Abstract
Recently it has been proposed that di-methylation of histone H3 on lysine 4 (H3K4me2) acts as an epigenetic memory to maintain transcriptional patterns in developing tissues. This model suggests that there may be a requirement to reprogram this modification in the germline to prevent transcriptional memory from being inappropriately transmitted to the next generation. We asked if SPR-5, the C. elegans ortholog of the H3K4me2 demethylase LSD1/KDM1, plays a role in epigenetically reprogramming H3K4me2. We show that spr-5 mutants exhibit progressive sterility over many generations due to defects in oogenesis and spermatogenesis. These defects correlate with a progressive failure to erase H3K4me2 in the primordial germ cells, resulting in the misregulation of spermatogenesis-expressed genes due to the transgenerational accumulation of H3K4me2 at these loci. These results suggest that H3K4me2 can serve as an epigenetic memory and that LSD1/KDM1 demethylases play a critical role in the reprogramming of this memory in the germline, preventing inappropriate epigenetic information from being propagated from one generation to the next.
Keywords: epigenetics, reprogramming, chromatin, germline, germ cells, C. elegans, histone demethylase, LSD1, SPR-5, KDM1
Introduction
The epigenetic state of the genome undergoes extensive reprogramming in the germline between generations, both in the gametes and the early embryo. For example, CpG methylation is high in differentiated tissues but low in oocytes and at the blastocyst stage of development in mice (Jaenisch, 1997; Li, 2002). However, despite advances in our understanding of epigenetic reprogramming, the mechanisms of resetting histone modifications in the germline and the role that the resetting of particular histone modifications plays in epigenetic reprogramming as a whole, remain poorly understood. A clear illustration of this can be seen in two landmark reprogramming studies. Over 10 years ago Ian Wilmut’s group showed that nuclear transfer could be used to clone Dolly the sheep (Campbell et al., 1996). Recently this work was extended by the Yamanaka group, who showed that the addition of four factors could successfully reprogram a mouse somatic cell into an induced pluripotent stem cell (Takahashi and Yamanaka, 2006). Yet in both cases, despite the tremendous advances provided by the work, we still know very few details about the molecular mechanisms involved or how these processes occur naturally in the germline (Nishikawa, 2007).
The modification of histone H3 by di-methylation of lysine 4 (H3K4me2) is associated with active genes in eukaryotes and was originally proposed to play a role in gene activation (Li et al., 2007). However, recent data have suggested that H3K4me2 may be acquired during transcriptional elongation. For example, the H3K4 methyltransferase Set1p is present in a complex with RNAPOLII in S. Cerevisiae and H3K4me2 distribution directly correlates with RNAPOLII occupation at active genes in Drosophila cells (Mito et al., 2005; Ng et al., 2003). In addition, in a Drosophila pgc (polar granule component) mutant, when transcription is ectopically activated in the primordial germ cells (PGCs), robust H3K4me2 is acquired (Hanyu-Nakamura et al., 2008). Since H3K4me2 may be deposited during transcriptional elongation, it is possible that H3K4me2 acts as an epigenetic memory facilitating the maintenance of transcriptional patterns during tissue differentiation, rather than in de novo transcriptional activation.
Evidence that H3K4me2 may act as an epigenetic memory can be found at the bithorax complex in Drosophila, where Hox genes are transcriptionally active through multiple cell divisions despite the absence of activating transcription factors (Ringrose and Paro, 2004). This epigenetic memory is dependent upon trithorax, an H3K4 methyltransferase, suggesting that H3K4 methylation plays a role in maintaining transcriptional patterns during development (Petruk et al., 2001; Poux et al., 2002). Such a model makes particular sense in the development of somatic tissues where genes that are activated may need to remain on following mitotic divisions. However, if H3K4me2 levels can be heritably maintained, this creates an intriguing dilemma for the germline. Genes such as those required for meiosis and gametogenesis, that are highly active during the germline program, would acquire significant levels of H3K4me2 in their chromatin. If this methylation were to be propagated to the next generation it could potentially result in the misregulation of gamete- and meiosis-specific genes in the zygote. As a result, there may be a requirement to remove H3K4me2 that is acquired during transcriptional elongation in the germline to prevent this epigenetic memory from being inappropriately transmitted from one generation to the next.
Is there any evidence for a requirement to reset H3K4me2? Recently Ng et al. demonstrated that inappropriate expression of endoderm genes is detected in embryos derived from differentiated endoderm nuclei transplanted into Xenopus eggs (Ng and Gurdon, 2008). This effect is dependent upon lysine 4 of the histone variant H3.3 that is incorporated during endodermal transcription (Ng and Gurdon, 2008). This implies that the complete reprogramming of a somatic nucleus may require efficient erasure of epigenetic information at H3K4. Thus, during normal transmission through the germline, it may be necessary to reset methylation at this residue.
The C. elegans germline provides an excellent model for trying to understand the role of histone modifications in epigenetic reprogramming. During early embryogenesis in C. elegans, the germline (P lineage) undergoes four asymmetric cell divisions in which both somatic and germline daughter cells are produced. Following these cell divisions, the P4 germline blastomere divides symmetrically to produce the two primordial germ cells (PGCs), Z2 and Z3, that remain mitotically quiescent through the rest of embryogenesis (Seydoux and Dunn, 1997; Seydoux et al., 1996; Sulston et al., 1983). Once the embryo hatches, the PGCs activate mitotically to populate the larval gonad and ultimately produce all of the sperm and the eggs in the adult. The chromatin in the germline P blastomeres preceding the PGCs initially contains high levels of H3K4me2 (Schaner et al., 2003). Once the P4 blastomere divides symmetrically to give rise to the PGCs and these cells are commited to the germ cell fate, the PGC chromatin uniquely and rapidly loses H3K4me2 (Schaner et al., 2003). The absence of H3K4me2 appears to be a conserved characteristic of PGCs as Drosophila pole cells also lack this modification (Schaner et al., 2003). This absence of H3K4me2 in the PGCs may be necessary for protecting and maintaining PGC fate through chromatin-based transcriptional repression (Schaner et al., 2003). However, direct evidence for this remains elusive.
The erasure of H3K4 methylation may require the activity of a histone demethylase. Shi et al. demonstrated that the mammalian amine oxidase LSD1/KDM1, a component of CoREST transcriptional repressor complexes, can specifically demethylate H3K4me2 (Shi et al., 2004). In addition, LSD1/KDM1 associates with the androgen receptor (AR). As a member of the CoREST complex, LSD1/KDM1 is thought to contribute to repression of neuronal targets in non-neuronal cell types by demethylating H3K4me2 (Ballas et al., 2001; Chong et al., 1995; Lee et al., 2005; Shi et al., 2005). In contrast, when bound to the AR it contributes to the derepression of AR targets by demethylating the heterochromatin-associated modification, H3K9me2 (Metzger et al., 2005; Wissmann et al., 2007). C. elegans has three lsd1/kdm1 homologs encoded by the genes spr-5, amx-1 and T08D10.2. Likely functional null mutations exist in each of these genes. The amx-1(ok659) and T08D10.2 mutations are deletion alleles while spr-5(by101) is a Tc3 transposon insertion in the middle of the conserved amine-oxidase catalytic domain (Eimer et al., 2002). This transposon insertion is predicted to abolish the demethylase activity of the amine oxidase domain. We took advantage of these mutant strains to examine the role of H3K4me2 in the C. elegans germline and to ask whether the resetting of this mark may be required to prevent inapropriate epigenetic memory from being transmitted from one generation to the next.
