showing preaxial polydactyly with six digits. (T–W) E16.5 skeletal preparations for wild-type forelimb (T) and hitchhiker mutants (U–W) demonstrating preaxial polydactyly with a range of phenotypes. Limbs are oriented with anterior to right. Scale bars: A–D and H: 2.5 mm; E–G: 0.5 mm; H and I: 5 mm; J and K: 1 mm; L–O: 4 mm; P and Q: 1 mm; R and S: 1.1 mm; T–W: 1.2 mm.
The hitchhiker line was named after a limb defect, in which the first digit exhibited an abnormal backward-bent position (Fig. 1P and Q). More often, mutants exhibited preaxial polydactyly on both fore- and hindlimbs, with six or occasionally seven digits (Fig. 1R and S). Skeletal preparations revealed a spectrum of defects, including apparent bifurcation or duplication of digit one, or full or partial duplication of another digit (Fig. 1T–W and data not shown).
The observed phenotypes were all partially penetrant and occurred in any combination: 68% of fetuses exhibited polydactyly (n = 73; ≥E12.5); 65%, spina bifida aperta; 58%, oedema; 37%, exencephaly; 8%, spina bifida occulta and 5% no detectable defect (n = 126, ≥E10.5). Approximately 6% of heterozygotes exhibited a mild head misshaping or slight caudal oedema.
hitchhiker carries a splice site mutation in Tulp3 causing a strongly hypomorphic function
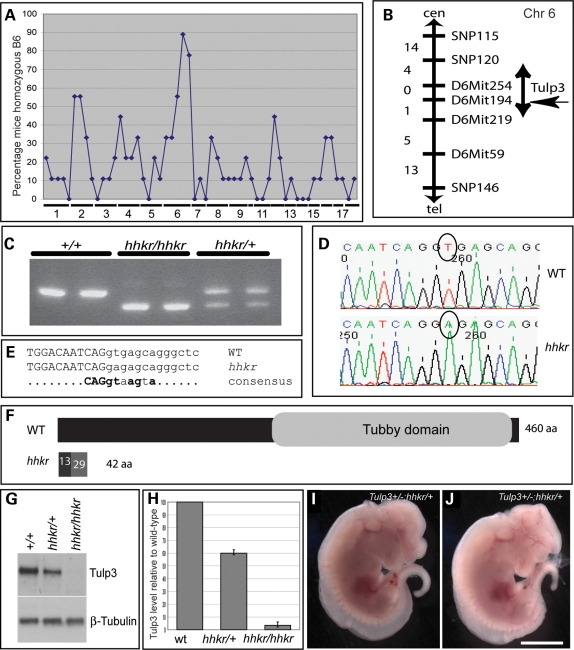
Genetic mapping using a genome-wide marker panel and nine affected individuals revealed linkage of hitchhiker to Chromosome 6 (Fig. 2A), and analysis with additional markers and individuals refined the interval to 12 Mb (Fig. 2B). Within this region lies Tulp3; since a targeted allele of Tulp3 exhibits neural tube defects similar to hitchhiker (58), we examined Tulp3 in our mutant. RT–PCR analysis revealed a reduced size for one Tulp3 amplicon (Fig. 2C), and sequencing revealed a deletion of 52 bp corresponding precisely to exon 2 (data not shown). Genomic DNA analysis identified a single nucleotide substitution at the highly invariant position 2 of the splice donor site immediately 3’ to exon 2 (Fig. 2D and E) (59).
Figure 2.
hitchhiker carries a splice site mutation in Tulp3 resulting in severely reduced protein expression and function. (A) Genome-wide scan with 55 SNP markers on nine affected fetuses, showing percentage of mice typing as homozygous C57BL/6 at each marker; hitchhiker shows linkage to Chromosome 6. (B) Genetic map following analysis with 67 affected fetuses; numbers on left indicate genetic separation of markers (in centimorgan). hitchhiker maps to 12 Mb (5 cM) region between SNP120 and D6Mit219, a region containing Tulp3. (C) Gel of RT–PCR products of Tulp3 amplified between exons 1 and 3, from wild-type, homozygous and heterozygous mutants, showing reduced amplicon size in hhkr. (D) Sequence trace from genomic DNA revealed single nucleotide substitution, T in wild-type to A in hhkr (circled). (E) Comparison with consensus splice sequence (59) reveals hhkr mutation occurs in the splice junction immediately 3’ to exon 2. Uppercase: exon, lowercase: intronic sequence, bold: invariant. (F) Schematic representation of Tulp3 protein in wild-type and predicted truncation following exon 2 deletion in hitchhiker. (G) Western blot analysis on total cell lysates from whole E9.5 embryos demonstrating reduced Tulp3 protein in heterozygous embryos and almost complete absence in homozygous mutant embryos, relative to wild-type littermates. (H) Quantitation of Tulp3 expression in heterozygous and mutant embryos, normalized to β-tubulin, shown as a percentage of wild-type; n = 3 blots, E9.5 or E11.5 whole embryos. (I and J) Complementation test between hitchhiker and the Tulp3-targeted allele, Tulp3tm1Jmg (58) generated double-mutant embryos with exencephaly, spina bifida and oedema, E12.5. Scale bar: I and J: 2.5 mm.
Deletion of exon 2 creates a frameshift that is predicted to cause premature truncation of the protein (Fig. 2F) and a null phenotype. Western blots confirmed a dramatic reduction in Tulp3 protein in hitchhiker homozygotes, although long exposures detected Tulp3 at ∼4% of the wild-type level (Fig. 2G and H), suggesting that hitchhiker is a strong hypomorph of Tulp3 rather than a complete null. Tulp3 protein was reduced to 60% wild-type level in heterozygotes (Fig. 2G and H). To further verify that Tulp3 mutation is responsible for the hitchhiker phenotype, we intercrossed hitchhiker heterozygotes with mice heterozygous for the Tulp3-targeted allele, Tulp3tm1Jng (58), with both strains on a C3H background. This yielded embryos exhibiting the range of phenotypes displayed by either homozygous mutant (Fig. 2I and J; n = 8), indicating a failure of complementation. The phenotype of both the targeted allele homozygotes and the double heterozygous embryos appears to be slightly more severe than that of hitchhiker homozygotes, with more extensive oedema and defects in eye development. Together, these data indicate that hitchhiker is a strong hypomorph of Tulp3.
Neural tube defects in hitchhiker result from a failure of brain and PNP closure
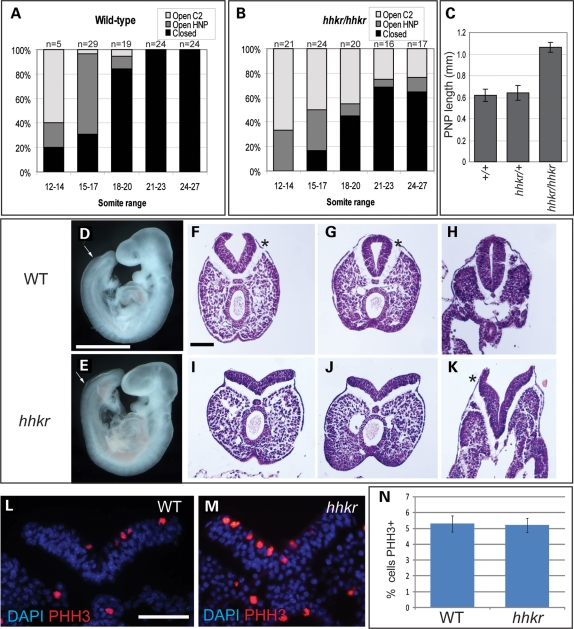
Following identification of the mutant gene, we conducted detailed morphological and histological examination of mutant embryos during the stages of neurulation in order to determine the cause of the neural tube defects. In the cranial region, mutant embryos exhibited normal de novo closure at the future cervical region (Closure 1) and the rostral extent of the forebrain (Closure 3) (data not shown). In contrast, the occurrence of Closure 2, at the forebrain–midbrain boundary, was delayed or failed: 22% of hitchhiker embryos exhibited failure of Closure 2 even at the 24–27 somite stage, while a further 15% had achieved Closure 2 but retained an open hindbrain neuropore at this stage, in contrast to the complete head closure seen in wild-type embryos by the 21 somite stage (Fig. 3A and B). The exencephalic phenotype observed in 37% of fetuses at later stages, therefore, corresponds precisely to the earlier failure of either Closure 2 or hindbrain neuropore closure.
Figure 3.
Exencephaly and spina bifida in hitchhiker result from a failure of neural tube closure. (A and B) Proportion of embryos demonstrating completely closed cranial neural tube (black bar), initiation of head closure at Closure 2 with open hindbrain neuropore (dark grey) or open cranial region with failure of Closure 2 (light grey), within the somite ranges marked, in wild-type (A) and hitchhiker (B) embryos. Number of embryos within each group is shown at the top of each column. (C) Length of PNP in wild-type, heterozygote and hitchhiker mutant embryos, at 22–24 somite stage, in millimetre; mean ± SEM. (D and E) Lateral view of E9.5 (20 somite) wild-type (D) and hitchhiker (E) embryos demonstrating enlarged PNP in mutants; anterior limit of PNP is marked with arrows. (F–K) Transverse H&E-stained sections through the caudal end of 24 somite wild-type (F–H) and hitchhiker (I–K) embryos, within the PNP (F, G, I and J), or closed neural tube (H). Wild-type embryos exhibit Mode 2 neurulation with clear DLHPs (asterisks), whereas hitchhiker embryos have reduced DLHPs that are apparent only adjacent to the closed neural tube (K, asterisk). (L and M) Immunostaining for phosphorylated histone H3 to examine cell proliferation in the caudal neuroepithelium of E9.5 wild-type (L) and hitchhiker (M) embryos. Red; phosphorylated histone H3; blue, DAPI. (N) Quantitative analysis of cell proliferation in the neuroepithelium, displayed as percentage of cells stained for phosphorylated histone H3, revealing no significant difference in percentage proliferation in hitchhiker caudal ends. Scale bar: D and E: 1 mm; F–M: 100 µm.
In the spinal region, embryos displayed a defect in PNP closure. From around the 17-somite stage, hitchhiker mutants exhibited a significantly enlarged PNP, compared with stage-matched wild-type and heterozygous littermates (Fig. 3C–E). In wild-types and heterozygotes, the PNP remained open in only 25% of embryos by the 27 somite stage (n = 4 of 20), whereas 72% of hitchhiker mutants exhibited an open PNP at this stage (n = 13 of 18). These figures correspond closely to the spina bifida observed in later embryos, both in the incidence (65%) and the region affected; at later stages, hitchhiker embryos exhibited an open neural tube corresponding to the region between somites 24 and 34. More caudally, the spinal neural tube appeared normal, suggesting that secondary neurulation proceeds unhindered in hitchhiker. Thus, in the spinal region as in the cranial region, we have demonstrated that neural tube defects occur as a primary defect in neural tube closure.
