Abstract
Mutations identified in a cohort of patients with atrioventricular septal defects as a part of Ellis van Creveld syndrome (EvC syndrome) led us to study the role of two non-homologous genes, EVC and LBN, in heart development and disease pathogenesis. To address the cause of locus heterogeneity resulting in an indistinguishable heart–hand phenotype, we carried out in situ hybridization and immunofluorescence and identified co-localization of Evc and Lbn mRNA and protein. In the heart, expression was identified to be strongest in the secondary heart field, including both the outflow tract and the dorsal mesenchymal protrusion, but was also found in mesenchymal structures of the atrial septum and the atrioventricular cushions. Finally, we studied the transcriptional hierarchy of EVC and LBN but did not find any evidence of direct transcriptional interregulation between the two. Due to the locus heterogeneity of human mutations predicted to result in a loss of protein function, a bidirectional genomic organization and overlapping expression patterns, we speculate that these proteins function coordinately in cardiac development and that loss of this coordinate function results in the characteristics of EvC syndrome.
INTRODUCTION
Congenital cardiovascular malformations, disproportionate limb dwarfism, post-axial polydactyly and ectodermal dysplasia are all clinical features of recessively inherited Ellis van Creveld Syndrome (EvC syndrome, OMIM #225500) (1–3). Family-based genetic studies identified human mutations in two previously unknown genes, EVC and LBN (EVC2), which are located head-to-head on chromosome 4p16.2 (4–6). Disruption of either of these non-homologous genes results in an indistinguishable heart–hand phenotype, but the role of EVC and LBN in development and disease pathogenesis remains largely unknown.
Cardiovascular malformations occur in ∼60% of patients with EvC syndrome (2). Most associated cardiac anomalies are variants of atrioventricular septal defects (AVSD) (7), which involve malformation of valvuloseptal structures. Proper atrioventricular (AV) septal development requires contributions from the muscular and mesenchymal atrial septum, the endocardial cushions, and extracardiac tissues (8,9). Though AVSDs have been referred to as endocardial cushion defects, invoking causation as improper morphogenesis of the AV cushions, recent evidence reveals that improper signaling in the secondary heart field (SHF) contributes to pathogenesis (9). Little is known about the role of EVC and LBN in either normal valvuloseptal development or AVSD pathogenesis. In the absence of an overt cardiac phenotype in the Evc knockout mouse model (10,11), other approaches need to be taken to unravel the mechanism underlying this human heart malformation. Therefore we employed genetic, developmental and biochemical techniques to facilitate understanding of the pathogenesis of AVSDs associated with EvC syndrome.
Genetic analysis of a small cohort of patients supported a mechanism of complete loss of EVC or LBN function in EvC syndrome and identified a novel missense mutation in the LBN gene. Developmental studies in mouse during valvuloseptal morphogenesis showed that Evc and Lbn mRNA and protein are coexpressed in the SHF, including both the outflow tract and the dorsal mesenchymal protrusion (DMP), but are also found in mesenchymal structures of the atrial septum and the AV cushions. This suggests that the two gene products work together to carry out their function. In addition, cilia were identified in cardiac structures that are malformed in EvC syndrome. Biochemical experiments eliminated hierarchal transcriptional interregulation as a mechanism for locus heterogeneity. Taken together, these studies support a role for EVC and LBN in development of cardiac AV structures and provide initial insight to the molecular basis for locus heterogeneity of EvC syndrome.
RESULTS
Mutational analysis of EvC syndrome cohort
Table 1 summarizes the phenotypic data from six patients with a clinical diagnosis of EvC syndrome characterized by incomplete AVSD, polydactyly and short stature. Patient ethnicity, race and gender varied even in our small cohort, which included three males and three females of African American (one patient), Caucasian (five patients) and Hispanic (one patient) heritage. Pathogenic mutations were identified in EVC (two patients) and LBN (three patients). No mutations were identified in the remaining patient. Though three of these individuals had homozygous mutations, haplotyping studies indicated that the parents were not related by blood (data not shown). The remaining two patients had compound heterozygous mutations.
Table 1.
Summary of phenotypic data in patients with EvC syndrome
| Patient | Race/Ethnicity | Gender | Phenotype |
|---|---|---|---|
| 1–12 | Caucasian | Female | Common atrium/partial AV canal, persistent superior left vena cava, short stature, polydactyly |
| 4–12 | African American | Female | Common atrium/partial AV canal, unroofed coronary sinus, persistent superior left vena cava, progressive AV block, short stature, polydactyly |
| 7–11 | Caucasian | Male | Common atrium/partial AV canal, large patent ductus arteriosis, short stature, polydactyly |
| 8–3 | Caucasian | Male | Common atrium/partial AV canal, coarctation of the aorta, short stature, polydactyly |
| 10–3 | Caucasian, Hispanic | Male | Common atrium/partial AV canal, short stature, polydactyly |
| 11–3 | Caucasian | Female | Common atrium/partial AV canal, short stature, polydactyly |
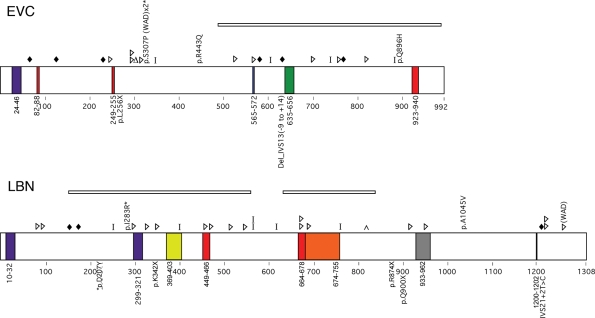
Mutations were considered to be pathogenic if they predicted a detrimental effect on the protein, altered a highly conserved residue, were previously reported to be disease causing and/or were absent in 100 control chromosomes. Most mutations were predicted to result in a premature stop codon (four mutations) or a splicing error (two mutations). However, in one case (Patient 11–3) a novel missense mutation was identified in a residue conserved in chicken and mouse. This mutation, which was not present in 100 control chromosomes, occurred in a patient with a point mutation on the transallele that is predicted to result in protein truncation. Figure 1 is a schematic diagram of the predicted protein effects from mutations identified in this study and those previously reported (4–6,12). Table 2 summarizes the pathogenic mutations found in this analysis. Other variants identified in our cohort are listed in Supplementary Material, Table S1.
Figure 1.
Summary of expected EVC and LBN protein changes based on previously reported mutation data (top of protein) or those identified in this study (bottom of protein) for patients with EvC syndrome. Colored boxes indicate putative domains. Purple, transmembrane; Red, nuclear localization signal; Green, leucine zipper; Blue, ATP/GTP binding site; Yellow, Flagellar hook associated protein; Orange, Helix-loop-helix; Gray, IQ motif; Black, RGD Cell attachment sequence. Shapes indicate predicted protein changes. Diamonds, splicing error; Triangles, insertion or deletion resulting in a frameshift; Delta sign, deletion of a single amino acid; Bar, truncation mutation; Caret, duplication. Deletions are indicated by rectangles over the deleted area. Asterisks denote missense mutations that occur in an amino acid conserved in mouse and chicken. WAD, Weyers Acrodental Dysostosis.