We find that the loss of the LSD1/KDM1 demethylase spr-5 results in a germline mortality phenotype in which the incidence of sterility increases across generations. This sterility correlates with the misreregulation of spermatogenesis-expressed genes due to the stable accumulation of H3K4me2 at these loci. This accumulation leads to the inappropriate retention of H3K4me2 in the PGCs as well as defective oogenesis and spermatogenesis. From these data we conclude that H3K4me2 provides an epigenetic memory and that the LSD1/KDM1 demethylase spr-5 plays a critical role in reprogamming this memory in the germline.
Results
SPR-5 Is an H3K4me2 Demethylase
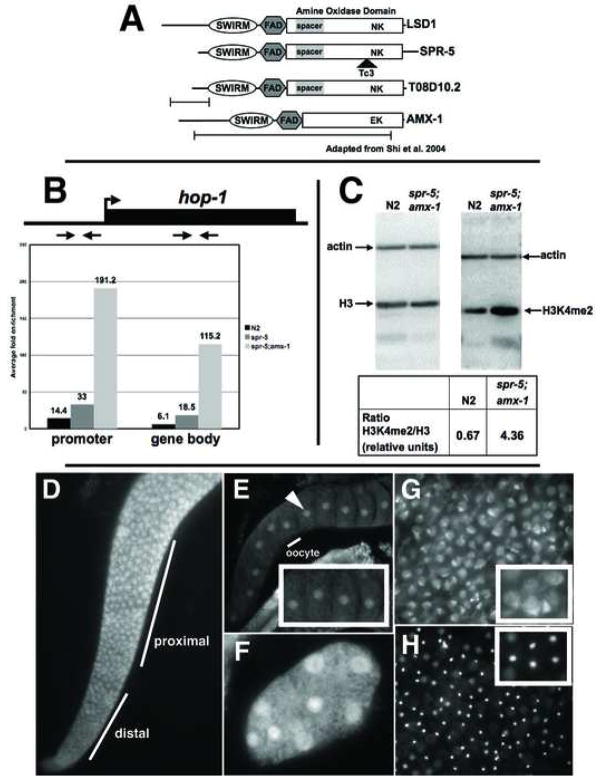
The worm protein SPR-5 is 27% identical (45% similar) to human LSD1/KDM1 and binds, like its mammalian counterpart, to the CoREST protein SPR-1 suggesting that it is the C. elegans ortholog of LSD1/KDM1 (Figure 1A) (Eimer et al., 2002; Jarriault, 2002). In addition, the critical catalytic residue (K661), within the highly conserved amine oxidase domain is conserved in SPR-5 (Figure 1A). To confirm that SPR-5 is an H3K4me2 demethylase in vivo we assayed for changes in H3K4me2 levels in spr-5 mutants. The presenilin-like gene hop-1 is a known SPR-5 target, since hop-1 transcription increases over 20-fold in the spr-5(by101) mutant (Eimer et al., 2002; Jarriault, 2002). We used ChIP to analyze H3K4me2 levels at the hop-1 locus in wild-type and spr-5 mutant animals. We observed a 2-fold (33/14.4) enrichment in H3K4me2 in spr5(by101) mutants compared to wild-type and a 13-fold (191.2/14.4) enrichment in spr5(by101);amx-1(ok659) double mutants compared to wild-type in the hop-1 promoter (Figure 1B). In addition, we observed a 3-fold (18.5/6.1) enrichment in H3K4me2 in spr5(by101) mutants compared to wild-type and a near 20-fold (115.2/6.1) enrichment in spr5(by101);amx-1(ok659) double mutants compared to wild-type in the hop-1 gene body (Figure 1B). Finally, western blot analysis revealed that total histone extracts made from spr-5;amx-1 mutants had approximately 6-fold (4.36/0.67) higher levels of H3K4me2 compared to wild-type when normalized for total histone H3, indicating that the defect is not restricted to the hop-1 locus (Figure 1C). Together these results strongly suggest that SPR-5 acts in vivo as a demethylase to affect H3K4me2 levels genome-wide.
Figure 1.
spr-5 is a H3K4me2 demethylase. A, Alignment of the C. elegans LSD1/KDM1 orthologs with human LSD1/KDM1. The key catalytic residues (N660 and K661) are shown at their relative position within the amine oxidase domain and the location of the mutations are shown beneath each gene. B, ChIP of H3K4me2 at hop-1 in wild-type (N2), spr-5(by101) and spr-5;amx-1 mutants. The fold enrichment (of the percentage of input precipitated) in H3K4me2 Ab over no Ab, with the positions of the primer sets in the promoter and gene body indicated above. C, Western blots of wild-type (N2) and spr-5;amx-1 protein extracts probed with actin, histoneH3 and H3K4me2 (positions marked). The ratio of H3K4me2/histoneH3 normalized to actin is shown below. Immunofluorescence with SPR-5 N (D,G,H) and C terminal (E,F) Ab’s showing the adult gonad (D), oocytes (E), an early embryo (F), sperm (G), and the corresponding DAPI image of sperm (H). The inset in panels E,F and G are zoomed in on part of the panel. The identical staining pattern was observed with both N and C terminal Ab’s.
Lack of Demethylase Activity Results in Germline Mortality
spr-5 is expressed in all stages but is enriched in the adult and highly expressed in the gonad indicating that, in addition to possible somatic expression, it is likely that spr-5 mRNA is maternally deposited in the germline (Eimer et al., 2002; nematode.lab.nig.ac.jp/). Consistent with this, immunofluorescence with two SPR-5 antibodies detected SPR-5 protein in the nuclei of the adult germline where it accumulates during meiosis (Figure 1D). In addition, we observed SPR-5 staining in sperm cytoplasm as well as in the nuclei of oocytes and the early embryo (Figure 1E–H). No SPR-5 staining was seen in somatic cells outside of the early embryo. This expression pattern is consistent with a potential role in resetting H3K4me2 between generations. Therefore, to determine if spr-5 plays a role in the germline we examined the phenotype of spr-5 mutant animals.
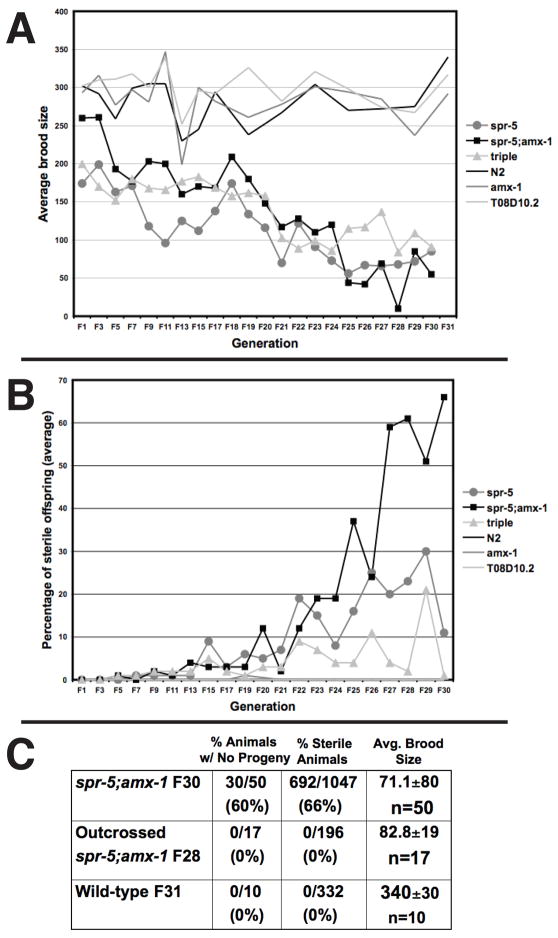
spr-5 mutants have a slight retention of embryos within the worm, or egg-laying defective (Egl) phenotype (Supplemental Figure 1B) and at early generations have a slightly reduced brood size compared to wild-type, amx-1 or T08D10.2 single mutants (Figure 2A). Despite this, all spr-5 mutants are initially fertile and we observed no significant early increase in the percentage of sterile animals, which can be scored under the light microscope (Figure 2B). Over 28 generations, however, the spr-5 mutants exhibited a germline mortality phenotype in which the brood size steadily declined with each successive generation (Figure 2A and Supplemental Figure 2). This decline occurred despite the fact that selecting fertile animals for passage inherently biased against this phenotype (see methods). In addition, after 20 generations we observed a sharp increase in the percentage of sterile worms, with the increasingly rare fertile animals producing few progeny (Figure 2B and Supplemental Figure 2). In contrast wild-type, amx-1 or T08D10.2 single mutants did not exhibit increased sterility (Figure 2A and 2B).
