Abstract
Tubby-like protein 3 (TULP3) is required for proper embryonic development in mice. Disruption of mouse Tulp3 results in morphological defects in the embryonic craniofacial regions, the spinal neural tube and the limbs. Here, we show that TULP3 functions as a novel negative regulator of Sonic hedgehog (Shh) signaling in the mouse. In Tulp3 mutants, ventral cell types in the lumbar neural tube, which acquire their identities in response to Shh signaling, are ectopically specified at the expense of dorsal cell types. Genetic epistasis experiments show that this ventralized phenotype occurs independently of Shh and the transmembrane protein Smoothened, but it is dependent on the transcription factor Gli2. The ventralized phenotype is also dependent on the kinesin II subunit Kif3A, which is required for intraflagellar transport and ciliogenesis. In addition, TULP3 is required for proper Shh-dependent limb patterning and for maintaining the correct balance between differentiation and proliferation in the neural tube. Finally, the localization of TULP3 to the tips of primary cilia raises the possibility that it regulates the Hedgehog pathway within this structure.
INTRODUCTION
Sonic hedgehog (Shh) is a member of the Hedgehog (Hh) family of secreted ligands, which control a number of developmental processes in vertebrates (1–4). In adults, dysregulated Hh signaling leads to cancers of the brain, skin and digestive tract (5). Despite the importance of this pathway in human development and disease, the mechanism of mammalian Hh signal transduction remains unclear.
Hh signal transduction is best understood in Drosophila (reviewed in 6). In flies, Hh acts by binding to its transmembrane receptor Patched (Ptc), preventing Ptc from antagonizing a second transmembrane protein, Smoothened (Smo) (7). Inhibition of Ptc allows Smo to transduce signals through a protein complex containing the atypical kinesin Costal-2, Fused kinase and the transcription factor Cubitus interruptus (Ci) (8). Hh signaling blocks the proteolytic cleavage of Ci, whose truncated form represses target gene expression. In the presence of ligand, full-length Ci translocates to the nucleus and activates Hh target genes (9).
Characterization of vertebrate homologs of Drosophila Hh pathway components suggests broad conservation of the mechanism. However, recent studies point to several differences between the pathways acting in mice and flies (10,11). For example, it appears that mouse fused and costal-2 homologs do not play roles analogous to those in flies (12,13) and the vertebrate pathway has been shown to require a number of components, such as Rab23, MIM/BEG4, Hip1, SIL, Gas1, Talpid3 and β-arrestin 2, which do not appear to have conserved functions in the Drosophila pathway (14–20). Recent work has also demonstrated that a number of factors participating in intraflagellar transport (IFT), a process essential for ciliogenesis, are also required for Hh signaling in mice (21–25), suggesting that the primary cilium is a key signaling center for the mammalian Hh pathway. The Gli proteins and a Gli antagonist, Suppressor of Fused (Sufu), localize to the tips of primary cilia (26). In addition, Patched1 (Ptch1) and Smo differentially localize to cilia in mammalian cells depending on exposure to Shh ligands (27–29). However, the steps in signal transduction downstream of Smo regulating the activity of the Gli transcription factors in the mouse are not understood.
Shh signaling plays a key role in the growth and patterning of many vertebrate tissues such as the limb and neural tube (reviewed in 30,31). In the developing limb, Shh, produced by a patch of cells in the posterior mesenchyme called the zone or polarizing activity, regulates gene expression and digit identity along the anterior–posterior (A–P) axis. In the developing neural tube, Shh patterns cell fates along the dorsal–ventral (D–V) axis. Shh ligand is produced from the notochord lying ventral to the neural tube and is distributed in a ventral-to-dorsal gradient (32,33). Progenitor cells are exposed to varying concentrations of Shh and they respond by adopting distinct identities. For example, cells closest to the source of Shh experience the highest level of signaling and are specified as floor plate, whereas more distant cells experience lower levels and develop into ventral interneuron and motor neuron progenitors (34).
Several novel Shh signaling components have been discovered in mice as a result of neural tube patterning defects in mutants. We have identified Tubby-like protein 3 (TULP3) as one such component. Tulp3 was originally identified through sequence homology to Tubby (Tub), a gene whose disruption results in obesity and sensory neurodegeneration in the adult mouse (35,36). Deletion of the Tubby domain of mouse Tulp3 is embryonic lethal, resulting in severe defects such as impaired neural tube closure and craniofacial abnormalities (37). These defects are also observed in other mouse mutants that exhibit upregulated Shh signaling (14,25,38), raising the possibility that TULP3 functions in Shh signaling. Here we show that TULP3 is required to antagonize the Shh pathway and we provide evidence that TULP3 inhibits Gli2 activity in an IFT-dependent manner at a step downstream of Shh and Smo. Findings similar to ours have been made for a strong hypomorphic allele of Tulp3, Tulp3hhkr (Patterson et al., submitted for publication). In addition, the subcellular localization of TULP3 protein suggests that it acts within the primary cilium to restrict Gli2 activity in the absence of Shh ligand.
RESULTS
TULP3 regulates cell identity in the spinal neural tube
Tulp3tm1Jng/ tm1Jng mutant embryos arrest on or before embryonic day 14.5 (E14.5) and exhibit defects including exencephaly, spina bifida, defective eye development and enlarged branchial arches (Fig. 1A, 37). Tulp3 mutants also displayed anterior outgrowth of the autopods indicative of polydactyly; cartilage staining confirmed the presence of one to two additional digits (Fig. 1B). These features are similar to those arising from mutations such as Rab23opb2 or Thm1aln, which exhibit inappropriate activation of the Shh signaling pathway (14,25), suggesting that Tulp3 is also required to inhibit this pathway in mice.
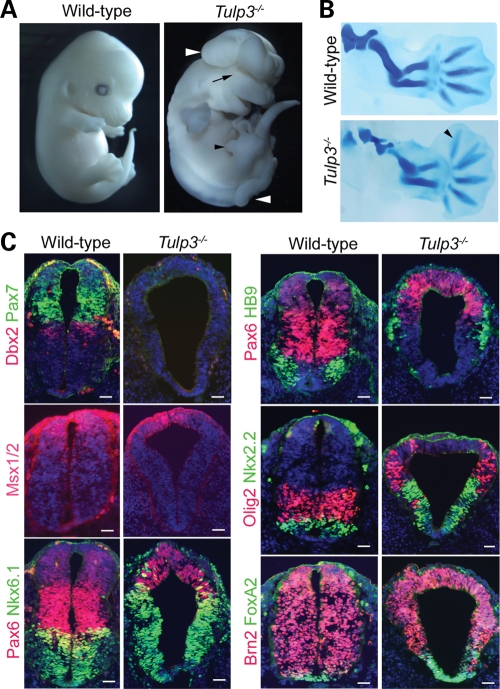
Figure 1.
The Tulp3 mutant phenotype. (A) E13.5 wild-type and Tulp3 mutants. The mutants exhibited neural tube closure defects (white arrowheads), eye defects (black arrow) and abnormal anterior outgrowth in the limbs (black arrowhead). (B) Alcian blue staining of E13.5 forelimbs revealed an additional anterior digit in the mutant limb (arrowhead). (C) E10.5 posterior neural tube sections from wild-type and Tulp3 embryos stained for Pax7, Dbx2, Msx1/2, Pax6, Nkx6.1, HB9, Olig2, FoxA2 and Brn2. Sections were counterstained with DAPI (blue). Scale bars are 50 µm.
Because Shh signaling has been extensively characterized in D–V patterning of the neural tube, we investigated this aspect in Tulp3 mutant embryos. In the lumbar neural tube of E10.5 Tulp3 mutants, Shh-dependent ventral cell types were ectopically specified at the expense of lateral and dorsal cell types (Fig. 1C). Specifically, FoxA2, Nkx2.2, Nkx6.1 (markers of floor plate, p3 and p3/pMN/p2 progenitors, respectively) showed enlarged expression domains expanding into comparatively more dorsal regions. Similarly, the expression domains of markers for differentiating motor neurons and their progenitors (HB9 and Olig2, respectively) were expanded into lateral regions. Shh signaling also inhibits the specification of lateral and dorsal neural cell types. In Tulp3 mutants, markers for these cell types were either strongly downregulated (Dbx2, Pax7) or were expressed in dorsally restricted domains (Pax6, Msx1/2). As in mouse embryos with mutations in Thm1, Rab23, Fkbp8 and Prkac (25,39–41), these effects were limited to the lumbar neural tube; D–V patterning at the level of the forelimbs appeared roughly similar in wild-type and Tulp3 mutants (data not shown). These changes are consistent with elevated Shh signaling in the lumbar neural tubes of Tulp3 mutants.
