Abstract
Autism spectrum disorders (ASDs) constitute a group of severe neurodevelopmental conditions with complex multifactorial etiology. In order to explore the hypothesis that submicroscopic genomic rearrangements underlie some ASD cases, we have analyzed 96 Spanish patients with idiopathic ASD after extensive clinical and laboratory screening, by array comparative genomic hybridization (aCGH) using a homemade bacterial artificial chromosome (BAC) array. Only 13 of the 238 detected copy number alterations, ranging in size from 89 kb to 2.4 Mb, were present specifically in the autistic population (12 out of 96 individuals, 12.5%). Following validation by additional molecular techniques, we have characterized these novel candidate regions containing 24 different genes including alterations in two previously reported regions of chromosome 7 associated with the ASD phenotype. Some of the genes located in ASD-specific copy number variants act in common pathways, most notably the phosphatidylinositol signaling and the glutamatergic synapse, both known to be affected in several genetic syndromes related with autism and previously associated with ASD. Our work supports the idea that the functional alteration of genes in related neuronal networks is involved in the etiology of the ASD phenotype and confirms a significant diagnostic yield for aCGH, which should probably be included in the diagnostic workup of idiopathic ASD.
INTRODUCTION
Autism spectrum disorders (ASDs) (OMIM: 209850) are a group of severe neurodevelopmental conditions, referred to a broader extent as pervasive developmental disorders, characterized by a triad associating impairments in social interactions, communication deficits and restricted repetitive and stereotyped behaviors and interests with an onset in infancy or early childhood (before 3 years). The estimated prevalence of ASD was 2–5/10 000 with a ratio four times higher in males than in females (1). In the last decades, a significant increase (6–10-fold) of prevalence has been noticed, partially explained by improvements in case ascertainment, making ASD a public health priority (2).
There is strong evidence for a genetic etiology of ASD given that the concordance rates in monozygotic twins are ∼90% for ASD, whereas concordance rates in dizygotic twins are ∼10% (3). ASD is considered to have a complex multifactorial etiology involving many genes. These genetic factors would contribute to the neurobiological alterations responsible for the final array of autistic phenotypes.
ASD is found in association with other conditions in ∼10% of cases, including known genetic disorders such as fragile-X syndrome, tuberous sclerosis and cytogenetic abnormalities, mostly duplications at 15q11.13 on the maternally inherited chromosome (Prader–Willi/Angelman syndrome region) and terminal deletions of chromosomes 2q or 22q (4,5). Genome-wide linkage and candidate–gene association studies have found evidence for autism susceptibility loci on more than 20 different chromosome regions including 1p, 2q, 5q, 7q, 15q, 17q 19p and Xq, although only a few of them have been replicated (6). Single-gene mutations in SHANK3 (gene ID: 85358) (22q13.3), NLGN3/NGLN4 (gene ID: 54413/57502) (Xq13/Xp22.33) and NRXN1 genes (gene ID: 9378) (2p16) have been described as causative of ASD in a small number of patients. Additional studies and homozygosity mapping in inbred families have also revealed genes with strong susceptibility to autism, such as the neurexin family member CNTNAP2 (gene ID: 26047) among others (7–13). Recent reports showed that submicroscopic copy number variants (CNVs) can also have a relevant role in autism (14,15) with de novo germline variants as a significant risk factor in sporadic forms of ASD (16). A specific CNV at 16p11.2 has been found in ∼1% of the ASD patients as well as in individuals with mental retardation (17–19). In addition, the reciprocal duplication of several deletion syndromes, dup7q11.23, dup17p11.2 and dup22q11, can lead to a phenotype of mental retardation and ASD (20–22). All these data further indicate the genetic heterogeneity of ASD with different modes of inheritance and different contribution of de novo and inherited variants in individual families.
Given that the cause of ASD in the majority of patients is still unknown and the fact that many genomic regions have been associated with the disorder, genome-wide screening methods seem to be indicated for the identification of affected regions and specific CNVs that could be associated to the disease. We have analyzed 96 Spanish patients with idiopathic ASD after ruling out most known genetic causes, using a homemade BAC microarray for comparative genomic hybridization (aCGH) (23). With this approach, we have identified 13 novel ASD-specific copy number changes including two rearrangements in genomic regions previously associated with the ASD phenotype, such as 7q11.22 and 7q31.33–32.1. Interestingly, a significant proportion of the 24 different genes located in these 13 loci are strong functional candidates for ASD, involved in common or related neuronal signaling networks.
RESULTS
We selected for the study 96 Spanish patients (12 females and 84 males, ratio: 1/7) with idiopathic ASD after extensive clinical and laboratory screening. ASD was confirmed in all cases using the Autism Diagnostic Interview-Revised (ADI-R). All patients had been examined by neurologists and clinical geneticists and also had an extensive negative laboratory workup, including standard karyotyping, fragile X molecular testing, subtelomeric and targeted multiplex ligation probe amplification (MLPA) screening (probes for the regions 1p36, 2q37, 7q11.23, 15q11–q13, 16p11.2, 17p11.2, 22q11.2 and 22q13.3), as well as metabolic studies in some cases when clinically indicated (Gener et al., submitted).
Autism-specific CNVs
We detected gains and losses of a total of 238 BACs in the ASD samples with the hot-spot-BAC-array (HSBA) aCGH: 194 of these CNVs had been previously described as putatively polymorphic in the population by other authors (http://projects.tcag.ca/variation) and 18 more were also detected in our control samples (n = 52). We considered that two CNVs could be the same when they overlapped in >70% of their predicted genomic content. The remaining 26 variants covering 21 genomic regions (5 deletions, 15 amplifications and 1 detected as both) were present specifically in the ASD population and were then considered as possibly pathogenic (Supplementary Material, Table S1). These CNVs ranged in size from 89 kb to 2.4 Mb. On average, we detected 9.8 CNVs per individual in ASD samples and 12.4 in controls, with no significant difference.
