Abstract
Tumor-associated macrophages (TAMs) invade the tumor stroma in many cancers, yet their role is incompletely understood. To visualize and better understand these critical cells in tumor progression, we screened a portfolio of rationally selected, injectable agents to image endogenous TAMs ubiquitously in three different cancer models (colon carcinoma, lung adenocarcinoma, and soft tissue sarcoma). AMTA680, a functionally derivatized magneto-fluorescent nanoparticle, labeled a subset of myeloid cells with an “M2” macrophage phenotype, whereas other neighboring cells, including tumor cells and a variety of other leukocytes, remained unlabeled. We further show that AMTA680-labeled endogenous TAMs are not altered and can be tracked noninvasively at different resolutions and using various imaging modalities, e.g., fluorescence molecular tomography, magnetic resonance imaging, and multiphoton and confocal intravital microscopy. Quantitative assessment of TAM distribution and activity in vivo identified that these cells cluster in delimited foci within tumors, show relatively low motility, and extend cytoplasmic protrusions for prolonged physical interactions with neighboring tumor cells. Noninvasive imaging can also be used to monitor TAM-depleting regimen quantitatively. Thus, AMTA680 or related cell-targeting agents represent appropriate injectable vehicles for in vivo analysis of the tumor microenvironment.
Introduction
Tumor-associated macrophages (TAMs) are bone marrow-derived mononuclear phagocytes that invade the tumor stroma in many cancers, participate in tumor invasion, angiogenesis and metastasis formation [1–5], and can modulate tumor-specific T-cell immunity [6,7]. The accumulation of TAMs is therefore generally associated with a poor prognosis [8], whereas their removal in animal models can lead to tumor regression [9]. Tumor-associated macrophages rely on migration and cell-cell contact for information transfer, yet most of our knowledge on the function mediated by these cells have been obtained ex vivo through inference from static snapshots. Also, surrogate in vitro culture models do not faithfully reproduce the behavior of immune cells in tissues [10]. Novel in vivo imaging technologies have recently made it possible to quantify and catalog the behavior and function of some immune cells that control tumor growth [11]. For example, elegant work on tumor cell invasion in animal models of breast cancer has revealed that perivascular TAMs recruit motile tumor cells and promote their exit into blood vessels [3,12]. In vivo studies have also shown that TAMs undergo long-lasting interactions with tumor-infiltrating lymphocytes [13] and thus may supply a variety of immunosuppressive signals that attenuate antitumor T cell activity [14,15]. These and other studies have used genetically modified mice expressing fluorescent reporters (e.g., enhanced green fluorescent protein [EGFP]) that can be tracked with appropriate optical imaging technologies. The genetic reporter approach is particularly useful for the study of cells at microscopic resolution; however, currently, it does not allow adequate imaging in larger fields of view (e.g., at mesoscopic and macroscopic scales), at increased depths (e.g., >600 µm), or in human patients with cancers.
Injectable multimodal imaging agents that are specific for molecular targets are an attractive alternative because they offer the advantage of being usable in both experimental animals and humans, they can carry multiple reporters for imaging at different resolutions and depths (e.g., fluorochromes, quantum dots, [super]paramagnetic particles, radionuclides), and they can combine diagnostic and therapeutic interventional capabilities [16–23]. Such agents exist for a small minority of targets of interest in cancer and are highly suitable for in vitro analysis, but their in vivo behavior (e.g., target selectivity, biocompatibility) is generally not fully characterized. Here, we developed and validated injectable imaging agents that can be used to target endogenous TAMs selectively. Such agents may be valuable for understanding the biology of these cells and may have important prognostic and therapeutic implications in cancer and other inflammatory diseases.
We previously tested a library of 146 nanoparticles decorated with different synthetic small molecules [18] and several protease-targeted optical sensors [24] for their capacity to target macrophages in vitro. Here, we rationally selected and extensively tested four lead candidates for their ability to selectively label TAMs in vivo. Two of these agents are cross-linked iron oxide (CLIO) magnetofluorescent nanoparticles (CLIO680 and AMTA680) with affinity for TAMs [18]. AMTA680 is decorated with N-terminal glycines and was identified among a library of modified CLIO nanoparticles with high affinity for activated macrophages in vitro [18]. The two other agents are enzyme-targeted optical sensors (ProSense680 and MMPSense680) that are activated by cathepsins and matrix metalloproteinases, respectively, expressed by TAMs [24,25]. We evaluated these agents in three animal models: a subcutaneously injected mouse colon carcinoma [26] and two genetically inducible mouse models that recapitulate either human lung adenocarcinoma or soft tissue sarcoma. Mice with conditional mutation in the endogenous Kras gene (LSL-KrasG12D mice) develop lung adenocarcinoma after intranasal instillation of an adenovirus expressing Cre recombinase [27], whereas mice with conditional mutations in the endogenous Kras and p53 genes (LSL-KrasG12D;LSL-p53) develop soft tissue sarcoma after intramuscular adenovirus injections [28]. The technique presented here profiles the cellular uptake of imaging agents in vivo, identifies one of them with specificity for a subset of TAMs within the tumor microenvironment, and demonstrates the efficient detection of endogenous cells by means of complementary microscopic, mesoscopic, and macroscopic imaging approaches. Labeling and imaging of endogenous TAMs allowed us to monitor TAMs-tumor cells interactions and to topograph TAMs infiltrates in the tumor microenvironment.