We next analyzed the expression of Ptch1 and Gli1 in Tulp3 mutant and wild-type neural tubes at E9.5 and E10.5 (Fig. 2A and Supplementary Material, Fig. S1A and B). In the wild-type neural tube, these targets are expressed ventrally in response to Shh signaling. In Tulp3 mutants, Ptch1 and Gli1 were inappropriately expressed in dorsal and lateral regions of the neural tube. The ventral border of the Gli1 expression domain in the mutants was shifted dorsally in comparison with wild-type. Although Gli1 is induced by Shh signaling, its expression is excluded from the wild-type floor plate and is repressed in the neural tubes of Patched1 mutants (42), suggesting that Gli1 is repressed ventrally by very strong Shh signaling in the Tulp3 mutant neural tube. Expression of Gli3 is normally restricted to dorsal regions by Shh signaling and this gene showed a more dorsally restricted expression pattern in Tulp3 mutants (Fig. 2A). The expression domain of Gli2 was roughly similar between genotypes, although it did not extend as far ventrally in the mutants. These data indicate that Tulp3 mutants experience elevated levels of Shh pathway activity.
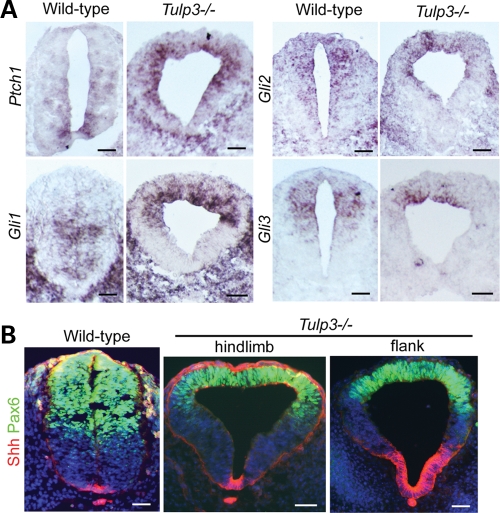
Figure 2.
Expression of Shh and Shh pathway components. (A) In situ hybridization for Patched1, Gli1, Gli2 and Gli3 in E10.5 wild-type and Tulp3 mutant posterior neural tube sections. The Shh targets Ptch1 and Gli1 were ectopically expressed in dorsal regions, and Gli3 expression was dorsally restricted in Tulp3 mutants. (B) Immunostaining for Shh (red) and Pax6 (green) in wild-type and Tulp3 mutants (shown at levels of the hindlimb and flank). Dorsal restriction of Pax6 was seen at both levels of the mutant neural tube, whereas expansion of the Shh+ domain was seen only at more anterior levels (flank). Scale bars are 50 µm.
It is possible that such an elevation stems from increased Shh ligand production. In fact, in the E10.5 mutant neural tube at the level just anterior to the hindlimbs (flank), we observed a broader domain of Shh protein expression (Fig. 2B). However, this was not the case at the level of the hindlimbs, where Shh expression was confined to its normal domain. We observed normal Shh expression in the mutant neural tube at E9.5 (Supplementary Material, Fig. S1A), although D–V patterning defects were evident by this stage. As Shh expression in the neural tube is driven by activation of the Shh signaling pathway (43), the variable expansion of the Shh expression domain at E10.5 is likely to be a consequence, rather than a cause, of increased pathway activity.
TULP3 controls Shh-dependent gene expression in the developing limbs
In addition to the neural patterning defects, Tulp3 mutant embryos exhibited limb preaxial polydactyly. This is obvious at E12.5 when the digit rays are outlined by the expression of Sox9 in the mutant limb buds (Fig. 3). The presence of abnormally long digit rays at the anterior limb bud raised the possibility that A–P patterning and Shh signaling are affected in the mutant limb buds. To address this, we examined the expression of Shh and its target genes in limb buds (Fig. 3). At E11.5, the expression of direct transcriptional targets of the Shh pathway, Gli1 and Ptch1, expanded to include the distal and anterior mesenchyme in mutant limb buds, whereas their expression was confined to posterior regions in normal limb buds (Fig. 3). Similarly, the Hoxd12 expression domain expanded into the anterior mesenchyme in E11.5 Tulp3 mutant limb buds, suggesting that A–P polarity is perturbed. These changes were accompanied by misexpression of Shh in anterior regions of mutant limb buds.
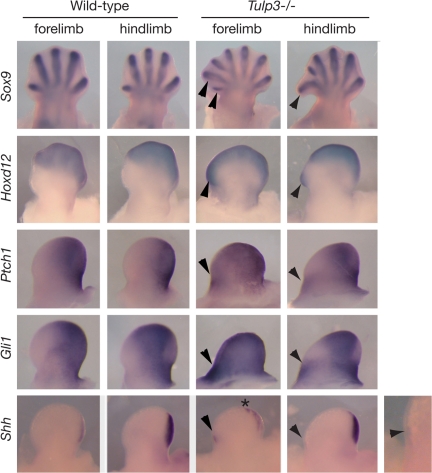
Figure 3.
Expression of Sox9, Hoxd12, Ptch1, Gli1 and Shh in limb buds. Expression in both forelimb and hindlimb buds is shown (anterior to the left). E12.5 Tulp3 mutant limbs show an increased number of digit rays as outlined by Sox9 expression, and disruption of A–P polarity as indicated by expansion of Hoxd12 expression into the anterior mesenchyme (arrowheads). In E11.5 mutant limb buds, both Ptch1 and Gli1 were expanded anteriorly in forelimb buds and ectopically expressed in a separate domain in the anterior mesenchyme of hindlimb buds (arrowheads). In mutant limb buds, weak Shh expression was detected in an ectopic anterior domain (arrowheads) separate from its normal domain in the posterior mesenchyme. In addition, Shh expression in the posterior domain of mutant forelimb buds was slightly expanded anteriorly (asterisk).
Anterior misexpression of Shh could be the cause or the consequence of ectopic Hoxd12 expression, as Hoxd12 and Shh control each other’s expression through positive feedback (44). Alternatively, inappropriate activity of the Shh signaling pathway could drive misexpression of both genes. One day earlier in development (E10.5), Tulp3 mutant forelimb buds were somewhat small and misshapen, although the hindlimb buds appeared normal. Nevertheless, both the mutant forelimb and hindlimb buds showed misexpression of Gli1 and Ptch1 in the anterior regions (Supplementary Material, Fig. S2). However, neither Shh nor Hoxd12 was misexpressed in the anterior domains at this stage, suggesting that their later misexpression is secondary to defects in the Shh signal transduction pathway. In both wild-type and mutant limb buds at E10.5, dHand and Gli3 were expressed in posterior and anterior domains, respectively (Supplementary Material, Fig. S2), indicating that limb bud prepattern (45) is retained independently of Tulp3. Collectively, these data indicate that the anterior regions of the Tulp3 limb buds experience inappropriate activation of the Shh pathway, eventually leading to polydactyly.
TULP3 regulates Shh signal transduction
We next investigated whether Shh pathway activation in Tulp3 mutants is Shh ligand-dependent. If TULP3 negatively regulates Shh ligand expression or distribution, neural patterning in Shh/Tulp3 double mutants should resemble that in Shh single mutants. However, if TULP3 antagonizes the pathway downstream of Shh, the pathway should remain active in the absence of ligand. We generated Shh/Tulp3 double mutants and found that their size and morphology resembled those of Tulp3 single mutants (Supplementary Material, Fig. S3A). In the Shh null mutant neural tube, specification of ventral FoxA2+ floor plate, Nkx2.2+ V3 interneuron progenitors and HB9+ motor neurons does not occur, whereas Pax6 and Pax7 expression expands throughout the ventral neural tube (14,46). In contrast, FoxA2+, Nkx2.2+ and HB9+ ventral cell types were restored in Shh/Tulp3 double mutants, which also showed dorsal restriction of Pax6 expression and repression of Pax7 (Fig. 4). Furthermore, Ptch1 and Gli1 were expressed in Shh/Tulp3 mutants in a pattern similar to that in Tulp3 single mutants (Supplementary Material, Fig. S1B). Shh ligand appeared to contribute relatively little to total pathway activation in the Tulp3 mutant posterior neural tube, as neural patterning was similar in Tulp3 and Shh/Tulp3 double mutants. However, the expansion of the FoxA2+ domain seen in Tulp3 single mutants was not observed in Shh/Tulp3 mutants, indicating that Tulp3 mutant neural progenitors are at least partially responsive to Shh.
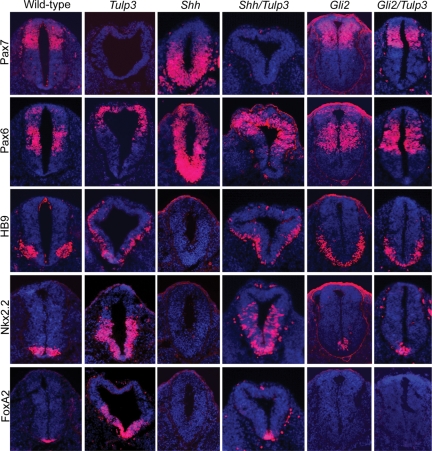
Figure 4.