Table 2.
Summary of mutations identified in this study
| Patient (Het/Homo) | Exon | Sequence change | Protein effect |
|---|---|---|---|
| EVC mutations | |||
| 4–12a (Homo) | 14 | Del_IVS13 (−9 to +14) | Predicted splice error |
| 10–3a (Homo) | 6 | c.770T > A | p.L256X |
| LBN mutations | |||
| 7–11a (Homo) | 9 | c.1024A > T | p.K342X |
| 8–3 (Compound Het) | 15 | c.2698G > T | p.Q900X |
| 21 | IVS21 + 2T > Cb | Predicted splice error | |
| 11–3 (Compound Het) | 5 | c.619G > T | p.D207Yc,d |
| 15 | c.2620C > T | p.R874X | |
aParents unrelated based on haplotyping studies.
bPreviously reported.
cConserved in mouse and chicken.
dChange not present in 100 control chromosomes.
Evc and Lbn mRNAs are present in developing valvuloseptal structures
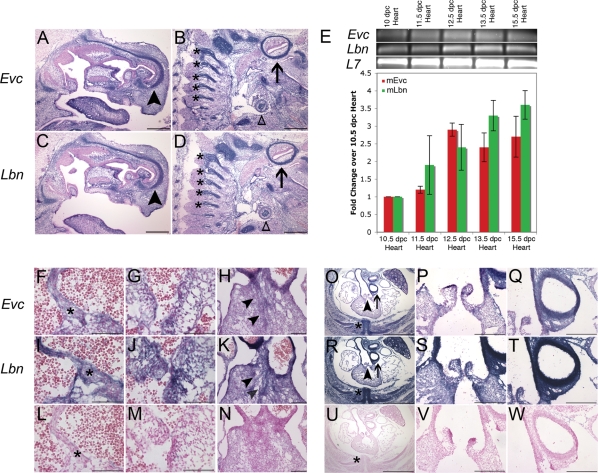
Based on the head-to-head genomic organization of Evc and Lbn and locus heterogeneity resulting in the same disease phenotype, we predicted tissue colocalization of the gene products. In situ hybridization on sagittal sections of 15.5 days post coital (dpc) embryo confirmed the presence of Evc mRNA in the embryonic cartilage primordium of the nasal bone, the vertebrae and other cartilaginous structures (Fig. 2A and B). Sister sections revealed the presence of Lbn message in overlapping anatomy (Fig. 2C and D). Sections of the developing mouse whole embryo at 15.5 dpc identified mRNA expression of both genes in structures affected by EvC syndrome including the ribs (Fig. 2O and R), heart (Fig. 2F–W) and limbs (data not shown).
Figure 2.
Expression of Evc and Lbn mRNA in the developing mouse embryo during AV septation. (A–D) Evc and Lbn mRNA expression in a sagittal section of a 15.5 dpc mouse whole embryo. Transcript for both genes is found in the cartilage primordium of the nasal bone (arrowhead), the vertebrae (astericks) and other cartilagenous structures like the primordium of the clavicle (open triangle) and the temporal bone (arrow). The scale bars in (A–D) represent 500 µM. (E) Quantitative RT–PCR confirms the presence of Evc and Lbn message in the developing murine heart from 10 dpc to 15.5 dpc. There is no statistically significant difference in the levels of Evc and Lbn mRNA transcript (P = NS). In situ hybridization in transverse sections of the heart at 13.5 dpc pinpoints co-expression at the tip of the primary atrial septum (asterisks in F and I), in the AV cushions (G and J) and in the connective tissue of the outflow tract (H and K). Sense controls are provided in (L–N). Transverse sections at 15.5 dpc reveal Evc and Lbn expression in structures affected in Evc syndrome including the heart (O–T) and the ribs (asterisks in O and R). Co-expression in the heart is strongest in outflow tract structures (O–T). The areas pointed out by the arrowhead in (O and R) are magnified in (P and S), while the arrows in (O and R) indicate the areas magnified in (Q and T). Sense controls are provided in (U–W). The scale bar represents 200 µM in all images from (F) to (W) except for (Q), (R) and (U), where they represent 1 mm.
To further investigate the EvC syndrome cardiac phenotype, we examined the expression of Evc and Lbn in the mouse heart over a developmental time course. Quantitative RT–PCR was used to assess mRNA presence in the developing heart (Fig. 2E) and expression in specific cardiac structures was identified by in situ hybridization (Fig. 2F–W). Both genes are expressed during time points related to AV septation and AV valve development. Relatively low levels of expression are identified at 10.5 dpc when the primary atrial septum is beginning to form and the DMP is just joining AV mesenchymal structures. As the primary atrial septum evaginates and merges with the DMP and the AV cushions between 11.5 and 13.5 dpc, mRNA for both genes increases. Co-expression of Evc and Lbn persists at 15.5 dpc when the endocardial cushions are remodeling to form the valves.
In situ hybridization of the heart at 13.5 dpc revealed broad expression of Evc and Lbn in the atrial septum (Fig. 2F and I) and in the AV cushion (Fig. 2G and J). Intriguingly, Evc and Lbn are strongly expressed in the connective tissue of the outflow tract (Fig. 2H and K). This coexpression continues at 15.5 dpc when Evc and Lbn mRNAs are identified in the cushions of the forming pulmonary valve and the muscular walls of the pulmonary and aortic trunks (Fig. 2O–T). Coexpression in embryos implicates a role for both genes in development, and together with head-to-head genomic orientation suggests coordinate regulation by a bidirectional promoter.