To define the experimental variability of the hybridization signal of the BACs detecting CNVs, we performed a boxplot analysis of the log2 ratios in all control samples; 7 out of 26 BACs showed high signal dispersion, suggesting possible false-positive results (Supplementary Material, Fig. S1).
In order to validate, better define and determine whether the novel rearrangements were inherited or de novo, we then targeted these 21 putative pathological variants with additional molecular technologies: MLPA assays with specific synthetic probes and oligo aCGH (Agilent 44k or 244K) or SNP array (Illumina 370). Using these tools, we confirmed the CNVs in 13 regions affecting 12 patients, and we defined the size and nature of the rearrangement, being complex in some cases (Table 1, Fig. 1). A total of eight CNVs detected with the BAC array were not confirmed by the other methods, six of them corresponding to single BACs with high dispersion of the signal (Supplementary Material, Fig. 1). The MLPA assays confirmed that the rearrangements were inherited in the five patients for whom both parental samples were available. Unfortunately, samples from one or both parents of the remaining seven patients were unavailable and we could not determine whether their CNVs were de novo or inherited (Table 1).
Table 1.
Description of the confirmed CNVs affecting each of the 12 patients with ASD, including the BAC ID, chromosomal location, size, CNV type, validation methods and gene content
| Sample | Gender | BAC ID | Cytoband | CNV | Validation | Start | End | Length (kb) | Origin | MLPA probe | Gene content |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUT21 | F | RP11-125A7 | chr13q14.11 | Gain | MLPA + Illumina | 41 003 320 | 41 418 753 | 415.433 | MAT | KIAA0564 | KIAA0564 |
| AUT24 | M | RP11-55L3a | chr4q26 | Gain | MLPA + Agilent | 117 191 242 | 119 614 346 | 2423.104 | MAT | TRAM1L1 | NDST3, PRSS12, TRAM1L1 |
| RP11-458P15 | |||||||||||
| RP11-61N14 | |||||||||||
| RP11-335K15a | |||||||||||
| RP11-778G8a | |||||||||||
| RP11-510D4a | |||||||||||
| RP11-750O22a | |||||||||||
| RP11-366I1a | |||||||||||
| CTD-201L19a | |||||||||||
| RP11-630F19a | |||||||||||
| AUT31 | M | RP1-117N3 | chr1p35.1 | Loss | Agilent | 33 158 375 | 33 263 299 | 104.924 | NA | RNF19B, AK2 | |
| AUT42 | M | RP11-140C4 | chr2q33.3 | Gain | MLPA + Illumina | 208 759 831 | 208 849 748 | 89.917 | NA | PIP5K3 | C2orf80, IDH1, PIP5K3 |
| AUT45 | F | RP11-266G18 | chr21q21.2 | Gain | Illumina | 25 518 184 | 25 708 178 | 189.994 | NA | No genes | |
| AUT84 | M | RP11-510O20a | chr7q31.33–q32.1 | Gain | MLPA + Agilent | 125 672 999 | 126 255 215 | 582.216 | NA | GRM8 | GRM8 |
| RP11-105E3 | |||||||||||
| RP11-432H11 | |||||||||||
| RP11-21K15 | |||||||||||
| RP11-475H14a | |||||||||||
| CTD-210M22a | |||||||||||
| RP11-290P11a | |||||||||||
| AUT91 | M | RP11-99M10 | chr18q12.1 | Gain | MLPA + Illumina | 10 127 382 | 10 594 362 | 466.98 | NA | NAPG | APCDD1, NAPG |
| AUT96 | M | RP11-21N8 | chr3q21.3 | Gain | MLPA + Agilent | 131 845 264 | 131 974 345 | 129.081 | NA | PIK3R4 | COL6A6, PIK3R4 |
| RP11-575M4 | chr7q11.22 | Gain | MLPA + Agilent | 69 630 362 | 69 955 721 | 325.359 | AUTS2 | AUTS2 | |||
| AUT138 | M | RP11-281H14 | chr11q14.1 | Loss | MLPA + Illumina | 84 032 216 | 84 276 593 | 244.377 | NA | DLG2 | DLG2 |
| AUT150 | M | RP11-47L17a RP11-158F10 RP11-194K9 |
chr5q35.3 | Gain | MLPA + Agilent | 177 501 908 | 177 688 820 | 186.912 | PAT | AGXT2L2 | COL23A1, AGXT2L2, GMCL1L, HNRNPAB, NOLA2, RMND5B |
| AUT186 | M | RP11-239E10 | chr1q41 | Loss | MLPA + Illumina | 221 401 766 | 221 501 748 | 99.982 | PAT | SUSD4 | SUSD4 |
| AUT195 | M | RP11-107B3 | chr3q22.3 | Gain | MLPA + Illumina | 139 934 042 | 140 070 771 | 136.729 | PAT | PIK3CB | PIK3CB |
The final CNV size estimate is based on oligo/SNP array data.
F, female; M, male; NA, not available; MAT, maternally inherited; PAT, paternally inherited.
aBACs overlapping with polymorphic CNVs described in controls.
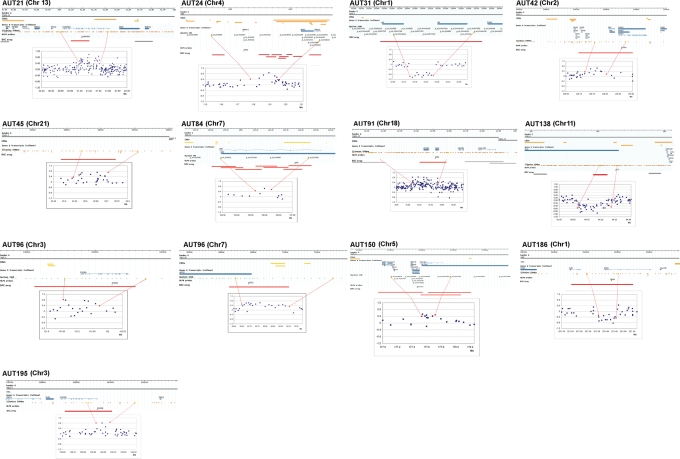
Figure 1.