Materials and Methods
Animal Models
Three murine tumor models were used for these studies.
Lung adenocarcinoma: Mice with conditional mutation in the endogenous Kras gene (LSL-KrasG12D mice) received intranasal instillation of an adenovirus expressing Cre recombinase to generate a temporally and spatially restricted model of lung adenocarcinoma [27].
Soft tissue sarcoma: Mice with conditional mutations in the endogenous Kras and p53 genes (LSL-KrasG12D;LSL-p53) received intramuscular injections of the adenovirus mentioned previously to generate a model of soft tissue sarcoma [28].
Syngeneic tumor model: Female BALB/c mice, 6 to 8 weeks old, received 1 x 106 CT26 or CT-26-GFP colon carcinoma cells subcutaneously into the upper side of the hind paw in 50-µl saline or intradermally in a dorsal skin window chamber in 20-µl saline as previously described [29].
The Massachusetts General Hospital Subcommittee on Research Animal Care approved all in vivo protocols and procedures.
Imaging Agents
From a library of 146 nanoparticles decorated with different synthetic small molecules [18] and protease-targeted optical sensors [24,30], four agents were selected because of their TAMs imaging potential and because they are close to clinical testing. These agents are ProSense680, MMPSense680, CLIO680, and AMTA680. All contain the fluorochrome VT680. AMTA680 is a derivative of CLIO680 decorated with N-terminal glycines conferring a negative Z-potential (ZAMTA = -24.5, ZCLIO = 6.38). We injected 2 nmol of ProSense680 or MMPSense680 (VisEn Medical, Woburn, MA). The probes generate far-red fluorescence when cleaved by cathepsins or matrix metalloproteinases, respectively. We also injected CLIO680 and AMTA680 (12.5-mg iron/kg), which are dextran-coated iron oxide core nanoparticles coupled with VT680. All probes were injected 24 hours before imaging unless otherwise stated. We also used AngioSense (VisEn Medical) for visualization of blood vessels, and CLIO, AMTA, and VT680 NHS ester [29] were used as control agents.
Flow Cytometry
Footpad, lung, and sarcoma tumors were excised and digested for 30 minutes at 37°C in RPMI medium completed with 0.2 mg/ml collagenase IV (BoehringerMannheim, Indianapolis, IN) and 0.04 mg/ml DNAse1 (Roche Molecular Biochemicals, Indianapolis, IN). Spleens were manually homogenized. The digested tumors and the processed spleens were filtered through a 70-µm cell strainer to obtain single-cell suspensions and were treated with an ACK buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2-EDTA, pH 7.3) to lyse red blood cells. Cells were either labeled with anti-CD11b-APC alone or together with CD90-PE, B220-PE, CD49b-PE, NK1.1-PE, and Ly-6G-PE (BD Biosciences, San Jose, CA) for 30 minutes at 4°C as described [24,31] and analyzed using an LSRII or sorted using FACSAria (both from BD Biosciences). Excitation and detection of the VT680 fluorochrome used a red helium-neon 635 nm laser and a filter combination of 685/LP and a 695/40BP. For comparison of fluorescence intensity measurements across studies, we calibrated the flow cytometer with RFP-30-5A rainbow particles (Spherotech, Lake Forest, IL) before each acquisition. Aliquots of 50,000 cells were sorted and morphologically characterized after staining with HEMA 3 (Biochemical Sciences, Bridgeport, NJ). Trypan blue exclusion staining was used to estimate the number of living cells.
Microscopic Imaging
We used a dorsal skin fold chamber model [29] for in vivo monitoring of AMTA680 uptake at microscopic resolution. After 7 days of tumor growth, AngioSense-584 and AMTA680 were administered intravenously, and mice were analyzed immediately by multiphoton microscopy (Prairie Technologies ULTIMA IV equipped with a 20x/0.95 NA objective lens and two Spectra-Physics Ti-S laser). For fluorescence excitation, the coaxially aligned beams of a DeepSee and a MaiTai HP Ti:Sapphire laser (both Spectra-Physics/Newport, Mountain View, CA) were individually tuned to achieve optimal excitation of all fluorochromes of the particular experiment. Typically, the excitation wavelengths used were 845 and 920 nm. Settings were optimal for dual observation of AMTA680 and tumor cells (EGFP) as described previously [29]. Stacks of 10 optical sections were acquired every 30 seconds for 4 hours to provide image volumes of 50 µm in depth and 500 µm in width and length for four-dimensional recordings of AMTA680 uptake. Some mice were also imaged by confocal microscopy using a Biorad Radiance 2100 multichannel laser scanning microscope (Bio-Rad, Hercules, CA) installed on a Nikon Eclipse E600 microscope (Nikon Instruments, Melville, NY) with a 20x water immersion objective (Nikon Fluor 20x/0.5 NA) and the Lasersharp 2000 software. Individual channels were scanned line by line separately to avoid bleed through between channels. A four-line Argon laser (457, 477, 488, and 514 nm) and a red laser diode (637 nm) were used for EGFP and AMTA680 excitation. The 560 DCLP and 650 DCLP dichroic mirrors and HQ515/30 (EGFP) and HQ660LP (VT680) emission filters were used to collect light from different channels. Four-dimensional recording were made by accumulating z-stacks of 11 images at 4-µm intervals along the z-axis at 15-second intervals for 45 minutes. z-Stacks were projected into a single plane using maximum-intensity projection for the final movie. Images are 256 x 256 pixels, and movies are shown at 20 images/sec, which corresponds to 300 times faster speed.