Figure 2.
The germline mortality phenotype. A, The average brood size of spr-5, spr-5;amx-1, spr-5;amx-1;T08D10.2 (triple), wild-type (N2), amx-1 and T08D10.2 strains in progressive generations are shown. B, The percentage of sterile animals in the same experiment as (A). The complete data from each strain are shown individually in Supplemental Figure 2. C, The percentage of animals with no progeny, the percentage of animals with a sterile phenotype and the average brood size of outcrossed spr-5;amx-1 (f28) worms compared to the corresponding (f30) non-outcrossed and wild-type generations.
All spr-5(by101)-containing mutant combinations and spr-5(by101) itself exhibited the germline mortality phenotype, suggesting that this phenotype is largely due to the spr-5(by101) allele. However, the spr-5;amx-1 double had a slightly more severe sterility defect than spr5(by101) alone, and the triple mutant including T08D10.2 was a little less severe. These data hint that amx-1 might be redundant with spr-5 while T08D10.2 may play an antagonistic role. Interestingly, microarray analysis demonstrated that the expression of amx-1 is elevated in spr-5 mutants (Supplemental Figure 7). This suggests that the activity of amx-1 may slightly compensate for the spr-5 mutant phenotype and may explain why the spr-5;amx-1 double mutant has a slightly more severe phenotype.
Although the number of generations required to reach the highest levels of sterility varied slightly between repetitions of the experiment, typically 20–30 generations were required. In addition, similar numbers of generations were required during multiple repetitions of the germline mortality phenotype at 16°C, despite the fact that worms develop more slowly at this temperature. This indicates that the number of generations, rather than absolute time or developmental time, is the most critical aspect of this phenotype. Additionally, forcing highly sterile spr-5(by101) populations to go through the alternate Dauer developmental pathway partially reset the germline mortality phenotype (Supplemental Figure 1D). Although it is possible that this is due to some type of survival selection, it is interesting to note that global chromatin changes associated with Dauer larvae formation have been observed(Jones et al., 2001) (and J. Waddle, personal communication).
Although progressive sterility is the predominant phenotype of the spr-5 mutants, a mutant in spr-5 was previously isolated in a screen for sel-12 suppressor mutants in the vulval pathway (Eimer et al., 2002; Jarriault, 2002). This suggests that spr-5 mutants may have somatic effects in addition to the effect on fertility. During the course of the experiments, we observed the sporadic appearance of a number of somatic phenotypes (e.g. rolling, dumpy and protruding/multivulval worms). The sporadic appearance of these low penetrance phenotypes is particularly intriguing in that these phenotypes were not heritable. For example, out of 119 progeny from 2 rolling worms (Rol phenotype), we observed 0 Rol offspring.
Mutations in mrt-2, a conserved DNA damage checkpoint protein, have also been shown to cause germline mortality (Ahmed and Hodgkin, 2000). In this mutant, the generational increase in sterility is associated with genetic defects, including telomere shortening and chromosome fusions (Ahmed and Hodgkin, 2000). We found no evidence of chromosome fusions in spr-5 mutants, suggesting that the germline mortality defect in these mutants is distinct from that of mrt-2 (Supplemental Figure 3). In addition, we did not observe any substantial increase in embryonic lethality or other heritable defects in the spr-5 mutants, indicating the absence of accumulating DNA damage (Supplemental Figure 4A and data not shown). We also did not observe any change in the percentage of XO male progeny suggesting that there are no gross defects in chromosomal segregation in these mutants (Supplemental Figure 4B). Finally, through multiple repetitions of the experiment we always observed the same germline and developmental phenotypes (see below). Taken together, these data suggest that an epigenetic defect, rather than the accumulation of random genetic mutations is more likely to be the cause of the germline mortality phenotype.
Germline Mortality Is Associated with a Defect in Oogenesis and Spermatogenesis
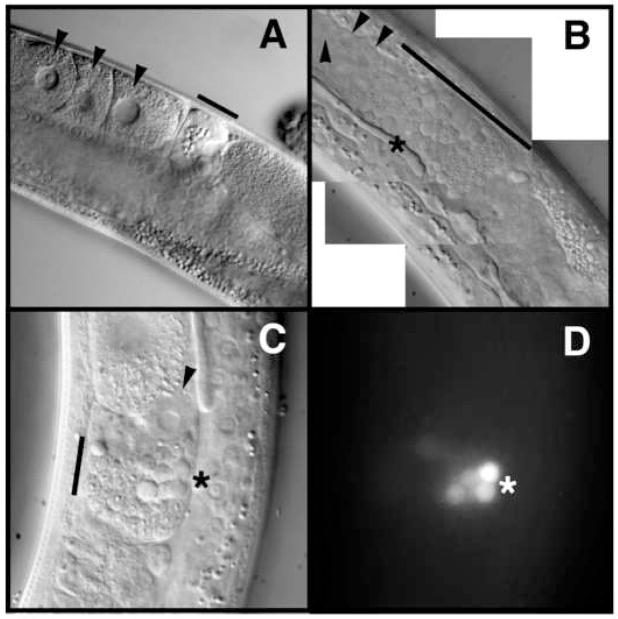
In order to determine why the spr-5 mutants become progressively sterile, we further analyzed the morphological characteristics of the severely sterile late-generation adults. By DIC, DAPI and phospho-histone H3 staining, proliferation of the mitotic germ cells appeared normal in sterile animals (Supplemental Figure 5). However, oogenesis was highly defective as many animals had either oocytes with abnormal morphology or no oocytes at all (Figure 3B and data not shown). In addition, we observed spermatids abnormally located in the proximal gonad where the oocytes normally would reside, amid many acridine orange staining residual bodies (Figure 3B–D and data not shown). In C. elegans hermaphrodites, spermatogenesis is normally completed in L4 larvae. Thus, the appearance of residual bodies, a by-product of spermatogenesis, in adult hermaphrodites suggests that spr-5 mutants also have a delay in spermatogenesis progression.
Figure 3.
Oogenesis and spermatogenesis defects in spr-5;amx-1 mutants. DIC microscopic imaging of the proximal gonad from wild-type (A) and severely sterile spr-5;amx-1 (B) adult worms. Black arrowheads point to oocytes (A) and defective looking oocytes (B and C). Black bars denote mature sperm in the spermatheca (A) as compared to early spermatogenic stages in the proximal gonad (B) and spermatheca (C). DIC imaging (C) and corresponding acridine orange staining (D) showing residual bodies (*).