Specific rearrangements detected in ASD patients. Each subfigure shows, under the patient’s identifier and chromosome number, a scheme of the genomic region containing the CNV. From top to bottom: (i) chromosomal scale (Mb), (ii) G-band of the chromosome, (iii) regional CNVs described in public databases (http://projects.tcag.ca/variation), (iv) genes in the interval, (v) probes of the Illumina or Agilent arrays, (vi) MLPA probes, (vii) probes of the BAC array and (viii) plot with the results of the oligo or SNP array hybridization showing the deleted (below 0.4) or duplicated (above 0.4) intervals flanked by red dots.
We then screened our entire cohort of autistic samples (n = 215) and controls (n = 120) with a homemade MLPA panel containing probes targeting each of the CNVs found by aCGH. We did not find rearrangements in any additional patient or control individual. We also searched the database of Genomics Variants (http://projects.tcag.ca/variation) for similar rearrangements, using the data from controls (n> 1800) that had been analyzed by high-density oligo arrays with enough probe coverage in the regions (more than 10 probes per locus) (24–26). Although there was some partial overlap at three loci with CNVs reported in population controls (Supplementary Material, Table S1), none was of the size and gene content of those found in ASD patients. Thus, the CNVs found are unique in each family and very rare or absent in the general population.
More detailed phenotype data for the 12 ASD patients with specific rearrangements are shown in Table 2.
Table 2.
Phenotype data of the 12 patients with autism spectrum and specific rearrangements
| Patient | Sex | Diagnosis | Mental retardation | Dysmorphism | Seizures | Aggressiveness | Other features |
|---|---|---|---|---|---|---|---|
| AUT21 | F | Autism | Moderate | No | No | No | |
| AUT24 | M | Autism | Moderate | No | No | No | Neurosensorial deafness (65 dB); diaphagmatic hernia (Bochdalek) |
| AUT31 | M | Autism | Severe | No | No | No | |
| AUT42 | M | Autism | Severe | No | No | Yesa | |
| AUT45 | F | Autism | Severe | No | Yes | Yes | Subcortical brain atrophy |
| AUT84 | M | Autism | Severe | No | Yes | No | Hyperekplexia or startle disease |
| AUT91 | M | Autism | Severe | No | No | No | |
| AUT96 | M | Autism | Severe | No | No | Yesa | |
| AUT138 | M | Autism | Severe | No | No | No | Hypermetropy (+5d), short stature (−2DS) |
| AUT150 | M | Autism | Moderate | No | No | No | Obsessive-compulsive disorder |
| AUT186 | M | PDD | Mild | Yes | No | No | Retrognatia |
| AUT195 | M | Autism | Mild | No | No | No | Unilateral neurosensorial deafness |
PDD, pervasive developmental disorder.
aOccasional self-injurious behavior.
Candidate ASD genes and pathways
We considered as candidates all genes located in the ASD-unique rearranged regions that were validated with additional molecular techniques—a total of 24 genes (Supplementary Material, Table S2). Only 2 out of 24 were completely deleted (AK2 and RNF19B) (gene IDs: 204/127544), 11 out of 24 completely duplicated and the remaining 11 out of 24 partially altered or disrupted.
Two of the detected alterations lie in regions of chromosome 7 previously related with ASD, 7q11.22 and 7q31.3, containing or disrupting a single candidate gene per rearrangement, AUTS2 (gene ID: 282553) and GRM8 (gene ID: 2918), respectively (Fig. 1). The remaining 11 CNV regions detected in ASD patients affected novel regions.
We then analyzed the data set of genes in ASD-related CNVs, using different computational resources [Ingenuity Pathway Analysis (IPA), ConsensusPathDB, KEGG], to obtain a general overview of their most relevant functions. Among the different functions encoded by those genes, the inositol and phosphatidylinositol-3-OH kinase (PI3K) signaling pathways were significantly overrepresented (P = 1.64E + 06) (Supplementary Material, Table S3). Interestingly, in addition to the two genes directly involved in the PI3K pathway (PIK3CB and PIP5K3) (gene IDs: 5291/200576), at least four other genes included in the ASD-specific CNVs encode proteins that act either upstream or downstream of PI3K: PIK3R4, DLG2, AGXT2L2 and GRM8 (Fig. 2 and Supplementary Material, Table S2), in related pathways such as the toll-like receptor signaling, the regulation of autophagy or the neuroactive ligand–receptor interaction. Therefore, at least 6 of the 13 ASD-specific CNVs affect genes with a role in related pathways, whereas only 4 of the remaining 212 non-ASD-specific CNVs contained genes in those pathways (significant Z-test with a 99% confidence interval). This finding strongly suggests a potential contribution of the identified ASD-specific CNVs to the ASD phenotype.
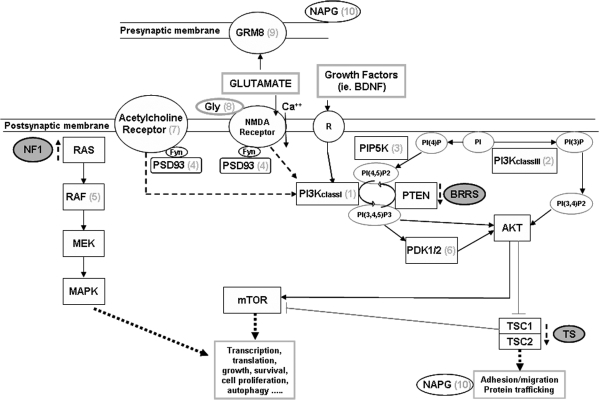
Figure 2.