Previous detailed analysis of spinal neural tube closure has documented three ‘modes’ of closure, characterized by the precise morphology of the neural folds (6). Histological sections through the PNP revealed abnormal morphology in hitchhiker embryos. At the 24 somite stage, wild-type and heterozygous embryos exhibited prominent DLHPs, characteristic of mode 2 neurulation (Fig. 3F–H). In contrast, hitchhiker mutants exhibited a reduction in this dorso-lateral bending, with small DLHP seen only in the sections immediately adjacent to the closed neural tube and straight neural folds observed more caudally (Fig. 3I–K). The rates of cell proliferation and cell death in the neuroepithelium of the caudal neural tube at E9.5 were not significantly different between hitchhiker mutants and wild-type littermates (Fig. 3L–N and data not shown). These results suggest that the abnormal neural fold morphology may be causative for the delay in PNP closure in hitchhiker mutants.
hitchhiker mutants exhibit increased Shh pathway activity during spinal neural tube closure
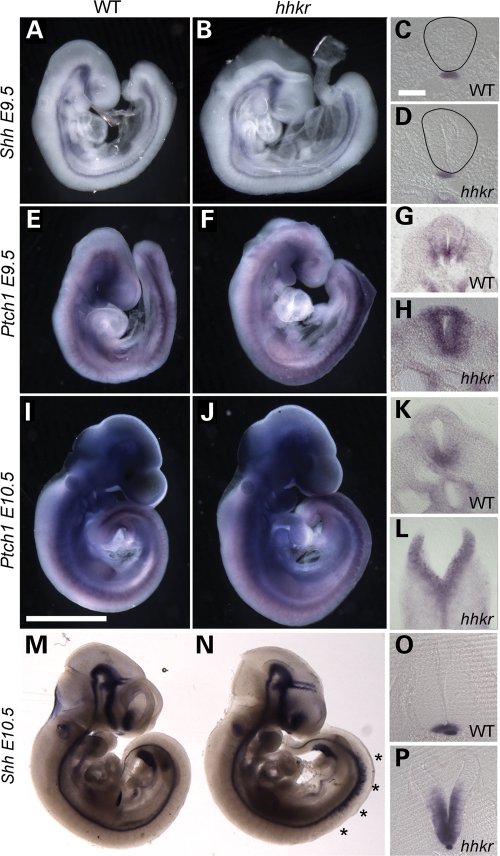
To investigate the cause of the neuroepithelial defects, we examined the expression of Shh, a key molecule involved in the regulation of neural tube closure. Strong expression of Shh in the notochord represses DLHP formation during mode 1 neurulation, whereas weaker Shh activity during mode 2 neurulation permits DLHPs to form (13). By whole mount in situ hybridization, Shh expression appeared comparable between hitchhiker mutants and wild-type embryos at E9.5 (Fig. 4A and B), and transverse sections through the caudal region revealed similar intensities of expression in the notochord (Fig. 4C and D). At this stage and axial level, Shh expression is not yet induced in the wild-type floor plate, and Shh is similarly absent from the hitchhiker floor plate (Fig. 4C and D). To investigate the activity of the Shh pathway, as opposed to the expression of Shh ligand, we examined the expression of the transcriptional target of Shh signalling, Ptch1. Whole mount in situ hybridization revealed a dramatic up-regulation of Ptch1 expression in the caudal region of the neural tube in hitchhiker mutants (Fig. 4E and F). Transverse sections through these embryos demonstrated an expanded domain of Ptch1 expression throughout the DV extent of the caudal neural tube in hitchhiker, compared with wild-type (Fig. 4G and H). Quantitative analysis by real-time RT–PCR (qRT–PCR) detected a 1.65-fold increase in Ptch1 expression in the caudal third of E9.5 embryos, compared with wild-type (Supplementary Material, Fig. S1A). In addition, a similar qRT–PCR analysis of Gli1, another transcriptional target of the Shh pathway, detected a 1.5-fold increase in Gli1 expression in hitchhiker (Supplementary Material, Fig. S1B; P < 0.05). These data suggest that the activity of the Shh pathway is increased in hitchhiker mutants, at E9.5, independently of a change in Shh expression. We suggest that this may inhibit the formation of DLHPs.
Figure 4.
hitchhiker mutants exhibit increased Ptch1 expression prior to increased Shh expression. E9.5 (A–H) and E10.5 (I–P) embryos following wholemount in situ hybridization with riboprobes for Shh (A–D and M–P) or Ptch1 (E–L). (A–D) At E9.5, Shh expression is comparable in wild-type (A) and hitchhiker (B) embryos, and transverse sections reveal expression is confined to the notochord (C and D). (E–H) Ptch1 expression at E9.5 is increased in hitchhiker mutants (F) compared with wild-type (E), in the caudal half of the embryo. Transverse sections demonstrate up-regulated and expanded expression in the caudal neural tube of mutant (H) compared with wild-type (G). (I–L) At E10.5, Ptch1 expression remains greatly enhanced at the caudal end of hitchhiker embryos (J and L) compared with wild-type (I and K). (M–P) At E10.5, ectopic Shh expression is evident in hitchhiker embryos along approximately one-third of the spinal cord, anterior to the hindlimb (N, asterisks, compare with M). Transverse sections reveal greatly expanded Shh expression domain within the caudal neural tube of hitchhiker (P) compared with wild-type (O). Scale bars: A, B, E, F, I, J, M and N: 1 mm; C, D, G, H, K, L, O, P: 100 µm.
Further analysis at E10.5 revealed continued overexpression of Ptch1 in hitchhiker mutants, compared with wild-type littermates. At this stage as at E9.5, the most dramatic up-regulation of Ptch1 expression was observed in the caudal neural tube, but ectopic expression was also evident in the caudal somites (Fig. 4I–L). Although Shh expression looked normal at E9.5, by E10.5 Shh is ectopically expressed in hitchhiker mutants, in approximately one-third of the spinal cord anterior to the hindlimb bud (Fig. 4M and N; asterisks). Transverse sections through the lumbar neural tube demonstrated a dramatic increase in the Shh expression domain. Shh expression extends throughout the ventral half of the neural tube in hitchhiker mutants, whereas it is restricted to the floor plate in wild-type embryos (Fig. 4O and P). The spatial and temporal relationship between the onset of Ptch1 and Shh overexpression suggest that in hitchhiker mutants, activation of the Shh pathway occurs independently of increased Shh ligand, which then subsequently (and indirectly) induces the ectopic expression of Shh within the neural tube.
hitchhiker mutants exhibit ventralization of the caudal spinal cord
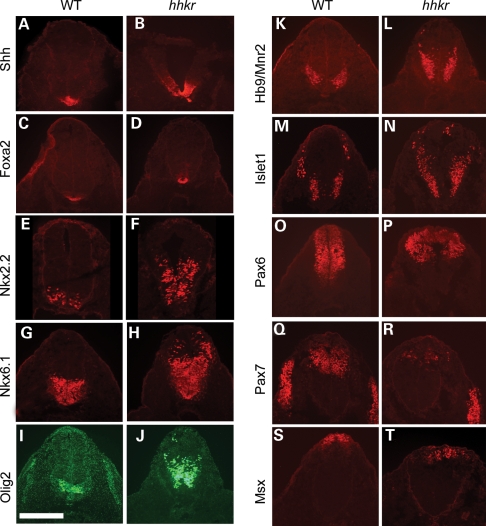
Activity of the Shh pathway is a key regulator of DV patterning within the neural tube, and the observed changes in Ptch1 and Shh expression in hitchhiker mutants prompted us to examine the expression of markers of DV patterning at E10.5. As anticipated from the expansion of Shh mRNA expression, we detected dorsal expansion of both Shh and Foxa2 protein expression, markers of floor plate, in the lumbar neural tube of hitchhiker mutants (Fig. 5A–D), although the region that exhibits the columnar morphology characteristic of floor plate appears to be unaltered. In addition, Nkx2.2, which marks the progenitors of the V3 ventral interneurons adjacent to the floor plate, and Nkx6.1, and Olig2 which mark other ventral and ventro-lateral neurons, are all expanded dorsally in hitchhiker mutant embryos (Fig. 5E–J). HB9/MNR2 and Islet1/2, markers of differentiating motorneurons, were also ectopically expressed in dorsally extended domains in hitchhiker mutants (Fig. 5K–N). Correspondingly, markers of dorso-lateral and dorsal neurons, Pax6, Pax7 and Msx, exhibited smaller expression domains in hitchhiker mutants, with a dorsal shift in their ventral limit of expression (Fig. 5O–T). Patterning of the neural tube at more cranial spinal levels was largely unaffected in hitchhiker mutants (Supplementary Material, Fig. S2). Thus, our data reveal ventralization of the caudal neural tube in hitchhiker mutants, consistent with Tulp3 acting as a negative regulator of the Shh pathway.
Figure 5.
hitchhiker mutants exhibit ventralization of the caudal spinal cord. Immunostaining of transverse sections through the neural tube immediately anterior to the hindlimb bud with antibodies against Shh (A and B), Foxa2 (C and D), Nkx2.2 (E and F), Nkx6.1 (G and H), Olig2 (I and J), HB9/MNR2 (K and L), Islet1 (M and N), Pax6 (O and P), Pax7 (Q and R) and Msx (S and T), in wild-type (A, C, E, G, I, K, M, O, Q and S) and hitchhiker (B, D, F, H, J, L, N, P, R and T) embryos at E10.5. Ventral and ventro-lateral markers (Shh, Foxa2, Nkx2.2, Nkx6.1, Olig2, HB9/MNR2 and Islet1) exhibit expanded expression domains in hhkr while dorsal markers (Pax6, Pax7 and Msx) are reduced. Scale bar: 300 µm.
Genetic analysis reveals Shh pathway activation in hitchhiker occurs independently of Shh ligand
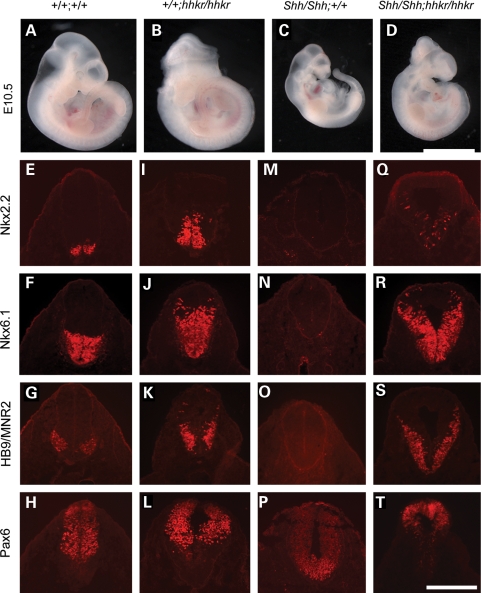
The expression changes observed at E9.5 suggested that up-regulation of the Shh pathway in hitchhiker mutants occurs independently of a change in Shh expression. We hypothesized that mutation of Tulp3 leads to loss of repression of the Shh pathway, downstream of Shh ligand. To test this genetically, we intercrossed hitchhiker with the Shh-targeted null allele (60). Doubly heterozygous (Shh/+;hhkr/+) mice were viable and fertile, and intercrosses yielded E10.5 embryos with the expected Mendelian ratio of genotypes. Although hhkr homozygotes often exhibited exencephaly and caudal oedema (Fig. 6B), Shh null mutants were characterized by holoprosencephaly and reduced growth (Fig. 6C, compare with A), as described previously (60). Compound homozygous Shh/Shh;hhkr/hhkr embryos exhibited exencephaly, caudal oedema and spina bifida (Fig. 6D). These phenotypes are characteristic of hitchhiker and distinct from those of the Shh null, demonstrating that hitchhiker is epistatic to Shh.
Figure 6.
Genetic analysis of epistasis places Tulp3 downstream of Shh. (A–D) E10.5 embryos generated from intercrossing Shh/+;hhkr/+ doubly heterozygotes, demonstrating phenotypes of wild-type (A), hitchhiker (B), Shh (C) and Shh/Shh;hhkr/hhkr doubly homozygous (D) embryos. Double mutants grossly resemble hitchhiker. (E–T) Immunostaining of transverse sections through the neural tube immediately anterior to the hindlimb bud with antibodies against Nkx2.2 (E, I, M and Q), Nkx6.1 (F, J, N and R), HB9/MNR2 (G, K, O and S) and Pax6 (H, L, P and T) in wild-type (E–H), hitchhiker (I–L), Shh (M–P) and Shh/Shh;hhkr/hhkr doubly homozygous (Q–T) embryos. Immunostaining of double mutants is similar to hitchhiker. Scale bar: A–D: 2.5 mm; E–T: 300 µm.