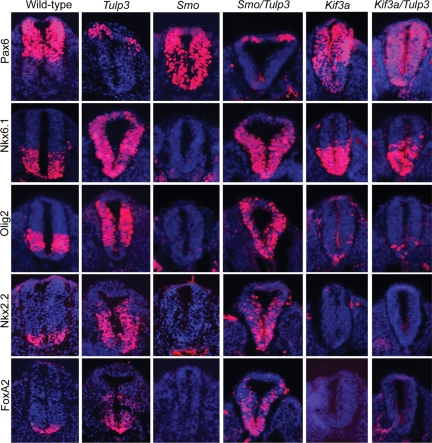
Neural tube patterning in Shh/Tulp3 and Gli2/Tulp double mutants. Sections through the neural tubes (lumbar level) of E10.5 wild-type, Tulp3−/−, Shh−/−, Gli2−/−, Shh−/−Tulp3−/− and Gli2−/−Tulp3−/− mutants stained for markers of dorsal/lateral (Pax7 and Pax6) and ventral (HB9, Nkx2.2, FoxA2) neural cell types, which are inhibited or induced by Shh signaling, respectively. Sections were counterstained with DAPI (blue).
This phenomenon was also evident in the limb buds of Shh/Tulp3 double mutants (Supplementary Material, Fig. S4). We found that, although Gli1, Ptch1 and Hoxd12 were expressed at lower levels in double mutant limb buds compared with those of wild-type or Tulp3 single mutants, their expression was maintained despite the absence of Shh. Shh mutant limbs buds fail to express Ptch1 and Gli1 and exhibit only very weak expression of Hoxd12 in hindlimbs (47). Furthermore, the double mutant limb bud size was intermediate between that of Tulp3 mutants and Shh mutants (not shown). These findings suggest that, although the total level of Shh pathway activity in Tulp3 mutant limbs depends heavily on signaling by Shh ligand, a low level of pathway activity also occurs in this tissue independently of Shh.
The Tulp3 neural patterning phenotype is Gli2-dependent
Although the data indicate that TULP3 controls neural patterning by inhibiting the Shh signal transduction pathway, it is still possible that this protein acts through a parallel pathway. To distinguish between these possibilities, we asked whether the effects of Tulp3 disruption were dependent on a downstream component transducing Shh signals, Gli2. In the Gli2 mutant neural tube, FoxA2+ cells are absent, Nkx2.2+ cells are reduced in number and the HB9+ domain expands across the ventral midline (48,49). Importantly, the ventralized phenotype of Tulp3 single mutants was suppressed in Gli2/Tulp3 double mutants, whose neural patterning and morphology closely resembled those of Gli2 single mutants (Fig. 4). The expression patterns of Shh, Ptch1 and Gli1 were also similar in Gli2 and Gli2/Tulp3 double mutants, although some weak expression of Ptch1 and Gli1 could be detected in more dorsal regions in the double mutant neural tubes (Supplementary Material, Fig. S1B). These data indicate that TULP3 functions in D–V neural patterning mainly through antagonizing Gli2 activity at a step downstream of Shh. However, slight differences in gene expression between Gli2 and Gli2/Tulp3 double mutants suggest that TULP3 functions to a minor extent through a separate mechanism, such as the regulation of Gli3.
We next considered how TULP3 might restrain Gli2 activity. This regulation is unlikely to be at the transcriptional level, as the expression of Gli2 mRNA in the Tulp3 mutant neural tube was comparable to that of wild-type (Fig. 2A). Recent studies indicate that the control of Gli2 protein stability by PKA-, CK1-, GSK3β- and DYRK2-mediated phosphorylation is important for the proper regulation of the Hh pathway (50,51). However, western blotting revealed comparable total steady-state Gli2 protein levels in Tulp3 mutant and wild-type embryos, and no clear differences in Gli2 electrophoretic mobility (indicative of differential posttranslational modification) were detected between genotypes (Supplementary Material, Fig. S5A). PKA-, CK1- and GSK3β-mediated phosphorylation events also control the activity of Gli3 (52). In the resting state, full-length 190 kDa Gli3 is phosphorylated and proteolytically processed into an 83 kDa transcriptional repressor (Gli3R); Shh signaling prevents this proteolysis allowing unprocessed Gli3 (Gli3A) to accumulate. The extent of Gli3 processing in E9.5 whole-embryonic extracts was comparable in wild-type and Tulp3 embryos, suggesting that phosphorylation and proteasome-dependent processing of Gli3 occur normally in Tulp3 mutants (Supplementary Material, Fig. S5B). Gli3 processing may be affected to some extent in E11.5 Tulp3 limb buds (Supplementary Material, Fig. S5C), but experimental variability made this difficult to confirm and we could not exclude potentially indirect effects of ectopic Shh expression (Fig. 3) on Gli3 processing in the limbs. Because we did not detect a physical interaction between TULP3 and Gli2 by co-immunoprecipitation (Supplementary Material, Fig. S5D and E), antagonism of Gli2 by TULP3 is likely to be indirect, although a small fraction of Gli2 and TULP3 could be physically associated in the cell. Moreover, the loss of TULP3 had no obvious effects on the association between Gli2 and Sufu proteins in the embryo (Supplementary Material, Fig. S5F). Thus, TULP3 appears to regulate Gli2 through a mechanism distinct from that controlling Gli stability and processing.
Subcellular localization of TULP3
Since several Shh pathway components (Ptch1, Smo, the Gli transcription factors, Sufu) localize to the primary cilium and genetic data indicate that IFT components are required for proper Hh signaling in mice (reviewed in 53), we considered the possibility that TULP3 controls the Shh pathway within the primary cilium. We assayed the ability of serum-starved primary mouse embryonic fibroblasts (pMEFs) from wild-type and Tulp3 mutant embryos to generate cilia and to properly localize ciliary proteins. Immunostaining of acetylated α-tubulin revealed that mutant fibroblasts generated primary cilia with no obvious defects in axonemal lengths (Supplementary Material, Fig. S6A and D). Mutant fibroblasts also showed normal ciliary localization of Ift88 and Gli2 (Supplementary Material, Fig. S6A and C). Ift88 staining of neural tube sections indicated that Tulp3 mutant neural progenitors properly generated cilia (Supplementary Material, Fig. S6B). The absence of left–right patterning defects in Tulp3 mutants (data not shown) suggested that mutant nodal cilia are able to generate leftward fluid flow (54). Thus, Tulp3 does not appear to regulate global aspects of ciliogenesis. Nevertheless, Tulp3 may have a role in the ultrastructure of the cilium or it may regulate the activities of Hh signal transduction components that reside within primary cilia.
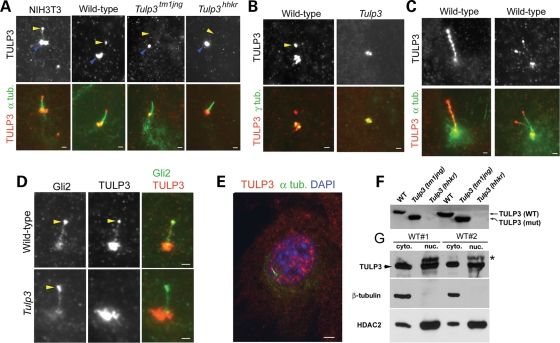
To determine where TULP3 may function in the cell to regulate Shh signaling, we performed immunostaining for TULP3. We found positive staining at the tips of cilia from wild-type pMEFs and NIH3T3 fibroblasts (Fig. 5A, yellow arrowheads). This localization pattern was somewhat variable as we observed it in most, but not all, cilia from wild-type cells (65%, n = 62). Staining at cilia tips was specific for TULP3 because it was not observed in cilia from Tulp3tm1jng mutant cells (n = 68), nor from cells mutant for a second allele, Tulp3hhkr (n = 45), which produce nearly undetectable levels of TULP3 protein (Fig. 5F; J. Murdoch, personal communication). Our antiserum also showed strong staining around the centrosome (Fig. 5B), but this appeared to be nonspecific, as it was also seen in mutant cells and was observed with our preimmune serum (not shown). We could not reliably detect TULP3 staining on cilia of neural progenitors in vivo (not shown). However, TULP3 protein may be rapidly transported into and out of cilia, resulting in very low steady-state levels within cilia that are continually being assembled and disassembled in dividing cells. Thus, TULP3 may accumulate only to detectable levels within stable primary cilia generated by quiescent cells. Consistent with the possibility that TULP3 undergoes transport in cilia, we observed strong punctate staining along the axonemes of longer cilia (>5 µm, Fig. 5C), suggesting that continuous transport through such long cilia occasionally results in local accumulation. Interestingly, the ciliary localization of TULP3 resembled that of endogenous Gli2 protein, although the two proteins did not show complete colocalization at this site (Fig. 5D).
