Abstract
Absence epilepsy is a common form of idiopathic generalized epilepsy whose etiology is poorly understood due to genetic and phenotypic heterogeneity. The inbred mouse strain C3H/He exhibits spontaneous absence seizures characterized by spike and wave discharges (SWD) on the electroencephalogram concomitant with behavioral arrest. Previous studies using the C3H/HeJ (HeJ) substrain identified a mutation in the Gria4 gene as a major susceptibility locus. In the present study we found that two closely-related substrains C3H/HeOuJ (OuJ) and C3H/HeSnJ, which have a similar SWD incidence as HeJ, do not contain the Gria4 mutation. Further analysis of backcross mice segregating OuJ and C57BL/6J alleles shows that, unlike the HeJ substrain, OuJ does not have a major locus for SWD but has suggestive loci at best that would explain only a fraction of the phenotypic variance. These results illustrate how the genetic etiology of a common neurological disorder can differ between substrains with similar phenotypes. We infer that all C3H strains are sensitized to SWD and that additional mutations affecting SWD arose or were fixed independently in the years since the substrains diverged.
Keywords: absence seizure, inbred mouse strains, complex trait, substrain divergence, AMPA receptor
Introduction
Absence epilepsy is a non-convulsive form of idiopathic generalized epilepsy that occurs frequently in childhood and adolescence. An absence seizure is defined by a sudden and brief arrest of normal activity that is accompanied by rhythmic and synchronous spike-wave discharges (SWD) on the electroencephalogram (EEG). Clinical and experimental studies indicate a central role of the thalamocortical circuitry in the genesis of SWD. The molecular pathogenesis, however, remains unclear because the inheritance is usually polygenic or multifactorial (Futatsugi & Riviello, 1998, Huguenard & Mccormick, 2007). Several mutant and inbred rodents strains, exhibiting SWD with behavioral and pharmacological features similar to absence seizure in humans, have long contributed to the field. For example, tottering, stargazer, ducky, and lethargic mutant mice, resulting from mutations in Ca2+ channel subunit genes, have been well accepted as absence seizure models despite their ataxic phenotype (Burgess et al., 1997, Cox et al., 1997, Fletcher et al., 1996, Letts et al., 1998, Barclay et al., 2001). More recently the mouse model harboring a GABAA γ2 missense mutation that segregated in a large family with childhood absence epilepsy and febrile seizures, recapitulated the primary seizure phenotype observed in patients and its absence seizure phenotype was very similar to prior mouse models (Tan et al., 2007). C3H is a very commonly used inbred mouse strain with no obvious neurological disease phenotypes, other than retinal degeneration (Fox, 1997). In 2005, EEG recording revealed that the C3H/HeJ (HeJ) substrain exhibited a modest incidence of 7–8-Hz SWD lasting about 2.5 seconds, concomitant with behavioral arrest (Frankel et al., 2005). Segregation tests using a backcross with the C57BL/6J (B6) strain indicated an unusual form of inheritance, with backcross mice showing a much higher incidence of SWD than HeJ itself. Nevertheless, a major HeJ-derived allele for SWD (spkw1) was genetically mapped to centromeric Chromosome 9. More recent work identified an intronic proviral insertion in the Gria4 gene encoding the ionotropic AMPA receptor subunit GRIA4 (also known as GLUR4), very likely to be synonymous with spkw1 (Beyer et al., 2008). The mutation resulted in the reduction of GRIA4 expression in HeJ mice. The cause-effect relationship between Gria4 mutation and seizure phenotype was confirmed by use of mice carrying a targeted mutation.
Other substrains of C3H/He, such as C3H/HeOuJ (OuJ) and C3H/HeSnJ (SnJ), have a similar incidence and form of SWD as HeJ. This suggested the hypothesis that susceptibility genes for SWD (e.g. Gria4) in HeJ, might be fixed in all C3H/He substrains. In the present study, however, we report that OuJ and SnJ substrains did not have a Gria4 mutation. The SWD incidence in backcross with OuJ and B6 appeared to show a more continuous distribution reflective of the parental strains, than that seen in crosses with HeJ. In OuJ we did not detect any significant loci associated with SWD, suggesting the genetic component is much more complex. Large-scale quantitative trait locus (QTL) analysis or alternative strategies will be required to further investigate the molecular basis of SWD in OuJ and other substrains of C3H/He.
Materials and Methods
Animals
All mice used in this study were obtained originally from The Jackson Laboratory (Bar Harbor, ME) and housed in the Research Facility at The Jackson Laboratory. All animal procedures were approved by Institutional Animal Care and Use Committee (IACUC).
Cortical EEG recordings
Mice that were between 6 and 10 weeks of age were tested for spontaneous SWD activity as previously described (Beyer et al., 2008). Briefly, mice were anesthetized with tribromoethanol (400mg/kg i.p.) and four Teflon-coated silver wires were implanted on both sides of the skull (1 mm anterior to the bregma, 2 mm posterior to the bregma and 2 mm lateral to the midline). At least 48 hours later, EEG activity of mice was measured twice for two hours on separate days (four hours total) using the Grass EEG Model 12 Neurodata Acquisition System and PolyViewPro software program (Grass-Telefactor, Inc.) or the Stellate Harmonie system (Stellate, Inc.). The mean values of SWD frequency and cumulative length per hour of the two sessions were calculated for individual mouse, with very good correspondence between sessions (r=0.62463; p=0.7282).