Schematic representation of the glutamatergic synapse and the postsynaptic neuronal signaling pathways relevant to ASD, with the putative location of the proteins encoded by the genes affected by the CNVs detected [(1) PIK3CB, (2) PIK3R4, (3) PIP5K3, (4) DLG2, (5) RAF1, (6) PDPK1, (7) CHRNA7, (8) AGXT2L2, (9) GRM8 and (10) NAPG]. The interrupted arrows show the abnormal function in three monogenic disorders associated with ASD: NF1, BRRS and TSC.
To explore whether the same pathways were also overrepresented in disease-specific CNVs detected in ASD samples by other studies (15,16,27,28), we downloaded the genes in those CNVs and analyzed their functional annotation by the ConsensusPathDB. We obtained a total of 2901 genes from the available data. Using the pathway-based sets, we observed the same pathways clearly overrepresented among those genes, being on top the toll-like receptor signaling, the regulation of autophagy and the mTOR signaling (P < 0.001), among others (Supplementary Material, Table S4). PI3K signaling is key in the mTOR upstream activation process (Fig. 2).
Non-specific CNVs
We also analyzed the CNVs found in our ASD patients that had also been described as rare variants in controls (reported in <1% of controls) for the presence of genes belonging to the same or related pathways. Interestingly, three patients showed a gain-type CNV in the 15q13.3 region that included the CHRNA7 gene (gene ID: 1139). Given that the flanking BACs in the array gave normal signal, this duplication likely corresponds to a ∼500 kb interval (29, 8–30, 3 Mb of chromosome 15) located between segmental duplications that has also been found in ∼1% of the general population (http://projects.tcag.ca/variation). In addition, an autistic patient showed a gain-type CNV, including the PDPK1 gene (gene ID: 5170), and another case had a gain-type CNV, including the RAF1 gene (gene ID: 5894). By MLPA analysis, we found that the CNVs in these five cases were inherited from unaffected progenitors. The RASSF5 gene (gene ID: 83593) as well as five RAB family members related to the same signaling pathway were also present in non-ASD exclusive CNVs.
DISCUSSION
The use of high-throughput genomic techniques has demonstrated to be a powerful strategy for the detection of genomic abnormalities associated with ASD (15,16,27). In our hands, the use of a BAC aCGH has allowed the identification of specific regions altered in copy number in a significant proportion of our autistic population, 12 out of 96 cases (12.5%), being inherited from a normal parent in all five cases with parental samples available. Some of these regions were already reported to harbor ASD genes by either linkage and association studies or by the finding of chromosomal translocation breakpoints in ASD patients (11,29–31). Other altered regions in our autistic population that could be linked to the pathological phenotype have also been found as rare variants in relatives and/or non-autistic population. The incomplete penetrance of these rare inherited CNVs could be due to several factors including interaction with other genetic or epigenetic alterations, environmental modifiers or the unmasking of recessive alleles. This is probably the expected situation in a multifactorial and complex disease such as ASD, a group of different conditions with overlapping phenotype generated each from a single or, more frequently, a combination of genetic changes with possible environmental contribution as well. Therefore, genetic studies should search for the different multifactorial combinations.
The CNVs found in ASD patients could be merely passenger changes or have a driver contribution to the autistic phenotype should they lead to disruption or deregulation of relevant genes. We initially focused our interest in the regions that were altered only in our autistic population and had not been found in controls. We detected and confirmed 13 chromosomal alterations affecting 12 of our patients. Since we had excluded from the study ASD patients with non-idiopathic ASD after previous extensive clinical and laboratory screening with targeted MLPA analyses for subtelomeric and known rearrangements, the yield of the aCGH in detecting novel CNVs potentially related with the ASD phenotype is quite significant—12.5%. Only 3 were genomic losses, whereas 10 changes were genomic gains.
Two of the ASD-related CNVs identified map to regions of strong linkage with ASD on chromosome 7, 7q11.22 and 7q31.3, and contain genes already proposed as candidates, AUTS2 and GRM8, respectively. The AUTS2 gene, encoding a large protein of unknown function, was found disrupted at the 7q11.2 breakpoint of different balanced translocations and inversion in ASD patients, as well as in unrelated patients with severe mental retardation (11,29,30). Genetic variants in the GRM8 gene, coding for the glutamatergic receptor 8, have also shown significant association with ASD (31). GRM8 is a strong functional candidate given that GRM8 overactivity in the lateral amygdala leads to the inhibition of synaptic transmission impairing learned fear acquisition, a feature present in ASD (32). It has also been shown that the lateral amygdala has an abnormal growth pattern and significantly fewer neurons in autistic patients (33). Interestingly, the patient with the rearranged GRM8 gene presented, in addition to ASD and severe mental retardation, an abnormal startle response to tactile stimuli and a diagnosis of hyperekplexia (OMIM: 149400). Dysfunction of the glutamatergic synapse in ASD is also supported by the finding of mutations in additional genes somehow involved in glutamatergic transmission, such as NLGN3, NLGN4, NRXN1, SHANK3, CNTNAP2 and even FMR1 (gene ID: 2332), the gene responsible for the fragile X syndrome (FXS, OMIM: 300624).
Out of the genes affected by the remaining ASD-related CNVs, we have highlighted some genes with causative potential considering their biological role. The most striking finding is that two of those genes directly participate in the PI3K signaling pathway (PIP5K3 and PIK3CB) and a third gene (PIK3R4) is highly related. Signaling by phosphorylated species of phosphatidylinositol regulates diverse cellular processes including membrane trafficking, cytoskeletal reorganization (34) and sex-dependent synaptic patterning (35), having also a role in glutamatergic and nicotinic neurotransmission and mTOR activation (36). The putative association of this pathway with the autistic phenotype has been previously proposed by other reports (31,37,38). Transmission disequilibrium test and haplotype analyses of regions of previous linkage to autism demonstrated that polymorphisms in the INPP1, PIK3CG and TSC2 genes (gene IDs: 3628/5294/7249), all in the PI3K pathway, are in linkage disequilibrium with autism (39).