Figure 5.
Subcellular localization of TULP3. (A) TULP3 staining (red) is seen at the tips of cilia labeled with α-acetylated alpha tubulin (green) in NIH3T3 fibroblasts and wild-type pMEFs, but not in pMEFs derived from embryos mutant for the tm1Jng1 or hhkr alleles of Tulp3. Cilia tips are indicated in top panels by yellow arrowheads. Nonspecific pericentrosomal staining is indicated by blue arrowheads. (B) Double labeling of γ-tubulin and TULP3 in wild-type and Tulp3 mutant pMEFs. Tip staining is indicated by a yellow arrowhead. (C) TULP3 immunostaining of unusually long cilia (left) or rare double cilia (right) from wild-type pMEFs. (D) TULP3 and Gli2 show partial colocalization at the tips of wild-type cilia (yellow arrowheads). (E) TULP3 (red) and acetylated alpha tubulin (green) immunostaining of a wild-type pMEF showing TULP3 staining in the nucleus (labeled with DAPI). (F) Western blotting of pMEF extracts reveals a truncated form of TULP3 (TULP3 mut) in the Tm1jng allele and very low levels of TULP3 in the hhkr allele. (G) TULP3 was found in nuclear and cytoplasmic factions from E9.5 embryos. HDAC2, which is enriched in nuclear fractions, and β-tubulin, which is limited to the cytoplasmic fractions, serve as controls. The slower migrating species (asterisk) in the nuclear fractions appears to be nonspecific, as it is also seen in Tulp3 mutant nuclear extracts (not shown). Scale bars are 1 µm in A–D and 3 µm in E.
Previous studies of Tubby family members raised the possibility that they might function in the nucleus. Tubby family members contain putative nuclear localization sequences, and TULP proteins have been observed in the nucleus (55–57). Boggon et al. (58) have suggested that the N- and C-terminal domains of Tubby are capable of transcriptional activation and double stranded DNA binding. However, there is no apparent sequence specificity in Tubby DNA binding, and transcriptional targets for Tubby proteins have not been identified. TULP3 antiserum variably showed punctate staining in the cytoplasm and nuclei of cultured fibroblasts (Fig. 5E). To address this issue, we performed whole-embryo subcellular fractionation. As observed for Tubby (58), we found TULP3 protein in cytoplasmic, nuclear and membrane fractions from such embryos (Fig. 5G; data not shown). This raises the possibility that TULP3 regulates the Hh pathway at sites other than the cilium, such as the nucleus. However, TULP3 does not appear to control Gli2 nuclear localization, as steady-state levels of Gli2 in nuclear fractions of Tulp3 mutants were comparable to those of wild-type (Supplementary Material, Fig. S5A).
Activation of the Shh pathway in Tulp3 mutants is Kif3a-dependent and Smo-independent
If TULP3 were to regulate the Hh pathway within primary cilia, the effects of disrupting Tulp3 on the Hh pathway should be suppressed in cells lacking these structures. Kinesin II is the primary molecular motor used for anterograde IFT in vertebrates, and cells lacking the kinesin II subunit Kif3A are defective in ciliogenesis (59,60). We investigated whether the Tulp3 mutant phenotype is dependent on kinesin II by generating Kif3a/Tulp3 double mutants. The general morphology of Kif3a/Tulp3 double mutants resembled that of Kif3a single mutants, which are smaller than wild-type embryos and arrest around E9.5 (Supplementary Material, Fig. S3B). The neural patterning phenotype of Kif3a mutants was strongly dorsalized at E9.5, although some neural cell types requiring low levels of Shh signaling for their specification persisted (Fig. 6). This is presumably due to diminished Gli3 repressor function (22,61). Cell types requiring the highest levels of Shh signaling, floor plate and p3 progenitors marked by FoxA2 and Nkx2.2, were absent in Kif3a embryos. In addition, Olig2+ cells were markedly reduced in number, and Pax6 was expressed in cells throughout the neural tube. Importantly, neural patterning in Kif3a/Tulp3 double mutant closely resembled that in Kif3a single mutants. The suppression of the Tulp3 phenotype by disruption of Kif3a was also demonstrated by the expression patterns of Shh, Ptch1 and Gli1 in Kif3a/Tulp3 double mutants (Supplementary Material, Fig. S1A). Thus, the activation of the Hh pathway resulting from Tulp3 disruption depends on kinesin II. These data are consistent with a model in which Tulp3 and kinesin II each regulate the Hh pathway at the level of the primary cilium. However, TULP3 and kinesin II also localize to other sites within the cell, such as on cellular membranes (62), and kinesin II is known to play many roles in the cell beyond IFT (63). Therefore, alternative models for Tulp3 function in the control of Hh signaling outside the cilium are possible.
Figure 6.
Neural tube patterning in Smo/Tulp3 and Kif3a/Tulp3 double mutants. Sections through posterior (lumbar) neural tubes of E9.5 wild-type, Tulp3−/−, Smo−/−, Kif3a−/−, Smo−/−Tulp3−/− and Kif3a−/−Tulp3−/− mutants stained for Pax6, Nkx6.1, Olig2, Nkx2.2 and FoxA2.
Recent data indicate that Smo localizes to the primary cilium upon exposure to Hh ligands and may act there to regulate signal transduction (27,29). We considered the possibility that TULP3 may function to inhibit Smo translocation to the cilium or Smo biochemical activity. To test this genetically, we generated Smo/Tulp3 double mutants. Smo null mutants lack all Hh signaling and exhibit a completely dorsalized neural patterning phenotype; Olig2, Nkx6.1, Nkx2.2, FoxA2, Shh, Ptch1 and Gli1 expression are suppressed in the Smo mutant neural tube, and Pax6 is not repressed in ventral cells (Fig. 6 and Supplementary Material, Fig. S1A). In contrast, the expansion of ventral identity markers and dorsal restriction of Pax6 occurred in Smo/Tulp3 mutants as in Tulp3 single mutants. The expression of Shh, Ptch1 and Gli1 in the Smo/Tulp3 double mutant neural tube was also similar to that in Tulp3 single mutants (Supplementary Material, Fig. S1A), and the growth retardation defect caused by the disruption of Smo was mostly suppressed in Smo/Tulp3 mutants (Supplementary Material, Fig. S3B). Thus, if TULP3 acts in the primary cilium to inhibit Gli2 function, this function is mostly, if not completely, independent of Smo.
The balance between neural proliferation and differentiation is disrupted in Tulp3 mutants
The morphology of the Tulp3 mutant neural tube is clearly abnormal; its shape is roughly triangular and the neuroepithelium appears thin (Fig. 1C). Although increased Shh signaling is generally thought to result in increased proliferation, the Tulp3 neural tube did not show signs of overgrowth. Nevertheless, the abnormal morphology of the mutant neural tube could be caused by changes in neural proliferation and differentiation.
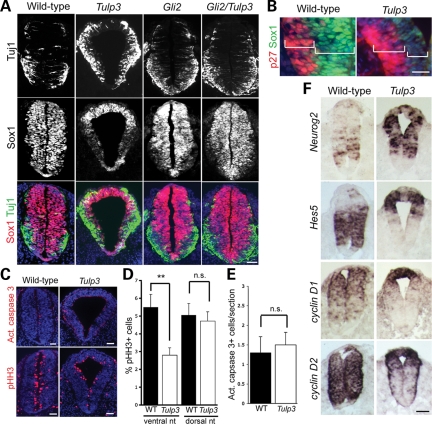
To investigate this, we examined the expression of the progenitor marker Sox1 and the postmitotic neuronal marker Tuj1 (class III beta-tubulin) in E10.5 wild-type and Tulp3 mutant posterior neural tubes. We observed clear downregulation of Sox1 expression in the ventral two-thirds of the mutant neural tube, whereas cells in the dorsal one-third retained robust expression of this marker (Fig. 7A). Conversely, many Tuj1+ cells were observed in the ventral and lateral regions of the mutant neural tube, whereas Tuj1+ cells were largely confined to the motor neuron domain in the wild-type. This effect was suppressed in Gli2/Tulp3 double mutants, whose expression of Sox1 and Tuj1 resembled that of Gli2 single mutants. This indicates that the changes in Sox1 and Tuj1 expression in Tulp3 mutants result from increased activity of the Shh pathway.
Figure 7.