Peptide antibodies generated to EVC and LBN are specific
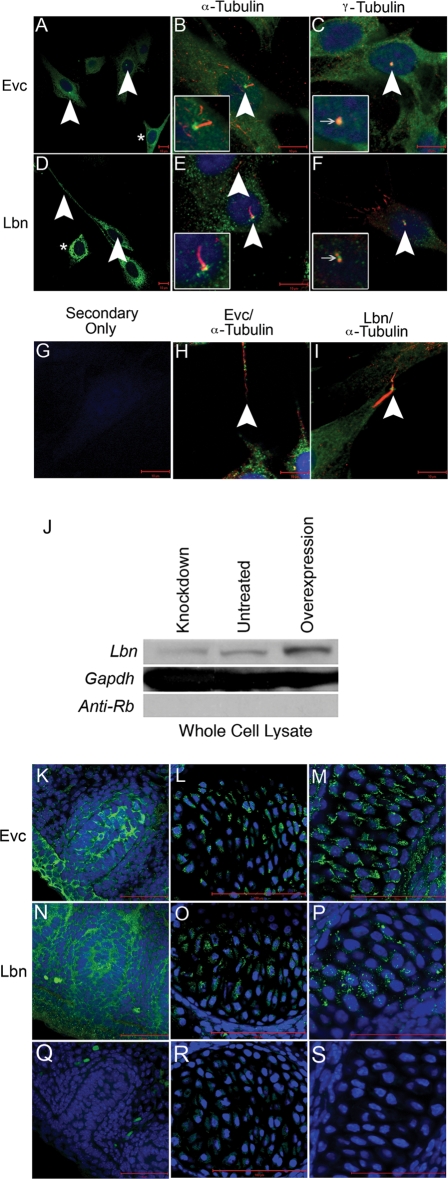
To confirm the mRNA results and study the degree of protein colocalization within tissues and cells, we generated peptide-specific antibodies to EVC and LBN. Prior to use in cardiac sections, we confirmed specificity of the EVC antibody using previously reported cell and tissue data generated by Ruiz-Perez et al. (10). In NIH3T3 cell culture, we identified coexpression of EVC with acetylated and gamma tubulin, confirming EVC’s presence at the base of cilia in cell culture (Fig. 3A–C). We further identified LBN in the area of the ciliary transition zone of NIH3T3 cells (Fig. 3D–F). Though we were able to show overlap of EVC and LBN with acetylated and gamma tubulin, expression was not restricted to the base of the cilia but extended throughout the cell in a peri-nuclear fashion (see asterisks in Fig. 3A and D). Both proteins could be found in cellular extensions characterized by acetylated tubulin expressions (Fig. 3H and I).
Figure 3.
Novel peptide-specific EVC antibodies confirm localization of Evc protein and co-staining with the newly generated LBN antibody reveals these proteins are expressed in overlapping structures. Expression of EVC alone (A) and co-staining with acetylated tubulin (B) or gamma tubulin (C) in NIH3T3 cells confirms antibody specificity by identification of EVC at the base of cilia. Expression of LBN alone (D) and co-staining with acetylated tubulin (E) or gamma tubulin (F) reveals an overlap of EVC and LBN protein in the cilia. Asterisks in (A and D) show protein presence in the cytoplasm of the cell outside of cilia structures. (G) serves as a negative control while (H and I) reveal the presence of EVC and LBN overlapping with acetylated tubulin-based structures that are not cilia. All scalebars in (A–I) represent 10 µM. Western blot for LBN protein (J) identified a band of the estimated size, 148 kDa. Compared with untreated cultures, the intensity of the LBN band in whole cell lysates decreased when treated with Lbn siRNA (Knockdown) and increased upon transfection with Sport6-pCMV-mLbn (Overexpression). GAPDH is used as a loading control and anti-Rb secondary antibody is a negative control for the western blot protocol. These findings confirm LBN antibody specificity. EVC antibody specificity is further confirmed in previously reported in vivo structures including the vibrissae (K), the vertebrae (L) and the cartilage primordium of the nasal bone (M) in a 15.5 dpc sagittal section of an embryo. LBN immunofluorescence reveals overlapping expression in these structures (N–P). Negative controls (Q–S) show minimal background. All scalebars in images (K–S) represent 50 µM.
Since no specific localization of LBN has been previously reported, we used western blotting to show specificity of this antibody. Our assay identified a band of the predicted size, 148 kDa (13), which increased in intensity after LBN overexpression studies and decreased after siRNA treatment in NIH3T3 cells (Fig. 3G).
To confirm antibody specificity of EVC in vivo, we carried out immunofluorescence on sagittal sections of 15.5 dpc embryos. We confirmed the presence of EVC in the vibrissae (Fig. 3K), the developing vertebrae (Fig. 3L) and the cartilage primordium of the nasal bone (Fig. 3M) as previously reported by Ruiz-Perez et al. (10). LBN protein was also identified in these structures (Fig. 3N–P) when compared with control sister sections with secondary antibody only (Fig. 3Q–S). Based on these results indicating antibody specificity and colocalization of the proteins, we analyzed EVC and LBN protein in the heart.
EVC and LBN coexpressed at the tip of the atrial septum primum
Immunofluorescence was used to examine EVC and LBN protein expression during valvuloseptal development. In Supplementary Material, Figure S1, we have independently stained for EVC and LBN in a transverse section of a 13.5 dpc heart to identify co-localization at the tip of the atrial septum and in the AV valves.
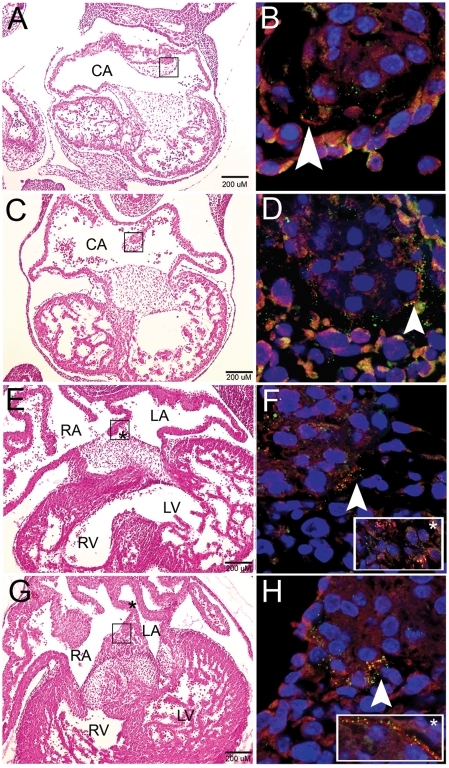
In Figure 4, the left column orients to the cardiac anatomy in transverse sections of the mouse embryonic heart at 10.5–13.5 dpc. The boxed atrial septum is magnified in the right column, where arrowheads indicate coexpression at the tip of the atrial septum. Though there were areas of independent expression, EVC and LBN were often in close proximity and sometimes overlapped. EVC and LBN proteins were consistently at the distal tip of the primary atrial septum during this period of AV septation. This tissue is continuous with the DMP after 10.5 dpc. Co-expression extended to the AV cushions after the primary atrial septum merged with this mesenchymal tissue at 12.5 dpc (Fig. 4E, inset). Further co-expression occurs along the edge of the atrial septum, in a location that overlaps with the SHF and is marked by the Mef2c-AHF-Cre;R26R mouse (9) (Fig. 4F, inset). Proximity of EVC and LBN protein in structures that are malformed in EvC syndrome implies coordinate function of EVC and LBN for proper cardiac morphogenesis.
Figure 4.
EVC and LBN protein colocalize at the tip of the primary atrial septum during murine cardiac development. Development of the atrial septum at stages 10.5–13.5 dpc (A,C,E and G). (B,D,F and H) are higher magnification images of the boxed area in the left column. The primary atrial septum shows colocalization of EVC (Red) and LBN (Green) at the tip of the developing septum as it dives down from the common atrial wall (10.5 and 11.5 dpc) and merges with the AV cushions (12.5 and 13.5 dpc). After the atrial septum joins the endocardial cushions, co-expression broadens to these AV structures (inset, F). The inset box in (H) represents overlap of EVC and LBN proteins along the edge of the atrial septum (location indicated by asterisk in G). Arrowheads are used to pinpoint areas of colocalization at the tip of the atrial septum. Blue color indicates nuclear staining with Topro-3. Scalebars represent 200 µM in (A,C,E and G). CA, common atrium; RA, right atrium; LA, left atrium; R, right ventricle; LV, left ventricle.