Genotyping
Genomic DNA was isolated from tails of the backcross progeny (N2) or intercross progeny (F2) that were generated between C3H/HeOuJ and C57BL/6J. Microsatellite markers distinguishing C3H and B6 alleles were amplified with standard PCR techniques and analyzed using agarose gel electrophoresis. N2 mice were genotyped with the total of 47 markers covering all mouse chromosomes including Chr X and F2 mice with nine markers based on the initial screening.
Data analysis
Marker regression analysis and interval mapping were performed using MapManagerQTXb20 (http://www.mapmanager.org/). Genome-wide significance thresholds were calculated using the permutation test, for 1,000 permutations. Calculated thresholds were suggestive linkage LRS > 5.0, significant linkage LRS > 8.6, and highly significant linkage LRS > 12.2. Further evaluations of association between genotypes and phenotypes were determined by one-way ANOVA and contingency test using JMP software (SAS Institute, Cary, NC)
Genomic PCR
The intracisternal A-particle (IAP) insertion site found in Gria4 in C3H/HeJ was amplified using genomic DNAs isolated from excised tail tips of C3H/HeJ, HeOuJ, HeSnJ, C3HeB/FeJ and C57BL/6J with the primers used in the previous study, which were designed in genomic regions located on either side of the mutation sites or within 5′ end of IAP long terminal repeat region (Beyer et al., 2008).
Western blot analysis
Protein lysates were processed using cerebellum of adult mice as previously reported (Beyer et al., 2008). Rabbit polyclonal antibodies for GRIA4 carboxyl terminus (1:200, Chemicon, CA) and for Calbindin D-28-K (1:5000, Swant, Switzerland) were used for primary antibodies. Horseradish peroxidase-conjugated secondary antibody (PerkinElmer, MA) was used. Protein bands were detected using the ECL-plus chemiluminescent reagent (Amersham Biosciences).
Results
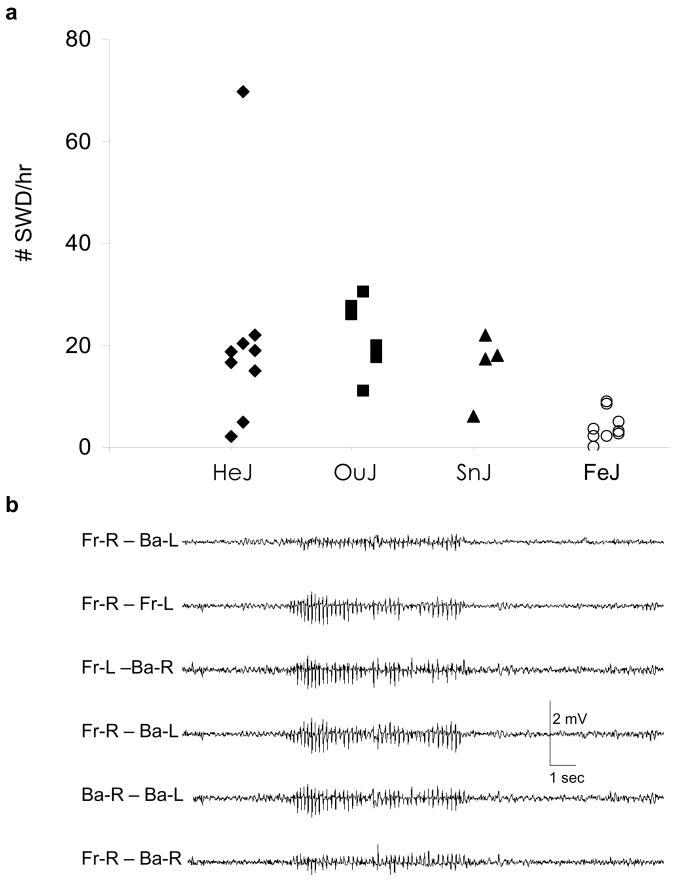
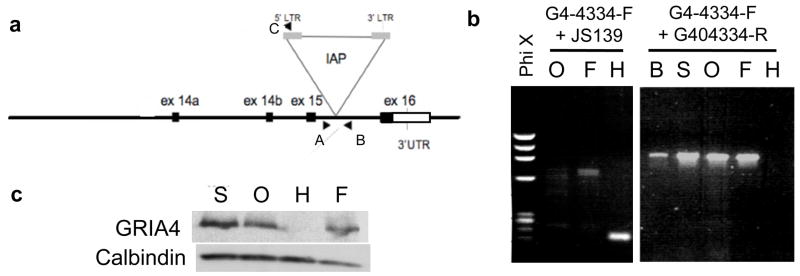
The C3H/He substrains: HeJ, OuJ and SnJ exhibit a modest incidence of SWD while another major substrain, C3HeB/FeJ, exhibits a only low incidence (Figure 1) (Frankel et al., 2005). Previous work revealed that the major genetic basis for SWD in HeJ mice is the IAP-insertion mutation in Gria4 (Beyer et al., 2008). Since the genetic background was expected to be very close among HeJ, OuJ and SnJ substrains, and the features of SWD were observed to be similar, we assumed that SWD in OuJ, SnJ and HeJ would have the same genetic basis. Genomic PCR with primer sets designed to distinguish between the proviral insertion mutation and wild type, and subsequent sequencing, however, showed OuJ and SnJ had the wild-type preintegration site, indicating these two substrains did not have the same allele of Gria4 as HeJ (Figure 2). Further, while we could not assess whether C3H/HeN mice exhibit SWD, examination of genomic DNA showed the wild–type allele at Gria4 (data not shown). Western blot analysis further revealed that OuJ and SnJ expressed the GRIA4 protein (Figure 2). These findings suggested that the seizure in both OuJ and SnJ are caused by genetic mutations other than that found in HeJ. This fact prompted us to further investigate the genetic basis of SWD in the C3H/He substrains, focusing on OuJ.