TULP3 is required for proper neural differentiation. (A) Neural tube sections from E10.5 wild-type, Tulp3−/−, Gli2−/− and Gli2−/−Tulp3−/− mutants stained for Tuj1 and Sox1. (B) Sox1+ progenitor and p27Kip1+ postmitotic zones in the ventral region of the E10.5 wild-type and Tulp3 mutant posterior neural tubes. Sox1 was weakly expressed by cells in the ventricular zone of Tulp3 mutants. (C) E10.5 wild-type and Tulp3 mutant sections stained with α-active caspase 3 and α-phosho-Histone H3 (pHH3) antibodies. (D) Percentage of pHH3+ cells in the ventral and dorsal halves of neural tube sections. Differences between genotypes for the ventral halves were significant (**P < 0.001, Student’s t-test). Differences for the dorsal halves were not significant (n.s., P > 0.2). (E) The number of apoptotic cells, marked by active caspase 3 staining, was comparable between genotypes. (F) Expression of Neurog2, Hes5, cyclin D1 and cyclin D2 in posterior neural tube sections from E9.5 wild-type and Tulp3 mutants. Scale bars are 50 µm in A, C and F, and 20 µm in B.
Thinning of the Tulp3 mutant neuroepithelium in ventral posterior regions was caused by the reduction of the ventricular (Sox1-expressing progenitor) zone, as this layer was noticeably reduced in Tulp3 mutants (Fig. 7B). In contrast, the thickness of the postmitotic, p27Kip1+ marginal zone (64) was slightly expanded in Tulp3 mutants. Although increased apoptotic cell death was previously described in the hindbrain region of Tulp3 mutants (37), the reduced thickness of the ventricular zone in the posterior neural tube was not associated with increased cell death (Fig. 7C and E). We did, however, observe fewer cells expressing the late G2/M-phase marker phospho-Histone H3 (pHH3) in the ventral half of the Tulp3 mutant neural tube in comparison with wild-type (Fig. 7C and D), which is consistent with diminished progenitor self-renewal and increased cell-cycle exit.
To investigate the cause of these defects, we focused our attention 1 day earlier in the development (E9.5), when Sox1 and Tuj1 expression was normal in Tulp3 mutants (data not shown). Altered patterns of proliferation and differentiation may be explained by changes in cell identity. For example, Shh signaling controls the expression of Pax6 and Olig2, which, in turn, control the expression of the neurogenic bHLH factor Neurogenin 2 (Neurog2) (65–68). In addition, recent data suggest that Shh signaling may directly regulate early Neurog2 expression through the Gli transcription factors (69). At E9.5, the Olig2 expression domain was expanded within the ventral two-thirds of the mutant neural tube, and Pax6 was confined to dorsal regions (Fig. 6). This was accompanied by increased Neurog2 expression throughout much of the neural tube (Fig. 7F). In addition, Hes5 negatively regulates Neurog2 expression in the spinal neural tube (70). At E9.5, Hes5 is expressed strongly in the ventral/lateral regions of the wild-type posterior neural tube, but its expression domain was dramatically shifted to the dorsal regions in the Tulp3 mutant (Fig. 7F). Thus, the expansion of the Olig2+ domain, ventral/lateral repression of Hes5 expression and direct activation by the Gli transcription factors could collectively account for the increased Neurog2 expression in Tulp3 mutants.
Restricting the activity of the D-type cyclins can also contribute to cell-cycle exit in the CNS (71–73). We found that cyclin D1 expression and D2 expression were markedly downregulated in the ventral and lateral regions of the E9.5 Tulp3 mutant neural tube in comparison with wild-type, yet these genes were strongly expressed in dorsal regions of the mutant neural tube (Fig. 7F). These changes may contribute to more efficient cell-cycle exit ventrally and continued proliferation dorsally. Thus, it is likely that the thinning of the Tulp3 mutant neuroepithelium by E10.5 stems from the misregulated expression of factors promoting or inhibiting neuronal differentiation at E9.5.
DISCUSSION
TULP3 antagonizes Shh signaling in the mouse neural tube
Tubby family members have not been previously linked to the control of mammalian Hh signaling. Here we have shown that one member of this family, TULP3, is required in the embryo to restrict Shh pathway activity in the absence of ligand. This is supported by the observation that Tulp3 mutants show a neural patterning phenotype resembling those of mutants with reduced or abolished function of Shh pathway antagonists such as Rab23, Thm1, Ptch1 and PKA (14,25,38,41). Moreover, direct Shh transcriptional target genes (e.g. Ptch1 and Gli1) are ectopically expressed in the Tulp3 mutant neural tube and limb buds, indicating that the Shh pathway is inappropriately activated in the absence of TULP3.
D–V patterning of the neural tube is driven primarily by the dorsalizing effects of BMP signaling and the ventralizing effects of Shh signaling (74). The specification of ventral neural cell types (such as motor neurons) is not a default state. Although dorsalizing signals such as BMPs and Wnts can attenuate the effects of Shh in gain-of-function experiments, blockade or removal of dorsalizing signals does not result in increased specification of ventral neural cell types, nor can the lack of such signals bypass the requirement for Shh (75–78). Thus, although signaling by dorsalizing factors may be impaired in Tulp3 mutants, it alone cannot explain the ectopic specification of ventral neural cells types in Shh/Tulp3 or Smo/Tulp3 double mutants. Since hyperactivation of Shh targets in Tulp3 mutants still depends on downstream Hh pathway components (Kif3a and Gli2), TULP3 must act within the context of Shh signal transduction.
The relationship between Tubby family proteins and primary cilia
The localization of TULP3 to cilia is interesting given the importance of primary cilia in mammalian Hh signaling. However, Tulp3 mutant cells do not show obvious defects in ciliogenesis. Nevertheless, several observations suggest a link between Tubby family proteins and processes dependent on cilia. For example, mice mutant for several Tubby family members collectively share phenotypic features with those of patients with Bardet–Biedl syndrome (BBS), such as obesity, retinal degeneration, polydactyly and hearing loss (53). BBS proteins play key roles in the function and/or assembly of primary cilia and basal bodies (79).
Some clues have emerged with respect to how Tubby family members function at the cellular level. For example, Tulp1 is expressed in ciliated photoreceptor cells of the retina, where it is required for their survival (80). In these cells, TULP1 localizes to inner segments and connecting cilia (81), and this protein has been implicated in vesicular trafficking of rhodopsin through the connecting cilium to outer segments (81–83). Tubby family members have also been implicated in cilia-related processes in other organisms. In Caenorhabditis elegans, the Tubby homolog TUB-1 is expressed in ciliated neurons and undergoes transport within cilia (84). In worms, TUB-1 plays an essential function in fat storage, chemotaxis and life span regulation, all processes that depend on proper assembly and function of cilia (84–87). It is also interesting that the expression of C. elegans tub-1 and that of Drosophila tulp are regulated by RFX transcription factors, which regulate genes encoding ciliary proteins (88,89) and that Tulp2 is strongly and rapidly upregulated during flagellar regeneration in Chlamydomonas reinhardtii (90).
The function of TULP3 in Shh signaling
Epistasis experiments indicate that TULP3 is required to prevent Gli2 from activating Shh target genes in the absence of Hh ligands. Smo is required for all Hh signaling, but how Smo controls Gli activity in mammals is unknown. TULP3 appears to participate in this step of the pathway. Gli2 nuclear localization and posttranslational modification of Gli2 appear normal in Tulp3 mutants, and we failed to detect an interaction between TULP3 and Gli2 proteins. Thus, regulation of the Hh pathway by TULP3 appears to represent a novel arm of the pathway. A better understanding of TULP3 function will rely on the identification and functional characterization of TULP3-associated proteins and the investigation of ciliary ultrastucture in Tulp3 mutants. We are currently investigating these aspects.
It is possible that TULP3 participates in membrane/vesicular trafficking. Consistent with this possibility, we found TULP3 in membrane fractions as described previously (55). Indeed, several Tubby family proteins exhibit association with cellular membranes (55,82), and mouse TULP1 and C. elegans TUB-1 appear to be involved in membrane trafficking (81,83,84,91). Recent studies suggest a close relationship between membrane trafficking and cilia. For example, the functions of several members of the small GTPase Rab and Arl families are tied to cilia-related processes. These include the small GTPases IFT27 and IFTA-2, Rab 8 and Arl6 and Arl13b (reviewed in 92). In addition, IFT components associate with Rab8 and Rabaptin5 (93), and BBS proteins function through the Rab8 GEF Rabin-8 (94). Interestingly, Arl13b regulates the mouse Shh pathway and is required for normal ciliary axonemal structure (95).
On the basis of our findings and those of others, we favor a model whereby TULP3 participates in trafficking events necessary for the transport of membrane-associated Hh regulatory components into or away from primary cilia. However, it is important to note that TULP3 also localizes to sites in the cell outside the primary cilium. Thus, it remains possible that TULP3 functions in the Shh pathway in a manner unrelated to primary cilia, possibly through the control of membrane trafficking within the cytoplasm or through direct transcriptional control within the nucleus.
TULP3 in neural proliferation and differentiation
In addition to neural cell identity, Shh signaling regulates proliferation and survival of neural progenitors (31). Shh is generally thought to stimulate cell proliferation in different regions of the developing CNS (96–98). However, Hh signaling can also promote cell-cycle exit and terminal differentiation in some contexts (99). For example, in the zebrafish retina, Shh accelerates the cell cycle in neural progenitor cells but, by doing so, it also advances the timing of cell-cycle exit and terminal differentiation (100–102).