A Late Larval Growth Delay Correlates with Increased Sterility
During the germline mortality experiment, we observed a severe delay in development that correlated with the onset of increasing sterility in the spr-5 mutants. We wondered if this delay could be linked to the defects in oogenesis and spermatogenesis. To examine this delay more closely, we selected wild-type and spr-5;amx-1 mutant embryos at the 2-cell stage and monitored the timing of development from embryogenesis through adult. At both early and late generations, we observed a very slight delay in embryogenesis and early larval stages in the spr-5;amx-1 double mutant (data not shown). However, beginning around generation 20, we observed an extended developmental delay in late larval stages in the spr-5;amx-1 mutants compared to wild-type. spr-5;amx-1 mutants were delayed at the L4 larval stage, and reached adulthood over 30hrs later than wild-type (Supplemental Figure 6). In C. elegans after spermatogenesis is completed in L4 larvae, oogenesis begins at the transition from L4 larvae to adult. Thus the extended developmental delay at the L4 larval stage may be linked to our observed defects in oogenesis and spermatogenesis.
Sterility of spr-5 Mutants Correlates with Retention of H3K4me2 in Z2/Z3
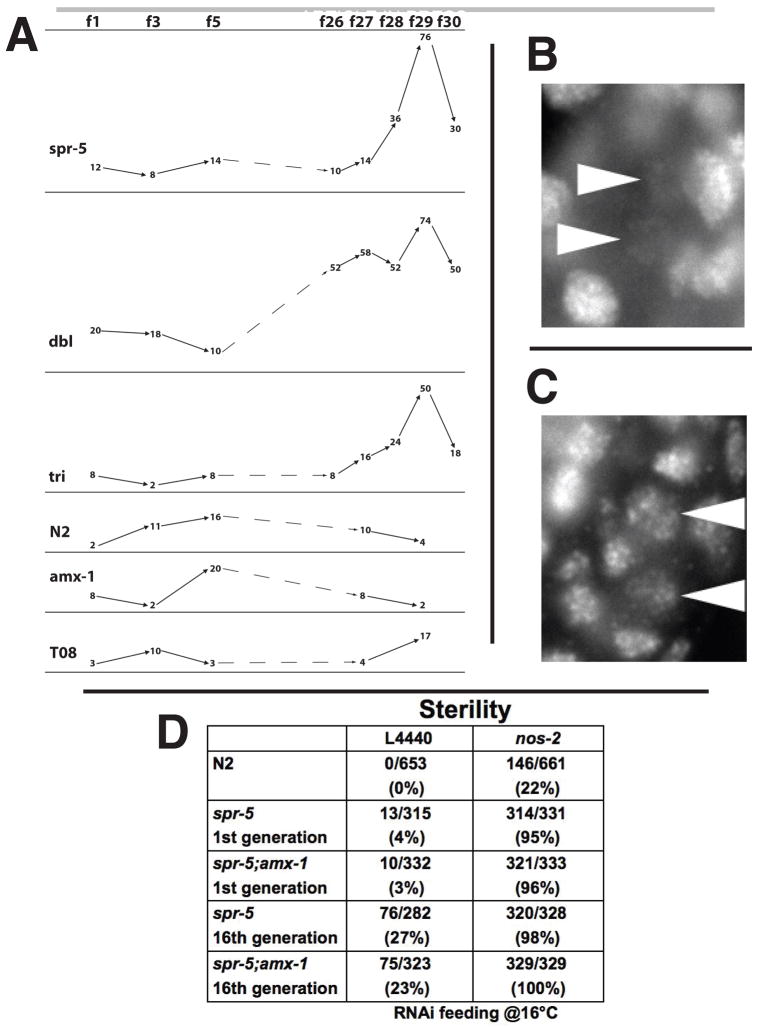
Why does the lack of spr-5 demethylase activity result in the observed defects in oogenesis and spermatogenesis? One possibility is that progressive and heritable accumulation of H3K4me2 through many cycles of the germline results in the inappropriate retention of H3K4me2 in the PGCs, Z2 and Z3. To test this hypothesis we examined H3K4me2 retention in the PGCs by immunofluorescence microscopy over the course of the generations in which the germline mortality was observed. In early generations, wild-type and all mutant combinations exhibited normal erasure of H3K4me2 in the PGCs (Figure 4A and 4B). This suggests that there is an spr-5 independent mechanism of chromatin remodeling acting directly in the PGCs. In addition we could not detect any increase over time in H3K4me2 levels in somatic tissues or in the adult germline, where H3K4me2 levels are already high in wild-type animals (data not shown). However, after many generations we observed a significant increase in PGC H3K4me2 retention in spr-5 mutants throughout the genome (Figure 4A and 4C). This increase correlated with generations exhibiting the lowest brood sizes and the sharpest rise in the percentage of sterile animals (Figure 2A and 2B). This indicates that although SPR-5 activity is not immediately required for the H3K4me2 erasure that occurs in the PGCs, its absence across multiple generations results in the inappropriate retention of H3K4me2 in the PGCs.
Figure 4.
Increased H3K4me2 retention in Z2/ Z3 correlates with sterility. A, Embryos from the experiment in Figure 2 were assayed for PGC H3K4me2 levels by immunofluorescence. Each number represents the percentage of embryos scored positive for H3K4me2 retention out of 50 total embryos for that generation. Examples of wild-type (B) and retained (C) H3K4me2 levels in Z2/Z3. Arrowheads indicate Z2 and Z3. D, The percentage of sterile offspring in spr-5, spr-5;amx-1, nos-2(RNAi), nos-2(RNAi);spr-5 and nos-2(RNAi);spr-5;amx-1 assayed at generations 1 and 16. L4440 is the vector only RNAi control.
Nanos and the spr-5 Mutants are Synthetically Sterile
To test whether the observed sterility may be due to a defect occurring in the PGCs, we performed a synthetic sterility experiment. Nanos is a highly conserved factor with essential germline functions in many metazoans (Kobayashi et al., 1996; Koprunner et al., 2001; Subramaniam and Seydoux, 1999; Tsuda et al., 2003). Two of the C. elegans nanos proteins, NOS-1 and NOS-2, are present exclusively in Z2/Z3 and are redundantly required for PGC survival as nos-1;nos-2 double mutants are 99% sterile in the first generation (Subramaniam and Seydoux, 1999). A nos-2 mutation alone results in 35% sterility (Subramaniam and Seydoux, 1999). If both NANOS and SPR-5 affect PGCs, partial loss of activity in both pathways could have a synergistic effect within these cells on sterility. We performed nos-2 RNAi on spr-5 mutants. nos-2(RNAi) or spr-5 mutants alone each had low levels of sterility (Figure 4D). However, a strong synthetic sterility phenotype (>95%) was observed in spr-5(by101) and spr-5;amx-1 animals with nos-2(RNAi) (Figure 4D). This suggests that the observed sterility in spr-5 mutants may be due to a defect in the PGCs.