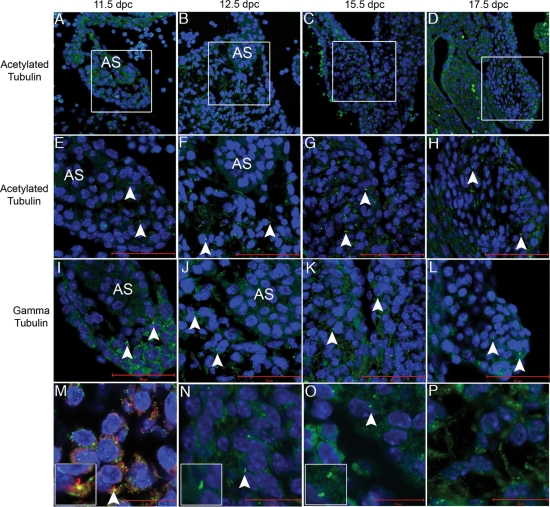
Acetylated tubulin expression indicates presence of cilia during cardiac morphogenesis
Tubulin has been identified as a marker of cilia (14). Limited studies exist on cardiac cilia and tubulin expression has not been reported in the developing murine heart. Based on the presence of EVC and LBN at the base of cilia in NIH3T3 cells, we examined the expression patterns of tubulin-based structures during cardiac morphogenesis. We carried out immunofluorescence with antibodies to acetylated and gamma tubulin in the embryonic mouse heart. We identified discrete tubulin stain in all stages assayed (Fig. 5). Tubulin was present in the developing atrial septum (Fig. 5 A, E and I), the AV cushions (Fig. 5 B,F and J) and the remodeling AV valves (Fig. 5C,D,G,H,K and L). The presence of cilia in the AV cushion was further supported by double staining for both acetylated tubulin and gamma tubulin in the same section (Fig. 5M). Magnified staining of acetylated tubulin (Fig. 5N) and gamma tubulin (Fig. 5O) show expected morphology for the ciliary axoneme and basal bodies, respectively. Acetylated tubulin staining in the AV cushion reveals the presence of tubulin-based structures that are not cilia (Fig. 5P). In Supplementary Material, Figure S2, the atrial septum of an 11.5 embryo serves as a negative control (Anti-MS 488 Only). At this time, we have been unable to identify an overlap of EVC and LBN with these markers of tubulin in these developmental time points and structures. However, identification of acetylated and gamma tubulin indicates the presence of cilia in the developing heart.
Figure 5.
Acetylated tubulin, a marker of cilia that colocalizes with EVC in cell culture, is present in the developing heart. Acetylated tubulin (seen here in green) is identified in the primary atrial septum (A,E and I), the AV junction (B,F and J) and the developing AV valves (C,D,G,H,K and L). Boxes in (A,B,C and D) indicate areas of higher magnification in (E–L). (M) displays co-staining of both acetylated and gamma tubulin confirming the presence of cilia. In images (E–N), the scale bars depict 50 µM while the scale bar represents 20 µM in (M–P). (N) is a higher magnification of acetylated tubulin revealing the cilia axonene and (O) is a higher magnification of gamma tubulin, which marks the basal bodies. (P) shows evidence of acetylated tubulin outside the cilia in mesenchymal structures of the heart. Blue Topro-3 staining marks the nuclei. Arrowheads indicate individual cilia.
EVC and LBN proteins are coexpressed but there is no hierarchal transcriptional interregulation
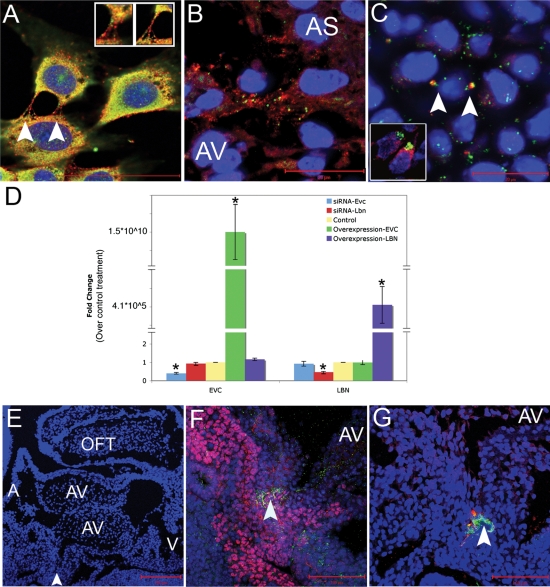
Gene mutations resulting in loss of protein function among members of a hierarchical pathway can result in similar disease phenotypes, as evidenced by the Ras pathway syndromes (15). Therefore, we used biochemical assays in cell culture to determine whether over-expression or knockdown of either EvC syndrome gene could affect the transcript levels of the other disease-causing gene. Figure 6 demonstrates results of immunofluorescent studies in cultured NIH3T3 cells (Fig. 6A), in cells at the tip of the atrial septum (Fig. 6B) and in chondrocytes of the 15.5 dpc developing rib (Fig. 6C). The bone cells showed overlapping expression in distinct projections from the cell, which was similar to the pattern seen in a 13.5 dpc AV cushion (Fig. 6C, inset). EVC and LBN showed adjacent expression in NIH3T3 and atrial septal tissue. Based on this localization and the hypothesis that EVC and LBN participate in subsequent transcriptional steps of a biochemical pathway, we sought a functional relationship between the two proteins. To test this, we independently transfected full length Evc and Lbn cDNAs or gene specific siRNAs, then used quantitative RT–PCR to measure transcription of the other gene. Transfection efficiency for overexpression and knockdown assays were 26 and 67%, respectively (data not shown). Overexpression of EVC in cell culture did not affect endogenous levels of Lbn transcript (Fig. 6D). Similarly, overexpression of LBN using the same methodology, did not affect levels of endogenous Evc transcript. Therefore, neither EVC nor LBN protein is sufficient to induce transcription of the other head-to-head gene. The complementary experiment using siRNA to knockdown each gene independently found that decrease of either Evc or Lbn did not impact the transcript levels of the other gene (Fig. 6J). Figure 3J provides evidence for protein knockdown and overexpression for LBN. Based on these studies, we concluded that reciprocal regulation does not occur at the level of transcription.
Figure 6.