We found that the balance between proliferating progenitors and differentiating neurons was shifted in favor of differentiation in the ventral and lateral regions of the Tulp3 mutant neural tube, although this balance was generally maintained in dorsal regions. These effects appear to result from the hyperactivation of the Shh-signaling pathway because the balance was restored in Gli2/Tulp3 double mutants. A similar phenomenon has also been observed in the neural tubes of Fkbp8 mutants, which also exhibit hyperactive Shh signaling (103). Thus, we propose that although Shh signaling is required for proliferation and cell survival in the spinal neural tube (46,98,104) and modest elevation of signaling can lead to overgrowth (105), high levels of Shh signaling favor cell-cycle exit and terminal differentiation in the posterior neural tube.
We investigated the mechanistic basis for this defect at E9.5, before any differentiation phenotype was evident. The bHLH protein Neurog2 promotes terminal differentiation of motor neurons and ventral interneurons (66,106,107). In the spinal neural tube, the expression of Neurog2 is positively regulated by Shh signaling directly through Gli transcription factors (69) and indirectly through Olig2 and Pax6 (65,67,70,108). Neurog2 expression is also negatively regulated by Hes5 (70). Olig2 and Pax6 also inhibit terminal differentiation at a step downstream of Neurogenin 2 protein. Thus, Neurogenin 2 levels must accumulate to sufficient levels to override the inhibitory effects of Olig2 and Pax6. At E9.5, Neurog2 was robustly expressed in a broad domain in the Tulp3 mutant neural tube, presumably resulting from increased Gli transcriptional activity, dorsal expansion of Olig2 expression and loss of ventral Hes5 expression. Because Pax6 was excluded from the ventral and lateral neural tube in the mutant, high levels of Neurogenin 2 would be free to promote extensive cell-cycle exit in the ventral two-thirds of the neural tube. Downregulation of cyclin D1 and cyclin D2 expression in these regions may also contribute to enhanced cell-cycle exit. Thus, decreased cyclin D expression, increased Neurog2 expression and repression of Pax6 in the ventral and lateral neural tube may synergize in promoting terminal differentiation. In contrast, the strong expression of cyclin D genes and Pax6 in the dorsal neural tube may be sufficient to maintain proliferation and restrict differentiation in this region.
In summary, we have demonstrated that TULP3 functions in the mid-gestation mouse embryo to antagonize the Shh signaling pathway at a step that is largely independent of the Shh ligand and Smo, but is dependent on Gli2. In the neural tube, this role is essential for proper patterning of progenitor cell identity and for controlling the balance between progenitor proliferation and neuronal differentiation. Owing to its intimate involvement in human development and disease, elucidating the mechanism of mammalian Hh signal transduction is of great clinical importance. The identification and functional characterization of novel factors controlling the Hh pathway, such as TULP3, will be crucial in this regard.
MATERIALS AND METHODS
Mouse strains
Heterozygous mice harboring the Tulp3tm1Jng allele were obtained from the Jackson Laboratory and bred into a C3Heb/FeJ background. The Tulp3tm1Jng allele generates a truncated form of mouse TULP3 (lacking the C-terminal Tubby domain) fused to GFP (37). Genotyping of Tulp3tm1Jng allele was performed as described previously (37). Double-mutant analysis was performed using Smotm1Amc, Shhtm1Chg, Kif3Atm1Gsn and Gli2tm1Alj mutant alleles, which were genotyped as described (4,46,59,109). Yolk sac DNAs were used for genotyping embryos. Noon of the day on which a vaginal plug was found was considered embryonic day 0.5 (E0.5).
Immunohistochemistry
Immunohistochemistry was carried out on cryosections as described previously (39). Antibodies against HB9/MNR2, Nkx2.2, Nkx6.1, FoxA2/Hnf3β, Pax7, Pax6, Msx1/2 and Shh were obtained from Developmental Studies Hybridoma Bank. Additional antibodies included α-Pax6 (Covance Research Products), α-Olig2 (R&D Systems), α-Brn2 (Santa Cruz Biotechnology), α-class III β-tubulin (Tuj1, Covance Research Products), α-Sox1 (R&D Systems), α-Dbx2 (Abcam), α-p27Kip1 (Santa Cruz Biotechnology), α-phospho-Histone H3 (pHH3, Upstate Cell Signaling) and α-active caspase 3 (Promega). Cy2- and Cy3-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. Sections were counterstained with DAPI (Sigma-Aldrich). Images were obtained using a fluorescent microscope (E800, Nikon) with digital camera (Princeton Instruments), using MetaMorph software (Universal Imaging Corporation). Image intensity and contrast were adjusted using Photoshop software (Adobe). Quantitation of the percentage of pHH3+ cells in the ventral and dorsal halves of the neural tube was performed by counting pHH3+ and total (DAPI+) nuclei from cryosections (nine sections from three embryos per genotype). The number of activated caspase 3+ cells/section was averaged from a similar set of nine sections per genotype.
In situ hybridization and skeletal preparation
In situ hybridization was performed on cryosections as described previously (110). Riboprobes were generated for mouse Ptch1 (111), Gli1 (112), Gli2 (112), Gli3 (40), Shh (113), Neurogenin 2 (114), Hes5 (115), Cyclin D1 (116) and Cyclin D2 (117). The mouse Hoxd12, Sox9 and dHand probes were generous gifts from D. Duboule, V. Lefebvre and K. Ligon, respectively. For skeleton preparations, embryos were dissected at E13.5, fixed, stained with Alcian blue and cleared as described (118). Images were acquired using a Leica stereomicroscope with digital camera (Optronics).
Primary MEF cell culture and staining
Embryos were dissected at E11.5 and wild-type and Tulp3tm1Jng mutant pMEFs were prepared as described (119). Tulp3hhkr mutant pMEFs were a gift from Dr Jennifer Murdoch. To generate ciliated NIH3T3 cells and pMEFs, cells were grown to ∼90% confluency on coverslips and then incubated with Opti-MEM (Gibco) for 48–72 h. Coverslips were washed; fixed with ice-cold acetone, air-dried, rehydrated and blocked with goat or horse serum in PBS. Primary antibodies included α-TULP3 (see what follows), α-acetylated-alpha tubulin and α-gamma tubulin (Sigma-Aldrich), α-Ift88 (generous gift from Dr Gregory Pazour) and α-mouse Gli2 antiserum (103).
Generation of TULP3 antiserum
Full-length mouse TULP3 cDNA was cloned into pET28A (Novagen) to generate recombinant protein. Purification was performed under denaturing conditions using urea and Ni-NTA agarose (Qiagen). Recombinant protein was mixed with Complete Freund’s Adjuvant and injected into New Zealand white rabbits. Affinity purification from bulk serum was performed using N-terminal recombinant TULP3 protein (residues 1–255) conjugated to CNBr-activated sepharose 4B from GE Healthcare Life Sciences according to the manufacturer’s instructions. Antisera from two rabbits gave similar results.
Immunoprecipitation
Embryos were dissected at E9.5 and disrupted by pipette in lysis buffer [50 mm Tris–Cl, pH 8.0, 150 mm NaCl, 1 mm NaF, 1 mm sodium orthovanadate, 1% NP-40, 1 mm PMSF, Complete Protease Inhibitor (Roche Applied Science)]. Immunoprecipitation was performed using protein G- or protein A-conjugated agarose beads (Invitrogen) as recommended by the manufacturer. Precleared lysates were incubated overnight with primary antibodies (α-Gli2 or α-TULP3) at 4°C, incubated with protein G/A beads and centrifuged. Beads were washed in ice-cold PBS, pelleted and then boiled in Laemmli sample buffer to release immunoprecipitated complexes.
Nuclear fractionation
Embryos were dissected at E9.5 and Dounce-homogenized in hypotonic buffer; nuclei were separated from crude cytoplasmic fractions by pelleting at 1500g for 5 min and then washed twice in hypotonic buffer (55). Nuclear protein was isolated by incubating fractions with hypertonic buffer for 30 min at 4°C, followed by sonication. Insoluble material was pelleted by high-speed centrifugation at 14 000g for 15 min; the supernatant was concentrated by TCA precipitation and resuspended in alkaline Laemmli sample buffer. Five micrograms of total protein per sample was separated through SDS–PAGE and transferred to nitrocellulose membrane according to standard protocols; quantification of loading and quality of fractionation were assessed through western blotting.