To investigate this further, we assessed the DV patterning in the caudal neural tube of double mutant embryos. Immunostaining on transverse sections through the caudal spinal cord of Shh mutants revealed a dorsalized phenotype, distinct from both the wild-type expression pattern (Fig. 6E–H) and ventralized phenotype of hhkr embryos (Fig. 6I–L). In Shh mutants, Nkx2.2, Nkx6.1, Olig2, HB9 and Islet1 are absent while Pax6, Pax7 and Msx are expanded ventrally (Fig. 6M–P and data not shown), as reported previously (45). In the Shh/Shh;hhkr/hhkr double mutants, Nkx2.2, Nkx6.1, Olig2, HB9 and Islet1 were expressed throughout a dorsally extended domain in double mutants, in a pattern similar to hhkr (Fig. 6Q–S and data not shown). The ventral limits of Pax6, Pax7 and Msx expression domains were shifted dorsally (Fig. 6T and data not shown), similar to hitchhiker. Thus, doubly homozygous Shh/Shh;hhkr/hhkr embryos revealed ventralization of the neural tube similar to that in hitchhiker mutants, and dissimilar to the dorsalization seen in Shh mutants. In addition, analysis of Ptch1 and Gli1 expression demonstrated increased expression in Shh/Shh;hhkr/hhkr embryos, similar to that observed in hhkr (Supplementary Material, Fig. S3), and in striking contrast to the absence of Ptch1 expression in Shh mutant embryos. These data further support our conclusion that Tulp3 acts downstream of Shh, and indicates that mutation of Tulp3 leads to activation of Shh pathway targets in the absence of Shh ligand.
hitchhiker is genetically epistatic to Smoothened
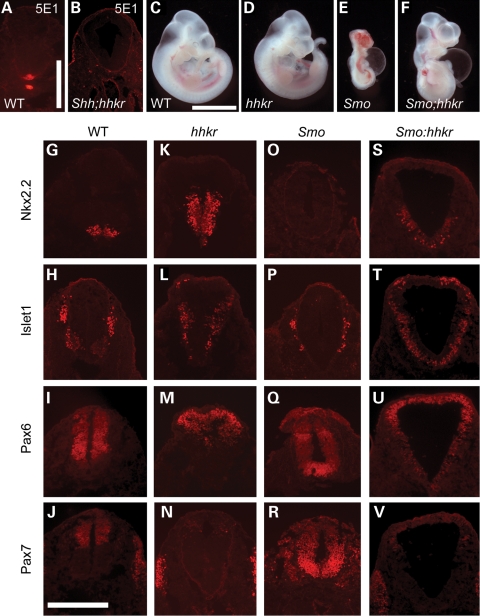
Since the hedgehog pathway can be activated by ligands other than Shh, we sought to test whether the pathway activation observed in hitchhiker mutants could be caused by increased expression of another ligand, such as Indian hedgehog or Desert hedgehog. Immunostaining with the 5E1 antibody that was raised against Shh (61), but which detects all hedgehog ligands (62), detected no observable signal in the notochord, neural tube or adjacent tissue of Shh/Shh;hhkr/hhkr double mutants, suggesting this is not the case (Fig. 7B, compare with A). For a more formal test, we intercrossed hitchhiker with the Smo-targeted null allele (63) to test for dependence on Smoothened, an obligate component of all hedgehog signalling. Doubly heterozygous (Smo/+;hhkr/+) mice were viable and fertile, and intercrosses yielded E10.5 embryos with the expected Mendelian ratios of genotypes. Smo null mutants were characterized by cyclopia, holoprosencephaly, retarded growth and pericardial oedema (Fig. 7E), as described previously (63). Compound homozygous Smo/Smo;hhkr/hhkr fetuses exhibited a phenotype more closely reminiscent of hhkr homozygotes than Smo null embryos, with expansion of the caudal spinal cord and a relatively normal head and growth, although pericardial oedema was also observed (Fig. 7F, compare with C–E).
Figure 7.
Ventralization of the spinal cord in hitchhiker occurs independently of Smo. (A and B) Immunostaining of wild-type and Shh/Shh;hhkr/hhkr double homozygous embryos with 5E1 antibody to detect all Hedgehog ligands. (C–F) E10.5 embryos generated from intercrossing Smo/+;hhkr/+ double heterozygotes, demonstrating phenotypes of wild-type (C), hitchhiker (D), Smo (E) and Smo/Smo;hhkr/hhkr doubly homozygous (F) embryos. Double mutants more closely resemble hitchhiker than Smo. (G–V) Immunostaining of transverse sections through the neural tube immediately anterior to the hindlimb bud with antibodies against Nkx2.2 (G, K, O and S), Islet1 (H, L, P and T), Pax6 (I, M, Q and U) and Pax7 (J, N, R and V) in wild-type (G–J), hitchhiker (K–N), Smo (O–R) and Smo/Smo;hhkr/hhkr (S–V) doubly homozygous embryos. Scale bar: A and B: 500 µm, C–F: 2.5 mm; G–V: 300 µm.
Immunostaining on transverse sections through the caudal spinal cord of doubly homozygous (Smo/Smo;hhkr/hhkr) embryos revealed ventralization of the neural tube similar to that seen in hitchhiker mutant embryos, and distinct to the dorsalized phenotype observed in Smo mutants (Fig. 7O–R). Nkx2.2 and Islet1 were expressed throughout a dorsally extended domain (Fig. 7S and T), whereas the ventral limits of Pax6, Pax7 and Msx expression domains were shifted dorsally (Fig. 7U and V and data not shown), similar to hitchhiker (Fig. 7K–N). These data reveal that Tulp3 acts downstream of Smo, and indicates that mutation of Tulp3 leads to activation of the Shh pathway independently of Smo receptor.
hitchhiker limb buds exhibit ectopic anterior activation of Shh signalling and a genetic interaction with Shh and Gli3
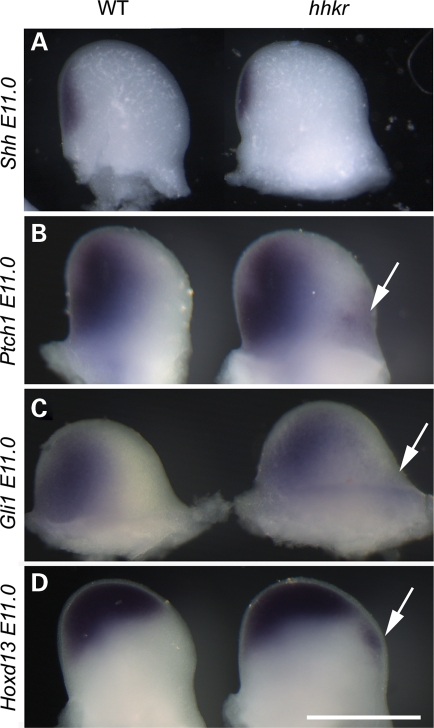
The hitchhiker mutants exhibit preaxial polydactyly, and we hypothesized that this might be caused by ectopic activation of the Shh pathway, similar to that observed in the neural tube. To address this, we examined the expression of targets of the Shh pathway in the developing limb buds. Shh expression appeared normal in hitchhiker limb buds, with a single domain of expression in the ZPA at E11.0 (Fig. 8A). In contrast, ectopic expression of Ptch1 was observed in the anterior region of mutant limb buds at E11.0, whereas expression appeared normal in the posterior region (Fig. 8B). We also observed ectopic expression of Gli1 in the anterior domain (Fig. 8C), although this was less robust than for Ptch1, and Hoxd13 (Fig. 8D) another target of Shh pathway activation. Thus, hitchhiker mutants show normal expression patterns of markers in the posterior region of the limb buds but appear to have ectopic activation of Shh pathway targets in the anterior region of the limb, in the absence of ectopic Shh ligand.
Figure 8.
hitchhiker mutants exhibit ectopic expression of Shh pathway targets in the anterior region of the limb buds. (A–D) Forelimb buds from E11.0 wild-type (left) or hitchhiker (right) embryos following wholemount in situ hybridization with riboprobes for Shh (A), Ptch1 (B), Gli1 (C) and Hoxd13 (D); anterior to right. Shh expression looks similar in wild-type and mutant limbs, whereas Ptch1, Gli1 and Hoxd13 show normal posterior expression but a small region of ectopic expression in the anterior domain (arrows). Scale bar: 1 mm.
To determine whether Tulp3 acts downstream of Shh in the limb (as well as in the neural tube), we examined limb morphology in Shh/Shh;hhkr/hhkr double mutants. Although hitchhiker mutants exhibited preaxial polydactyly with six digits on both forelimbs and hindlimbs (Fig. 9B, compare with A), Shh mutants are characterized by shortened and narrowed limbs bearing only a single digit (Fig. 9C), as described previously (64). The Shh/Shh;hhkr/hhkr double homozygotes reproducibly demonstrated an intermediate phenotype, with two digits on the forelimbs and four digits on hindlimbs (Fig. 9D). Although the limbs of double homozygotes do not resemble either single mutant, the mutation of Tulp3 can partially rescue the severe limb defects of Shh mutants, consistent with Tulp3 acting genetically downstream of Shh.
Figure 9.
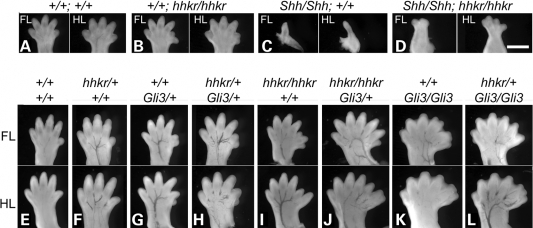
Genetic analysis shows hitchhiker acts downstream of Shh and interacts with Gli3 in limb development. (A–D) E12.5 embryos generated from intercrossing Shh/+;hhkr/+ double heterozygotes, demonstrating phenotypes of the forelimbs (FL) and hindlimbs (HL) of wild-type (A), hitchhiker (B), Shh (C) and Shh/Shh;hhkr/hhkr double mutant fetuses (D); anterior to right. (E–L) FL and HL of E15.5 embryos generated from intercrossing hhkr/+;Gli3/+ double heterozygotes; positioned with anterior to the right. Wild-type (E) and hhkr/+ (F) embryos have normal limbs with five digits; Gli3/+ (G) and hhkr/+;Gli3/+ (H) display six digits with a small additional preaxial digit; hhkr/hhkr (I) demonstrates six or seven digits; hhkr/hhkr;Gli3/+ (J) exhibits seven digits; Gli3/Gli3 (K) has polysyndactyly with seven or eight digits, plus an extra postaxial nubin; hhkr/+;Gli3/Gli3 (L) exhibits polysyndactyly with seven or eight digits, plus an extra postaxial nubin. Scale bar: 1 mm.