EVC and LBN proteins are coexpressed in cell culture and in cells crucial for cardiac AV septation but there is no hierarchal transcriptional interregulation. EVC and LBN proteins are coexpressed in a similar pattern in NIH3T3 cells (A) and in vivo at the tip of the atrial septum (B) of a 12.5 dpc mouse. Arrows indicate overlapping expression of EVC and LBN in bone (C) and in the developing AV valve at 13.5 dpc (inset in C). Knockdown and overexpression of EvC-associated genes reveals a lack of hierarchial transcriptional interregulation (D). Manipulation of gene expression in cell culture revealed that changes in expression of EVC did not impact the levels of Lbn transcript and changes in the expression of LBN did not affect Evc transcription. The blue bars indicate successful knockdown of Evc after treatment with siRNA, but no change in Lbn gene expression. The red bars show Lbn could be knockeddown but the treatment did not affect Evc transcription. The yellow bar represents transfection with a control plasmid or a control siRNA. The green bar indicates successful overexpression of EVC with no affect on levels of Lbn transcription. The purple bar shows transfection of LBN does not alter the levels of Evc in the cell. Asterisks indicate statistical significance based on the Bonferroni adjusted P-value <0.0125. (E–G) are sagittal sections of a mouse heart at 10.5 dpc. AS, atrial septum; AV, atrioventricular cushions; A, atria; V, ventricles; OFT, outflow tract. The arrowhead in (E) indicates the area of zoom in (F) and (G). (F) shows overlap of ISL-1 (pink) and LBN (green) while G shows overlap of EVC (red) and LBN (green) in the DMP.
Studies in the Mef2C-AHF-Cre;R26R mouse (9) revealed cardiac developmental expression that overlaps with our findings for Evc and Lbn. In this mouse model, cre-recombination takes place in the outflow tract, the right ventricle, the atrial septum, the AV cushion and the DMP. The DMP is an extracardiac, non-endothelially derived mesenchyme that acts as glue between the tip of the primary atrial septum and the AV cushions during development (9,16,17). Based on the suspected importance of the DMP in AV development, we carried out immunohistochemistry on sections of the DMP at 11.5 dpc. We were able to identify overlap of LBN with ISL-1, a marker of the DMP (Fig. 6F). In an adjacent section, we identified overlap of Evc and Lbn in the same location. Based on these studies, Evc and Lbn are co-expressed in the DMP (Fig. 6G).
DISCUSSION
In this study, we found co-expression of EVC and LBN in structures affected by EvC syndrome. We identified novel mutations, which are predicted to result in protein loss of function. By in situ hybridization, quantitative RT–PCR and immunofluorescence, we showed co-expression of Evc and Lbn gene products during heart morphogenesis in key valvuloseptal structures that are malformed in EvC syndrome. These findings coupled with the head-to-head orientation of EVC and LBN suggest that the proteins function coordinately during cardiac development and that loss of this coordinate function provides reasoning for EvC syndrome locus heterogeneity. However, our studies conclude that reciprocal regulation based on a transcriptional hierarchy between EVC and LBN is not the mechanism of such coordinate function.
While little is known about the pathogenic mechanisms of EvC syndrome mutations, we and others (5,6,12) have identified missense mutations, splicing errors and nonsense mutations that are predicted to result in truncation or loss of function of EVC or LBN proteins. A microdeletion identified in this study is predicted to overlap with a splicing error described by Ruiz-Perez et al. (4) in the initial Amish pedigree. Interestingly, these splicing errors would be predicted to occur just prior to a previously predicted leucine zipper domain (4), suggesting this putative domain may be important for protein function. In the LBN gene, we identified a missense mutation that changed conserved residue 207 from a smaller Aspartic Acid to a Tyrosine with a bulky benzene group. Ruiz-Perez et al. have previously reported a point mutation that affects nearby conserved residue 283. This suggests a crucial region of the protein that is not tolerant to even a single amino acid change. We report a second patient who has an LBN mutation that is expected to truncate the C-terminus of the protein. This lends support to the presence of a functional domain distal to the 21st exon of LBN. Mutational analysis is useful in the continued identification of putative domains that hint at protein function. Future studies of cohorts enriched for cardiac malformations could aid in the identification of genotype/phenotype correlations.
The conserved arrangement of EVC and LBN as head-to-head genes sharing common 5′ genomic DNA (5,6) may underlie the mechanism for gene regulation. This bi-directional genomic arrangement most commonly results in co-expression of the genes (18), and can be important for transcripts necessary in overlapping biological pathways, expression of proteins in a stoichiometric relationship or other coordinated responses (18). Given the presence of both Evc and Lbn message in the same embryonic tissues, we speculate that Evc and Lbn are coordinately regulated and have synergistic protein function in cardiac development.
Previous experiments have identified monocilia in endocardial tissues of the embryonic chicken (19) but similar studies have not been applied to mice. Based on staining with tubulin markers, we were able to identify cilia in the developing atrial septum, AV junction and later in the maturing AV valve. The role of primary cilia in cardiac development remains unknown at this time, but may include signal transduction and/or mechanosensation. This and earlier studies (10) have revealed the presence of EVC and LBN at the base of cilia in cell culture, suggesting these proteins may be important for cilia function. Loss of Evc in a knockout mouse model did not disrupt the formation of cilia (10), indicating that Evc’s role in cilia is more likely to be functional and not structural. It is unclear at this time whether EVC and LBN are involved in ciliary signaling during heart development. While we have not shown overlap of the proteins with markers of cilia in the developing heart, we have shown that all the players are present, and a closer look at developmental timepoints and structures is warranted. One possibility is that cilia are present in or adjacent to tissues of the SHF, particularly the DMP. Sonic Hedgehog (SHH) signaling has been shown to be important for DMP migration (9) and cilia are known to be associated with SHH signaling (20). Further work is necessary to determine whether there is a role for SHH, EVC and LBN in cilia of the developing heart.
In an effort to understand the basis of locus heterogeneity associated with EvC syndrome, we performed biochemical analyses. Considering that mutation in either gene results in an identical human phenotype and that gene products can be identified in the same structures during development, these proteins are most likely functioning coordinately. We investigated hierarchal interregulation at the transcriptional level but found that this was not a plausible mechanism for coordinate function. Future biochemical studies investigating the nature of this coordinate function, perhaps protein–protein interaction, will shed light on a potential disease mechanism by which mutation in either gene results in an indistinguishable patient phenotype.
Emerging investigations of valvuloseptal development focus on the tissues that contribute to AV septation and valve formation. AVSDs have been thought to be caused entirely by failure of the superior and inferior AV cushions to merge during AV morphogenesis. However, more recent work has shown that mouse models with AVSDs due to Trisomy 16 (equivalent to human Trisomy 21) and fetuses with Down syndrome actually lack discrete mesenchymal structures that have origins outside the AV cushion (21,22). Recently, considerable excitement has developed over reports of the DMP, an extracardiac Isl-1 positive, non-endothelially derived mesenchyme that fills the gap between the mesenchymal atrial septum and the endocardial cushions in the embryonic 10.5 dpc heart (16,23). The DMP and the anterior heart field (AHF) are marked by recombination in the Mef2C-AHF-Cre;Rosa26 reporter mouse model (9). Loss of crucial signals using this cre line result in AVSD due to improper differentiation and migration of the DMP, indicating that this tissue is crucial for AV septation (9). Here, we show that both EVC and LBN are present in cells of the DMP. This provides the first example of gene expression tied to human disease pathogenesis associated with the DMP. We also identify EVC and LBN in the SHF and in other mesenchymal structures including the mesenchymal tip of the atrial septum and later in development of the AV endocardial cushions.