Macroscopic Imaging
Mice received a nonfluorescent/reduced manganese diet (Harlan-Teklad, Indianapolis, IN) 5 days before imaging and were shaved and depilated. Imaging was performed 24 hours after probe administration, unless otherwise stated. Fluorescence molecular tomography (FMT) used an FMT or an FMT2500 system (VisEn Medical) for excitation and emission scans at 670 and 700 nm, respectively. Anesthetized mice were maintained in a fixed position within an imaging cartridge to allow point-based image fusion. Reconstruction of three-dimensional maps of fluorochromes was done as described [32], and data were expressed as mean fluorescent concentration. Magnetic resonance imaging (MRI) used a BioSpin 7T (Bruker BioSpin, Billerica, MA) with T2-weighted RARE sequence, echo time = 48 milliseconds, repetition time = 2000 milliseconds, and number of averages = 8. Eighteen coronal images were acquired with a matrix size of 192 x 256 and 0.195 x 0.260 x 1-mm voxels. Baseline control images for MRI and FMT were acquired before AMTA680 injection. The FMT-MRI image fusion used the Tudor DICOM ImageJ plugin (http://imagejdocu.tudor.lu/imagej-documentation-wiki/plugins/the-tudor-dicom-toolkit) to produce reflectance-fluorescence fused FMT DICOM images, dicomlib (http://dicomlib.swri.ca/dicomlib.html) to generate adequate headers, and Osirix 3 (The Osirix Foundation, Geneva, Switzerland) for point-based registrations. Quantification of cell motility and interaction time was calculated as described [33].
Immunohistochemistry
CT26 tumors excised on day 7 were frozen in OCT compound (Sakura Finetek, Torrance, CA) and sectioned in 5-µm slices. Adjacent sections were incubated with 0.3% hydrogen peroxide to inhibit endogenous peroxidase activity and incubated with rat antimouse CD11b (BD Pharmingen, San Diego, CA) or NIMP-R14 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) antibody. Sections were washed with PBS, labeled with biotinylated antirat immunoglobulin G antibody (Vector Laboratories, Inc, Burlingame, CA) and with an avidin-peroxidase complex (Vectastain ABC Kit; Vector Laboratories). Reactions were visualized with 3-amino-9-ethyl carbazole substrate (AEC; Sigma Chemical, Co, St. Louis, MO). Sections were counter-stained with Mayer's hematoxylin solution (Sigma) and mounted. Images were captured on a digital camera (Nikon DXM1200-F; Nikon Inc) with ACT-1 imaging version 2.63 software. Fluorescence microscopy visualized simultaneously 1) AMTA680, 2) NIMP-R14+ cells or CD11b+ cells labeled with fluorescein-conjugated streptavidin (GE Healthcare Bio-Sciences, Piscataway, NJ), and 3) 4′-6′-diamidino-2-phenylindole (DAPI; Vector Laboratories). Images were captured and processed with an epifluorescence microscope (Eclipse 80i; Nikon Instruments) equipped with a cooledCCDcamera (Cascade; Photometrics, Tucson, AZ).
Real-time Polymerase Chain Reaction
RNA extraction was performed using the RNAeasy Kit (Qiagen, Valencia, CA) and with flow-sorted cells from spleen or tumor on day 14 after tumor implantation or from spleen of tumor-free mice. Complementary DNA preparation and quantitative polymerase chain reactions were performed using TaqMan gene expression assays according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). Mean normalized gene expression (“deltaCt”) was calculated from two independent experiments using 18s RNA as a reference gene and expression values for TAMs were set to 1. Assays used interleukin 4 (IL-4) Mm00445259_m1, IL-10 Mm00439616_m1, Tie2 Mm00443242_m1, VEGFa Mm00437304_m1, and EGF Mm00438696_m1.
Statistics
Experiments were performed at least in triplicate (except for sarcoma) and representative data are shown. Results were tested for statistical significance using Student's t test withWelch's correction when two variables were measured, whereas three variables or more were tested with one-way analysis of variance with subsequent Tukey tests of each pairs with GraphPad Prism version 4.0a software (Graph Pad Software, La Jolla, CA).