The downstream effectors of the Shh pathway are the three Gli-family transcription factors, with Gli3 playing the major role in limb patterning (30–32). To investigate the relationship between Gli3 and Tulp3 genetically, we intercrossed hitchhiker mutants with mice heterozygous for the Gli3 null allele, Gli3XtJ (65). Doubly heterozygous hhkr/+;Gli3/+ mice were viable and fertile, and intercrosses yielded embryos of all the expected genotypes except for surviving doubly homozygous mutants, at E15.5. Analysis of digit number in the different genotypic classes revealed an additive genetic interaction between hhkr and Gli3 (Table 1). Heterozygous hitchhiker mice exhibited normal limbs (Fig. 9F, compare with E), whereas Gli3/+ fetuses exhibited an extra preaxial digit (Fig. 9G). The hhkr/+;Gli3/+ double heterozygotes appeared almost phenotypically indistinguishable to single Gli3/+ heterozygotes, with most limbs exhibiting a single extra digit (Fig. 9H). The hhkr/hhkr single homozygotes exhibited preaxial polydactlyly with six or seven digits (Fig. 9I). Introduction of one Gli3 mutant allele onto the homozygous hhkr/hhkr genotype appears to exacerbate the hitchhiker phenotype, with more severe polydactyly than observed in either hhkr/hhkr single mutants or Gli3/+ heterozygotes, with often seven or eight digits (Fig. 9J; compare with I and G; Table 1). Homozygous Gli3/Gli3 embryos exhibited severe polysyndactyly with often seven to nine digits (Fig. 9K), as described previously (65). Introduction of one hhkr mutant allele onto the Gli3/Gli3 genotype (hhkr/+;Gli3/Gli3) appears to make no difference to the limb defect, which is almost indistinguishable to that observed in Gli3/Gli3 single mutants, with seven to nine digits formed (Fig. 9L, compare with K; Table 1). Doubly homozygous hhkr/hhkr;Gli3/Gli3 embryos did not survive to E15.5. Attempts to collect embryos at E12.5 yielded only one double mutant in which limb morphology was abnormal, but retarded development meant digit number could not be reliably ascertained. The absence of doubly homozygous mutant data precludes a formal assessment of epistasis. However, the analysis of intermediate genotypes demonstrates that there is a genetic interaction between the two loci, and suggests that Tulp3 and Gli3 may act in concert during limb morphogenesis.
Table 1.
Genetic interaction between hitchhiker and Gli3, determined by analysis of digit number
| hhkr genotype | +/+ | +/+ | +/+ | hhkr/+ | hhkr/+ | hhkr/+ | hhkr/hhkr | hhkr/hhkr |
| Gli3 genotype | +/+ | Gli3/+ | Gli3/Gli3 | +/+ | Gli3/+ | Gli3/Gli3 | +/+ | Gli3/+ |
| Number of fetuses examined | 10 | 10 | 5 | 21 | 24 | 7 | 7 | 9 |
| Average number of digits | 20.0 | 24.0 | 28.8 | 20.0 | 24.2 | 29.0 | 23.9 | 27.0 |
| Percentage of limbs with no extra digits | 100 | 0 | 0 | 100 | 0 | 0 | 14 | 0 |
| Percentage of limbs with one extra digit | 0 | 100 | 20 | 0 | 96 | 14 | 75 | 36 |
| Percentage of limbs with two extra digits | 0 | 0 | 45 | 0 | 2 | 50 | 11 | 53 |
| Percentage of limbs with three extra digits | 0 | 0 | 25 | 0 | 2 | 32 | 0 | 11 |
| Percentage of limbs with four extra digits | 0 | 0 | 10 | 0 | 0 | 4 | 0 | 0 |
Gli2 and Gli3 abundance and Gli3 processing appear unaltered in hitchhiker mutants
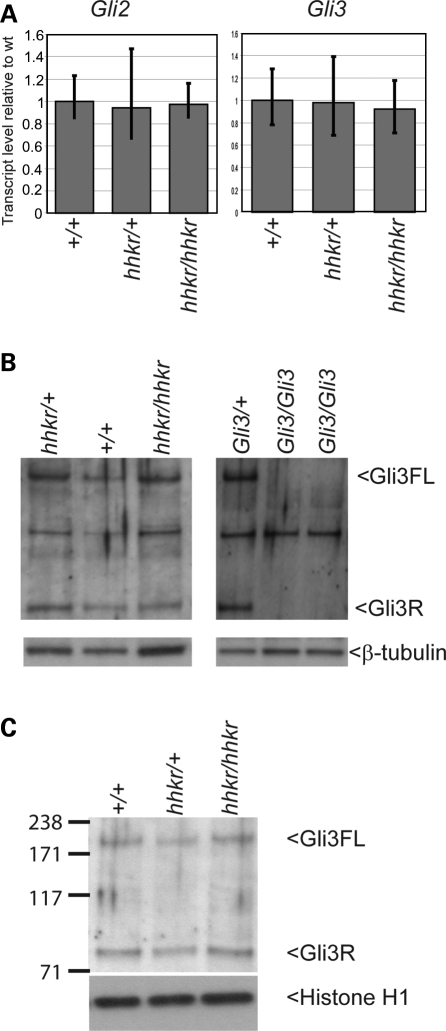
Shh pathway activity is mediated by the stimulation of Gli activators and inhibition of Gli repressors. Since we have demonstrated that Tulp3 acts genetically downstream of Shh and Smo, and genetically interacts with Gli3, we sought to determine whether Gli2 and Gli3 expression are altered in hitchhiker mutants. By qRT–PCR, the expression of Gli2 and Gli3 was not detectably altered in the caudal end of E9.5 mutant embryos, compared with wild-type (Fig. 10A). Other mutants that affect Shh signalling demonstrate dramatic differences in Gli3 processing, as seen, for example, in the ftm, IFT88/polaris and Dnchc2 mutants (66–69), prompting us to look for similar changes in the abundance of the Gli3 isoforms in hitchhiker mutants. Since the Gli3 Ab we used detects multiple bands, we performed western blots with protein extracts from Gli3XtJ/XtJ mutants, which lack Gli3 protein (65,70), in order to confirm the correct band identity (Fig. 10B). Experiments with forelimb buds isolated from E9.5 embryos, thus at a stage before changes in gene expression are apparent, revealed no observable change in the abundance of either the full length or cleaved Gli3 isoforms compared with wild-type or heterozygous littermates (Fig. 10B). In addition, Westerns performed with nuclear extracts from E10.5 embryos detected no change in the abundance of either full length or cleaved isoforms, in hitchhiker mutants, suggesting that Gli3 entry into the nucleus is not affected (Fig. 10C). From these data, we conclude that the action of Tulp3 does not appear to be mediated through regulation of Gli expression, processing or nuclear localization, although we cannot rule out the possibility that small differences might exist that would not be easily detectable due to experimental variability.
Figure 10.
Gli2 and Gli3 expression and Gli3 processing are not altered in hitchhiker mutants. (A) Quantitative RT–PCR analysis with primers for Gli2 and Gli3 on RNA from caudal ends of E9.5 wild-type, heterozygous and hitchhiker mutant embryos, showing no change in expression. (B) Western blot analysis on total cell lysates from E9.5 limb buds demonstrate no apparent change in abundance of the full length 190 kDa Gli3 isoform (Gli3FL) nor the 83 kDa cleaved repressor isoform (Gli3R), in hitchhiker embryos compared with wild-type or heterozygous littermates. Western blots with extracts from Gli3/+ and Gli3/Gli3 mutants confirm correct Gli3 band identify. β-Tubulin was used as a loading control. (C) Western blots with nuclear extracts from E10.5 caudal ends demonstrate similar abundance of the 190 and 83 kDa Gli3 isoforms in hitchhiker embryos compared with wild-type or heterozygous littermates; histone H1 was used as a loading control.
Expression of Rab23, Fkbp8, Thm1 and Sufu, activity of PKA and morphology of cilia are unaltered in hitchhiker
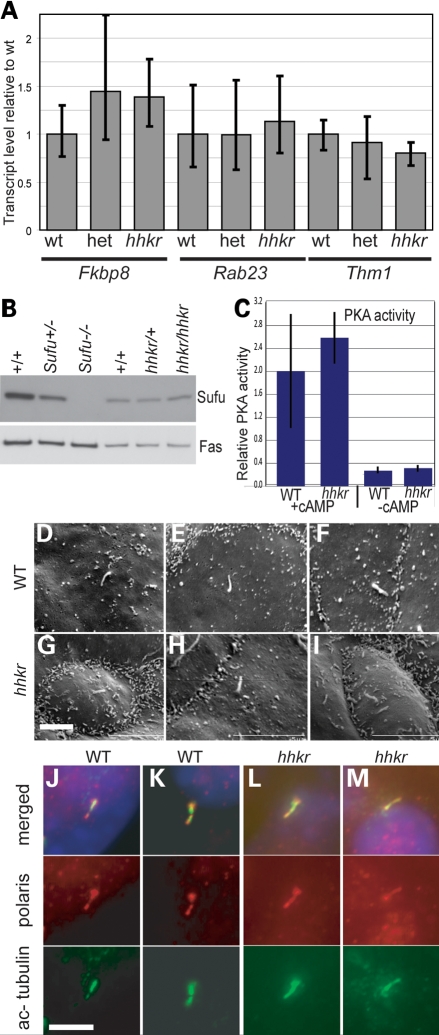
Since tubby family proteins may function as transcription factors (71), we hypothesized that Tulp3 might act by regulating the expression of another negative regulator. By qRT–PCR, we detected no significant change in Rab23, Fkbp8 or Thm1 (Ttc21b) expression in hitchhiker homozygotes compared with wild-type or heterozygous littermates, in the caudal thirds of E9.5 embryos, where the phenotype is apparent (Fig. 11A). Western blotting detected no change in Sufu expression in hitchhiker mutants compared with wild-type embryos at E9.5 (Fig. 11B). Assays for PKA activity in protein extracts from the caudal third of E9.5 embryos detected no significant difference in PKA activity between hitchhiker mutants and wild-types (Fig. 11C). We conclude that Tulp3 does not regulate the expression of Rab23, Fkbp8, Thm1 or Sufu, nor the activity of PKA.
Figure 11.
Evaluation of changes in downstream molecules and cilia structure. (A) Quantitative RT–PCR analysis with primers for Fkbp8, Rab23 and Thm1 (Ttc21b) on RNA from caudal ends of E9.5 wild-type, heterozygous and hitchhiker mutant embryos, showing no significant change in expression. (B) Western blot analysis on total cell lysates from E10.5 limb buds for Sufu expression revealed no change in hitchhiker mutants; fatty acid synthase (Fas) was used as a loading control. (C) PKA activity with or without cAMP addition, in protein extracts from E9.5 caudal ends of wild-type and hitchhiker embryos; no significant difference is detected. (D–I) SEM of E11.5 forelimb buds from wild-type (D–F) and hitchhiker mutant (G–I) embryos revealing cilia of normal length and morphology. (J–M) Immunostaining of MEFs from wild-type (J and K) and hitchhiker embryos (L and M) with anti-acetylated tubulin (green), anti-polaris (red) antibodies and DAPI (blue) demonstrate similar staining in mutants and wild-type cilia. Scale bars: 2 µm.
Recent evidence indicates that cilia play a major role in Shh pathway activity. Although most mutants affecting cilial morphology or function lead to a decrease in Shh pathway function, such as seen in the IFT52, IFT57, IFT88/polaris, IFT172 and Kif3a mutants (66–69,72,73), at least one mutant that affects retrograde intraflagellar transport results in activation of the Shh pathway (42). This prompted us to examine cilial morphology in hitchhiker mutants. Scanning electron microscopy (SEM) on limb buds from E11.5 embryos revealed the presence of cilia on both wild-type and hitchhiker mutant cells, with no overt difference in their length or morphology (Fig. 11D–I). Immunofluorescence with anti-acetylated tubulin and anti-polaris on cultures of mouse embryonic fibroblasts or limb bud cells confirmed the presence of cilia in both hitchhiker mutant and wild-type cells with no detectable difference in polaris distribution (Fig. 11J–M and data not shown). Thus, no gross defect in cilial morphology was observed in hitchhiker homozygotes.