Figure 1. Spike and wave discharge (SWD) in C3H substrains.
(a) Distribution of SWD per hour in C3H/HeJ (HeJ), C3H/HeOuJ (OuJ), C3H/HeSnJ (SnJ) and C3HeB/FeJ (FeJ). HeJ and FeJ data were replotted from Beyer et al., 2008. (HeJ: 20.9 ± 6.51 SWD/hr (n=9), OuJ: 22.1 ± 2.96 SWD/hr (n=6), SnJ: 15.9 ± 3.43 SWD/hr (n=4), FeJ: 3.96 ± 1.00 SWD/hr (n=9). Mean ± S.E.M.)
(b) Representatives of EEG trace recorded with the OuJ mouse. EEG was obtained from electrodes placed over the right front (Fr-R), left front (Fr-L), right back (Ba-R), and left back (Ba-L) surface of the cerebral cortex. SWD were characterized by a high amplitude, synchronous, bilateral and generalized signal.
Figure 2. The IAP insertion mutation in Gria4 in HeJ is not present in other C3H substrains.
(a) Schematic map of the 3′ end of the Gria4 gene (modified from Beyer et al, 2008). The IAP proviral insertion was located within intron 15 in the C3H/HeJ substrain. Arrow heads represent the location of primers used in genomic PCR; A: G4-4334-F, B:G4-4334-R, C:JS139.
(b) 250 bp of the IAP junction fragment in HeJ is amplified with the genomic primer G4-4334-F and the IAP primer JS139, but this fragment was not present in lanes O and F (left panel). The genomic primer G4-4334-F and G4-4334-R amplified an 800 bp fragments from all the strains except for HeJ (right panel). The fragment length for HeJ was expected to be 5 kb, too large to be amplified under the PCR conditions of the present study. The absence of the IAP insertion mutation in OuJ and SnJ was also confirmed by genomic sequence (data not shown). (H: C3H/HeJ, O: C3H/HeOuJ, S: C3H/HeSnJ, F: C3H HeB/FeJ, B: C57B6/6J)
(c) Expression of the GRIA4 protein. Western blot analysis of cerebellum was performed using anti-GRIA4 and anti-calbindin for loading control. OuJ, FeJ and SnJ mice expressed GRIA4 as well as FeJ but no GRIA4 expression was detected in HeJ.
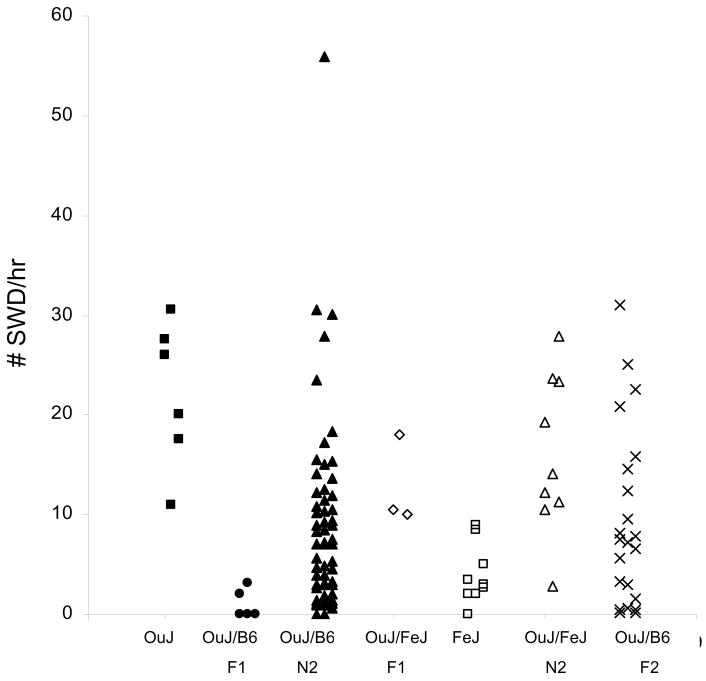
We crossed OuJ to B6 or FeJ to determine the mode of inheritance. Of five OuJ x B6 F1 hybrid mice tested, SWD were observed at low incidence in two animals and no SWD in others, suggesting that inheritance was mostly recessive (Figure 3). We then crossed the F1 hybrid mice to OuJ mice and assessed SWD in the resultant backcross generation. Almost all progeny showed some SWD, the frequency of which spanned the entire range defined by parental strains, with only one mouse exhibiting much higher frequency (56 SWD/hr) (Figure 3); this is in contrast with previous results from HeJ x B6 where a large number of backcross mice had SWD at a much higher rate than HeJ itself (Frankel et al., 2005). About one third of the current backcross showed modest frequency similar to OuJ (10–30 SWD/hr) and the rest showed lower frequency (<10 SWD/hr). Those findings further suggested that the inheritance was recessive but non-Mendelian in this OuJ and B6 cross. On the other hand, in F1 hybrids between OuJ and FeJ substrains of C3H/He, all mice exhibited a similar frequency to OuJ, and the backcross progeny to OuJ also showed a modest frequency of SWD except for one mouse with a low incidence (Figure 3). We infer that the major genes underlying SWD in OuJ are dominant or semidominant when on the C3H background - showing further that background appears to influence the mode of inheritance of the seizure phenotype of OuJ.