Results
A Screening Method for Evaluating Injectable Imaging Agents
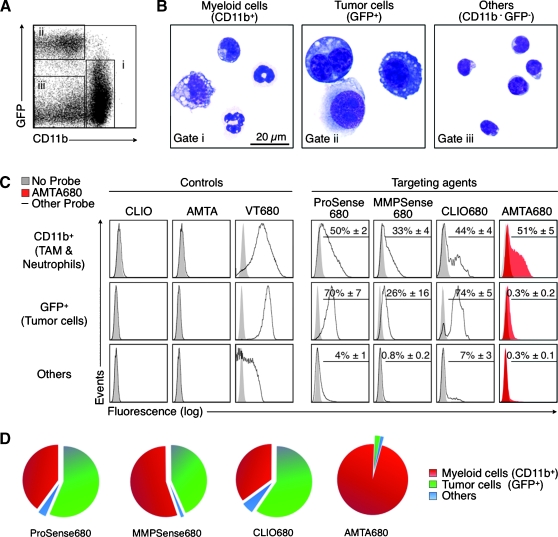
From an initial library of ∼150 injectable imaging agents [18,24,30], we rationally selected ProSense, MMPSense, CLIO, and AMTA based on their capacity to target macrophages in vitro and tested their cellular uptake in vivo in BALB/c mice bearing carcinoma-derived CT26-GFP tumor cells. All agents contained the far-red fluorochrome VT680 [29]. We administered each agent intravenously in separate mice 6 days after tumor inoculation and used flow cytometry on day 7 to analyze single-cell suspensions obtained from excised tumors. Myeloid cells were defined as CD11b+GFP-, tumor cells as CD11b-GFP+, and “other” cells as CD11b-GFP- (Figure 1A). Microscopic examination of flow-sorted cell subsets confirmed their morphological features: CD11b+ cells showed heterogeneous myeloid-like phenotypes typical of neutrophils or TAMs; GFP+ cells showed large, polymorphic, and sometimes polynucleated attributes distinguishing tumor cells; CD11b-GFP- cells consisted mainly of smaller cells with round nuclei suggesting lymphocytes (Figure 1B). Among the four agents tested, AMTA680 was the only one with specificity for myeloid cells versus tumor cells (P < .001). ProSense680 and CLIO680 showed higher uptake in tumor cells than in myeloid cells (P < .05 and P < .01, respectively), whereas we detected similar uptake of MMPSense680 by tumor cells and myeloid cells, although we noted highly variable fluorescence intensity in tumor cells (Figure 1C). Nonfluorescent nanoparticles (CLIO and AMTA) served as negative controls. The NHS-ester VT680 alone (i.e., fluorochrome not attached to a nanoparticle) served as an additional control (Figure 1C), as it is known to covalently label virtually all cells nonspecifically [29]. A pie chart representation of the percentage of each probe-positive population shows the difference of cellular uptake pattern for AMTA680 compared with the other imaging agents (Figure 1D). Our data also indicate that AMTA680 is biocompatible because myeloid cells in mice injected with the agent remained viable, in same numbers, and retained their forward and side scatter profiles and overall surface phenotype (Figure W1). Similar profiling in genetically induced lung adenocarcinoma and soft tissue sarcoma confirmed the specificity of AMTA680 for myeloid cells (Figure 2E). AMTA680 also labels macrophages in tumor-free tissues, e.g., the spleen (Figure W2).
Figure 1.
In vivo targeting of endogenous tumor-associated myeloid cells with AMTA680. (A) Flow cytometric analysis of cell suspensions obtained from excised tumors and stained with anti-CD11b mAb (tumor cells stably express EGFP) identifies myeloid cells (i), tumor cells (ii), and “other” cells (iii), n = 9. (B) Morphological analysis of sorted cells. (C) Analysis of tumor cell suspensions 24 hours after intravenous administration of “control” (CLIO, AMTA, VT680) or “targeting” agents (ProSense680, MMPSense680, CLIO680, and AMTA680). Each histogram shows a representative example of the fluorescence intensity (uptake) of each agent for each cell type. Figures in the upper right of the histograms show the percentage (mean ± SEM) of cells labeled with the agent when compared with the corresponding cells in mice that did not receive any agent. All agents but AMTA680: n = 3; AMTA680: n = 27. (D) Distribution of the targeting agents into different cell types that are present in the tumor environment. The analysis shows the specificity of AMTA680 (but not of the other agents) for tumor-associated myeloid cells.
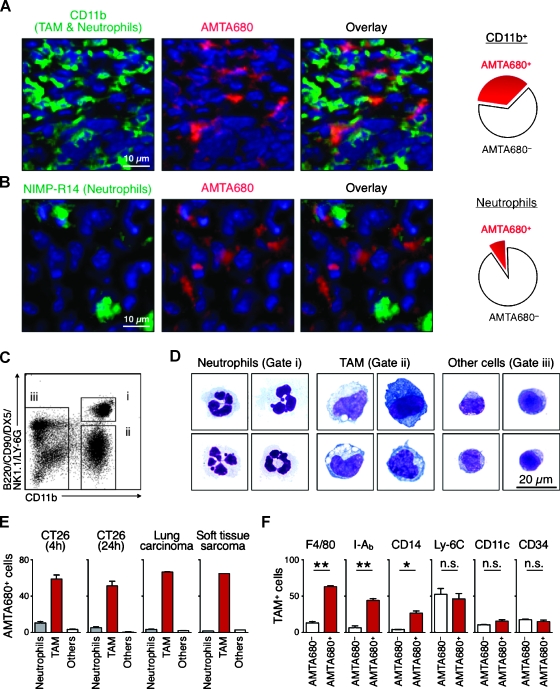
Figure 2.
AMTA680 preferentially labels TAMs with an “M2-like” phenotype. Immunofluorescence analysis of frozen sections from nonfluorescent CT26 tumors labeled with anti-CD11b (A) or NIMP-R14 (B) mAb. DAPI is shown in blue. During image acquisition, the signal integration times required to detect AMTA680 in (B) were an order of magnitude lower than in (A) suggesting that the few AMTA680-positive neutrophils contain less AMTA680 than TAMs, n = 3. (C) Flow cytometry analysis of cell suspensions of excised tumors identifies neutrophils (i), TAMs (ii), and other cells (iii). (D) Morphological analysis of sorted cells, n = 3. (E) Percentage of different cell types labeled with AMTA680 24 hours after administration of the agent in mice with subcutaneous CT26 (n = 4), lung carcinoma (n = 3), or soft tissue sarcoma (n = 1). Mice with subcutaneous CT26 were also tested after 4 hours (n = 4). (F) Surface phenotype of AMTA680+ and AMTA680- TAMs obtained from CT26 tumors and analyzed by flow cytometry 24 hours after administration of the agent. *P < .005, **P < .0005, n = 3.