DISCUSSION
In this study, we have identified a novel ENU-induced strongly hypomorphic allele of Tulp3 which exhibits ventralization of the caudal neural tube, neural tube defects and polydactyly. We demonstrate ectopic expression of Shh pathway targets, independently of a change in Shh expression, and show genetically that Tulp3 functions downstream of Shh and Smo. We also demonstrate that Tulp3 genetically interacts with Gli3 in regulating limb morphogenesis, but does not noticeably affect Gli3 expression or processing. We propose that Tulp3 functions to negatively regulate the Shh pathway in the caudal neural tube and limb buds.
Tulp3 functions as a negative regulator of Shh signalling in neural tube and limb
The targeted allele of Tulp3 has previously demonstrated a requirement for Tulp3 in neural tube closure (58); however, Tulp3 has not previously been associated with DV patterning of the neural tube nor with limb defects. In this study, we have demonstrated that Tulp3 functions as a negative regulator of the Shh pathway in the developing neural tube and limbs. This conclusion is based on three sets of experimental results, which are the established criteria for identifying novel components of the Shh pathway (45,47,74). First, we observed increased/ectopic expression of genes activated by Shh signalling in the hitchhiker mutants, in the caudal spinal cord and anterior limb domains. Shh is necessary and sufficient for the expression of ventral neural tube cell types including floor plate, motor neurons, V2 and V3 interneurons (60,61,75), and markers of these neurons/progenitors were expressed in expanded domains in the caudal neural tube of hitchhiker mutants. Neural tube patterning involves both the ventralizing signal of Shh from the notochord and floor plate, and the dorsalizing signal of BMPs and Wnts from the roof plate and surface ectoderm, with modulation of patterning by FGFs and retinoic acid (14–18,76). Although antagonism of BMP signalling in neural explants leads to a dorsal-to-ventral switch in the neuronal progenitor cell identity induced by a set concentration of Shh (77), embryos in which BMP or Wnt signalling is abrogated exhibit disruption of the dorsal interneuron cell populations yet no defects in ventral neuronal patterning (78–80) arguing against a role for Tulp3 in activating BMP or Wnt signalling. Moreover, four of the overexpressed genes (Foxa2, Nkx2.2, Ptch1 and Gli1) are direct transcriptional targets of Shh signalling (81,82). Secondly, genes that are normally repressed by Shh signalling are shifted dorsally in the hitchhiker neural tube. Thirdly, analysis of Shh/Shh;hhkr/hhkr and Smo/Smo;hhkr/hhkr double mutants shows that the activation of Shh target genes in hitchhiker mutants is independent of both Shh and Smo, and that substantial rescue of neural tube patterning and limb development occur in double mutants. Although BMP and Wnt signals can modulate the cellular response to Shh, the specification of ventral cell fates does not occur in the absence of hedgehog signalling, even without a ‘repressive’ effect of the dorsalizing signals (77,83). These defects observed in hitchhiker are established criteria for identifying negative regulators of the Shh pathway, as used in the characterization of Fkbp8 and Rab23 (45,47).
We have shown that the DV patterning of the neural tube in Shh/Shh;hhkr/hhkr and Smo/Smo;hhkr/hhkr double mutants is more closely similar to hitchhiker homozygotes than either Shh or Smo mutants, indicating that the DV markers are expressed independently of Shh and Smo. However, the expression patterns are not identical between double mutants and hitchhiker homozygotes. Nkx2.2 and Islet1 are expressed throughout the most ventral (floor plate) domain in double mutants, whereas they are excluded from this region in wild-type and hitchhiker homozygote. Foxa2 is expressed in an expanded domain in hitchhiker homozygotes, but appears to be absent from double mutants (Supplementary Material, Fig. S3). In addition, low-intensity Pax6 expression was evident throughout the ventral neural tube of Smo/Smo;hhkr/hhkr mutants, while it is excluded from this region in wild-types and hitchhiker homozygotes. These results indicate that the activation of Foxa2 expression and the repression of Nkx2.2, Islet1 and Pax6 from the ventral domains remain dependent on functional Shh and Smo activity. Foxa2 induction and Nkx2.2, Islet1 and Pax6 repression require very high levels of Shh activity (84–86), suggesting that down-regulation of Tulp3 in hitchhiker mutants is not sufficient to induce high-level activation of the pathway. A similar situation seems to occur in the developing limbs, whereby the intermediate phenotype of double mutants suggests that, in the absence of Shh, mutation of Tulp3 is not sufficient to induce full activation of the Shh pathway. Intriguingly, Shh−/−;Gli3+/+ mice typically form limbs with two to four digits (31), similar to the phenotype observed in Shh/Shh;hhkr/hhkr double homozygotes. Although Tulp3 mutation can lead to activation of some markers in a Shh-independent manner, the level of Shh pathway activation does not reach the highest levels that normally occur and, therefore, some residual Shh-dependent patterning remains.
Some regulators of Shh activity are also transcriptional targets of the Shh pathway. By western blotting, we found no substantial change in the abundance of Tulp3 protein in Shh, Smo or Gli3 mutants (data not shown), suggesting that Tulp3 is not itself a transcriptional target of the Shh pathway. Involvement of Tulp3 as a negative regulator of the Shh pathway suggests that down-regulation of Tulp3 activity might be required for Shh signalling. Tulp3 has several putative sites for phosphorylation, myristoylation and glycosylation (87). It is possible that regulation of Tulp3 activity might occur through effects on post-translational modification.
Possible functions of Tulp3 in the Shh pathway
The tubby family consists of four proteins, Tubby and Tulps1–3, which are characterized by the C-terminal ‘tubby’ domain (88,89). Disruption of other tubby proteins has been associated with a range of phenotypes, including retinal and cochlear degeneration following progressive cell apoptotis, and maturity-onset obesity (90–94). No tubby member has previously been linked to Shh signalling.
We have demonstrated that hitchhiker mutants exhibit a phenotype consistent with Tulp3 acting as a negative regulator of Shh signalling, and we have positioned Tulp3 genetically downstream of Shh and Smo. Many regulators of the Shh pathway have recently been discovered, and several have been shown to act genetically downstream of Shh and Smo. These include proteins involved in the formation or maintenance of cilia and intraflagellar transport, such as Arl13b/hennin, IFT88/polaris/flexo, IFT172/wimple, Kif3a, Rpgrip1l/Fantom and Thm1 (38,42,66,68,69,72,95). Disruption of IFT or cilia usually results in reduced Shh pathway activity in the neural tube, evidenced by the absence of Shh-dependent ventral cell types, as seen in mutants for IFT88/polaris/flexo, IFT172/wimple, Kif3a, Rpgrip1l/Fantom, Dync2h1/Dnchc2, IFT52/Ngd5, IFT57/hippi and Ofd1 (67,68,72,96,97). However, mutation of Arl13b/hennin results in constitutive activation of the Shh pathway at an intermediate level (95), whereas loss of Thm1 causes increased Shh activity and expansion of ventral markers (42). The Gli-mediated transcriptional response to Shh and the effects of the negative regulators Sufu and PKA are abrogated in cells mutant for IFT components (98). The negative regulators Rab23 and Fkbp8 function genetically between Shh/Smo and Gli2 (45,47,99,100), and their activity is dependent on functional IFT (66,100). Rab23 is a member of the Rab-GTPase family of vesicular transport proteins and may regulate vesicular transport within (or dependent on) cilia, although the targets of Rab23 trafficking have not been identified (55,99,101). Fkbp8 is a member of the FK506-binding protein family (immunophilins), and may also play a role in membrane-associated proteasome function and trafficking (56,102).
The importance of cilia is further underlined by the finding that several Shh pathway components are localized within the cilia, including Smo, Ptch, Sufu and Gli proteins (38–41,67,98). Smo becomes enriched in cilia following treatment of cells with Shh (39,40,98), and this ciliary localization is necessary (39) but not sufficient (98) for pathway activation. Tulp3 protein can be detected at the tips of cilia (37), and evidence from other members of the tubby family suggests that Tulp3 might play a role in vesicular trafficking. Tulp1 interacts with several proteins implicated in the formation and movement of vesicles including F-actin, dynamin-1, clathrin heavy chain, dynein intermediate chain and tubulin (103,104). Tulp1 mutants exhibit accumulation of rhodopsin-containing vesicles in the interphotoreceptor matrix, consistent with loss of polarized trafficking between photoreceptor segments (91,105). However, examination of cilial morphology in hitchhiker mutants has detected no overt cilial defect, whereas immunofluorescence studies revealed no apparent difference in localization of polaris/IFT88. Thus, there is no evidence to suggest that cilial morphology or transport is grossly disrupted in hitchhiker mutants, although more subtle structural or functional defects would not have been detected.
An alternative hypothesis for the role of Tulp3 is as a transcriptional regulator. The tubby proteins are characterized by the C-terminal ‘tubby’ domain that binds to double-stranded DNA, while the N-terminal region can function as a transcriptional activator (71). The tubby proteins also encode nuclear localization signals and can be detected in the cell nucleus (87,93,106). Experiments suggest tubby proteins translocate from the plasma membrane into the nucleus in response to Gαq-coupled receptor signalling (107). The phenotype of hitchhiker mutants might be consistent with transcriptional down-regulation of another negative regulator of the Shh pathway. Indeed, it is striking that hitchhiker bears many similarities with the Rab23opb mutant, including gross phenotypic effects (exencephaly, dorsal expansion of the caudal spine, small and dorsally misplaced dorsal root ganglia, polydactyly, rib bifurcations and splayed vertebrae) (108,109), similar patterning defects in the caudal neural tube (45,99,110) and positioning genetically downstream of Shh and Smo (45,99). The disruption of Fkbp8 or partial loss of function of PKA (Pka-plof) also yield partial ventralization of the neural tube, similar to that seen in hitchhiker (46,47,100). However, we have shown that expression of Rab23, Fkbp8, Thm1, Gli3 and Sufu and activity of PKA are not detectably altered in hitchhiker mutants. Thus, although we cannot rule out a transcriptional role for Tulp3 on some other yet undiscovered negative regulator, we have excluded transcriptional modulation of the known negative modulators of Shh signalling.
Modulation of Gli protein activity is the downstream consequence of the Shh pathway, since the presence of Shh ligand acts to inhibit cleavage of Gli3 (30) and suppress the processing and degradation of Gli2 (111). Many mutants that exhibit down-regulation of the Shh pathway show impaired Gli3 processing, with a decrease in the amount of Gli3 repressor relative to the amount of full-length Gli3 protein, as seen in mutants for Dync2h1/Dnchc2, IFT88/polaris, IFT172/wimple, Kif3a, Rpgrip1l/fantom and the chicken talpid3 mutant (38,67–69,72,73,112). Gli3 processing is also disrupted in the Thm1 mutant, which has activation of Shh signalling (42). The negative regulator Sufu binds to the Gli proteins (113,114) and may act in part by sequestering Gli1 and Gli2 in the cytoplasm (49,53,114,115), a function that involves CRM1-dependent nuclear export of Gli1 but not Gli2 (54). We have shown that Tulp3 genetically interacts with Gli3 in regulating limb development. However, we observed no change in the expression levels of Gli2 or Gli3, and we detected no substantial change in the abundance of the Gli3 isoforms in hitchhiker. This suggests that Tulp3 function is not associated with Gli expression or processing. In addition, we observed no alteration in abundance of Gli3 within the nucleus, suggesting that Tulp3 does not act to regulate entry into the nucleus.