Although the functional consequences of these three CNVs are still unknown, we propose that they all might lead to the upregulation of the PI3K pathway. The first CNV includes a partial duplication of PIK3CB affecting the N terminal part of the gene that contains the p85-negative regulatory subunit-binding site and the RAS-binding site but not the catalytic C terminal domain. A deletion mutant of PIK3CB lacking the entire p85-binding domain has been previously described that efficiently activated PI3K signaling (40). The second amplification-type CNV includes the promoter region of PIP5K3. It is logical to propose that overfunction might be related to ASD, since heterozygous hypomorphic mutations of this gene cause the Francois–Neetens fleck corneal dystrophy (OMIM: 121850) (41), a phenotype confined to the cornea that was not observed in our patient. A third CNV contains a complete duplication of the PIK3R4 gene, coding for a highly conserved protein from yeast to humans that interacts with PIK3C3 in vivo to regulate the protein trafficking required for PI3K activity (42).
Interestingly, we found additional genes related to the same pathway in CNVs detected in ASD patients; initially discarded because they had been reported in normal controls as rare variants. The duplication-type CNV containing the PDPK1 gene in one patient might somehow alter the PI3K signaling pathway since PDPK1 directly phosphorylates AKT (Fig. 2). The duplication-type CNV observed in three patients at 15q13.3 containing the CHRNA7 gene which codes for the alpha7 nicotinic acetylcholine receptor could also contribute to pathway disregulation because the cytoplasmatic signal transmission of this receptor involves PI3K (36). Microdeletion and microduplication of a larger interval in this region have recently been described as a novel syndrome with incomplete penetrance and variable phenotype including mental retardation, autistic features, epilepsy and other neuropsychiatric disorders (43).
We sought further evidence of genetic variation in the PI3K pathway in previously published data of autistic populations (15,16,27,28). Three other genes directly related to this pathway were found in ASD-associated rearrangements: five ASD patients had a loss of GAB2 (gene ID: 9846), coding for a PI3K-negative regulator in the TCR signaling pathway, one case had a gain of PIK3C2G (gene ID: 5288) and one case had a PIP5K1B (gene ID: 8395) gene disruption by a balanced translocation (15).
Additional evidence supporting a role of the PI3K signaling pathway in autism comes from several well-characterized genetic diseases frequently comorbid with ASD, such as tuberous sclerosis (TSC, OMIM: 191100), neurofibromatosis 1 (NF1, OMIM: 162200), Bannayan–Riley–Rubalcaba syndrome (BRRS, OMIM: 153480) and FXS (44). The prevalence of autism in patients with TSC is higher than in any other condition (43–86%) (45). The TSC genes, TSC1 and TSC2, code for proteins that act downstream the PI3K signaling pathway downregulating mTOR (Fig. 2). As an integrator of external stimuli, a tight regulation of mTOR activity promoting cell growth, survival and proliferation is required for neural development (46,47). BRRS patients with PTEN mutations may also show ASD-like phenotype. In addition, Pten KO mice show macrocephaly, socialization problems and exaggerated responses to sensory stimuli, along with abnormal neurons with dendritic hypertrophy (37). PTEN acts as a negative regulator of the PI3K signaling pathway, and overactivity of PI3K could overcome the PTEN-negative control (Fig. 2). NF1 has also been associated with high risk for ASD, suggesting related etiologies for both disorders (48). NF1 is due to a reduction of neurofibromin activity, a tumor-suppressor protein with RAS GTPase activity that attenuates the mitogen-activated protein kinase and PI3K pathways (49). Mice that lack neurofibromin in cortical neurons and astrocytes fail to form cortical barrels in the somatosensory cortex (50). RAS also activates the RAF1 kinase, included as a gain-type CNV in one of our ASD patients and his mother, as well as in 2 out of 506 controls (24). Finally, 33% of FXS patients fulfill criteria for ASD. FXS is caused by the silencing of FMR1, coding for FMRP, a negative regulator of mRNA translation. The loss of FMRP in FXS leads to mGluR-dependent LTD increase, which, at the same time, is sensitive to PI3K inhibitors (51).
The PI3K signaling pathway is one of the postsynaptic transductors of glutamate neurotransmission. The glutamatergic neurotransmission has been repeatedly implicated in some of the pathogenic mechanisms of ASD (52–54). In addition to the evidence coming from the patient with the GRM8 alteration, another ASD case showed a disruption of the DLG2 gene which codes for PSD93, a protein that mediates tyrosine-phosphorylation of the N-methyl-d-aspartic acid (NMDA) receptors by Fyn (55). This phosphorylation upregulates NMDA receptor (NMDAR) function and is also needed for its interaction with PI3K (56). Therefore, DLG2 deletion might cause a reduction in NMDAR ability to transduce signaling through PI3K. Another putative candidate is the duplication of the NAPG gene coding for the N-ethylmaleimide-sensitive factor attachment protein gamma. NAPG interacts with syntaxin 8 and is required for vesicular transport between the endoplasmic reticulum and the Golgi apparatus to control membrane fusion (57). Finally, the gain of AGXT2L2 gene copy number, seen in one of our patients (AUT150), also has a potential role perturbing neurotransmission. This gene codes for a glycine biosynthesis enzyme. Glycine, in addition to its direct role as neurotransmitter, is a necessary cofactor for glutamate action through the NMDAR (58).