AMTA680 Labels TAMs with an “M2” Phenotype
The screening described above identified AMTA680 as a myeloid cell-targeting agent. Histological analysis of CT26 tumor tissue sections confirmed that AMTA680 accumulates in CD11b+ myeloid cells but not in tumor cells (Figure 2A) and further identified that CD11b+ NIMP-R1+ neutrophils do not accumulate the imaging agent to a significant amount (Figure 2B). Control experiments included frozen sections frommice that did not receive AMTA680 and unstained frozen sections (data not shown). To quantify precisely the distribution of AMTA680 within various myeloid cells, we used a flow cytometry approach (Figure 2C) previously validated for simultaneous analysis of neutrophils (CD11b+ (B220/CD90/DX5/NK1.1/Ly-6G)hi cells) versus macrophages and dendritic cells (CD11b+ (B220/CD90/DX5/NK1.1/Ly-6G)lo cells) [24]. Microscopic examination of flow-sorted cell subsets confirmed the morphological features of neutrophils and TAMs, whereas the “other cells” were predominantly lymphocytes (Figure 2D). Analysis by flow cytometry revealed that AMTA680 accumulates much more efficiently in TAMs than in lymphocytes (P < .001) and neutrophils (P < .001). AMTA680 had accumulated in TAMs at 4 and 24 hours after injection and in all models tested (e. g., CT26 tumors as well as genetically induced lung adenocarcinoma and soft tissue sarcoma; Figures 2E and W3). Immunofluorescence analysis in the last two models also confirmed the specificity of AMTA680 for TAMs but not for tumor cells (Figure 4E for soft tissue sarcoma, and data not shown for lung adenocarcinoma). Ex vivo analysis of TAMs purified from implanted CT26 tumors showed selective AMTA680 labeling of a subset of TAMs with a F4/80hi I-Abhi CD14lo phenotype, whereas AMTA680- cells were mostly F4/80lo I-Ablo CD14- (Figure 2F). In addition, the labeled cells were mostly CD34- and CD11c- and thus contained neither stem cells nor dendritic cells. The TAMs also showed increased messenger RNA levels of proangiogenic (Tie-2, vascular endothelial growth factor [VEGF], endothelial growth factor [EGF]) and immunosuppressive (IL-4, IL-10) genes, when compared with splenic monocytes purified from non-tumor- or tumor-bearing mice (Figure W4). We obtained comparable results for TAMs purified from mice with lung adenocarcinoma (data not shown). Thus, AMTA680-labeled cells represent mostly F4/80hi “M2-like” tissue macrophages but not “M1” macrophages or more immature myeloid-derived suppressor cells [1].
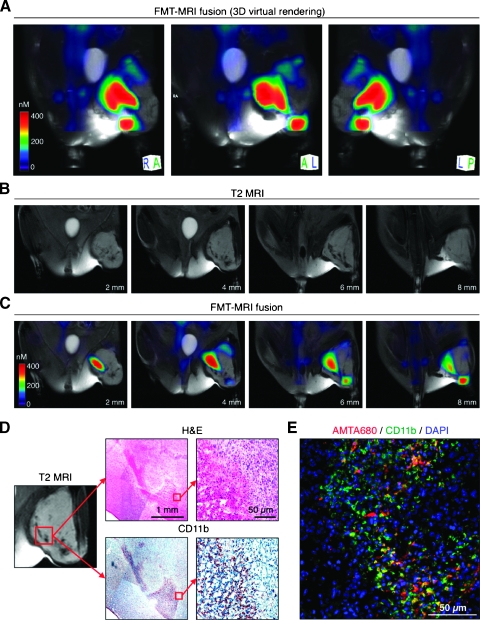
Figure 4.
Macroscopic imaging of TAMs distribution. A mouse with soft tissue sarcoma in the left crural muscle received AMTA680 intravenously and was analyzed 24 hours later with the following imaging modalities: (A–C) FMT, MRI, and FMT-MRI fusion. (A) Three-dimensional virtual rendering of TAMs using FMT-MRI fusion. The color-coded optical images use advanced algorithms [32] to reconstruct and quantitate three-dimensional maps of AMTA680. (B) Different sections of T2-weighted MRIs generated by the iron oxide superparamagnetic core of AMTA680. (C) Corresponding FMT-MRI fused sections. (D) Hematoxylin/eosin (H&E) and anti-CD11b staining in an AMTA680 rich region of the tumor as identified by MRI (and FMT). (E) Immunofluorescence analysis for colocalization of AMTA680+ and CD11b+ cells.