spr-5 Mutants Exhibit Transgene Desilencing in the Germline
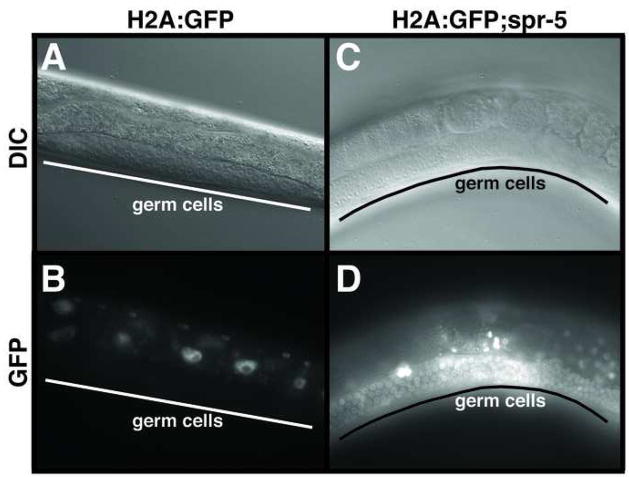
To determine if increased H3K4me2 in the PGCs leads to increased transcription throughout the germline, we performed a transgene desilencing experiment. In C. elegans, multi-copy transgenes are normally silenced in the germline(Kelly and Fire, 1998). For example, an integrated multi-copy histone H2A:GFP transgene is expressed at high levels throughout the soma, but is not expressed in the germline (Figure 5A and 5B). To ask if the increased H3K4me2 in spr-5 mutants can cause germline desilencing, we crossed the H2A:GFP transgene into spr-5 mutants. A dramatic increase in germline expression of the H2A:GFP transgene was observed in many of the spr-5(by101) animals (Figure 5C and 5D). Interestingly, the spr-5 mutation had no affect on the desilencing of an extrachromosomal multi-copy let-858 transgene that can be desilenced by mutations in both transcriptional and post-transcriptional silencing mechanisms (Kelly and Fire, 1998) (and data not shown). Although these results were obtained using different reporter genes, since a major difference between these two transgenes is that one is extrachromosomal while the other is integrated, this finding hints that SPR-5 may only demethylate histones in a chromosomal context.
Figure 5.
Increased H3K4me2 affects transcription. DIC (A,C) and GFP (B,D) imaging of H2A:GFP transgene expression in wild-type (A,B) and spr-5 (C,D) animals.
Microarray Analysis Suggests the Misregulation of Spermatogenesis-enriched Genes
If H3K4me2 can function as an epigenetic memory in the C. elegans germline and SPR-5 demethylation activity resets this memory, then genes that are expressed in the germline could accumulate H3K4me2 and exhibit a transgenerational increase in expression in spr-5 mutants. To look for evidence of this, we performed expression microarray analysis to compare RNA made from mixed-stage populations taken from generation 1, 13 and 26 spr-5 mutant animals as well as wild-type animals.
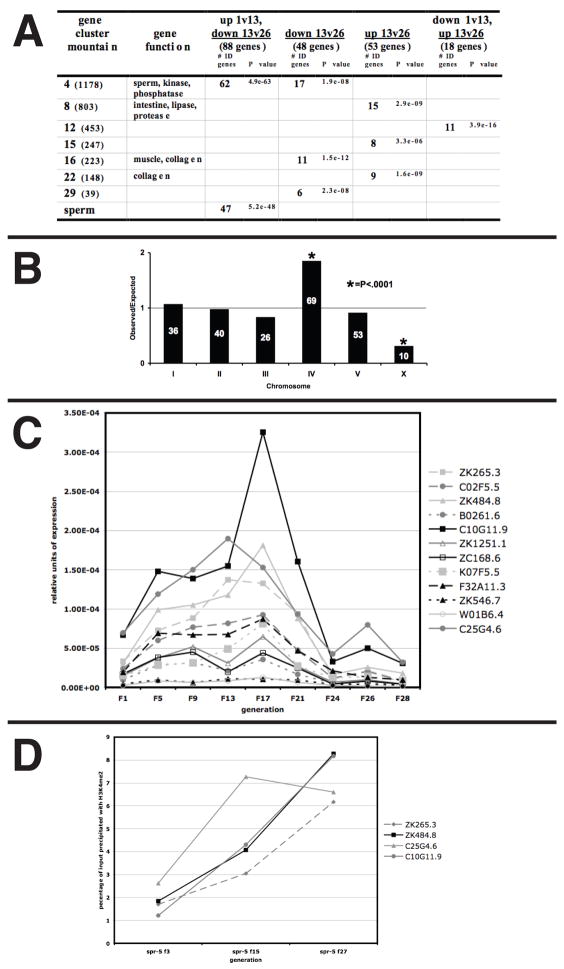
Using strict cutoffs for signal intensity and fold-expression change, the microarray analyses identified a class of 88 genes whose expression coordinately increased from generations 1 to 13 and then decreased from generations 13 to 26 in spr-5(by101) mutants (Figure 6A). Remarkably, 53% of these genes are genes that are expressed during spermatogenesis (Figure 6A) (Reinke et al., 2004). No other significant categories of genes that are coordinately regulated were identified (Figure 6A). Based on the striking group of coordinately regulated spermatogenesis-expressed genes, we examined the microarray expression data from all of the genes that are annotated as being expressed during spermatogenesis. We find that most spermatogenesis-enriched genes also showed a similar pattern of expression, though they fall slightly below the strict cutoffs that were employed (Supplemental Figure 7 and data not shown). In addition, our microarray analysis demonstrated that the distribution of the class of the 234 total regulated genes (>2 fold change in any comparison) was remarkably similar to the chromosomal distribution of spermatogenesis genes, including significant under-representation of X-linked genes and significant over-representation of genes on chromosome IV (Reinke et al., 2004) (Figure 6B). Taken together, these results suggest that nearly all spermatogenesis-expressed genes are coordinately misregulated in spr-5 mutants. Thus, a failure to reset spermatogenesis-acquired H3K4me2 may result in the progressive sterility that is observed in spr-5 mutants.
Figure 6.
The trans-generational accumulation of H3K4me2 and expression at spermatogenesis genes. A, Genes that were changed twofold or greater in comparisons between spr-5 f1,f13 and f26 were compared against the topomap gene function list from over 500 microarray experiments (http://elegans.uky.edu/gl/cgi-bin/gl_mod.cgi?action=compare2). Only over-representation P values of less than 1×10−6 are shown. The number of overlapping genes and the corresponding P value are listed. Mountains with no listed gene function have no known function. All genes that are listed under multiple regulated categories (ex. up1v13, down13v26) are not listed in the individual category (ex. down13v26). B, The chromosomal distribution of the 234 regulated genes, represented as the ratio of observed to expected. The number of genes on each chromosome is shown within the bar. (*)Indicates significant (P<0.001) deviation from the expected. C, Quantitative RT-PCR showing the relative expression of 12 spermatogenesis genes from the 88 coordinately expressed genes (Figure 6A, column 3) over 28 generations. The expression is normalized to actin (act-1). D, ChIP on 4 of the spermatogenesis genes from c, showing the percentage of input precipitated with an H3K4me2 Ab in spr-5 mutants from generations 3, 15 and 27. The primers used for ChIP are immediately downstream of the start site. The percentage precipitated with no Ab was <0.1 in all cases.
Heritable Accumulation of Spermatogenesis-enriched Expression in spr-5 Mutants
To validate the microarray analysis, we performed quantitative RT-PCR analysis on 12 of the spermatogenesis-expressed genes identified in the microarray experiment using RNA made from mixed stage populations from nine generations distributed throughout the germline mortality experiment. This analysis demonstrated that the expression of spermatogenesis-enriched genes increases stably over 20 generations (Figure 6C). This correlates directly with the slow decrease in brood size observed in spr-5 mutants (Figure 2A). After approximately 20 generations, the expression of spermatogenesis-enriched genes decreased (Figure 6C). This decrease coincides with the sharp increase in the percentage of sterile animals, the large increase in H3K4me2 retention in the PGCs, and the onset of the developmental delay (Figure 2B, 4A and Supplemental Figure 6). It is important to note that the observed developmental delay could lead to an increased representation of L4 larvae, in which sperm are produced, within the late generation population. However, we observed no increase in larval-expressed genes, either in the microarray results or by additional quantitative RT-PCR (data not shown). Furthermore, an increased representation of sperm-producing stages is inconsistent with the decline in sperm-enriched expression that we observed after 20 generations.