It is possible that Tulp3 may function as a co-factor in Gli activity or may affect the interaction of Gli with other regulators. Sufu can inhibit the transcriptional activator function of Gli1 and Gli2, independently of cytoplasmic sequestration (54). Sufu mutant MEFs exhibit constitutive Gli activity yet do not show accumulation of Gli1 in the nucleus (50). Sufu interacts with SAP18, Galectin3 and other nuclear proteins (116) as well as with Gli1 bound to DNA (53), and Sufu represses Gli-mediated transcription by recruiting the histone deacetylation machinery through interaction with SAP18 (117). Although we detected no change in Sufu expression level, it is possible that loss of Tulp3 might affect the interaction of Sufu with Gli; recent data have revealed that Cdc2l1 can act as a pathway activator, through binding to Sufu and relieving its inhibition on Gli (118). Alternatively, Tulp3 might act to regulate the expression or activity of other factors that regulate Gli protein function, such as MED12 (119), MIM/BEG4 (120), CBP (81), Ski (121), Dyrk1 (122), DYRK2 or MAP3K10 (123). The precise functional and molecular relationship between Gli3 and Tulp3 requires further analysis.
It is intriguing that the hitchhiker limb buds exhibit a localized domain of ectopic gene expression, at the anterior limit of the limb buds. This is distinct from the ectopic expression patterns observed in other mutants, in which there is an anterior expansion of the normal expression domains. Many other mutants with polydactyly have impaired processing of Gli3, and this may cause the limb defect. It is possible that Tulp3 acts in regulation of specific targets of Gli3 function, which then subsequently results in the region-specific activation of other molecules. Microarray analysis of transcriptional changes in Gli3 mutant limb buds identified a number of targets, including some that are expressed in localized anterior domains and which are down-regulated in Gli3 mutants; these include Pax9, Cdo and Id2 and novel transcript BG074838 (124). We might hypothesize that Tulp3 has a role in co-regulation of specific Gli3 targets, resulting in loss of an anterior-specific factor, rather than a general change in Gli3 effects in the limb.
hitchhiker mutants exhibit spina bifida as a result of a failure of neural tube closure
We have demonstrated that spina bifida in hitchhiker results from a primary failure of spinal neural tube closure. Closure of the PNP occurs with three ‘modes’ of closure, at different gestational ages and, therefore, at different axial levels, typified by the formation of MHPs (mode 1), DLHPs (mode 3) or both (mode 2) (6). The regulation of DLHP formation is modulated by Shh expression, with strong Shh expression from the notochord during mode 1 neurulation inhibiting DLHPs (12), through the noggin-mediated antagonism of Bmp signalling (13). Formation of DLHPs is critical for spinal closure, and failure to form DLHPs results in severe spina bifida as seen in the Zic2 mutant (13). In hitchhiker mutants, the formation of DLHPs appears to be partially suppressed, although they are not inhibited completely, and correspondingly, the spina bifida in hitchhiker is less severe than observed in Zic2 mutants. We have demonstrated increased activation of the Shh pathway at the caudal end of hitchhiker mutants, and this would be anticipated to repress DLHP formation. Indeed, the loss of other negative regulators of the Shh pathway can also result in spina bifida, including Sufu, Rab23, PKA and Ptch1 (45,46,48,109). However, we also observed defects in DV patterning of the neuroepithelium at E9.5 (data not shown). It is likely that PNP closure is inhibited by a combination of these effects.
Regional differences in the role of Tulp3
Our data reveal a role for Tulp3 in DV patterning of the neural tube, with a more important role in caudal than cranial regions of the spinal cord. Tulp3 is expressed ubiquitously (58), suggesting that the AP difference is not attributable to differential expression. It is notable that the role of other molecules in DV patterning also varies along the AP axis. For example, mutation of Rab23, Fkbp8 or PKA-deficiency affect only caudal spinal cord patterning (45–47,110), whereas the Dync2h1/Dnchc2 mutant exhibits a more severe patterning disruption in rostral than caudal regions of the spinal cord (72) and retinoic acid modulates patterning in the rostral spinal cord of quail embryos (125). Smo−/−;Gli3−/− embryos show reduced rescue of the ventral markers more caudally, and this is taken to indicate an AP difference in the requirement for Hh/Gli3 interactions in the specification of similar cell identities (75). The molecular basis of the regional differences and the relationship between the pathway regulators has yet to be fully elucidated.
The data presented here led to similar conclusions as a study performed with the Tulp3-targeted allele (Norman et al., submitted). However, there are minor variations between the phenotypes, such as the absence of Foxa2 expression in the Shh/Shh;hhkr/hhkr double mutant, compared with expression of this floor plate marker in double mutants with the Tulp3 null allele, and no significant change in proliferation in the caudal neural tube of hitchhiker, compared with decreased proliferation in the Tulp3 knockout. These qualitative differences likely reflect the difference in abundance of Tulp3 protein, in the two alleles, since the targeted allele is likely to be a true null, while we have demonstrated in hitchhiker that a small amount (∼4%) of wild-type protein remains.
In conclusion, correct regulation of the activity of the Shh pathway is essential for normal development and continued health in adults. Misregulation during embryonic development generates a spectrum of congenital abnormalities, including holoprosencephaly, neural tube defects and polydactyly. The continued identification of new components of the Shh pathway, such as Tulp3, coupled with further research to understand their molecular function, holds significant promise for the diagnosis and treatment of human disease.
MATERIALS AND METHODS
Mice and embryos
hitchhiker was identified during a three-generation (G3) recessive mutagenesis screen (details to be described elsewhere). Briefly, C57BL/6 males were injected with ENU and outcrossed to C3H/HeH; male F1 offspring were mated to C3H/HeH and F2 females backcrossed to their father. G3 embryos were examined at E13.5 for developmental abnormalities. Affected fetuses were used for genetic mapping with a 55-marker genome-wide SNP panel (sequences available on request), and additional microsatellite markers. The mutant line was maintained by backcrossing to C3H/HeH and, following gene identification, genotyped for the mutation by pyrosequencing. The MGI accession ID for this allele is MGI:3662473. Mice heterozygous for Shhtm1Chg (60) or Smotm1Amc (63) were maintained on 129S6/SvEv and genotyped by PCR (N. Warr, personal communication). Mice heterozygous for the Tulp3tm1Jng-targeted allele (58) on C3Heb/FeJ were obtained from J. Eggenschwiler (Princeton University). Mice heterozygous for Gli3 deletion (Gli3XtJ/+) on a mixed C57BL/6/C3H background were obtained from T. Theil (Edinburgh University) and genotyped as described (126). Animals were maintained following guidelines of the Medical Research Council and in accordance with the Animals (Scientific Procedures) Act, 1986.
Mice were maintained routinely on a 12 h light–dark cycle (dark from 19:00 to 07:00 h). Embryos were generated by overnight matings, with the day of finding a copulation plug designated as embryonic day (E) 0.5. Some mice were maintained on a reverse light–dark cycle (dark from 10:00 to 22:00 h), and litters from these animals were designated as E1.0 on the day of plugging. Embryos were dissected in PBS with 10% newborn calf serum and processed according to downstream application.
Sequencing
RNA was extracted from E13.5 fetuses using GenElute (Sigma) and reverse transcribed with MMLV-RT (Invitrogen). DNA and cDNA were amplified with intron or exon-specific Tulp3 primers (sequences available on request), purified using Qiaquick (Qiagen) then sequenced with BigDye reagent (ABI) and ABI3700.
Histology and in situ hybridization
Histology and skeletal preparations used standard protocols. Wholemount in situ hybridization was performed essentially as described (127), using digoxigenin-labelled riboprobes for Shh, Ptch1, Gli1, and Hoxd13. At least three embryos of each genotype were analysed with each probe and processed under identical conditions. Embryos were photographed on a Leica MZ16 stereomicroscope and vibratome sectioned at 35–50 µm as described (128). Slide in situ hybridization was performed using 10 µm cryosections as described elsewhere (129), with digoxigenin-labelled riboprobes for Ptch1 and Gli1.
Protein extraction, western blots and PKA assay
Total cell lysates were generated in RIPA buffer (PBS with 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS). Nuclear fractions were generated using the Proteoextract Subcellular Fractionation kit (Calbiochem). Proteins were quantitated using the DC assay (Biorad). Westerns used 1–5 µg protein per lane on 7% or 3–8% Tris-Acetate NuPAGE gels with SeeBluePlus2 or HiMark ladders (Invitrogen). Proteins were transferred onto Hybond ECL (GE Healthcare) and detected with antibodies against Tulp3 (1:300; gift from J. Eggenschwiler), Gli3 (1:1000; Santa Cruz sc-20688), Sufu (1:200; sc-10933), β-tubulin (1:5000; sc9104), histone H1 (1:1000, sc-8030) or fatty acid synthase (1:1000; sc-55580) with HRP-conjugated secondary antibodies (1:12 000; DAKO) and detection with ECL Advance (GE Healthcare). Extracts from Gli3XtJ/XtJ (65) and Sufu−/− (50) mutants were used to verify Gli3 and Sufu band identity, respectively, and the antibodies used have been published previously (70,130). Bands were quantitated from scanned images using Adobe Photoshop CS2; data are expressed as means ± standard errors, and significance tested using two-sample equal variance, two-tailed distribution Student’s t-test. PKA activity was measured on extracts from E9.5 caudal thirds using a non-radioactive assay (Calbiochem), following Bradford assay quantitation.
SEM, cell culture and immunofluorescence
Embryos were processed for SEM as described (38) and imaged on an Hitachi S-530 scanning electron microscope. Mouse embryonic fibroblast primary cultures were immunostained for N-acetylated tubulin (1:10 000; Sigma) or Polaris (1:1000; gift from B. Yoder) as described (38). Immunofluorescence on 10 µm cryosections was performed as described (131), using antibodies against Nkx2.2, Shh, Islet1, Msx1/2, Foxa2, HB9/MNR2, Nkx6.1, Pax6 and Pax7 (1:10; Developmental Studies Hybridoma Bank) and Olig2 (1:500, Abcam ab33427). Cell proliferation rates were determined by staining with antibody against phospho-Histone H3 (pHH3, Upstate Cell Signalling). Primary antibody staining was detected with appropriate secondary antibodies conjugated with AlexaFluor-488 or AlexaFluor-594 (1:250 dilution; Invitrogen), mounted in Vectashield with DAPI (Vector Laboratories) and imaged with a Zeiss Axiophot microscope. Quantitation of proliferation rates was performed by counting pHH3+ cells and total (DAPI+) cells within the caudal neuroepithelium of E9.5 (17–19 somite) embryos; the mitotic index was calculated as the average from 12 sections from each of six embryos of each genotype, with analysis from the region immediately posterior to the caudal extremity of the vitelline artery thus within the PNP.
Quantitative real-time RT–PCR
Total RNA was extracted from caudal thirds of E9.5 embryos and DNase treated using RNAeasy plus mini kit (Qiagen), quantified with a Nanodrop ND1000 and the integrity assessed by gel electrophoresis. Double-stranded cDNA was then synthesized from 1 µg RNA using High Capacity cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR was performed with 10 ng cDNA using TaqMan® gene expression assays on a 7500 Fast Real-Time PCR System (Applied Biosystems). The assays used were: Mm00494645_m1 (Gli1); Mm00487418_m1 (Fkbp8); Mm01293116_m1 (Gli2); Mm00492333_m1 (Gli3); Mm01545399_m1 (Hprt); Mm00436209 (Rab23); Mm00436026 (Ptch1) and Mm01270405 (Ttc21b). Samples were analysed in quadruplicate, using at least three embryos of each genotype, and calculations performed using the comparative CT method. The values were normalized to the endogenous reference gene (Hprt) and the transcript levels are presented as fold change relative to the wild-type sample in relative quantification (RQ) units. Error bars indicate the calculated maximum (RQMax) and minimum (RQMin) expression levels, with a 95% confidence level. Statistical significance was tested using two-sample equal variance, two-tailed distribution Student’s t-test and is also represented as non-overlapping error bars if the samples are significantly different (P < 0.05).