Microscopic In Vivo Imaging of TAMs
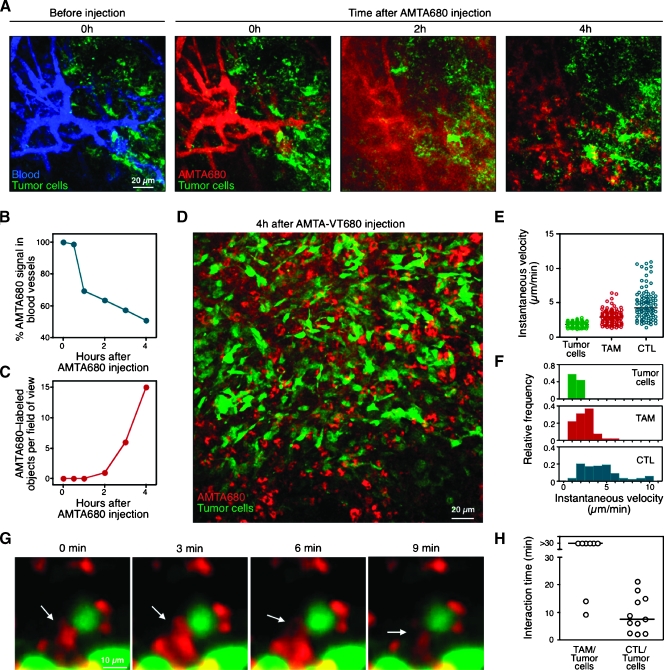
Having established that AMTA680 selectively labels endogenous TAMs, we set out in situ imaging approaches to study these cells in their native environment. Initially, we used multiphoton and confocal intravital microscopy to image the tumor microenvironment at different time points as a way to determine the distribution and cellular uptake of the agent over time. This involved surgical implantation in the dorsum of the animal of a tumor-window chamber containing CT26-GFP tumor cells [29], followed on day 7 by intravenous injection of AMTA680 and a blood pool agent. Immediately on intravenous injection, AMTA680 distributed in the tumor vasculature (Figure 3A), whereas within 4 hours, a significant fraction of the agent had spread into the tumor parenchyma (Figure 3B) and integrated in the cytoplasm of single cells (Figure 3, C and D). We could detect AMTA680-labeled TAMs efficiently by both multiphoton and confocal microscopy when using appropriate excitation and detection wavelengths (Materials and Methods). Next, we performed time-lapse intravital confocal microscopy to derive quantitative parameters of TAMs migration and their physical interactions with surrounding cellular partners. Measures of instantaneous velocities of cell centroids revealed that both tumor cells and TAMs are relatively immotile (Figure 3, E and F). We compared the velocity of endogenous TAMs to the one of HA-specific effector cytotoxic T lymphocytes (CTLs) that were also recruited to the tumor stroma and confirmed that CTLs are typically faster, as reported previously [10,13,34]. We also found that stationary TAMs polarize and present cytoplasmic protrusions that physically interact with neighboring tumor cells for extended periods (typically >30 minutes; Figure 3, G and H, see also Video W1).
Figure 3.
Microscopic imaging of TAMs behavior. (A) Time-course analysis of extravasation and cellular uptake of AMTA680. CT44-GFP tumor-bearing mice received intravenous injection of a blood pool agent (AngioSense) to delineate vessels (blue, left panel) and AMTA680 (red, adjacent panels). Multiphoton microscopy images of the same tumor region were recorded every hour. Tumor cells are also shown (green). (B and C) Change in the percentage of AMTA680 fluorescence in blood vessels (B) and in the number of AMTA680-labeled extravascular objects (C) over time. (D) Overview of a TAMs-rich region labeled with AMTA680 and recorded 4 hours after injection. (E) Instantaneous velocity of tumor cells, TAMs, and tumor antigen-specific CTLs in the tumor microenvironment. Cytotoxic Tlymphocyte specific for the HA antigen (that is expressed by CT44-GFP tumors) were stimulated in vitro with cognate peptide and IL-2 and injected intravenously as previously described [13]. Tumors: n = 11, TAMs: n = 109, CTLs: n = 91. (F) Instantaneous velocity of cells plotted in 1-µm/min bins. (G) Physical interactions between TAMs (red) and tumor cells (green). (H) Long-lasting physical interactions between TAMs and tumor cells (n = 8) in comparison to the ones observed between CTLs and tumor cells (n = 11).
Macroscopic In Vivo Imaging of TAMs
We then tested whether mesoscopic and macroscopic imaging modalities, e.g., FMT, MRI, and FMT-MRI fusion imaging, allow to assess the in vivo distribution of AMTA680-labeled TAMs at the whole tumor (or body) level. To this end, we used a LSL-KrasG12D-p53-/- mouse ∼25 weeks after initiation of soft tissue sarcoma in the left crural muscle [28] and imaged the mouse 24 hours after administration of AMTA680. Fluorescence molecular tomography allowed us to reconstruct and quantitate three-dimensional maps of AMTA680 and to demonstrate well-delimited signal foci within the tumor (Figure 4A). Magnetic resonance imaging also identified submillimeter foci of hyposignal (black) on T2-weighted images generated by the iron oxide superparamagnetic core of AMTA680 (Figure 4B and Video W2). Point-based fusion of FMT and MRI data sets revealed colocalization of the signals obtained from both modalities (Figure 4C).
Next we performed hematoxylin-eosin and CD11b staining in a section of the tumor matching the AMTA680 signal observed by FMT-MRI. The overall morphology of the lesion and presence of CD11b+ cells confirmed the presence of a dense infiltrate of myeloid cells where AMTA680 signal was detected (Figure 4D). Immunofluorescence analysis demonstrated further that AMTA680 accumulated selectively in the myeloid cell-rich regions (Figure 4E). Thus, the combination of AMTA680 with whole-body noninvasive imaging techniques such as FMT and MRI permits to assess the distribution of AMTA680-labeled TAMs noninvasively and in three dimensions in vivo. These macroscopic approaches for analysis of large fields of view and deep in tissue complement single-cell confocal and multiphoton microscopy techniques.