Local protein synthesis in neuronal dendrites is critical for synaptic plasticity. However, the signaling cascades that couple synaptic activation to dendritic protein synthesis in live neurons are not fully understood although various subtypes of glutamate receptors and the PI3K/mTOR signaling have been demonstrated to have a prevalent role in the control of synaptic activity-induced dendritic protein synthesis in hippocampal neurons. Our finding of multiple genes in these pathways affected in copy number in ASD patients further supports the hypothesis that ASD can be caused, at least in some cases, by perturbation of the regulatory mechanisms of protein synthesis in the dendrites.
The diagnosis of autism or ASD is still based on behavioral criteria that do not allow differentiating among the underlying pathologies. The finding of multiple uncommon ASD-related CNVs, each in a specific patient, further reflects the complexity and multifactorial nature of the ASD phenotype. The search for genetic and genomic variation along the whole genome is still badly needed to better ascertain the genetic background of autistic phenotypes. Given the detection yield of aCGH technologies in our and other’s experience, it should probably be included in the diagnostic workup of idiopathic ASD, despite that genetic counseling may be difficult since many of the rearrangements detected can also be present in asymptomatic progenitors. In addition, more focused studies on genetic and epigenetic variations of specific pathways using the available higher resolution genomic technologies could lead to a better definition of molecular signatures, being the basis for an improved diagnosis and the ultimate development of specific therapeutic targets. The glutamatergic neurotransmission and the PI3K signaling pathway appear among the candidates for this approach.
MATERIAL AND METHODS
Patients
We have studied 96 Spanish patients (74 children followed in the neurology clinic and 22 institutionalized mentally retarded adults) with a confirmed diagnosis of one of the categories of ASD listed in the Diagnosis and Statistical Manual of Mental Diseases (DSM-IV). All patients were studied using the ADI-R to define a specific category of ASD and the Wechsler Intelligence Scale for Children or Wechsler Adult Intelligence Scale. These assessments provide a measure of general, verbal and performance IQ as well as analysis of multiple factorial components of cognitive functioning. We also use the Leiter International Performance Scale-Revised and the Raven Progressive Matrices in the non-verbal patients. All patients had an extensive evaluation by neurologists and clinical geneticists along with an intensive laboratory workup including standard karyotyping, fragile X molecular testing, subtelomeric and targeted MLPA assays (homemade panel designed to detect genomic duplications/deletions of specific regions associated with ASD and mental retardation: 1p36, 2q37, 7q11.23, 15q11–q13, 17p11.2, 16p11.2, 22q11.2 and 22q13.3), as well as metabolic and brain image studies in some cases when clinically indicated (Gener et al. submitted). All subjects participated after informed consent was obtained from their families or other legal caregivers. The study was approved by the medical ethical committees of the centers involved. Blood samples were obtained under institutional review board-approved informed consent, and genomic DNA was extracted by the salting out method using the Puregene® DNA Purification Kit (Gentra Systems). Parental samples were also obtained from the available parents who gave informed consent.
Controls
DNA samples from 100 population control individuals matched for population ancestry (Spanish anonymous blood donors) were used to prepare reference pools (50 males and 50 females) for hybridization experiments. DNAs from 52 control individuals (27 males and 25 females) were used in order to define polymorphic changes in DNA dosage in aCGH experiments.
CGH arrays
We used three different microarray platforms. The first screening was performed using a homemade BAC array containing 5442 large insert DNA fragments (BACs) with a global coverage of 23% of the euchromatic genome and higher probe density in genomic regions presumed hot-spots for rearrangements, named HSBA (23) (see Supplementary Material, Table S5, for more detailed information). To confirm the different variants detected with HSBA, we used two commercial arrays, either an oligoarray (Agilent 44K or 244K) or an SNP array (Illumina 370K). We performed hybridization experiments and subsequent analyses of aCGH as described previously in detail (23), and we followed the manufacturer’s recommendations for the SNP array. We used the PennCNV (19 November 2008 version, sample option) and CNV partition software for the analysis of the Illumina 370K array data (59).
Multiplex ligation probe amplification
A total of 100 ng of genomic DNA from each sample was subjected to MLPA using synthetic probes designed to target the specific CNV detected by aCGH, at least one locus per CNV. Oligonucleotide sequences for MLPA at the analyzed loci are described in the Supplementary Material, Table S6. The MLPA reactions were performed essentially as described previously and products were analyzed on an ABI PRISM 3100 genetic analyzer according to manufacturers’ instructions. For quantitative analysis, trace data were retrieved using the accompanying software (GeneScan, Applied Biosystems). Each MLPA signal was normalized and compared with the corresponding mean peak height obtained from five control DNA samples.
Gene ontology analyses
To obtain a general overview of the most relevant key functions represented in our data set, the genes located in the identified CNVs were used for ontology and biofunction analyses. We used the IPA software (http://www.ingenuity.com), along with two additional resources freely available: the ConsensusPathDB (http://cpdb.molgen.mpg.de) and the KEGG Pathway Database (http://www.genome.ad.jp/kegg/pathway.html) (60). We performed a core analysis to categorize the genes on the basis of their locations and their reported or suggested biochemical, biological and molecular functions. In the ConsensusPathDB, we checked the functional annotation of the gene list by using the pathway-based set options.
Statistical analysis
Fisher’s exact test for count data and Pearson’s χ2 test with simulated P-value (based on 10 000 replicates) were applied when appropriate for measuring significant differences in gene copy number frequencies or pathway representation. In all cases, statistical significance was considered for corrected P-values <0.05. Differences were assessed with the χ2 test using resampling. The Z-test was used in inference to determine whether the number of genes affecting relevant pathways in ASD was statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
Spanish Ministry of Health (FIS PI076832 and RETIC G03/184), Genome Spain and the European Commission (FP6-2005-037627). Funding to pay the Open Access publication charges for this article was provided by FIS PI076832.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the ASD patients and their families for their support as well as the different pediatricians and neurologists for referring the patients.