Previously, bone growth plate experiments revealed a role for Evc in the mediation of Indian Hedgehog (IHH) signaling and suggested a role in other hedgehog signaling pathways (10). Further work by Goddeeris et al. (9) indicated the importance of SHH signaling in the SHF for proper AV septation. Based on the AVSD phenotype in EvC syndrome and our identification of Evc and Lbn in developing mouse SHF, we speculate that EVC and LBN have coordinate functions in a pathway related to SHH signaling during cardiac development. Since SHH signaling has been shown to occur at cilia surfaces (20), cilia may be involved with this signal transduction. Mutation in either EVC or LBN could interfere with this signaling function and/or prevent DMP contribution to AV septation during heart development. Presumably, these disruptions of coordinate function could occur at multiple time points affecting multiple facets of valvuloseptal development. Future experiments addressing these questions will be important in the further elucidation of the role of EVC and LBN in cardiac development and pathogenesis, and the contributions of the DMP to AV septation.
MATERIALS AND METHODS
Patient clinical evaluation and sample collection
Patients with EvC syndrome were evaluated by echocardiography for structural heart malformations, physical exam for stature and post-axial polydactyly and by medical record review. Family history, gender and ethnicity were also noted on a multi-generation pedigree. A 10 mL intravenous blood sample was collected from each participant in acid-citrate dextrose (ACD) tubes. Informed consent was obtained for all participants. This study was approved by the Institutional Review Board of Cincinnati Children’s Hospital Medical Center.
Genomic extraction and polymerase chain reaction
The Puregene DNA Isolation Kit (Gentra Systems, Mineapolis, MN, USA) was used to extract genomic DNA from whole blood. PCR was carried out using EVC (Supplementary Material, Table S2) and LBN primers (5) designed to amplify the coding sequence and flanking intronic sequence. PCR was carried out on the PTC-225 Peltier Thermal Cycler (Bio-Rad, Hercules, CA, USA) for 35 cycles of denaturation at 95°C for 30 s, primer annealing at 55–65°C for 30 s depending on the primer pair and extension at 72°C for 60 s. PCR products were electrophoresed on a 1–2% Agarose gel, and then purified with the QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA, USA). The BigDye Terminator Cycle Sequencing Kit (v3.0) was used to generate fluorescently labeled fragments for analysis on the Applied Biosystem’s 3100 Capillary Gel Electrophoresis DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were compared with consensus EVC (AF216184) and LBN (NM 147127) transcripts from the National Center for Biotechnology Information (NCBI). One mutation resulted in a 23 bp deletion and was verified by PCR followed by gel electrophoresis. All other mutations were verified by restriction enzyme digest. Mutation origin was traced in families when parental DNA was available. Identification of putative protein domains was based on previously reported domains (4), or Ensembl (24) or Motif Scan (25) analysis.
Genotyping with polymorphic short-tandem repeat markers
Five microsatellite markers, D4S412, D4S3023, D4S431, D4S2366 and D4S2983, encompassing ∼4.5 Mb of the EvC locus (4p16.2) were used to genotype participants in three families where the proband had a homozygous mutation. Genomic DNA was amplified with a 32P-labeled primer in a 10 µL reaction containing 4 pmols of each primer, 2 pmols dNTPs and 0.28 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). Samples were denatured at 94°C for 15 s, annealed at 57°C for 30 s and extended for 45 s at 72°C for 10 cycles, then denatured at 94°C for 15 s, annealed at 57°C for 30 s and extended at 72°C for 45 s for another 25 cycles. Final extension was for 10 min at 72°C. PCR products were loaded to a 6% polyacrylimide gel and electrophoresed for 2 h and 15 min, then exposed for 3 h on a phosphoimager cassette (Molecular Dynamics, Sunnyvale, CA, USA). Manually constructed haplotypes were then examined for evidence of homozygosity.
Quantitative RT–PCR analysis of gene expression in mice
Total RNA was extracted from NIH3T3 cells and embryonic hearts using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and chloroform. RNA was precipitated overnight in isopropanol and 5 µg of glycogen (Ambion, Austin, TX, USA) then quantified and diluted to 3 ng/µL. cDNA generation and PCR amplification was performed with the iScript One-Step RT–PCR Kit With SYBR Green (Bio-Rad, Hercules, CA, USA). Products were detected and quantified with the MJ Research Opticon 2 (Bio-Rad). qRT–PCR primers designed across exons to prevent amplification of genomic DNA were as follows: Evc 5′-AAGACGAAGGAGCTGCAGAAC-3′and 5′-AAGACGAAGGAGCTGCAGAAC-3′, Lbn 5′-AAGACGAAGGAGCTGCAGAAC-3′and 5′-AAGACGAAGGAGCTGCAGAAC-3′, and mL7 5′-GAAGCTCATCTATGAGAAGGC-3′ and 5′-AAGACGAAGGAGCTGCAGAAC-3′. Evc and Lbn gene expression levels were quantified with respect to control treatment using the Stealth RNAi Negative Control LO GC (Invitrogen) for gene knockdown experiments or an empty pcDNA3.1 (−) vector (Invitrogen) for overexpression studies. A standard curve was generated for all primer pairs using RNA extracted from untreated NIH3T3 cells. Standardized RNA was further normalized to amplification with the murine L7 primer pair to account for spectrophotometer error. Quantitative RT–PCR experiments were carried out three times, each time in triplicate.
Preparation of mouse tissues for in situ hybridization or immunohistofluorescence
Timed matings of C57Bl/6 mice were set up, with the morning of a vaginal mucous plug considered to be 0.5 dpc. Females were sacrificed by CO2 asphyxiation and the embryos were extracted between 9.5 and 17.5 dpc. Mouse embryos were dissected and fixed overnight in 4% paraformaldehyde/PBS at 4°C. In preparation for in situ hybridization, mouse embryos were snap frozen in Optimal Cutting Temperature (OCT) medium (Tissue-Tek, Torrance, CA, USA) and sectioned at 14 µM. Tissues to be used for immunofluorescence were dehydrated through a methanol/PBS gradient and stored in 100% MeOH at −20°C. Prior to staining, tissues were hydrated in an ethanol series, cleared in xylene and washed three times for 1 h in paraffin at 65°C, then paraffin embedded and sectioned at 8 µM. All animal procedures were carried out according to Institutional Animal Care and Use Committee (IACUC)-approved methods.