Western blotting
Embryos were processed for Gli3 protein analysis through western blotting as described previously (120,121). α-Beta actin (Abcam) was used as a loading control. Gli2, Sufu and TULP3 proteins in fibroblasts and embryonic extracts were analyzed by western blotting using standard techniques. Rabbit α-Sufu was obtained from Santa Cruz Biotechnology, Inc. α-Beta tubulin (Sigma) and α-HDAC2 (Upstate) were used as loading controls and for determining purity of fractions. Scanned blot images were used for the quantification of band intensity using ImageJ software (NIH).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the National Institutes of Health (HD050761, HD045522) (to J.T.E. and X.S., respectively) and by fellowships from the New Jersey Commission on Spinal Cord Research (to R.X.N. and H.W.K.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Akihiro Ikeda, Jürgen Naggert and Patsy Nishina for helpful discussions and for supplying the Tulp3tm1Jng allele. We also thank Kathryn Anderson and Rebecca Burdine for their comments on the manuscript, Yulian Lin and Jamie Verheyden for help with some experiments, and Jennifer Murdoch for communicating unpublished data and for supplying Tulp3hhkr mutant fibroblasts. We also thank G. Pazour, T. Li, S. Mackem, A. McMahon, A. Joyner, K. Anderson, R. Kageyama, P. Sicinski, Q. Ma, D. Duboule and M. Scott for providing reagents and mouse strains. Monoclonal antibodies developed by T.M. Jessell, S. Brenner-Morton, O.D. Madsen and A. Kawakami were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, The University of Iowa, Iowa City IA 52242, USA.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Marti E., Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- 2.Palma V., Lim D.A., Dahmane N., Sanchez P., Brionne T.C., Herzberg C.D., Gitton Y., Carleton A., Alvarez-Buylla A., Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeller R. It takes time to make a pinky: unexpected insights into how SHH patterns vertebrate digits. Sci. STKE. 2004;2004:e53. doi: 10.1126/stke.2592004pe53. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X.M., Ramalho-Santos M., McMahon A.P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 5.Mullor J.L., Sanchez P., Altaba A.R. Pathways and consequences: Hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562–569. doi: 10.1016/s0962-8924(02)02405-4. [DOI] [PubMed] [Google Scholar]
- 6.Jia J., Jiang J. Decoding the Hedgehog signal in animal development. Cell. Mol. Life Sci. 2006;63:1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- 8.Ruel L., Rodriguez R., Gallet A., Lavenant-Staccini L., Therond P.P. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 9.Methot N., Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- 10.Varjosalo M., Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., McMahon A.P., Allen B.L. Shifting paradigms in Hedgehog signaling. Curr. Opin. Cell Biol. 2007;19:159–165. doi: 10.1016/j.ceb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Varjosalo M., Li S.P., Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Merchant M., Evangelista M., Luoh S.M., Frantz G.D., Chalasani S., Carano R.A., van Hoy M., Ramirez J., Ogasawara A.K., McFarland L.M., et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol. Cell. Biol. 2005;25:7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggenschwiler J.T., Espinoza E., Anderson K.V. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 15.Callahan C.A., Ofstad T., Horng L., Wang J.K., Zhen H.H., Coulombe P.A., Oro A.E. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18:2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang P.T., McMahon A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 17.Izraeli S., Lowe L.A., Bertness V.L., Campaner S., Hahn H., Kirsch I.R., Kuehn M.R. Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis. 2001;31:72–77. doi: 10.1002/gene.10004. [DOI] [PubMed] [Google Scholar]
- 18.Martinelli D.C., Fan C.M. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey M.G., Paton I.R., Yin Y., Schmidt M., Bangs F.K., Morrice D.R., Smith T.G., Buxton P., Stamataki D., Tanaka M., et al. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–1377. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilbanks A.M., Fralish G.B., Kirby M.L., Barak L.S., Li Y.X., Caron M.G. Beta-arrestin 2 regulates zebrafish development through the hedgehog signaling pathway. Science. 2004;306:2264–2267. doi: 10.1126/science.1104193. [DOI] [PubMed] [Google Scholar]
- 21.Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 22.Liu A., Wang B., Niswander L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 23.May S.R., Ashique A.M., Karlen M., Wang B., Shen Y., Zarbalis K., Reiter J., Ericson J., Peterson A.S. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 24.Koyama E., Young B., Nagayama M., Shibukawa Y., Enomoto-Iwamoto M., Iwamoto M., Maeda Y., Lanske B., Song B., Serra R., et al. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–2169. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran P.V., Haycraft C.J., Besschetnova T.Y., Turbe-Doan A., Stottmann R.W., Herron B.J., Chesebro A.L., Qiu H., Scherz P.J., Shah J.V., et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haycraft C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., Yoder B.K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs J.J., Whalen E.J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., Lefkowitz R.J. {beta}-Arrestin-mediated localization of Smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 30.McGlinn E., Tabin C.J. Mechanistic insight into how Shh patterns the vertebrate limb. Curr. Opin. Genet. Dev. 2006;16:426–432. doi: 10.1016/j.gde.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Ulloa F., Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- 32.Chamberlain C.E., Jeong J., Guo C., Allen B.L., McMahon A.P. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135:1097–1106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- 33.Gritli-Linde A., Lewis P., McMahon A.P., Linde A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev. Biol. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- 34.Roelink H., Porter J.A., Chiang C., Tanabe Y., Chang D.T., Beachy P.A., Jessell T.M. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 35.Nishina P.M., North M.A., Ikeda A., Yan Y., Naggert J.K. Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics. 1998;54:215–220. doi: 10.1006/geno.1998.5567. [DOI] [PubMed] [Google Scholar]
- 36.Noben-Trauth K., Naggert J.K., North M.A., Nishina P.M. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda A., Ikeda S., Gridley T., Nishina P.M., Naggert J.K. Neural tube defects and neuroepithelial cell death in Tulp3 knockout mice. Hum. Mol. Genet. 2001;10:1325–1334. doi: 10.1093/hmg/10.12.1325. [DOI] [PubMed] [Google Scholar]
- 38.Milenkovic L., Goodrich L.V., Higgins K.M., Scott M.P. Mouse patched1 controls body size determination and limb patterning. Development. 1999;126:4431–4440. doi: 10.1242/dev.126.20.4431. [DOI] [PubMed] [Google Scholar]
- 39.Eggenschwiler J.T., Anderson K.V. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev. Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]
- 40.Bulgakov O.V., Eggenschwiler J.T., Hong D.H., Anderson K.V., Li T. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development. 2004;131:2149–2159. doi: 10.1242/dev.01122. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y., Roelink H., McKnight G.S. Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 2002;277:19889–19896. doi: 10.1074/jbc.M111412200. [DOI] [PubMed] [Google Scholar]
- 42.Motoyama J., Milenkovic L., Iwama M., Shikata Y., Scott M.P., Hui C.C. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev. Biol. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 43.Jeong Y., Epstein D.J. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development. 2003;130:3891–3902. doi: 10.1242/dev.00590. [DOI] [PubMed] [Google Scholar]
- 44.Knezevic V., De Santo R., Schughart K., Huffstadt U., Chiang C., Mahon K.A., Mackem S. Hoxd-12 differentially affects preaxial and postaxial chondrogenic branches in the limb and regulates Sonic hedgehog in a positive feedback loop. Development. 1997;124:4523–4536. doi: 10.1242/dev.124.22.4523. [DOI] [PubMed] [Google Scholar]
- 45.te Welscher P., Fernandez-Teran M., Ros M.A., Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002;16:421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 47.Chiang C., Litingtung Y., Harris M.P., Simandl B.K., Li Y., Beachy P.A., Fallon J.F. Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev. Biol. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 48.Bai C.B., Auerbach W., Lee J.S., Stephen D., Joyner A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 49.Matise M.P., Epstein D.J., Park H.L., Platt K.A., Joyner A.L. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 50.Pan Y., Bai C.B., Joyner A.L., Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varjosalo M., Bjorklund M., Cheng F., Syvanen H., Kivioja T., Kilpinen S., Sun Z., Kallioniemi O., Stunnenberg H.G., He W.W., et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 52.Wang B., Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc. Natl Acad. Sci. USA. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eggenschwiler J.T., Anderson K.V. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiratori H., Hamada H. The left–right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 55.Santagata S., Boggon T.J., Baird C.L., Gomez C.A., Zhao J., Shan W.S., Myszka D.G., Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 56.Ikeda A., Nishina P.M., Naggert J.K. The tubby-like proteins, a family with roles in neuronal development and function. J. Cell Sci. 2002;115:9–14. doi: 10.1242/jcs.115.1.9. [DOI] [PubMed] [Google Scholar]