Heritable Accumulation of H3K4me2 in spr-5 Mutants
The transgenerational increase in spermatogenesis-enriched expression that occurs in the absence of SPR-5 demethylase activity may be due to a heritable accumulation of H3K4me2 in these loci. To test this, we performed ChIP assays with an H3K4me2 antibody on four of the spermatogenesis genes exhibiting generational expression changes in the RT-PCR experiment (Figure 6C). The analysis was performed on mixed stage spr-5 mutants taken from generations 3, 15 and 27. H3K4me2 increases continuously in spr-5 mutants, suggesting that the generational increase in spermatogenesis expression is due to the heritable increase in H3K4me2 at these loci (Figure 6D). Importantly, despite the decrease in expression of spermatogenesis genes at late generations, H3K4me2 levels continue to increase at these genes (Figure 6D). This argues that the increase in H3K4me2 is the cause, not an effect, of the increased expression of spermatogenesis-enriched genes.
Outcrossing at Severely Sterile Generations Rescues the Sterility Defect
If SPR-5 erases H3K4me2 at each generation, then providing wild-type demethylase activity to spr-5 mutants should reset the spr-5 sterility phenotype. To test this we outcrossed spr-5;amx-1 mutants to a balancer worm strain at generation 28 of the germline mortality experiment. Following one passage through the germline with heterozygous demethylase activity, these worms were then returned to a homozygous mutant state and scored for sterility. Outcrossing decreased the frequency of sterile progeny to wild-type levels in the first generation after hemizygosity, suggesting that the most severely sterile worms in the population were completely rescued (Figure 2C). Interestingly, the lower brood size defect was only slightly reversed, indicating that complete epigenetic resetting could not be immediately restored within one generation of exposure to heterozygous SPR-5 activity (Figure 2C).
Discussion
During somatic tissue differentiation, transcriptional activation results in the acquisition of H3K4me2 in the gene bodies of activated genes (Li et al., 2007). This histone methylation may act as an epigenetic memory to facilitate the maintenance of the transcriptional program as cells divide to generate a tissue. Such a model is consistent with the gene regulation of the bithorax complex during differentiation in Drosophila (Ringrose and Paro, 2004). In addition, genome wide evidence of this is seen during the differentiation of mammalian ES cells, where bivalent domains consisting of H3K27me3 and H3K4me3 resolve into domains containing H3K4me3 upon gene activation (Bernstein et al., 2006). Furthermore, evidence for transcriptional memory has recently been provided by the nuclear transfer experiments performed in Xenopus by Ng et. al (Ng and Gurdon, 2008). Despite these observations, direct evidence of the epigenetic inheritance of any histone modification remains elusive.
It has long been suspected that epigenetic modifications could play a role in limiting the developmental potential of a tissue during the complex pattern of tissue differentiation (Jenuwein and Allis, 2001). This has led to the idea that a fundamental aspect of the difference between the “mortal” soma and the “immortal” germline may be the ability to reprogram the genome of the germ cells epigenetically in order to maintain totipotency. However, gaps remain in our understanding of the epigenetic reprogramming between generations. For example, DNA methylation undergoes resetting between generations, but the mechanism of erasing DNA methylation remains unknown (Jaenisch, 1997; Li, 2002). In addition, there is little evidence of the resetting of any histone modification between generations in the germline.
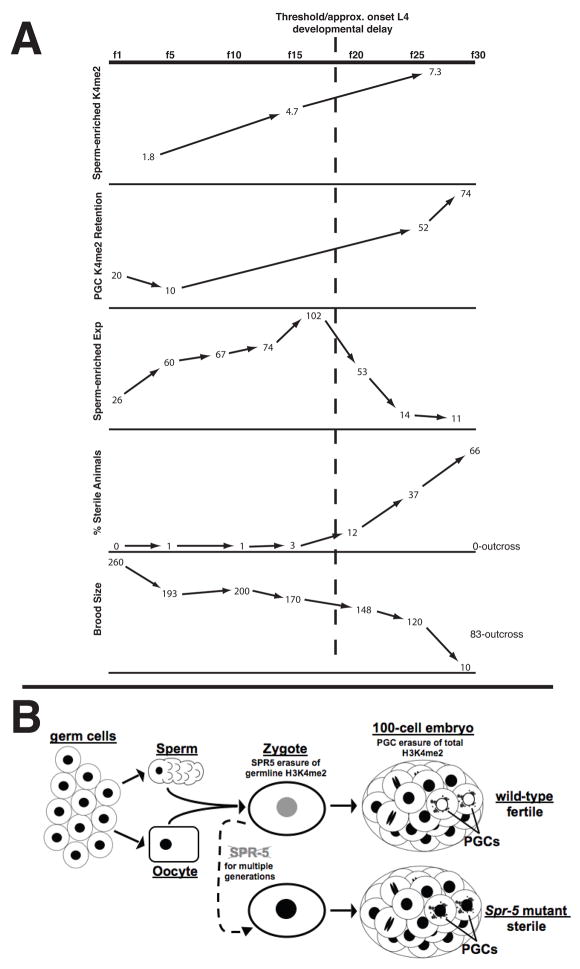
During germline transcription, H3K4me2 is acquired in activated genes. If this methylation acts as an epigenetic memory, there may be a requirement to reset this modification during gametogenesis or in the zygote to prevent this epigenetic memory from inappropriately transmitting from one generation to the next. Our results indicate that a lack of LSD1/KDM1 demethylase activity results in increasing sterility due to defects in oogenesis and spermatogenesis. Why does the lack of SPR-5 demethylation activity result in these sterility defects? During early generations, we observed normal H3K4me2 erasure in the PGCs of spr-5 mutants. This, along with other evidence in our lab (H. Furuhashi, W. Kelly, unpublished), demonstrates that there are spr-5 independent mechanisms required for this erasure. However, at late generations we observed the increasing accumulation of inappropriate H3K4me2 retention in the PGCs. This retention correlates directly with the generations exhibiting the highest degree of sterility (Figure 7A). In addition we find that the SPR-5 has a synergistic effect on sterility with NOS-2, which is present exclusively in the PGCs. Taken together these results argue that the absence of SPR-5 activity causes a transgenerational accumulation of H3K4me2 that eventually saturates the second mechanism of chromatin remodeling in Z2/Z3, resulting in the increased retention of H3K4me2 in the PGCs and increasing sterility (fig 7b.). This data provides the first direct evidence that the proper removal of H3K4me2 in the PGCs is essential for germline maintenance. Furthermore, after approximately 20 generations, we observed a developmental delay in spr-5 mutants at the L4 larval stage (Figure 7A). This delay occurs in somatic lineages in addition to the germline, but only at generations that are largely sterile. This implies that the germline defects in spr-5 mutants may activate a feedback between the germline and soma that can cause a developmental delay of the whole animal.
Figure 7.
The summary of data (A) and model (B). A, Graphs comparing the percentage of H3K4me2 precipitated by ChIP in spermatogenesis-expressed genes(Figure 6D), the percentage of H3K4me2 retention in the PGCs (Figure 4), the relative expression of spermatogenesis-enriched genes (Figure 6C), the percentage of sterile animals (Figure 2B) and the brood size (Figure 2A) over generations in spr-5 mutants. The trans-generational increase in H3K4me2 in spermatogenesis-enriched genes due to the failure to demethylate results in increased H3K4me2 retention in the PGCs and the continuous misexpression of spermatogenesis-enriched genes. This leads to an increasing percentage of sterile animals and decreasing brood size, which can be reset by outcrossing the mutation (Figure 2). B, Germ cells, sperm and oocytes have high levels (black nuclei) of H3K4me2 throughout the genome. SPR-5 erases the epigenetic memory of germline transcription by demethylating H3K4me2 at germline-expressed loci in the gametes or early embryo (grey nucleus). A second mechanism removes H3K4me2 throughout the genome in the PGCs (white nuclei). In spr-5 mutants, the failure to reset germline H3K4me2 over multiple generations results in the inappropriate retention of H3K4me2 in the PGCs (black nuclei) and sterility.