In situ hybridization
To generate antisense RNA probes, mouse Evc (AK148262) and Lbn (AK165051) products were first amplified from mouse whole embryo cDNA with the following primers: Evc 5′°-AGCTGTACTGTAGCACCATG-3′ and 5′-GACACTAGTGCTGGTCACAT-3′, and Lbn 5′-TACTCAGGATGGAATCCAGACT-3′ and 5′-CTCATCTCTAGGTCACACTCTG-3′. All primers were designed to produce amplicons crossing exons so only mRNA would be detected after hybridization. PCR products were cloned into the pCR 2.1-Topo Vector with the TOPO TA Cloning Kit (Invitrogen) then moved to pBluecript SK+ (Stratagene, La Jolla, CA, USA) where sequence accuracy was confirmed. Digoxigenin labeled antisense and sense probes were generated using the T7 and the T3 promoter, respectively (26). Both Evc and Lbn sense probes were used as controls. Figure 2L–N and U–W are Evc sense slides which are representative of all sense control slides.
In situ hybridization was carried out on formaldehyde-fixed, OCT-embedded tissues cut at 14 µM and mounted on Superfrost Plus microscope slides (Fisher Scientific, Pittsburg, PA, USA). Slides were brought to room temperature prior to running on the Ventana Discovery XT (Ventana Medical Systems Inc., Tuscon, AZ, USA). The colorimetric reaction was carried out using the ChromoMap Blue Kit (Ventana Medical Systems Inc). Slides were counterstained with Nuclear Fast Red. Signal was visualized with an Olympus BX60 microscope (Olympus Imaging American Inc., Center Valley, PA, USA).
Polyclonal antibody production
Mouse peptide-specific antibodies were generated to keyhole limpet hemocyanin (KLH)-conjugated oligopeptides representing either EVC or LBN (Zymed/Invitrogen Custom Antibody Production). Peptide sequences were identified based on the likelihood of the region to be on the protein’s exterior and avoidance of regions with high homology to other mouse and chicken proteins. The EVC peptide (mEvc: KLH-CKARRRQRETTRDED) was injected into goat, while the LBN peptide (mLbn: KLH-CENESSGEQAQAEQSKRRKH) was introduced to rabbits. The use of two different host species allowed for simultaneous staining of protein products.
Immunocytofluorescence
NIH3T3 cells were grown on cover slips, rinsed twice with PBS then fixed in 3.7% formaldehyde/PBS. Following fixation, cells were again rinsed with PBS then blocked for 1 h with 2% horse serum, 2%BSA and 1% NP-40 in PBS. Cells were incubated with anti-EVC (1:100 dilution), anti-LBN (1:100 dilution), anti-acetylated Tubulin (Sigma, St Louis, MO, USA) and/or anti-gamma-tubulin (Sigma) antibodies overnight at 4°C. Cells were washed five times for 5 min with 1% NP-40/PBS before secondary antibodies, Alexafluor 568 rabbit anti-goat and Alexafluor 488 donkey anti-rabbit (Invitrogen) were used to detect EVC and LBN, respectively. TO-PRO-3 iodide (Molecular Probes/Invitrogen) was used to counter stain the nucleus. Fluorescence was visualized with a Nikon inverted Eclipse TE 300 (Nikon Instruments Inc., Mellville, NY, USA).
Immunohistofluorescence
Paraffin-embedded heart and whole embryo sections were hydrated, rinsed with PBST, and boiled in antigen unmasking solution (Vector Labs, Burlingame, CA, USA) prior to blocking for 1 h in immunoblock (1% BSA, 0.1% cold water fish skin gelatin, 0.1% TWEEN-20 in PBS and 0.05% NaN3). After blocking, the primary antibody was added overnight at 4°C. Slides were rinsed in PBST, then incubated in the dark with the appropriate secondary AlexaFluor (Invitrogen) antibody for 1 h. After rinse, TO-PRO-3 Iodide was applied at a 1:1000 dilution for 20 min before final rinses and mounting with Vectashield Mounting Media for Fluorescence (Vector Labs). Confocal images were acquired with either the Nikon inverted Eclipse TE 300 (Nikon Instruments) or the Axio plane LSM 510 (Carl Zeiss MicroImaging, Thornwood, NY, USA). The Isl-1 antibody made by Thomas M. Jessell was obtained from the Developmental Studies Hybridoma Bank created under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences. Reactions were carried out with secondary only negative controls for Evc (Anti-Goat-488 Only) and Lbn (Anti-Rb-488 Only). Figure 3Q–S, which is Anti-Goat-488 only, displays representative negative control slides.
Obtaining cDNA of murine Evc and Lbn
pCMV-sport6-mLbn (MGC-47318) was purchased from ATCC (Manassas, VA). The Expand Long Template PCR system (Roche Scientific) was used to amplify Evc from mouse whole embryo cDNA with primers mEvcF_LR: TGAGCCCTCCATGACCTGCACTAAAGATGC and mEvcR_LR: GTCCCAAGAGGAAGTGAGGTGCGTGCAG. The PCR product was gel purified and cloned into the pCR 2.1-Topo Vector. Cloned Evc sequence was compared with accession number AK148262. The Evc open reading frame was then cut out of Topo-mEvc with EcoRV and XbaI and ligated to pCMV-sport6 with T4 DNA Ligase (Invitrogen, Carlsbad, CA, USA) to ensure similar degrees of overexpression in cell culture.
Transfections for overexpression
NIH3T3 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) modified with 4 mm l-glutamine and 4.5 g/L glucose (Gibco/Invitrogen), 1.5 g/L sodium bicarbonate and 10% fetal bovine serum. Media was changed to OptiMEM I Reduced Serum Medium (Invitrogen) prior to transfection with the Lipofectamine reagent (Invitrogen). Transfection was carried out according to the manufacturer’s guidelines. Briefly, plasmid purified with Qiafilter Plasmid Maxi Kit (Qiagen) was incubated with Optimem and Lipofectamine for 20 min prior to dropwise addition to cells. After 48 h, transfection efficiency was measured by counting the number of cells expressing pcDNA3.1-eGFP and dividing by the total number of cells in the field. Transcript overexpression was verified by quantitative RT–PCR.
siRNA transfections
The Stealth™ Select RNAi Card for Evc and Lbn were tested for efficacy and the following sequences were chosen for experimentation: Evc 5′-ACAAGCUGUGGAAGAAGCAAGAAUU-3′ and Lbn 5′-CCUCACAGAACAGAACUCAGCUAAA-3′. All siRNA transfections were carried out at a 100 nM concentration using Lipofectamine 2000 reagent (Invitrogen). Transfected cells were incubated for 48 h before collection in Trizol reagent. The transfection efficiency was determined by counting the number of cells containing the green labeled BLOCK-iT Fluorescent Oligo (Invitrogen) and dividing by the total number of cells in the same field. The percent transcript knockdown was determined by quantitative RT–PCR.
Statistical methods
Significance values were Bonferroni adjusted to account for the multiple testing strategy. To achieve statistical significance the P-value had to be <0.0125. Due to non-normal distribution, overexpression data were analyzed with the non-parametric Wilcoxon sign-rank. The remaining comparisons were made with a Student’s t-test.