Figure 3.
Spike and wave discharge incidence in OuJ mice, F1 hybrids, intercross F2, and backcross N2 progeny with B6 or FeJ mice. OuJ and FeJ data was replotted from Figure 1.
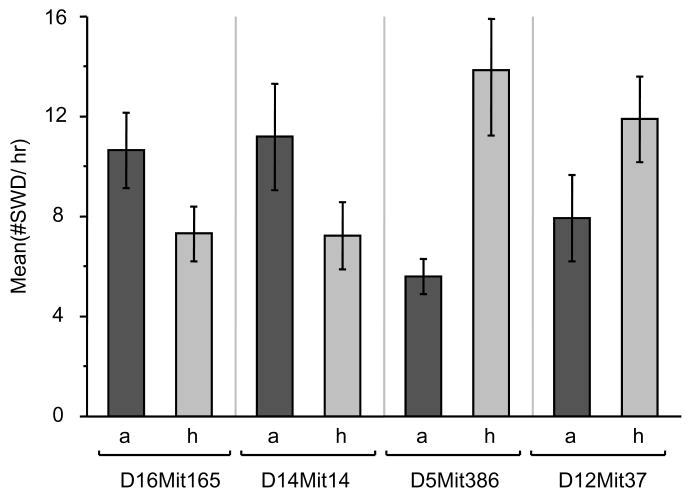
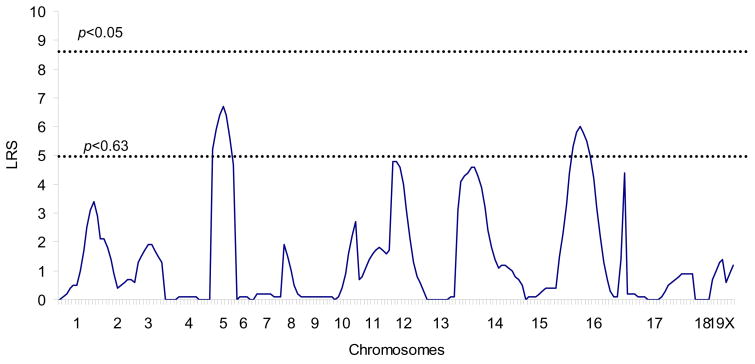
To explore the genetic basis of SWD in OuJ mice, we first conducted parametric analysis using 56 OuJ and B6 N2 backcross mice. The number of SWD per hour was determined and transformed into natural log to obtain a more normal distribution and to equalize the variance as much as possible; a genome scan was then performed. The preliminary marker regression analysis using 42 polymorphic microsatellite markers gave potential evidence that Chr 5, 12, 14 and 16 are associated with the phenotype (Table 1). The susceptibility alleles on Chr 14 and 16 would be derived from OuJ, but those on Chr 5 and 12 would be transgressive susceptibility alleles derived from the B6 strain (Figure 4). Subsequently, additional markers for potential OuJ susceptibility loci were genotyped and tested. As a result, suggestive associations between phenotype and genotype were detected at the centromere-proximal markers on Chr 16 (D16Mit165; Table 2 and Figure 4). We then performed interval mapping, and modest peaks were detected only within Chr 5 and Chr 16, surpassing the suggestive threshold as estimated by using a permutation test (Figure 5). Neither main effect loci nor pairwise interactions were found to be significantly associated with the phenotype, nor was there a significant effect of gender. Moreover, no association was seen in the Gria4 region of Chr 9, suggesting that Gria4 variants are not involved in SWD of OuJ mice (D9Mit67, p=0.759; D9Mit18, p=0.768).
Table 1.
SWD susceptibility loci detected by marker regression analysis a
| Chr | Locus | Position (cM) | LRS | Genetic variance explained (%) | p-value | Additive effect b | Susceptibility allele |
|---|---|---|---|---|---|---|---|
| 5 | D5Mit386 | 9.8 | 5.2 | 9 | 0.022 | 1.14 | B6 |
| 5 | D5Mit95 | 57.9 | 3.8 | 7 | 0.049 | 0.98 | B6 |
| 12 | D12Mit37 | 1.1 | 4.8 | 8 | 0.028 | 1.15 | B6 |
| 14 | D14Mit14 | 16.4 | 4.3 | 7 | 0.039 | −1.03 | OuJ |
| 16 | D16Mit152 | 45.9 | 4.6 | 8 | 0.032 | −1.09 | OuJ |
56 N2 progeny were genotyped with 42 polymorphic markers. Marker regression analysis was performed using MapManager QTXb20. (Markers associated at p<0.05 are shown.)