Conflict of Interest statement. L.A.P.-J. declares that he is founding partner and advisor of qGenomics S.L., a company involved in aCGH technology for diagnostic applications. The other authors declare that they have no competing interests.
REFERENCES
- 1.Smalley S.L. Genetic influences in childhood-onset psychiatric disorders: autism and attention-deficit/hyperactivity disorder. Am. J. Hum.Genet. 1997;60:1276–1282. doi: 10.1086/515485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fombonne E. The prevalence of autism. JAMA. 2003;289:87–89. doi: 10.1001/jama.289.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams B.S., Geschwind D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veenstra-Vanderweele J., Christian S.L., Cook E.H., Jr Autism as a paradigmatic complex genetic disorder. Annu. Rev. Genomics. Hum. Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- 5.Goizet C., Excoffier E., Taine L., Taupiac E., El Moneim A.A., Arveiler B., Bouvard M., Lacombe D. Case with autistic syndrome and chromosome 22q13.3 deletion detected by FISH. Am. J. Med. Genet. 2000;96:839–844. [PubMed] [Google Scholar]
- 6.Klauck S.M. Genetics of autism spectrum disorder. Eur. J. Hum. Genet. 2006;14:714–720. doi: 10.1038/sj.ejhg.5201610. [DOI] [PubMed] [Google Scholar]
- 7.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsater H., et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng J., Schroer R., Yan J., Song W., Yang C., Bockholt A., Cook E.H., Jr, Skinner C., Schwartz C.E., Sommer S.S. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci. Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Morrow E.M., Yoo S.Y., Flavell S.W., Kim T.K., Lin Y., Hill R.S., Mukaddes N.M., Balkhy S., Gascon G., Hashmi A., et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakkaloglu B., O’Roak B.J., Louvi A., Gupta A.R., Abelson J.F., Morgan T.M., Chawarska K., Klin A., Ercan-Sencicek A.G., Stillman A.A., et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arking D.E., Cutler D.J., Brune C.W., Teslovich T.M., West K., Ikeda M., Rea A., Guy M., Lin S., Cook E.H., et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alarcon M., Abrahams B.S., Stone J.L., Duvall J.A., Perederiy J.V., Bomar J.M., Sebat J., Wigler M., Martin C.L., Ledbetter D.H., et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquemont M.L., Sanlaville D., Redon R., Raoul O., Cormier-Daire V., Lyonnet S., Amiel J., Le Merrer M., Heron D., de Blois M.C., et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghebranious N., Giampietro P.F., Wesbrook F.P., Rezkalla S.H. A novel microdeletion at 16p11.2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am. J. Med. Genet. 2007;143A:1462–1471. doi: 10.1002/ajmg.a.31837. [DOI] [PubMed] [Google Scholar]
- 18.Ballif B.C., Hornor S.A., Jenkins E., Madan-Khetarpal S., Surti U., Jackson K.E., Asamoah A., Brock P.L., Gowans G.C., Conway R.L., et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2–p12.2. Nat. Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 19.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 20.Berg J.S., Brunetti-Pierri N., Peters S.U., Kang S.H., Fong C.T., Salamone J., Freedenberg D., Hannig V.L., Prock L.A., Miller D.T., et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams–Beuren syndrome region. Genet. Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 21.Keller K., Williams C., Wharton P., Paulk M., Bent-Williams A., Gray B., Ward A., Stalker H., Wallace M., Carter R., et al. Routine cytogenetic and FISH studies for 17p11/15q11 duplications and subtelomeric rearrangement studies in children with autism spectrum disorders. Am. J. Med. Genet. 2003;117A:105–111. doi: 10.1002/ajmg.a.10042. [DOI] [PubMed] [Google Scholar]
- 22.Mukaddes N.M., Herguner S. Autistic disorder and 22q11.2 duplication. World J. Biol. Psychiatry. 2007;8:127–130. doi: 10.1080/15622970601026701. [DOI] [PubMed] [Google Scholar]
- 23.Cusco I., del Campo M., Vilardell M., Gonzalez E., Gener B., Galan E., Toledo L., Perez-Jurado L.A. Array-CGH in patients with Kabuki-like phenotype: identification of two patients with complex rearrangements including 2q37 deletions and no other recurrent aberration. BMC Med. Genet. 2008;9:27. doi: 10.1186/1471-2350-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto D., Marshall C., Feuk L., Scherer S.W. Copy-number variation in control population cohorts. Hum. Mol. Genet. 2007;16(Spec no. 2):R168–R173. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 25.Zogopoulos G., Ha K.C., Naqib F., Moore S., Kim H., Montpetit A., Robidoux F., Laflamme P., Cotterchio M., Greenwood C., et al. Germ-line DNA copy number variation frequencies in a large North American population. Hum. Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 26.Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 27.Christian S.L., Brune C.W., Sudi J., Kumar R.A., Liu S., Karamohamed S., Badner J.A., Matsui S., Conroy J., McQuaid D., et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol. Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szatmari P., Paterson A.D., Zwaigenbaum L., Roberts W., Brian J., Liu X.Q., Vincent J.B., Skaug J.L., Thompson A.P., Senman L., Feuk L., et al. Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sultana R., Yu C.E., Yu J., Munson J., Chen D., Hua W., Estes A., Cortes F., de la Barra F., Yu D., et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 30.Kalscheuer V.M., FitzPatrick D., Tommerup N., Bugge M., Niebuhr E., Neumann L.M., Tzschach A., Shoichet S.A., Menzel C., Erdogan F., et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum. Genet. 2007;121:501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- 31.Serajee F.J., Zhong H., Nabi R., Huq A.H. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J. Med. Genet. 2003;40:e42. doi: 10.1136/jmg.40.4.