In Vivo Imaging of Anti-TAMs Treatment
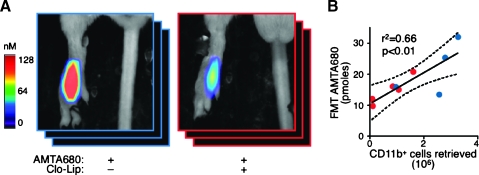
Imaging of TAMs can have important prognostic and therapeutic implications [3,35]. In addition, targeting TAMs is a possible strategy against cancer, and tools to measure the efficiency of TAMs depletion in vivo would be useful. Here, we tested the ability of AMTA680 and FMT to assess the depletion of endogenous TAMs in mice that received 0.1 ml of clodronate-loaded liposomes (Clo-Lip). CT26 tumor-bearing mice received Clo-Lip and AMTA680 on day 6 and were analyzed by FMT on day 7. Flow cytometry analysis revealed that treatment with Clo-Lip reduced the number of CD11b+ cells retrieved from the tumor site. Noninvasive FMT imaging also showed reduced TAMs-associated AMTA680 activity in lesions (Figure 5A and Video W3), and the number of retrieved CD11b+ cells positively and linearly correlated with in vivo AMTA680 activity (Figure 5B: blue dots, mice untreated; red dots, mice treated with Clo-Lip). Thus, FMT can noninvasively inform on quantitative alterations of TAMs populations over time.
Figure 5.
In vivo imaging of anti-TAMs treatment. (A) In vivo FMT imaging of AMTA680-derived signal from the tumor site of CT26 tumor-bearing mice that received Clo-Lip on day 6 (red) or not (blue). All mice received AMTA680 intravenously on day 6 and were imaged by FMT on day 7. Clo-Lip-treated mice: n = 5, control mice: n = 4. (B) Positive and linear relationship between FMT-based AMTA680-derived signal and the number of CD11b+ cells retrieved from excised tumors and analyzed by flow cytometry.
Discussion
Understanding the function and activity of endogenous immune cells within complex biological systems necessitates the development of in vivo imaging technologies that are cell-specific and quantitative. In this study, we set up a screening method of injectable imaging agents to assess cellular specificity and to quantify cellular uptake. We show that AMTA680, a surface-tuned magnetofluorescent nanoparticle, labels endogenous TAMs selectively and permits to track these cells in their native microenvironment. AMTA680 incorporates two reporter tags: a fluorescent dye for optical imaging and a superparamagnetic core for MRI. The VT680 fluorochrome emits at a far-red wavelength where autofluorescence and tissue absorbance are minimal and thus allows imaging in deep tissues. In addition, VT680 is adequate for multiscopic imaging because single-cell and whole-body information can be integrated from the same animal by intravital microscopy and FMT, respectively. The combination of microscopic and macroscopic imaging offers the possibility to evaluate cellular activity and biodistribution quantitatively and simultaneously. Such advances should foster the identification of novel biomarkers that, when used together, will facilitate a systems-based understanding of tumor-related cellular and molecular events.
Here, we have focused on TAMs because these immune cells are recruited to most tumors and are involved in crucial processes during tumor development. We have found that AMTA680 preferentially targets “M2-like” TAMs, which express high levels of F4/80 [1] and VEGF [24]. Together with the overexpression of the endothelial tyrosine kinase Tie-2 [36], these markers indicate previously described cells that have a profound influence on tumor angiogenesis [1,37]. Using MRI or FMT imaging, we identified that labeled TAMs mainly cluster in delimited foci within tumors and mostly in peripheral regions that are rich in newly formed blood vessels (data not shown). On the basis of their relatively low motility, these TAMs are likely confined to their microenvironment. Tumor-associated macrophages are also known to present antigens to T cells and secrete factors that suppress neighboring T cells. Interestingly, the latter cells, as shown here and in previous studies, have significantly higher motility and infiltrate the tumor from the periphery to the center [10,13,34,38]. It remains to be determined whether peripheral T cells, en route to the center of the tumor, interact with these TAMs and receive the immunosuppressive signals. At least, the TAMs undergo prolonged physical interactions with neighboring tumor cells, mostly through the extension of cytoplasmic protrusions. The quality of signals that are delivered through cell-cell interactions in vivo needs to be clarified, but this will require the generation of sophisticated imaging systems that can monitor fluorescence-based molecular reporters that are expressed at physiological levels.
Injectable imaging agents are now starting to uncover molecular and cellular processes in vivo. The development of agents with properties similar to those of AMTA680 and that are approved for injection into patients would allow in vivo noninvasive monitoring of the activity and location of biological indicators. The approach would be useful to diagnose tumor progression and to follow responses to therapeutic drugs. Tissue biopsies or blood samples could also be used to evaluate cell-targeting agents in patients. Therefore, the technology presented here is transposable to a variety of experimental approaches and potentially offers clinical translatability.
Supplementary Material
Acknowledgments
The authors thank Tyler Jacks and Monte Winslow (Koch Institute for Cancer Research, Massachusetts Institute of Technology, Cambridge) for providing the LSL-KrasG12D and LSL-KrasG12D;LSL-p53 mice and the adenovirus-expressing Cre-recombinase and Tyler Jacks, Richard Hynes (Koch Institute for Cancer Research, Massachusetts Institute of Technology, Cambridge), and Robert Weinberg (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, Cambridge) for helpful discussions.