Additive effect was calculated by MapManager program.
Figure 4. SWD effects for marker alleles with suggestive evidence or association in backcross mice.
The top of each bar represents mean ± S.E.M. The mean SWD/hr of homozygous alleles was larger than that of heterozygous alleles at D16Mit165 or D14Mit14. In contrast, the mean SWD/hr of homozygous alleles was smaller than that of heterozygous alleles at D5Mit386 or D12Mit37; a: C3H/HeOuJ homozygous alleles, h: heterozygous alleles. The number of animal (a, h) is: (33, 23) for D16Mit165, (29, 27) for D14Mit14, (31, 25) for D5Mit386 and (37, 19) for D12Mit37.
Table 2.
Likelihood ratio test statistics (LRS) scores of additional markers on Chr 16 and 14
| Chr | Locus a | Position (cM) | LRS | Genetic variance explained (%) | p-value |
|---|---|---|---|---|---|
| 16 | D16Mit165 | 10.9 | 6.0 | 10 | 0.014 |
| D16Mit4 | 25.1 | 0.1 | 0 | 0.786 | |
| D16Mit30 | 30.6 | 4.4 | 8 | 0.036 | |
| 14 | D14Mit10 | 3.3 | 3.1 | 5 | 0.076 |
| D14Mit102 | 37.2 | 1.1 | 2 | 0.287 |
Five markers were further tested by marker regression analysis with 56 N2 progeny.
Figure 5.
Genetic map indicating interval likelihood ratio test statistics (LRS) scores for linkage to loci associated with SWD in OuJ and B6 backcross progeny (n=56). LRS scores were determined with MapManager QTX20 using ln (#SWD) as the trait measure. Significance thresholds were estimated by permutation test.
As an alternative, we applied a non-parametric approach to detecting SWD loci, hypothesizing that an obligate interaction between several loci might be required for high seizure incidence. We noted that five N2 progeny showed a much higher incidence of SWD (>20 SWD) compared with the remaining animals (Figure 3). An obligate interaction between three recessive alleles would result in approximately 1/8 of all the backcross mice having a high seizure incidence – not significantly different from 1/10 observed. An allele frequency test between markers and phenotype was therefore conducted between these two groups. While none of markers on Chr 5, 12, 14 and 16, previously suggested by parametric analysis, showed a significant difference between groups, the non-parametric approach showed that all individuals in the high-incidence group had OuJ homozygous alleles at D4Mit54 and D18Mit120 (Fisher’s exact test: p<0.0257 and p<0.0445, respectively; Table 3).
Table 3.
Non-parametric analysis with high (>20 SWD/hr) and low (<20 SWD/hr) seizure groups
| N2 backcross | F2 intercross | |||||
|---|---|---|---|---|---|---|
| Genotype | High | Low | Genotype | High | Low | |
| D4Mit54 | A/A | 5 (8.9%) | 23 (41.1%) | A/A | 2 (9.1%) | 4 (18.2%) |
| A/B | 0 (0.0%) | 28 (50.0%) | A/B | 2 (9.1%) | 10 (45.5%) | |
| B/B | 0 (0.0%) | 4 (18.2%) | ||||
| Fisher’s test a | p=0.0257 | |||||
| D18Mit120 | A/A | 5 (8.9%) | 26 (46.4%) | A/A | 1 (4.5%) | 2 (9.1%) |
| A/B | 0 (0.0%) | 25 (44.6%) | A/B | 2 (9.1%) | 12 (54.6%) | |
| B/B | 1(4.5%) | 4 (18.2%) | ||||
| Fisher’s test a | p=0.0445 | |||||
A: OuJ allele, B: B6 allele
Contingency analysis using JMP software.
Although the evidence was weak for OuJ-derived SWD susceptibility loci obtained from either parametric or non-parametric analysis, we thought that it was still possible that major loci could be located on Chr 16 or 14 (the two chromosomes with the most promising OuJ-derived susceptibility loci), but perhaps their effects were masked by other complexities in the backcross. An intercross would provide a more diffuse population structure than the corresponding backcross, for example, breaking-up certain types of interactions that might interfere with the ability to detect major effects. Such complexities might include transgressive alleles – even minor ones - that are linked to Chr 16 or Chr 14, or epistasis between main effect loci, and recessive OuJ-derived modifiers in the background, which would be less prolific in an intercross than in a backcross.
To examine whether the suggestive associations on Chr 16 and Chr 14 could be detected in intercross mice, 40 F2 progeny of OuJ and B6 were generated and genotyped before examining phenotype. To perform the hypothesis test efficiently, EEG was ultimately recorded from 22 F2 mice that were homozygous for one or the other parental allele of Chr 16. As shown in Figure 3, one third (seven) of F2 mice exhibited the similar frequency of SWD (>10 SWDs/hr) to OuJ parental strain while one third showed fewer than five times per hour. Linear regression analysis was applied to Chr 16 markers to explore possible correlation between genotype and phenotype in the F2 population. Similarly, we tested for Chr 14. Consistent with the backcross results there was no evidence for major effects on either chromosome and only a persistent trend for OuJ-derived susceptibility loci (Table 4).
Table 4.