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid S., Fendt M. Effects of the mGluR8 agonist (S)-3,4-DCPG in the lateral amygdala on acquisition/expression of fear-potentiated startle, synaptic transmission, and plasticity. Neuropharmacology. 2006;50:154–164. doi: 10.1016/j.neuropharm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Schumann C.M., Amaral D.G. Stereological analysis of amygdala neuron number in autism. J. Neurosci. 2006;26:7674–7679. doi: 10.1523/JNEUROSCI.1285-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toker A., Cantley L.C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz J.M., Liang S.L., Thompson S.M., McCarthy M.M. Estradiol induces hypothalamic dendritic spines by enhancing glutamate release: a mechanism for organizational sex differences. Neuron. 2008;58:584–598. doi: 10.1016/j.neuron.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kihara T., Shimohama S., Sawada H., Honda K., Nakamizo T., Shibasaki H., Kume T., Akaike A. Alpha 7 nicotinic receptor transduces signals to phosphatidylinositol 3-kinase to block A beta-amyloid-induced neurotoxicity. J. Biol. Chem. 2001;276:13541–13546. doi: 10.1074/jbc.M008035200. [DOI] [PubMed] [Google Scholar]
- 37.Kwon C.H., Luikart B.W., Powell C.M., Zhou J., Matheny S.A., Zhang W., Li Y., Baker S.J., Parada L.F. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belmonte M.K., Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat. Neurosci. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 39.Serajee F.J., Nabi R., Zhong H., Mahbubul Huq A.H. Association of INPP1, PIK3CG, and TSC2 gene variants with autistic disorder: implications for phosphatidylinositol signalling in autism. J. Med. Genet. 2003;40:e119. doi: 10.1136/jmg.40.11.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J.J., Liu Z., Wang L., Shin E., Loda M.F., Roberts T.M. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc. Natl Acad. Sci. USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S., Tiab L., Jiao X., Munier F.L., Zografos L., Frueh B.E., Sergeev Y., Smith J., Rubin B., Meallet M.A., et al. Mutations in PIP5K3 are associated with Francois–Neetens mouchetee fleck corneal dystrophy. Am. J. Hum. Genet. 2005;77:54–63. doi: 10.1086/431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panaretou C., Domin J., Cockcroft S., Waterfield M.D. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J. Biol. Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- 43.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J., et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J. Med. Genet. 2008 doi: 10.1136/jmg.2008.059907. [Epub ahead of print November 26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geschwind D.H. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrison J.E., Bolton P.F. Annotation: tuberous sclerosis. J. Child. Psychol. Psychiatry. 1997;38:603–614. doi: 10.1111/j.1469-7610.1997.tb01687.x. [DOI] [PubMed] [Google Scholar]
- 46.Swiech L., Perycz M., Malik A., Jaworski J. Role of mTOR in physiology and pathology of the nervous system. Biochim. Biophys. Acta. 2008;1784:116–132. doi: 10.1016/j.bbapap.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Kwon C.H., Zhu X., Zhang J., Baker S.J. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl Acad. Sci. USA. 2003;100:12923–12928. doi: 10.1073/pnas.2132711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marui T., Hashimoto O., Nanba E., Kato C., Tochigi M., Umekage T., Ishijima M., Kohda K., Kato N., Sasaki T. Association between the neurofibromatosis-1 (NF1) locus and autism in the Japanese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;131B:43–47. doi: 10.1002/ajmg.b.20119. [DOI] [PubMed] [Google Scholar]
- 49.Lavery W., Hall V., Yager J.C., Rottgers A., Wells M.C., Stern M. Phosphatidylinositol 3-kinase and Akt nonautonomously promote perineurial glial growth in Drosophila peripheral nerves. J. Neurosci. 2007;27:279–288. doi: 10.1523/JNEUROSCI.3370-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lush M.E., Li Y., Kwon C.H., Chen J., Parada L.F. Neurofibromin is required for barrel formation in the mouse somatosensory cortex. J. Neurosci. 2008;28:1580–1587. doi: 10.1523/JNEUROSCI.5236-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antion M.D., Hou L., Wong H., Hoeffer C.A., Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol. Cell. Biol. 2008;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinohe A., Hashimoto K., Nakamura K., Tsujii M., Iwata Y., Tsuchiya K.J., Sekine Y., Suda S., Suzuki K., Sugihara G., et al. Increased serum levels of glutamate in adult patients with autism. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2006;30:1472–1477. doi: 10.1016/j.pnpbp.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 53.McDougle C.J., Erickson C.A., Stigler K.A., Posey D.J. Neurochemistry in the pathophysiology of autism. J. Clin. Psychiatry. 2005;66(Suppl. 10):9–18. [PubMed] [Google Scholar]
- 54.Rubenstein J.L., Merzenich M.M. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato Y., Tao Y.X., Su Q., Johns R.A. Post-synaptic density-93 mediates tyrosine-phosphorylation of the N-methyl-d-aspartate receptors. Neuroscience. 2008;153:700–708. doi: 10.1016/j.neuroscience.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hisatsune C., Umemori H., Mishina M., Yamamoto T. Phosphorylation-dependent interaction of the N-methyl-d-aspartate receptor epsilon 2 subunit with phosphatidylinositol 3-kinase. Genes Cells. 1999;4:657–666. doi: 10.1046/j.1365-2443.1999.00287.x. [DOI] [PubMed] [Google Scholar]
- 57.Subramaniam V.N., Loh E., Horstmann H., Habermann A., Xu Y., Coe J., Griffiths G., Hong W. Preferential association of syntaxin 8 with the early endosome. J. Cell Sci. 2000;113:997–1008. doi: 10.1242/jcs.113.6.997. [DOI] [PubMed] [Google Scholar]
- 58.Lester R.A., Tong G., Jahr C.E. Interactions between the glycine and glutamate binding sites of the NMDA receptor. J. Neurosci. 1993;13:1088–1096. doi: 10.1523/JNEUROSCI.13-03-01088.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.