SUPPLEMENTARY MATERIAL
FUNDING
Medical Research Council (Career Development Award to J.N.M); and the Genetics Society. Funding to pay the Open Access charge was provided by the Medical Research Council.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to J. Eggenschwiler (Princeton University) for anti-Tulp3 antibody, for providing targeted Tulp3 mice and for sharing unpublished information. We thank T. Theil (Edinburgh University) for providing Gli3Xt/+ mice; B. Yoder (University of Alabama) for anti-polaris antibody; V. Fotaki (Edinburgh University) for Gli3Xt/Xt protein extracts; S. Teglund (Karolinska Institutet) for Sufu−/− protein extracts; L. Niswander (University of Colorado Health Sciences Center) and C. Chiang (Vanderbilt University Medical Center) for plasmids. Monoclonal antibodies against Nkx2.2, Shh, Islet1, MNR2 (developed by Jessell), Msx1/2, Foxa2 (Jessell and Brenner-Morton), Nkx6.1 (Madsen), Pax6 and Pax7 (Kawakami) were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa. We thank S. Townsend, N. Powles-Glover, N. Warr, GEMS core and Harwell histology service for technical assistance; S. Wells and other Mary Lyon Centre staff for mouse husbandry; S. Polley and M. Cormack for support in mutagenesis; and C. Dean, D. Norris and N. Greene for helpful discussions and comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.European Surveillance of Congenital Anomalies. Final activity report. EUROCAT central registry. 2005 wwww.eurocat.ulster.ac.uk/pubdata/Annual-Reports .
- 2.Kasarskis A., Manova K., Anderson K.V. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Natl Acad. Sci. USA. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herron B.J., Lu W., Rao C., Liu S., Peters H., Bronson R.T., Justice M.J., McDonald J.D., Beier D.R. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- 4.Zohn I.E., Anderson K.V., Niswander L. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res. A Clin. Mol. Teratol. 2005;73:583–590. doi: 10.1002/bdra.20164. [DOI] [PubMed] [Google Scholar]
- 5.Copp A.J., Brook F.A., Estibeiro J.P., Shum A.S.W., Cockroft D.L. The embryonic development of mammalian neural tube defects. Prog. Neurobiol. 1990;35:363–403. doi: 10.1016/0301-0082(90)90037-h. [DOI] [PubMed] [Google Scholar]
- 6.Shum A.S.W., Copp A.J. Regional differences in morphogenesis of the neuroepithelium suggest multiple mechanisms of spinal neurulation in the mouse. Anat. Embryol. 1996;194:65–73. doi: 10.1007/BF00196316. [DOI] [PubMed] [Google Scholar]
- 7.Copp A.J., Greene N.D., Murdoch J.N. The genetic basis of mammalian neurulation. Nat. Rev. Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- 8.McMahon A.P., Ingham P.W., Tabin C.J. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 9.Varjosalo M., Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 10.Pasca D.M., Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 11.Wicking C., McGlinn E. The role of hedgehog signalling in tumorigenesis. Cancer Lett. 2001;173:1–7. doi: 10.1016/s0304-3835(01)00676-0. [DOI] [PubMed] [Google Scholar]
- 12.Ybot-Gonzalez P., Cogram P., Gerrelli D., Copp A.J. Sonic hedgehog and the molecular regulation of neural tube closure. Development. 2002;129:2507–2517. doi: 10.1242/dev.129.10.2507. [DOI] [PubMed] [Google Scholar]
- 13.Ybot-Gonzalez P., Gaston-Massuet C., Girdler G., Klingensmith J., Arkell R., Greene N.D., Copp A.J. Neural plate morphogenesis during mouse neurulation is regulated by antagonism of Bmp signalling. Development. 2007;134:3203–3211. doi: 10.1242/dev.008177. [DOI] [PubMed] [Google Scholar]
- 14.Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 15.Jacob J., Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson L., Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev. Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 17.Ulloa F., Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- 18.Briscoe J., Novitch B.G. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philos. Trans. R. Soc. Lond B Biol. Sci. 2008;363:57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlinn E., Tabin C.J. Mechanistic insight into how Shh patterns the vertebrate limb. Curr. Opin. Genet. Dev. 2006;16:426–432. doi: 10.1016/j.gde.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Robert B., Lallemand Y. Anteroposterior patterning in the limb and digit specification: contribution of mouse genetics. Dev. Dyn. 2006;235:2337–2352. doi: 10.1002/dvdy.20890. [DOI] [PubMed] [Google Scholar]
- 21.Tickle C. Making digit patterns in the vertebrate limb. Nat. Rev. Mol. Cell Biol. 2006;7:45–53. doi: 10.1038/nrm1830. [DOI] [PubMed] [Google Scholar]
- 22.Huangfu D., Anderson K.V. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., McMahon A.P., Allen B.L. Shifting paradigms in Hedgehog signaling. Curr. Opin. Cell Biol. 2007;19:159–165. doi: 10.1016/j.ceb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Riobo N.A., Manning D.R. Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem. J. 2007;403:369–379. doi: 10.1042/BJ20061723. [DOI] [PubMed] [Google Scholar]
- 25.Matise M.P., Epstein D.J., Park H.L., Platt K.A., Joyner A.L. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 26.Ding Q., Motoyama J., Gasca S., Mo R., Sasaki H., Rossant J., Hui C.C. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 27.Bai C.B., Auerbach W., Lee J.S., Stephen D., Joyner A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 28.Persson M., Stamataki D., Te W.P., Andersson E., Bose J., Ruther U., Ericson J., Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai C.B., Stephen D., Joyner A.L. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 30.Wang B., Fallon J.F., Beachy P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 31.Litingtung Y., Dahn R.D., Li Y., Fallon J.F., Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 32.Wang C., Ruther U., Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev. Biol. 2007;305:460–469. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterlund T., Kogerman P. Hedgehog signalling: how to get from Smo to Ci and Gli. Trends Cell Biol. 2006;16:176–180. doi: 10.1016/j.tcb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Varjosalo M., Taipale J. Hedgehog signaling. J. Cell Sci. 2007;120:3–6. doi: 10.1242/jcs.03309. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Garcia M.J., Eggenschwiler J.T., Caspary T., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Anderson K.V. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl Acad. Sci. USA. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caspary T., Anderson K.V. Uncovering the uncharacterized and unexpected: unbiased phenotype-driven screens in the mouse. Dev. Dyn. 2006;235:2412–2423. doi: 10.1002/dvdy.20853. [DOI] [PubMed] [Google Scholar]
- 37.Eggenschwiler J.T., Anderson K.V. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haycraft C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., Yoder B.K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 40.Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 41.Kovacs J.J., Whalen E.J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., Lefkowitz R.J. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran P.V., Haycraft C.J., Besschetnova T.Y., Turbe-Doan A., Stottmann R.W., Herron B.J., Chesebro A.L., Qiu H., Scherz P.J., Shah J.V., Yoder B.K., Beier D.R. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epstein D.J., Marti E., Scott M.P., McMahon A.P. Antagonizing cAMP-dependent protein kinase A in the dorsal CNS activates a conserved Sonic hedgehog signaling pathway. Development. 1996;122:2885–2894. doi: 10.1242/dev.122.9.2885. [DOI] [PubMed] [Google Scholar]
- 44.Goodrich L.V., Milenkovic L., Higgins K.M., Scott M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 45.Eggenschwiler J.T., Espinoza E., Anderson K.V. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y., Roelink H., McKnight G.S. Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 2002;277:19889–19896. doi: 10.1074/jbc.M111412200. [DOI] [PubMed] [Google Scholar]
- 47.Bulgakov O.V., Eggenschwiler J.T., Hong D.H., Anderson K.V., Li T. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development. 2004;131:2149–2159. doi: 10.1242/dev.01122. [DOI] [PubMed] [Google Scholar]
- 48.Cooper A.F., Yu K.P., Brueckner M., Brailey L.L., Johnson L., McGrath J.M., Bale A.E. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 49.Varjosalo M., Li S.P., Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 50.Svard J., Heby-Henricson K., Persson-Lek M., Rozell B., Lauth M., Bergstrom A., Ericson J., Toftgard R., Teglund S. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 51.Wong R.L., Wlodarczyk B.J., Min K.S., Scott M.L., Kartiko S., Yu W., Merriweather M.Y., Vogel P., Zambrowicz B.P., Finnell R.H. Mouse Fkbp8 activity is required to inhibit cell death and establish dorso-ventral patterning in the posterior neural tube. Hum. Mol. Genet. 2008;17:587–601. doi: 10.1093/hmg/ddm333. [DOI] [PubMed] [Google Scholar]
- 52.Tempe D., Casas M., Karaz S., Blanchet-Tournier M.F., Concordet J.P. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol. Cell Biol. 2006;26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kogerman P., Grimm T., Kogerman L., Krause D., Unden A.B., Sandstedt B., Toftgard R., Zaphiropoulos P.G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 54.Barnfield P.C., Zhang X., Thanabalasingham V., Yoshida M., Hui C.C. Negative regulation of Gli1 and Gli2 activator function by suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 55.Evans T.M., Ferguson C., Wainwright B.J., Parton R.G., Wicking C. Rab23, a negative regulator of hedgehog signaling, localizes to the plasma membrane and the endocytic pathway. Traffic. 2003;4:869–884. doi: 10.1046/j.1600-0854.2003.00141.x. [DOI] [PubMed] [Google Scholar]
- 56.Walker V.E., Atanasiu R., Lam H., Shrier A. Co-chaperone FKBP38 promotes HERG trafficking. J. Biol. Chem. 2007;282:23509–23516. doi: 10.1074/jbc.M701006200. [DOI] [PubMed] [Google Scholar]
- 57.Christ B., Huang R., Scaal M. Amniote somite derivatives. Dev. Dyn. 2007;236:2382–2396. doi: 10.1002/dvdy.21189. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda A., Ikeda S., Gridley T., Nishina P.M., Naggert J.K. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum. Mol. Genet. 2001;10:1325–1334. doi: 10.1093/hmg/10.12.1325. [DOI] [PubMed] [Google Scholar]
- 59.Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 61.Ericson J., Morton S., Kawakami A., Roelink H., Jessell T.M. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identify. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 62.Wang L.C., Liu Z.Y., Gambardella L., Delacour A., Shapiro R., Yang J., Sizing I., Rayhorn P., Garber E.A., Benjamin C.D., et al. Regular articles: conditional disruption of hedgehog signaling pathway defines its critical role in hair development and regeneration. J. Invest. Dermatol. 2000;114:901–908. doi: 10.1046/j.1523-1747.2000.00951.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X.M., Ramalho-Santos M., McMahon A.P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 64.Chiang C., Litingtung Y., Harris M.P., Simandl B.K., Li Y., Beachy P.A., Fallon J.F. Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev. Biol. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 65.Hui C.C., Joyner A.L. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 66.Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 67.May S.R., Ashique A.M., Karlen M., Wang B., Shen Y., Zarbalis K., Reiter J., Ericson J., Peterson A.S. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 68.Liu A., Wang B., Niswander L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 69.Vierkotten J., Dildrop R., Peters T., Wang B., Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- 70.Zaki P.A., Martynoga B., Delafield-Butt J.T., Fotaki V., Yu T., Price D.J. Loss of Gli3 enhances the viability of embryonic telencephalic cells in vitro. Eur. J. Neurosci. 2005;22:1547–1551. doi: 10.1111/j.1460-9568.2005.04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boggon T.J., Shan W.S., Santagata S., Myers S.C., Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 72.Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kolpakova-Hart E., Jinnin M., Hou B., Fukai N., Olsen B.R. Kinesin-2 controls development and patterning of the vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev. Biol. 2007;309:273–284. doi: 10.1016/j.ydbio.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiter J.F., Skarnes W.C. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20:22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wijgerde M., McMahon J.A., Rule M., McMahon A.P. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 2002;16:2849–2864. doi: 10.1101/gad.1025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lupo G., Harris W.A., Lewis K.E. Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- 77.Liem K.F., Jr., Jessell T.M., Briscoe J. Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development. 2000;127:4855–4866. doi: 10.1242/dev.127.22.4855. [DOI] [PubMed] [Google Scholar]