Western blot
Cells grown in a 100 mm dish were washed twice with 1× PBS and lysed in M-PER Mammalian Protein Extraction Reagent (Pierce, Rockford, IL, USA) plus Complete protease inhibitor (Roche Diagnostics, Indianapolis, IN). After three cycles of freeze–thaw, debris was removed by centrifugation at 18 000g for 10 min and protein was stored temporarily at −20°C. For western Blot, Laemmli sample buffer (Bio-Rad) plus ß-Mercaptoethanol was added to 50 µg of protein and loaded on a 4–12% Tris–Glycine SDS gel (Invitrogen, Carlsbad, CA, USA). Gels were run at room temperature for 90–120 min at 100 V. Membranes were blocked for 1 h in 5% powdered milk in PBST and antibody was added overnight at a 1:100 dilution. Membranes were rinsed repeatedly before addition of ECL Donkey Anti-Rabbit IgG Horseradish Peroxidase linked whole antibody (Amersham, Piscataway, NJ, USA). Amersham ECL Plus Western Blotting Detection System was used to detect HRP activity (Amersham, Piscataway, NJ, USA).
SUPPLEMENTARY MATERIAL
FUNDING
American Heart Association Pre-doctoral grant (0615186B) to K.L.S.; National Institutes of Health(HL074728, HL69712 to D.W.B.).
Supplementary Material
REFERENCES
- 1.Ellis R.W.B., van Creveld S. A syndrome characterized by ectodermal dysplasia, polydactyly, chondro-dysplasia and congenital morbus cordis: report of three cases. Arch. Dis. Child. 1940;15:65–84. doi: 10.1136/adc.15.82.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKusick V.A., Egeland J.A., Eldridge R., Krusen D.E. Dwarfism in the Amish I.The Ellis-van Creveld syndrome. Bull. Johns Hopkins Hosp. 1964;115:306–336. [PubMed] [Google Scholar]
- 3.McKusick V.A. Ellis-van Creveld syndrome and the Amish. Nat. Genet. 2000;24:203–204. doi: 10.1038/73389. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-Perez V.L., Ide S.E., Strom T.M., Lorenz B., Wilson D., Woods K., King L., Francomano C., Freisinger P., Spranger S., et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat. Genet. 2000;24:283–286. doi: 10.1038/73508. [DOI] [PubMed] [Google Scholar]
- 5.Galdzicka M., Patnala S., Hirshman M.G., Cai J.F., Nitowsky H., Egeland J.A., Ginns E.I. A new gene, EVC2, is mutated in Ellis-van Creveld syndrome. Mol. Genet. Metab. 2002;77:291–295. doi: 10.1016/s1096-7192(02)00178-6. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Perez V.L., Tompson S.W., Blair H.J., Espinoza-Valdez C., Lapunzina P., Silva E.O., Hamel B., Gibbs J.L., Young I.D., Wright M.J., et al. Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am. J. Hum. Genet. 2003;72:728–732. doi: 10.1086/368063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digilio M.C., Marino B., Ammirati A., Borzaga U., Giannotti A., Dallapiccola B. Cardiac malformations in patients with oral-facial-skeletal syndromes: clinical similarities with heterotaxia. Am. J. Med. Genet. 1999;84:350–356. doi: 10.1002/(sici)1096-8628(19990604)84:4<350::aid-ajmg8>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Webb S., Brown N., Anderson R. Formation of the atrioventricular septal structures in the normal mouse. Circ. Res. 1998;82:645–656. doi: 10.1161/01.res.82.6.645. [DOI] [PubMed] [Google Scholar]
- 9.Goddeeris M.M., Rho S., Petiet A., Davenport C.L., Johnson G.A., Meyers E.N., Klingensmith J. Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development. 2008;135:1887–1895. doi: 10.1242/dev.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Perez V.L., Blair H.J., Rodriguez-Andres M.E., Blanco M.J., Wilson A., Liu Y.N., Miles C., Peters H., Goodship J.A. Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development (Cambridge, England) 2007;134:2903–2912. doi: 10.1242/dev.007542. [DOI] [PubMed] [Google Scholar]
- 11.University, I.o.H.G.a.N. Professor Judith Goodship: Research Interests; Cilia and Development. 2008 [Google Scholar]
- 12.Tompson S.W., Ruiz-Perez V.L., Blair H.J., Barton S., Navarro V., Robson J.L., Wright M.J., Goodship J.A. Sequencing EVC and EVC2 identifies mutations in two-thirds of Ellis-van Creveld syndrome patients. Hum. Genet. 2007;120:663–670. doi: 10.1007/s00439-006-0237-7. [DOI] [PubMed] [Google Scholar]
- 13.Burland T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 14.Piperno G., Fuller M.T. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell. Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kratz C.P., Schubbert S., Bollag G., Niemeyer C.M., Shannon K.M., Zenker M. Germline mutations in components of the Ras signaling pathway in Noonan syndrome and related disorders. Cell Cycle. 2006;5:1607–1611. doi: 10.4161/cc.5.15.3128. [DOI] [PubMed] [Google Scholar]
- 16.Mommersteeg M.T., Soufan A.T., de Lange F.J., van den Hoff M.J., Anderson R.H., Christoffels V.M., Moorman A.F. Two distinct pools of mesenchyme contribute to the development of the atrial septum. Circ. Res. 2006;99:351–353. doi: 10.1161/01.RES.0000238360.33284.a0. [DOI] [PubMed] [Google Scholar]
- 17.Snarr B.S., Wirrig E.E., Phelps A.L., Trusk T.C., Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev. Dyn. 2007;236:1287–1294. doi: 10.1002/dvdy.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Heiden K., Groenendijk B.C., Hierck B.P., Hogers B., Koerten H.K., Mommaas A.M., Gittenberger-de Groot A.C., Poelmann R.E. Monocilia on chicken embryonic endocardium in low shear stress areas. Dev. Dyn. 2006;235:19–28. doi: 10.1002/dvdy.20557. [DOI] [PubMed] [Google Scholar]
- 20.Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science (New York), N.Y. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 21.Webb S., Anderson R.H., Lamers W.H., Brown N.A. Mechanisms of deficient cardiac septation in the mouse with trisomy 16. Circ. Res. 1999;84:897–905. doi: 10.1161/01.res.84.8.897. [DOI] [PubMed] [Google Scholar]
- 22.Blom N.A., Ottenkamp J., Wenink A.G., Gittenberger-de Groot A.C. Deficiency of the vestibular spine in atrioventricular septal defects in human fetuses with Down syndrome. Am. J. Cardiol. 2003;91:180–184. doi: 10.1016/s0002-9149(02)03106-5. [DOI] [PubMed] [Google Scholar]
- 23.Snarr B.S., O’Neal J.L., Chintalapudi M.R., Wirrig E.E., Phelps A.L., Kubalak S.W., Wessels A. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ. Res. 2007;101:971–974. doi: 10.1161/CIRCRESAHA.107.162206. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C.V., Hau J., Martin O., Kuznetsov D., Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yutzey K.E., Rhee J.T., Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development (Cambridge, England) 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.