Microarray analysis followed by quantitative RT-PCR analysis demonstrated that the expression of spermatogenesis-enriched genes steadily increases in spr-5 mutants from generation 1 through approximately generation 20 (Figure 7A). In addition, H3K4me2 ChIP analysis demonstrated that H3K4me2 increases continually at spermatogenesis-expressed genes (Figure 7A). Importantly, the ChIP assays were performed on mixed stage populations. In these populations, the cells that are undergoing spermatogenesis are a small fraction of the overall population of cells. This argues that the increase in H3K4me2 at these spermatogenesis-expressed genes may not be confined to spermatogenesis. Rather it is likely that this increase in H3K4me2 is occurring throughout the germline or even throughout the whole animal.
Taken together, our data suggest the following model for the spr-5 germline mortality phenotype (Figure 7). During spermatogenesis, H3K4me2 is acquired at spermatogenesis-expressed genes as they are activated. In the absence of SPR-5 demethylation, this histone methylation is transmitted to the subsequent generation and accumulates for many generations. Increasingly with each generation this accumulation overwhelms the chromatin remodeling mechanism(s) in the PGCs. This results in increased ectopic H3K4me2 in the PGCs and the continuous, inappropriate overexpression of spermatogenesis-expressed genes throughout the germline. Although it is possible that SPR-5 acts in the PGCs where it might be partially redundant with other H3K4me2 erasure mechanism(s) there, the fact that SPR-5 appears to be maternally deposited is also consistent with SPR-5 acting either in gametogenesis and/or in the early embryo. In either case, increasingly with each generation, the continuous misexpression of spermatogenesis-enriched genes in the germline results in delayed spermatogenesis and is incompatible with oogenesis. After approximately 20 generations, the RT-PCR analysis demonstrated that the expression of the spermatogenesis-enriched genes decreases. It is unclear exactly why this occurs. However, this correlates with the sharp increase in the percentage of sterile worms at late generations as well as the onset of the L4 developmental delay and the large increase in H3K4me2 retention in the PGCs (Figure 7A). Thus, it is possible that after many generations, the increase in H3K4me2 may reach a threshold where sperm development is more severely compromised (Figure 7A). This, in turn, may lead to the dramatic decrease in sperm expression.
If H3K4me2 in spermatogenesis-expressed genes is increasingly inappropriately inherited throughout the tissues of offspring, does this result in somatic phenotypes? In spr-5 mutants we observe the appearance, at a low frequency, of numerous somatic phenotypes such rolling worms, dumpy worms, protruding/multivulvals, etc. It is possible that these phenotypes are due to a requirement for SPR-5 activity in these tissues. Alternatively, it is possible that these phenotypes are due to the somatic expression of germline genes that is the result of the failure to reset germline H3K4me2 from the previous generation. It is unclear, if this is the case, why a higher penetrance of somatic phenotypes is not observed. However, although increased somatic H3K4me2 in germline genes could make these loci more permissive for transcription, it may be very difficult for the inappropriate expression of germline genes to overcome the robust somatic program. For example, in mep-1 mutants in C. elegans, germline granules are inappropriately segregated to somatic cells and some germline genes are inappropriately expressed there, but the somatic tissues are still specified (Unhavaithaya et al., 2002).
In our microarray analysis we observed dramatic changes in spermatogenesis expression but not changes in the expression of meiosis genes or oogenesis genes. One possible explanation for this result is that SPR-5 plays a specific role in demethylating H3K4me2 during spermatogenesis. Alternatively, SPR-5 may play a role in resetting germline H3K4me2 in general. The expression of SPR-5 throughout the adult gonad is consistent with a role in resetting germline H3K4me2 in general. In addition, the ectopic H3K4me2 staining in the PGCs is genome wide, suggesting that more than spermatogenesis-expressed genes are being affected. In C. elegans spermatogenesis occurs only during the brief L4 developmental stages. As a result, spermatogenesis genes are likely expressed at high levels in order to provide sufficient protein for the complex spermatogenesis differentiation program. This feature of brief, intensive transcription may make spermatogenesis genes particularly sensitive to the loss of SPR-5.
The data presented here provide the first evidence that H3K4me2 levels can be faithfully transmitted through many cell divisions and act as an epigenetic memory to affect transcription. In addition, this phenotype strongly suggests that SPR-5 functions in the epigenetic reprogramming of the germline by erasing H3K4me2 at numerous loci, preventing it from being inappropriately propagated to the next generation (Figure 7B). Thus we propose that the ability to reset H3K4me2 plays a critical role in distinguishing the “immortal” germline from the “mortal” soma by erasing the epigenetic memory of germline transcription at each generation. This is the first example of a mechanism of epigenetic reprogramming for which the enzymatic activity has been identified. Based on our results, we hypothesize that efficient LSD1/KDM1-mediated demethylation may be part of what is missing in the incomplete reprogramming that was observed in the nuclear transfer experiments performed by Ng et al. (Ng and Gurdon, 2008). In addition, it is possible that LSD1/KDM1 demethylation may also be part of the mechanism of somatic reprogramming that is induced by the four pluripotent factors in the experiments performed by Yamanaka (Takahashi and Yamanaka, 2006).
Mutations in Su(var)3-3, the Drosophila ortholog of LSD1/KDM1, result in sterility in both males and females due to defects in oogenesis and spermatogenesis (Rudolph et al., 2007; Szabad et al., 1988). In addition, Su(var)3-3 mutations result in ectopic H3K4me2 in the PGCs(Rudolph et al., 2007). These phenotypes occur in one generation rather than over multiple generations. However, despite these observations, it remains unclear how the lack of Su(var)3-3 demethylase activity results in the observed sterility defects in Drosophila. We propose that LSD1/KDM1 demethylation acts similarly in both C. elegans and Drosophila. In the case of Drosophila, the pole cells are set aside already lacking H3K4me2 and are maintained in this state (Schaner et al., 2003; Technau and Campos-Ortega, 1986; Underwood et al., 1980). This is in contrast to C. elegans where H3K4me2 is initially abundant in the P blastomere chromatin, but is rapidly erased from the genome once the germline is separated from the somatic lineages (Schaner et al., 2003). Thus the P blastomeres, unlike Drosophila pole cells, may have an epigenetic signature more similar to ES cells, with a subsequent requirement for the erasure of this signature once the PGCs are established. As a result, a second demethylase-independent chromatin erasure mechanism may be required in worms but not flies. We speculate that the sterility in the Drosophila Su(var)3-3 mutant is immediate, rather than requiring many generations, due to the lack of requirement for, and thus absence of a second chromatin erasure mechanism. Taken together, we believe that the phenotypes in flies and worms argue that the function of LSD1/KDM1 demethylation elucidated here may be the primordial function in metazoans. As such, this function may be conserved in mammals. Interestingly we note that should LSD1/KDM1 play a similar role in humans, defective LSD1/KDM1 regulation in the germline could result in the misregulation of multiple genes in the next generation. Thus, mutations in LSD1/KDM1 could result in complex diseases where the phenotype is due to the contribution of multiple loci.