Footnotes
This work was funded in part by the National Institutes of Health (NIH) U54-CA126515 (to R.W.), NIH 5P50 CA86355 (to R.W. and M.J.P.), NIH U24-CA092782 (to R.W.), NIH U54-CA119349 (to R.W.), and Swiss NSF PBLAB-11555 (to A.L.).
This article refers to supplementary materials, which are designated by Figures W1 to W4 and Videos W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 5.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 6.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 7.Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, Gabrilovich DI. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 9.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittet MJ, Mempel TR. Regulation of T-cell migration and effector functions: insights from in vivo imaging studies. Immunol Rev. 2008;221:107–129. doi: 10.1111/j.1600-065X.2008.00584.x. [DOI] [PubMed] [Google Scholar]
- 11.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 13.Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, Cavanagh LL, von Andrian UH, Ertl HC, Haydon PG, et al. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J Exp Med. 2006;203:2749–2761. doi: 10.1084/jem.20060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pittet MJ. Behavior of immune players in the tumor microenvironment. Curr Opin Oncol. 2009;21:53–59. doi: 10.1097/CCO.0b013e32831bc38a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zippelius A, Batard P, Rubio-Godoy V, Bioley G, Lienard D, Lejeune F, Rimoldi D, Guillaume P, Meidenbauer N, Mackensen A, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Res. 2004;64:2865–2873. doi: 10.1158/0008-5472.can-03-3066. [DOI] [PubMed] [Google Scholar]
- 16.Gupta AK, Gupta M. Synthesis and surface engineering of iron oxide nanoparticles for biomedical applications. Biomaterials. 2005;26:3995–4021. doi: 10.1016/j.biomaterials.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 19.Pittet MJ, Swirski FK, Reynolds F, Josephson L, Weissleder R. Labeling of immune cells for in vivo imaging using magnetofluorescent nanoparticles. Nat Protoc. 2006;1:73–79. doi: 10.1038/nprot.2006.11. [DOI] [PubMed] [Google Scholar]
- 20.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang L, Derfus AM, Yang M, Hoffman RM, Bhatia S, et al. Biomimetic amplification of nanoparticle homing to tumors. Proc Natl Acad Sci USA. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath JR, Davis ME. Nanotechnology and cancer. Annu Rev Med. 2008;59:251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radu CG, Shu CJ, Nair-Gill E, Shelly SM, Barrio JR, Satyamurthy N, Phelps ME, Witte ON. Molecular imaging of lymphoid organs and immune activation by positron emission tomography with a new [(18)F]-labeled 2′-deoxycytidine analog. Nat Med. 2008;14:783–788. doi: 10.1038/nm1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw SY, Westly EC, Pittet MJ, Subramanian A, Schreiber SL, Weissleder R. Perturbational profiling of nanomaterial biologic activity. Proc Natl Acad Sci USA. 2008;105:7387–7392. doi: 10.1073/pnas.0802878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 26.Chen ML, Pittet MJ, Gorelik L, Flavell RA, Weissleder R, von Boehmer H, Khazaie K. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci USA. 2005;102:419–424. doi: 10.1073/pnas.0408197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm J, Kirsch DG, Windsor SD, Kim CF, Santiago PM, Ntziachristos V, Jacks T, Weissleder R. Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc Natl Acad Sci USA. 2005;102:14404–14409. doi: 10.1073/pnas.0503920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 29.Swirski FK, Berger CR, Figueiredo JL, Mempel TR, von Andrian UH, Pittet MJ, Weissleder R. A near-infrared cell tracker reagent for multiscopic in vivo imaging and quantification of leukocyte immune responses. PLoS ONE. 2007;2:1075. doi: 10.1371/journal.pone.0001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ntziachristos V, Tung CH, Bremer C, Weissleder R. Fluorescence molecular tomography resolves protease activity in vivo. Nat Med. 2002;8:757–760. doi: 10.1038/nm729. [DOI] [PubMed] [Google Scholar]
- 31.Swirski FK, Libby P, Aikawa E, Alcaide P, Luscinskas FW, Weissleder R, Pittet MJ. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117:195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 33.Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, von Andrian UH. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 2006;25:129–141. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, Amigorena S. In vivo imaging of cytotoxic T cell infiltration and elimination of a solid tumor. J Exp Med. 2007;204:345–356. doi: 10.1084/jem.20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enochs WS, Harsh G, Hochberg F, Weissleder R. Improved delineation of human brain tumors on MR images using a long-circulating, superparamagnetic iron oxide agent. J Magn Reson Imaging. 1999;9:228–232. doi: 10.1002/(sici)1522-2586(199902)9:2<228::aid-jmri12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.De Palma M, Venneri MA, Galli R, Sergi Sergi L, Politi LS, Sampaolesi M, Naldini L. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell. 2005;8:211–226. doi: 10.1016/j.ccr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 37.De Palma M, Murdoch C, Venneri MA, Naldini L, Lewis CE. Tie2-expressing monocytes: regulation of tumor angiogenesis and therapeutic implications. Trends Immunol. 2007;28:519–524. doi: 10.1016/j.it.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Pittet MJ, Grimm J, Berger CR, Tamura T, Wojtkiewicz G, Nahrendorf M, Romero P, Swirski FK, Weissleder R. In vivo imaging of T cell delivery to tumors after adoptive transfer therapy. Proc Natl Acad Sci USA. 2007;104:12457–12461. doi: 10.1073/pnas.0704460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.