- 57.He W., Ikeda S., Bronson R.T., Yan G., Nishina P.M., North M.A., Naggert J.K. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Brain Res. Mol. Brain Res. 2000;81:109–117. doi: 10.1016/s0169-328x(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 58.Boggon T.J., Shan W.S., Santagata S., Myers S.C., Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 59.Marszalek J.R., Ruiz-Lozano P., Roberts E., Chien K.R., Goldstein L.S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl Acad. Sci. USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin F., Hiesberger T., Cordes K., Sinclair A.M., Goldstein L.S., Somlo S., Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl Acad. Sci. USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bananis E., Nath S., Gordon K., Satir P., Stockert R.J., Murray J.W., Wolkoff A.W. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for dynein and kinesin. Mol. Biol. Cell. 2004;15:3688–3697. doi: 10.1091/mbc.E04-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown C.L., Maier K.C., Stauber T., Ginkel L.M., Wordeman L., Vernos I., Schroer T.A. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 2005;6:1114–1124. doi: 10.1111/j.1600-0854.2005.00347.x. [DOI] [PubMed] [Google Scholar]
- 64.Gui H., Li S., Matise M.P. A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev. Biol. 2007;301:14–26. doi: 10.1016/j.ydbio.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bel-Vialar S., Medevielle F., Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev. Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Lee S.K., Lee B., Ruiz E.C., Pfaff S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Novitch B.G., Chen A.I., Jessell T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 68.Scardigli R., Baumer N., Gruss P., Guillemot F., Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- 69.Ribes V., Stutzmann F., Bianchetti L., Guillemot F., Dolle P., Le Roux I. Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev. Biol. 2008;321:470–481. doi: 10.1016/j.ydbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Holmberg J., Hansson E., Malewicz M., Sandberg M., Perlmann T., Lendahl U., Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135:1843–1851. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi M., Kojima M., Nakajima K., Suzuki-Migishima R., Takeuchi T. Functions of a jumonji-cyclin D1 pathway in the coordination of cell cycle exit and migration during neurogenesis in the mouse hindbrain. Dev. Biol. 2007;303:549–560. doi: 10.1016/j.ydbio.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 72.Lobjois V., Bel-Vialar S., Trousse F., Pituello F. Forcing neural progenitor cells to cycle is insufficient to alter cell-fate decision and timing of neuronal differentiation in the spinal cord. Neural Dev. 2008;3:4. doi: 10.1186/1749-8104-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lobjois V., Benazeraf B., Bertrand N., Medevielle F., Pituello F. Specific regulation of cyclins D1 and D2 by FGF and Shh signaling coordinates cell cycle progression, patterning, and differentiation during early steps of spinal cord development. Dev. Biol. 2004;273:195–209. doi: 10.1016/j.ydbio.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 74.Wilson L., Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev. Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 75.Wine-Lee L., Ahn K.J., Richardson R.D., Mishina Y., Lyons K.M., Crenshaw E.B., III Signaling through BMP type 1 receptors is required for development of interneuron cell types in the dorsal spinal cord. Development. 2004;131:5393–5403. doi: 10.1242/dev.01379. [DOI] [PubMed] [Google Scholar]
- 76.Liem K.F., Jr, Jessell T.M., Briscoe J. Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development. 2000;127:4855–4866. doi: 10.1242/dev.127.22.4855. [DOI] [PubMed] [Google Scholar]
- 77.Ikeya M., Lee S.M., Johnson J.E., McMahon A.P., Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 78.Robertson C.P., Braun M.M., Roelink H. Sonic hedgehog patterning in chick neural plate is antagonized by a Wnt3-like signal. Dev. Dyn. 2004;229:510–519. doi: 10.1002/dvdy.10501. [DOI] [PubMed] [Google Scholar]
- 79.Mykytyn K., Sheffield V.C. Establishing a connection between cilia and Bardet–Biedl Syndrome. Trends Mol. Med. 2004;10:106–109. doi: 10.1016/j.molmed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 80.Hagstrom S.A., North M.A., Nishina P.L., Berson E.L., Dryja T.P. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat. Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 81.Hagstrom S.A., Adamian M., Scimeca M., Pawlyk B.S., Yue G., Li T. A role for the Tubby-like protein 1 in rhodopsin transport. Invest. Ophthalmol. Vis. Sci. 2001;42:1955–1962. [PubMed] [Google Scholar]
- 82.Xi Q., Pauer G.J., Marmorstein A.D., Crabb J.W., Hagstrom S.A. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest. Ophthalmol. Vis. Sci. 2005;46:4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xi Q., Pauer G.J., Ball S.L., Rayborn M., Hollyfield J.G., Peachey N.S., Crabb J.W., Hagstrom S.A. Interaction between the photoreceptor-specific tubby-like protein 1 and the neuronal-specific GTPase dynamin-1. Invest. Ophthalmol. Vis. Sci. 2007;48:2837–2844. doi: 10.1167/iovs.06-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukhopadhyay A., Deplancke B., Walhout A.J., Tissenbaum H.A. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2005;2:35–42. doi: 10.1016/j.cmet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Perkins L.A., Hedgecock E.M., Thomson J.N., Culotti J.G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 86.Apfeld J., Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 87.Mak H.Y., Nelson L.S., Basson M., Johnson C.D., Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 88.Efimenko E., Bubb K., Mak H.Y., Holzman T., Leroux M.R., Ruvkun G., Thomas J.H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 89.Laurencon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007;8:R195. doi: 10.1186/gb-2007-8-9-r195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stolc V., Samanta M.P., Tongprasit W., Marshall W.F. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl Acad. Sci. USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukhopadhyay A., Pan X., Lambright D.G., Tissenbaum H.A. An endocytic pathway as a target of tubby for regulation of fat storage. EMBO Rep. 2007;8:931–938. doi: 10.1038/sj.embor.7401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oro A.E. The primary cilia, a ‘Rab-id’ transit system for hedgehog signaling. Curr. Opin. Cell Biol. 2007;19:691–696. doi: 10.1016/j.ceb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Omori Y., Zhao C., Saras A., Mukhopadhyay S., Kim W., Furukawa T., Sengupta P., Veraksa A., Malicki J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat. Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- 94.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 95.Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 96.Kenney A.M., Cole M.D., Rowitch D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 97.Britto J., Tannahill D., Keynes R. A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat. Neurosci. 2002;5:103–110. doi: 10.1038/nn797. [DOI] [PubMed] [Google Scholar]
- 98.Cayuso J., Ulloa F., Cox B., Briscoe J., Marti E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- 99.Neumann C.J. Hedgehogs as negative regulators of the cell cycle. Cell Cycle. 2005;4:1139–1140. doi: 10.4161/cc.4.9.1999. [DOI] [PubMed] [Google Scholar]
- 100.Masai I., Yamaguchi M., Tonou-Fujimori N., Komori A., Okamoto H. The hedgehog-PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development. 2005;132:1539–1553. doi: 10.1242/dev.01714. [DOI] [PubMed] [Google Scholar]
- 101.Locker M., Agathocleous M., Amato M.A., Parain K., Harris W.A., Perron M. Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shkumatava A., Neumann C.J. Shh directs cell-cycle exit by activating p57Kip2 in the zebrafish retina. EMBO Rep. 2005;6:563–569. doi: 10.1038/sj.embor.7400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho A., Ko H.W., Eggenschwiler J.T. FKBP8 cell autonomously controls neural tube patterning through a Gli2- and Kif3a-dependent mechanism. Dev. Biol. 2008;321:27–39. doi: 10.1016/j.ydbio.2008.05.558. [DOI] [PubMed] [Google Scholar]
- 104.Litingtung Y., Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 105.Jeong J., McMahon A.P. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–154. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- 106.Mizuguchi R., Sugimori M., Takebayashi H., Kosako H., Nagao M., Yoshida S., Nabeshima Y., Shimamura K., Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 107.Scardigli R., Schuurmans C., Gradwohl G., Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Q., Anderson D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 109.Mo R., Freer A.M., Zinyk D.L., Crackower M.A., Michaud J., Heng H.H., Chik K.W., Shi X.M., Tsui L.C., Cheng S.H., et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 110.Schaeren-Wiemers N., Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 111.Goodrich L.V., Johnson R.L., Milenkovic L., McMahon J.A., Scott M.P. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 112.Hui C.C., Slusarski D., Platt K.A., Holmgren R., Joyner A.L. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2 and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 113.Echelard Y., Epstein D.J., St-Jacques B., Shen L., Mohler J., McMahon J.A., McMahon A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 114.Ma Q., Fode C., Guillemot F., Anderson D.J. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ishibashi M., Ang S.L., Shiota K., Nakanishi S., Kageyama R., Guillemot F. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix–loop–helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 116.Sicinski P., Donaher J.L., Parker S.B., Li T., Fazeli A., Gardner H., Haslam S.Z., Bronson R.T., Elledge S.J., Weinberg R.A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 117.Sicinski P., Donaher J.L., Geng Y., Parker S.B., Gardner H., Park M.Y., Robker R.L., Richards J.S., McGinnis L.K., Biggers J.D., et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 118.Jegalian B.G., De Robertis E.M. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71:901–910. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- 119.Nagy A., Gertenstein M., Vintersten K., Behringer R. Manipulating the Mouse Embryo. A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2003. [Google Scholar]
- 120.Eggenschwiler J.T., Bulgakov O.V., Qin J., Li T., Anderson K.V. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev. Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 121.Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.