Linear regression analysis of the frequency of SWD on Chr 16 and 14 in F2 mice
| F | χ2 | p-value | SWD/hr (mean ±S.E.M.) | |||
|---|---|---|---|---|---|---|
| A/A (n) | A/B (n) | B/B (n) | ||||
| D16Mit9 | 1.56 | 0.07 | 0.23 | 11.4±3.2 (8) | 9.9±3.0 (9) | 4.8±4.0 (4) |
| D16Mit152 | 1.78 | 0.08 | 0.20 | 12.0±2.8 (10) | - | 7.0±2.5 (12) |
| D14Mit14 | 0.05 | 0.002 | 0.82 | 10.9±4.7 (4) | 9.4±3.0 (8) | 8.3±3.3 (10) |
| D14Mit102 | 0.74 | 0.03 | 0.40 | 11.4±3.3 (8) | 8.4±2.8 (11) | 6.8±5.3 (3) |
A: OuJ allele, B: B6 allele
With similar rationale, the F2 population was analyzed non-parametrically for Chr 4 and Chr 18. Four of the 22 F2 mice showed a relatively high seizure incidence compared to the remainder (>20 SWD). Although it is difficult to reconcile the N2 and F2 incidences with a similar epistatic model, all F2 progeny were nevertheless genotyped for D4Mit54and D18Mit120 and an allele frequency test was again performed. In this analysis, a suggestive trend was noted on Chr 4, where all high-incidence mice had OuJ homozygous or heterozygous allele. Together with the non-parametric results from N2 progeny, this implies a potential involvement of Chr 4 in seizure incidence. However, considering the two datasets together, the overall evidence for trait loci remains suggestive at best.
Discussion
In 2005, it was reported that C3H/He substrains are prone to absence seizures (Frankel et al., 2005). Subsequently, a retroelement insertion mutation in the Gria4 gene was discovered as the major cause of SWD in the HeJ substrain (Beyer et al., 2008). While HeJ, OuJ and SnJ are genetically very similar, the genetic basis for SWD in these other He substrains was not directly assessed. The present study revealed that OuJ and SnJ substrains do not have the Gria4 mutation associated with their seizure phenotype, and genetic analysis provided only suggestive evidence for susceptibility loci of any kind. Although our study was not powered to detect minor locus effects to a high degree of confidence, it was larger than that used to initially implicate Gria4 as a candidate gene and to detect an epistatic interaction in HeJ mice. Therefore, we would have detected a major effect locus if it did exist. From the results, however, we conclude that the etiology of SWD in OuJ is most likely to be genetically complex, with multiple susceptibility loci interacting to yield a modest level of SWD.
Indeed, we found no evidence that Gria4 variants were involved in the OuJ phenotype. OuJ and SnJ substrains expressed normal amount levels of GRIA4 protein, and marker-phenotype assessment in N2 progeny derived from OuJ and B6 mice did not indicate any correlations – even suggestive ones - between SWD phenotype and Chr 9 near Gria4. Lastly, unlike the previous study involving HeJ in which it was likely that unmasking of recessive SWD resistance loci in HeJ led to many Gria4-deficient backcross progeny having a much higher SWD incidence than either parent, in the present study the phenotype of most OuJ backcross progeny was well within the range of the parents (Beyer et al., 2008, Frankel et al., 2005). This suggests that the innate effect of Gria4 deficiency, in the absence of resistance loci, is quite profound. In contrast, in the equivalent backcross for OuJ, we saw only suggestive correlations at best on Chr 5, 12, 14 and 16, and Chr 4 for high SWD incidence. Acknowledging the limitation of our study to confidently detect trait loci of small effects, these results suggest that there are multiple variants which together contrive to cause SWD in OuJ. We may infer, but we do not know formally, that additional substrains of C3H/He such as SnJ and HeN have the same etiology as OuJ.
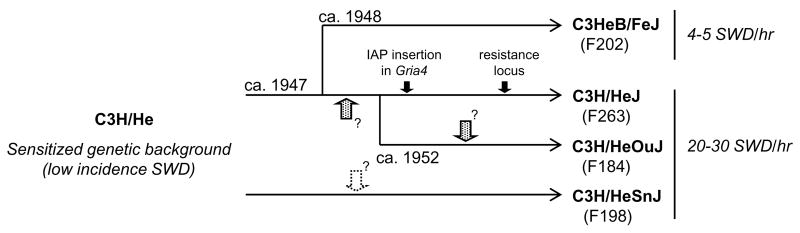
C3H/He is a general purpose inbred strain used in a wide variety of research areas. The HeJ substrain arose from progenitor C3H/He strain in 1947 and SnJ had been separated from C3H/He earlier than HeJ. HeJ and OuJ were separated in 1952 (Figure 6). Those substrains share the same genetic background and are well known to have a high incidence of mammary tumors caused by an exogenous mouse mammary tumor virus and also severe visual impairment in young adulthood caused by a retinal degeneration mutation (Fox, 1997., Bowes et al., 1993). On the other hand, several mutations which have become differentially fixed in these substrains have been reported. Perhaps the most famous is one that occurred in HeJ in 1960’s at the lipopolysaccharide response locus (in the toll-like receptor 4 gene, Tlr4lps) making HeJ mice endotoxin resistant while the other C3H strains are endotoxin sensitive (Poltorak et al., 1998). It seems likely that the progenitor strain already had a predisposition to SWD because at least four substrains, including C3HeB/FeJ, which was separated in 1948, exhibit at least some SWD. It is presumed that, during approximately 60 years (and some 200 generations) of divergence, the major-effect Gria4 mutation arose in HeJ and during the same period independent, but more minor-effect mutations arose in OuJ, resulting in the modest aggravation of its susceptibility for SWD. It is also possible that the minor mutations might occur before segregating of OuJ and are present in HeJ mice, their effects on phenotype being dwarfed by that of Gria4 deficiency, or that FeJ acquired resistance alleles that reduced SWD frequency. Regardless of the specifics, this study illustrates the general principle that even substrains with the same progenitor and with the same or very similar phenotypes, may have a different genetic basis.