Methods
Strains
The C. elegans strains spr-5(by101) and T08D10.2 were provided by R. Baumeister and V. Reinke. amx-1(ok659) was provided by the Caenorhabditis Genetics Center. The ht2g balancer strain was used for the outcross experiment.
Western and ChIP
The protein extracts were made as in Katz et al. except that worms were washed off of six 10cM plates with 500ul of PBS, flash frozen at −80°C for 30min before homogenization and centrifuged 3 times at 2,000rpms following homogenization. H3K4me2 Ab (Upstate), H3 Ab (Abcam), and Actin Ab (Chemicon) were used at 1/2000, 1/5000 and 1/1200 respectively in the westerns and the westerns were quantitated using a Bio-Rad Flour-S Max Multimager. The hop-1 ChIP was done as in Katz et al. except that worms from 20 10cm plates were bleached with 1M NaOH, 10% bleach to isolate embryos and hatched into L1 larvae overnight on 5 plates. In addition, worms were freeze-thawed before homogenization as above and sonicated for 200sec (4sec on/10sec off) at 20% in a Fisher Sonic Dismembrator 500. 10ul of H3K4me2 Ab (Upstate) was used for immunoprecipitaiton. For the spermatogenesis genes ChIP, mixed stage worms were washed off of 3 10cM plates before proceeding directly to the freeze-thaw step. The Bio-Rad iCycler PCR primers are shown in Supplemental Table 1.
Immunofluorescence
A methanol/acetone fixation procedure was used (Strome and Wood 1983) for embryo and gonad staining with 1/50 N and C terminal SPR-5 peptide Ab’s (Santa Cruz sc-68340-R, 68339-R), 1/250 H3K4me2 Ab (Upstate 07-030) and undiluted P-granule Ab (Strome and Wood, 1983).
Germline Mortality Experiment
During the germline mortality experiment, the worms were maintained at 20°C by picking three fertile adults on to another 6cm plate every 4th day. Because the worms are translucent, embryos can be visualized inside the adult worm, allowing us to select fertile adults. Prior to the experiment all mutants were balanced for a minimum of 3 generations and then re-homozygoused. The generation number was then counted from the 1st homozygous mutant generation. The wild-type (N2 Bristol), amx-1 and T08D10.2 strains were counted every other generation throughout the experiment. The spr-5(by101), spr-5;amx-1 and triple (spr-5;amx-1;T08D10.2) strains were counted every other generation for the first 17 generations and then every generation through generation 31. The average brood size of the spr-5(by101) and spr-5;amx-1 strains was calculated from the progeny of 10 worms through generation 18 and then out of 50 worms through the end of the experiment, while the average brood size of the triple strain was calculated from the progeny of 10 worms through generation 24 and then out of 50 through the end of the experiment. The percentage of males was counted out of the combined total brood while the percentage of embryonic lethality was taken from the first ~1000 embryo’s laid divided equally among the individual broods. The percentage of sterile animals for the spr-5(by101) and spr-5;amx-1 strains was counted out of >300 through generation 21 and then out of >1000 through generation 31, while the percentage of sterile animals for the triple strain was counted out of >300 for all generations except generation 26 where it was counted out of >1000. At all generations the percentage of sterile animals for the spr-5(by101), spr-5;amx-1 and triple strains was counted from the progeny of at least 5 adults.
Each percentage of H3K4me2 retention data point was calculated from ~50 different embryos of ~100 cells or greater from at least 2 slides. Special care was taken to try and avoid counting more than 5 embryos from any 1 animal. In each case Z2 and Z3, as marked by P-granule staining, were scored either positive or negative under exposure conditions where the surrounding cells had adequate levels of H3K4me2 staining. In cases where only Z2 or Z3 were positive, that embryo was scored as positive. All scoring was done blind to the genotype with all 6 genotypes and all generations that were scored intermixed.
RNA Interference
RNAi feeding was performed as in Kamath et al. (Kamath et al., 2001). All RNAi constructs were taken from the Ahringer RNAi library (Kamath and Ahringer, 2003).
Apoptosis Assays and the Larval Delay Experiment
To stain with acridine orange (AO) 500ul of 0.04mg/ml AO in M9 buffer was dropped on top of a 6cm seeded NGM plate of worms and incubated in the dark for 1hr at 20°C. Individual worms were then recovered for 1hr on a seeded plate without AO and then visualized by DIC imaging. To synchronized adults for the larval delay experiment, cloned out 2-cell embryos were allowed to develop on seeded plates and then visualized by DIC as above.
Microarray Analysis and Quantitative RT-PCR
To generate total RNA 3 10cM plates of worms were flash frozen in 500ul of PBS. Following freezing 1ml of Trizol reagent was added to the pellet and the pellet was thawed, vortexed and re-frozen 4 times. After 4 cycles, the sample was chloroform extracted, precipitated with isopropanol, washed with 75% ethanol and cleaned up with the Qiagen RNeasy kit. For the microarray analysis 8ug of total RNA was hybridized pair-wise, along with a dye flip, from N2f1 (wild-type) and spr-5(by101) f1, 13 and f26, for a total of 12 arrays. The microarray hybridizations were performed at the Washington University Microarray Core Facility (Gene Expression Omnibus Accession number GSE14432). Significantly differentially regulated genes were selected using a two-fold difference along with intensity values >1000. For RT-PCR, 5ug of total RNA (isolated as above) was reverse-transcribed using the Invitrogen First Strand Synthesis System with Oligo(dt)20 priming. The results were normalized to Actin (act-1). The quantitative PCR primers are shown in Supplemental Table 2.
Supplementary Material
Acknowledgments
We thank R. Baumeister for providing strains; A. Printz for help with data presentation; C. Bean, B. Lakowski, G. Reuter, T. Schedl, Y. Shi and J. Waddle for helpful discussions on the work; T. Caspary, J. Lucchesi and S. Tilghman for critical comments on the manuscript; and all of our laboratory members for assistance throughout. This work was supported by grants from the NIH to DJK and WGK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Eimer S, Lakowski B, Donhauser R, Baumeister R. Loss of spr-5 bypasses the requirement for the C. elegans presenilin sel-12 by derepressing hop-1. Embo J. 2002;21:5787–5796. doi: 10.1093/emboj/cdf561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Jarriault SGI. Genes Dev. 2002;16:2713–2728. doi: 10.1101/gad.1022402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods (San Diego, Calif. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nature genetics. 2005;37:1090–1097. doi: 10.1038/ng1637. nematode.lab.nig.ac.jp/ [DOI] [PubMed]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. Reprogramming by the numbers. Nat Biotechnol. 2007;25:877–878. doi: 10.1038/nbt0807-877. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Poux S, Horard B, Sigrist CJ, Pirrotta V. The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development. 2002;129:2483–2493. doi: 10.1242/dev.129.10.2483. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annual review of genetics. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Szabad J, Reuter G, Schroder MB. The effects of two mutations connected with chromatin functions on female germ-line cells of Drosophila. Mol Gen Genet. 1988;211:56–62. doi: 10.1007/BF00338393. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Technau G, Campos-Ortega J. Lineage Analysis of transplanted individual cells in embryos of Drosophila Melanogaster. III. Commitment and proliferation capabilities of pole cells and midgut progenitors. Rouxs Arch Dev Biol. 1986;195:489–498. doi: 10.1007/BF00375889. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Underwood EM, Caulton JH, Allis CD, Mahowald AP. Developmental fate of pole cells in Drosophila melanogaster. Dev Biol. 1980;77:303–314. doi: 10.1016/0012-1606(80)90476-5. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.