- 78.Ikeya M., Lee S.M.K., Johnson J.E., McMahon A.P., Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 79.Muroyama Y., Fujihara M., Ikeya M., Kondoh H., Takada S. Wnt signaling plays an essential role in neuronal specification of the dorsal spinal cord. Genes Dev. 2002;16:548–553. doi: 10.1101/gad.937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wine-Lee L., Ahn K.J., Richardson R.D., Mishina Y., Lyons K.M., Crenshaw E.B., III Signaling through BMP type 1 receptors is required for development of interneuron cell types in the dorsal spinal cord. Development. 2004;131:5393–5403. doi: 10.1242/dev.01379. [DOI] [PubMed] [Google Scholar]
- 81.Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 82.Vokes S.A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W.J., Davidson E.H., Wong W.H., McMahon A.P. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 83.Robertson C.P., Braun M.M., Roelink H. Sonic hedgehog patterning in chick neural plate is antagonized by a Wnt3-like signal. Dev. Dyn. 2004;229:510–519. doi: 10.1002/dvdy.10501. [DOI] [PubMed] [Google Scholar]
- 84.Marti E., Bumcrot D.A., Takada R., McMahon A.P. Requirement of 19K form of Sonic hedgehog for induction of distinct ventral cell types in CNS explants. Nature. 1995;375:322–325. doi: 10.1038/375322a0. [DOI] [PubMed] [Google Scholar]
- 85.Ericson J., Briscoe J., Rashbass P., Van Heyningen V., Jessell T.M. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harb. Symp. Quant. Biol. 1997;62:451–466. [PubMed] [Google Scholar]
- 86.Briscoe J., Sussel L., Serup P., Hartigan-O’Connor D., Jessell T.M., Rubenstein J.L.R., Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- 87.Nishina P.M., North M.A., Ikeda A., Yan Y., Naggert J.K. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics. 1998;54:215–220. doi: 10.1006/geno.1998.5567. [DOI] [PubMed] [Google Scholar]
- 88.Ikeda A., Nishina P.M., Naggert J.K. The tubby-like proteins, a family with roles in neuronal development and function. J. Cell Sci. 2002;115:9–14. doi: 10.1242/jcs.115.1.9. [DOI] [PubMed] [Google Scholar]
- 89.Carroll K., Gomez C., Shapiro L. Tubby proteins: the plot thickens. Nat. Rev. Mol. Cell Biol. 2004;5:55–63. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- 90.Noben-Trauth K., Naggert J.K., North M.A., Nishina P.M. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 91.Hagstrom S.A., Duyao M., North M.A., Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 92.Stubdal H., Lynch C.A., Moriarty A., Fang Q., Chickering T., Deeds J.D., Fairchild-Huntress V., Charlat O., Dunmore J.H., Kleyn P., Huszar D., Kapeller R. Targeted deletion of the tub mouse obesity gene reveals that tubby is a loss-of-function mutation. Mol. Cell Biol. 2000;20:878–882. doi: 10.1128/mcb.20.3.878-882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ikeda S., Shiva N., Ikeda A., Smith R.S., Nusinowitz S., Yan G., Lin T.R., Chu S., Heckenlively J.R., North M.A., et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum. Mol. Genet. 2000;9:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- 94.Bode C., Wolfrum U. Caspase-3 inhibitor reduces apototic photoreceptor cell death during inherited retinal degeneration in tubby mice. Mol. Vis. 2003;9:144–150. [PubMed] [Google Scholar]
- 95.Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 96.Ferrante M.I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dolle P., Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 97.Houde C., Dickinson R.J., Houtzager V.M., Cullum R., Montpetit R., Metzler M., Simpson E.M., Roy S., Hayden M.R., Hoodless P.A., Nicholson D.W. Hippi is essential for node cilia assembly and Sonic hedgehog signaling. Dev. Biol. 2006;300:523–533. doi: 10.1016/j.ydbio.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ocbina P.J., Anderson K.V. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev. Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eggenschwiler J.T., Bulgakov O.V., Qin J., Li T., Anderson K.V. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev. Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 100.Cho A., Ko H.W., Eggenschwiler J.T. FKBP8 cell-autonomously controls neural tube patterning through a Gli2- and Kif3a-dependent mechanism. Dev. Biol. 2008;321:27–39. doi: 10.1016/j.ydbio.2008.05.558. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y., Ng E.L., Tang B.L. Rab23: what exactly does it traffic? Traffic. 2006;7:746–750. doi: 10.1111/j.1600-0854.2006.00416.x. [DOI] [PubMed] [Google Scholar]
- 102.Nakagawa T., Shirane M., Iemura S., Natsume T., Nakayama K.I. Anchoring of the 26S proteasome to the organellar membrane by FKBP38. Genes Cells. 2007;12:709–719. doi: 10.1111/j.1365-2443.2007.01086.x. [DOI] [PubMed] [Google Scholar]
- 103.Xi Q., Pauer G.J., Marmorstein A.D., Crabb J.W., Hagstrom S.A. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest. Ophthalmol. Vis. Sci. 2005;46:4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xi Q., Pauer G.J., Ball S.L., Rayborn M., Hollyfield J.G., Peachey N.S., Crabb J.W., Hagstrom S.A. Interaction between the photoreceptor-specific tubby-like protein 1 and the neuronal-specific GTPase dynamin-1. Invest. Ophthalmol. Vis. Sci. 2007;48:2837–2844. doi: 10.1167/iovs.06-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hagstrom S.A., Adamian M., Scimeca M., Pawlyk B.S., Yue G., Li T. A role for the Tubby-like protein 1 in rhodopsin transport. Invest. Ophthalmol. Vis. Sci. 2001;42:1955–1962. [PubMed] [Google Scholar]
- 106.He W., Ikeda S., Bronson R.T., Yan G., Nishina P.M., North M.A., Naggert J.K. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Brain Res. Mol. Brain Res. 2000;81:109–117. doi: 10.1016/s0169-328x(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 107.Santagata S., Boggon T.J., Baird C.L., Gomez C.A., Zhao J., Shan W.S., Myszka D.G., Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 108.Gunther T., Struwe M., Aguzzi A., Schughart K. Open brain, a new mouse mutant with severe neural tube defects, shows altered gene expression patterns in the developing spinal cord. Development. 1994;120:3119–3130. doi: 10.1242/dev.120.11.3119. [DOI] [PubMed] [Google Scholar]
- 109.Gunther T., Sporle R., Schughart K. The open brain (opb) mutation maps to mouse chromosome 1. Mamm. Genome. 1997;8:583–585. doi: 10.1007/s003359900509. [DOI] [PubMed] [Google Scholar]
- 110.Eggenschwiler J.T., Anderson K.V. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev. Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]
- 111.Pan Y., Bai C.B., Joyner A.L., Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davey M.G., Paton I.R., Yin Y., Schmidt M., Bangs F.K., Morrice D.R., Smith T.G., Buxton P., Stamataki D., Tanaka M., et al. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–1377. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simon-Chazottes D., Paces-Fessy M., Lamour-Isnard C., Guenet J.L., Blanchet-Tournier M.F. Genomic organization, chromosomal assignment, and expression analysis of the mouse suppressor of fused gene (Sufu) coding a Gli protein partner. Mamm. Genome. 2000;11:614–621. doi: 10.1007/s003350010144. [DOI] [PubMed] [Google Scholar]
- 114.Dunaeva M., Michelson P., Kogerman P., Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 115.Shimokawa T., Tostar U., Lauth M., Palaniswamy R., Kasper M., Toftgard R., Zaphiropoulos P.G. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J. Biol. Chem. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Paces-Fessy M., Boucher D., Petit E., Paute-Briand S., Blanchet-Tournier M.F. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem. J. 2004;378:353–362. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng S.Y., Bishop J.M. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl Acad. Sci. USA. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Evangelista M., Lim T.Y., Lee J., Parker L., Ashique A., Peterson A.S., Ye W., Davis D.P., De Sauvage F.J. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci. Signal. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- 119.Zhou H., Kim S., Ishii S., Boyer T.G. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol. Cell Biol. 2006;26:8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Callahan C.A., Ofstad T., Horng L., Wang J.K., Zhen H.H., Coulombe P.A., Oro A.E. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18:2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dai P., Shinagawa T., Nomura T., Harada J., Kaul S.C., Wadhwa R., Khan M.M., Akimaru H., Sasaki H., Colmenares C., Ishii S. Ski is involved in transcriptional regulation by the repressor and full-length forms of Gli3. Genes Dev. 2002;16:2843–2848. doi: 10.1101/gad.1017302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mao J., Maye P., Kogerman P., Tejedor F.J., Toftgard R., Xie W., Wu G., Wu D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 123.Varjosalo M., Bjorklund M., Cheng F., Syvanen H., Kivioja T., Kilpinen S., Sun Z., Kallioniemi O., Stunnenberg H.G., He W.W., Ojala P., Taipale J. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 124.McGlinn E., van Bueren K.L., Fiorenza S., Mo R., Poh A.M., Forrest A., Soares M.B., Bonaldo M.F., Grimmond S., Hui C.C., Wainwright B., Wicking C. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 2005;122:1218–1233. doi: 10.1016/j.mod.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 125.Wilson L., Gale E., Chambers D., Maden M. Retinoic acid and the control of dorsoventral patterning in the avian spinal cord. Dev. Biol. 2004;269:433–446. doi: 10.1016/j.ydbio.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 126.Maynard T.M., Jain M.D., Balmer C.W., LaMantia A.S. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm. Genome. 2002;13:58–61. doi: 10.1007/s00335-001-2115-x. [DOI] [PubMed] [Google Scholar]
- 127.Wilkinson D.G. Oxford: IRL Press; 1992. In Situ Hybridisation: A Practical Approach. [Google Scholar]
- 128.Murdoch J.N., Eddleston J., Leblond-Bourget N., Stanier P., Copp A.J. Sequence and expression analysis of Nhlh1: a basic helix-loop-helix gene implicated in neurogenesis. Dev. Genet. 1999;24:165–177. doi: 10.1002/(SICI)1520-6408(1999)24:1/2<165::AID-DVG15>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 129.Murdoch J.N., Doudney K., Paternotte C., Copp A.J., Stanier P. Severe neural tube defects in the loop-tail mouse result from mutation of Lpp1, a novel gene involved in floor plate specification. Hum. Mol. Genet. 2001;10:2593–2601. doi: 10.1093/hmg/10.22.2593. [DOI] [PubMed] [Google Scholar]
- 130.Szczepny A., Hime G.R., Loveland K.L. Expression of hedgehog signalling components in adult mouse testis. Dev. Dyn. 2006;235:3063–3070. doi: 10.1002/dvdy.20931. [DOI] [PubMed] [Google Scholar]
- 131.Briscoe J., Pierani A., Jessell T.M., Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.