Figure 6. Schematic figure of the genealogical tree for C3H/He substrains.
The C3H/He progenitor strain arose in 1942 from original the C3H inbred strain which had been derived from a cross of a Bagg albino female with a DBA male. C3H/HeJ (HeJ) was introduced to The Jackson Laboratory in 1947 and C3HeB/Fe was developed in 1948 by transfer of fertilized ova of HeJ to C57BL/6. HeJ and C3H/HeOuJ (OuJ) substrains were separated in 1952. C3H/HeSnJ (SnJ) arose from C3H/He prior to the separation of HeJ. HeJ, OuJ and SnJ show an average of 20–30 SWD/hr while C3HeB/FeJ shows 4–5 SWD/hr. The present generation of each substrain was from JAX® Mice database (http://jaxmice.jax.org, Jan 2008). Black arrows represent events of mutations identified in HeJ in the previous study (Frankel et al., 2005). Sparkled and white arrows represent hypothetical events of mutations in OuJ and SnJ, respectively.
In recent years, genetic study of human families has provided strong evidence of a genetic etiology in idiopathic generalized epilepsy (IGE), common forms of which are childhood absence epilepsy (CAE), juvenile absence epilepsy (JAE) and juvenile myoclonic epilepsy. Several potential loci and genes associated with IGE, such as CLCN2 (chloride channel 2) and CACNA1H (T-type voltage-dependent calcium channel α1H subunit), as reported in linkage analyses and genetic association studies (Chen et al., 2003, Crunelli & Leresche, 2002, Haug et al., 2003, Steinlein, 2004, Wallace et al., 2001). Replication studies, however, are often unsuccessful because of strong phenotypic and genotypic heterogeneity. Genetic animal models are therefore powerful complementary tools for studying complex disorders such as epilepsy, to understand the molecular and physiological bases, and to guide the development of novel antiepileptic drugs (Frankel, 1999, Futatsugi & Riviello, 1998, Smith et al., 2007). C3H substrains seem a particularly attractive mouse model for absence epilepsy because they do not have any other neurological symptoms or gross morphological changes in the brain (their retinal degeneration is a defect of the peripheral CNS). On the other hand, in behavioral tests C3H should be used with care given their visual impairment in addition to their absence seizure episodes (Schmidt & Lolley, 1973).
Despite the suggestive nature of the SWD loci we detected, it is noteworthy that mouse Chr 16 has conserved synteny with human Chr 21q22 where the GRIK1 gene (encoding the ionotropic glutamate receptor GluR5) is located. It has been reported that a polymorphism of GRIK1 was associated with JAE in family-based association analysis (Sander et al., 1997). Chr 14 also has conserved synteny with human Chr 14q23 where suggestive evidence for IGE has been found previously (Sander et al., 2000).
In conclusion, C3H strains appear to harbor a variety of susceptibility and modifier genes involved in absence seizures. Their further study has the potential to contribute to understanding the etiology of absence epilepsy.
Supplementary Material
Acknowledgments
We thank Dr. Tatyana Golovkina for her gift of C3H/HeN DNA and Dr. Verity Letts for providing protein expression data. We also thank Drs. Greg Cox and Letts for comments on this manuscript and Mrs. Carolyne Dunbar for technical assistance. This work was supported by NIH grant NS31348 to WNF. ST was supported by a postdoctoral fellowship from The Jackson Laboratory training program.
References
- Barclay J, Balaguero N, Mione M, Ackerman SL, Letts VA, Brodbeck J, Canti C, Meir A, Page KM, Kusumi K, Perez-Reyes E, Lander ES, Frankel WN, Gardiner RM, Dolphin AC, Rees M. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. J Neurosci. 2001;21:6095–6104. doi: 10.1523/JNEUROSCI.21-16-06095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer B, Deleuze C, Letts VA, Mahaffey CL, Boumil RM, Lew TA, Huguenard JR, Frankel WN. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes C, Li T, Frankel WN, Danciger M, Coffin JM, Applebury ML, Farber DB. Localization of a retroviral element within the rd gene coding for the beta subunit of cGMP phosphodiesterase. Proc Natl Acad Sci U S A. 1993;90:2955–2959. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL, Jones JM, Meisler MH, Noebels JL. Mutation of the Ca2+ channel beta subunit gene Cchb4 is associated with ataxia and seizures in the lethargic (lh) mouse. Cell. 1997;88:385–392. doi: 10.1016/s0092-8674(00)81877-2. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lu J, Pan H, Zhang Y, Wu H, Xu K, Liu X, Jiang Y, Bao X, Yao Z, Ding K, Lo WH, Qiang B, Chan P, Shen Y, Wu X. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–243. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- Cox GA, Lutz CM, Yang CL, Biemesderfer D, Bronson RT, Fu A, Aronson PS, Noebels JL, Frankel WN. Sodium/hydrogen exchanger gene defect in slow-wave epilepsy mutant mice. Cell. 1997;91:139–148. doi: 10.1016/s0092-8674(01)80016-7. [DOI] [PubMed] [Google Scholar]
- Crunelli V, Leresche N. Childhood absence epilepsy: genes, channels, neurons and networks. Nat Rev Neurosci. 2002;3:371–382. doi: 10.1038/nrn811. [DOI] [PubMed] [Google Scholar]
- Fletcher CF, Lutz CM, O’Sullivan TN, Shaughnessy JD, Jr, Hawkes R, Frankel WN, Copeland NG, Jenkins NA. Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell. 1996;87:607–617. doi: 10.1016/s0092-8674(00)81381-1. [DOI] [PubMed] [Google Scholar]
- Fox RR, Witham Barbara A, Neleski Linda A. Handbook on genetically standardized JAX mice. Bar Harbor: ME Jackson Laboratory; 1997. [Google Scholar]
- Frankel WN. Detecting genes in new and old mouse models for epilepsy: a prospectus through the magnifying glass. Epilepsy Res. 1999;36:97–110. doi: 10.1016/s0920-1211(99)00044-3. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Beyer B, Maxwell CR, Pretel S, Letts VA, Siegel SJ. Development of a new genetic model for absence epilepsy: spike-wave seizures in C3H/He and backcross mice. J Neurosci. 2005;25:3452–3458. doi: 10.1523/JNEUROSCI.0231-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futatsugi Y, Riviello JJ., Jr Mechanisms of generalized absence epilepsy. Brain Dev. 1998;20:75–79. doi: 10.1016/s0387-7604(97)00107-1. [DOI] [PubMed] [Google Scholar]
- Haug K, Warnstedt M, Alekov AK, Sander T, Ramirez A, Poser B, Maljevic S, Hebeisen S, Kubisch C, Rebstock J, Horvath S, Hallmann K, Dullinger JS, Rau B, Haverkamp F, Beyenburg S, Schulz H, Janz D, Giese B, Muller-Newen G, Propping P, Elger CE, Fahlke C, Lerche H, Heils A. Mutations in CLCN2 encoding a voltage-gated chloride channel are associated with idiopathic generalized epilepsies. Nat Genet. 2003;33:527–532. doi: 10.1038/ng1121. [DOI] [PubMed] [Google Scholar]
- Huguenard JR, McCormick DA. Thalamic synchrony and dynamic regulation of global forebrain oscillations. Trends Neurosci. 2007;30:350–356. doi: 10.1016/j.tins.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett FS, 2nd, Mori Y, Campbell KP, Frankel WN. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Sander T, Hildmann T, Kretz R, Furst R, Sailer U, Bauer G, Schmitz B, Beck-Mannagetta G, Wienker TF, Janz D. Allelic association of juvenile absence epilepsy with a GluR5 kainate receptor gene (GRIK1) polymorphism. Am J Med Genet. 1997;74:416–421. [PubMed] [Google Scholar]
- Sander T, Schulz H, Saar K, Gennaro E, Riggio MC, Bianchi A, Zara F, Luna D, Bulteau C, Kaminska A, Ville D, Cieuta C, Picard F, Prud’homme JF, Bate L, Sundquist A, Gardiner RM, Janssen GA, de Haan GJ, Kasteleijn-Nolst-Trenite DG, Bader A, Lindhout D, Riess O, Wienker TF, Janz D, Reis A. Genome search for susceptibility loci of common idiopathic generalised epilepsies. Hum Mol Genet. 2000;9:1465–1472. doi: 10.1093/hmg/9.10.1465. [DOI] [PubMed] [Google Scholar]
- Schmidt SY, Lolley RN. Cyclic-nucleotide phosphodiesterase: an early defect in inherited retinal degeneration of C3H mice. J Cell Biol. 1973;57:117–123. doi: 10.1083/jcb.57.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Wilcox KS, White HS. Discovery of antiepileptic drugs. Neurotherapeutics. 2007;4:12–17. doi: 10.1016/j.nurt.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinlein OK. Genetic mechanisms that underlie epilepsy. Nat Rev Neurosci. 2004;5:400–408. doi: 10.1038/nrn1388. [DOI] [PubMed] [Google Scholar]
- Tan HO, Reid CA, Single FN, Davies PJ, Chiu C, Murphy S, Clarke AL, Dibbens L, Krestel H, Mulley JC, Jones MV, Seeburg PH, Sakmann B, Berkovic SF, Sprengel R, Petrou S. Reduced cortical inhibition in a mouse model of familial childhood absence epilepsy. Proc Natl Acad Sci U S A. 2007;104:17536–17541. doi: 10.1073/pnas.0708440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG, Williams DA, Sutherland GR, Mulley JC, Scheffer IE, Berkovic SF. Mutant GABA(A) receptor gamma2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet. 2001;28:49–52. doi: 10.1038/ng0501-49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.