Abstract
Lung cancer is the leading cause of cancer deaths worldwide, yet few genetic markers of lung cancer risk useful for screening exist. The let-7 family-of-microRNAs (miRNAs) are global genetic regulators important in controlling lung cancer oncogene expression by binding to the 3′UTRs (untranslated regions) of their target messenger RNAs (mRNAs). The purpose of this study was to identify single nucleotide polymorphisms (SNPs) that could modify let-7 binding and to assess the effect of such SNPs on target gene regulation and risk for non-small cell lung cancer (NSCLC). let-7 complementary sites (LCSs) were sequenced in the KRAS 3′UTR from 74 NSCLC cases to identify mutations and SNPs that correlated with NSCLC. The allele frequency of a previously un-identified SNP at LCS6 was characterized in 2433 people (representing 46 human populations). The frequency of the variant allele is 18.1–20.3% in NSCLC patients and 5.8% in world populations. The association between the SNP and the risk for NSCLC was defined in two independent case-control studies. A case-control study of lung cancer from New Mexico showed a 2.3-fold increased risk (C.I.= 1.1–4.6, p = 0.02) for NSCLC cancer in patients who smoked < 40 pack years. This association was validated in a second independent case-control study. Functionally, the variant allele results in KRAS over-expression in vitro. The LCS6 variant allele in a KRAS miRNA complementary site is significantly associated with increased risk for NSCLC among moderate smokers, and represents a new paradigm for let-7 miRNAs in lung cancer susceptibility.
Introduction
MiRNAs are recently identified gene regulators that are at abnormal levels and implicated in virtually all cancer subtypes studied.1 MiRNAs bind to the 3′UTRs of their target genes, regions which are evolutionarily highly conserved,2, 3 suggesting an important role for these regions in natural selection. Because miRNAs each regulate hundreds of mRNAs simultaneously,4 the potential of cellular transformation resulting from dysfunction of a single miRNA is high. The role of miRNA SNPs in disease is just being defined. For example, SNPs in miRNAs important in cancer have been identified; mir-125a, shown to be altered in breast cancer,5, 6 has a variant allele at a SNP in the mature miRNA sequence that decreases expression.7 There is also evidence that SNPs in miRNA binding sites could be associated with disease, for example, a point mutation identified in several Tourette’s syndrome patients in the 3′UTR of SLITRK1 disrupts the binding of miR-189.8 Furthermore, two recent papers report SNPs in miRNA target sites in human cancer genes9 and show that allele frequencies vary between cancerous and normal tissues.10 Finally, a SNP identified in a miRNA binding site in the kit oncogene was associated with increased gene expression in papillary thyroid cancer.11
The let-7 family of miRNAs appears to play a key role in lung cancer: they are at low levels in NSCLC;12, 13 their lower levels are biomarkers of a poor outcome;14, 15 they regulate multiple important lung cancer oncogenes, including RAS;12, 16 and they inhibit growth of lung cancer cell lines in vitro12, 14 and in vivo.17, 18 The purpose of this study was to identify SNPs that could potentially modify let-7 binding and to assess the effect of these SNPs on target gene regulation and the risk for NSCLC. Here we show that a variant allele at a SNP in a let-7 complementary site in the KRAS 3′UTR is associated with increased risk for NSCLC in moderate smokers. Furthermore, this variant allele leads to altered KRAS regulation in vitro, with higher KRAS expression in the presence of the variant allele. In addition, in tumors from patients harboring the variant allele, let-7s were lower than in non-variant allele tumors, suggesting the SNP might be associated with NSCLC with a poor prognosis. The finding that a SNP disrupting let-7 miRNA regulation of a known oncogene can affect cancer predisposition to NSCLC is a new paradigm, and supports further studies to identify similar SNPs in all cancer types.
Methods
Study Populations
Lung tissue samples from patients with a diagnosis of NSCLC were collected following Yale University Human Investigation Committee approval. Cases were chosen based on the availability of frozen stored tissue from lung tumor resections from 1994 through 2003, and from recent cases with extra tissue available. Tissue was collected from 87 patients. Seven patients were excluded due to other risk factors for lung cancer (e.g., immunosuppression, tuberculosis) and six were excluded due to their tumors being non-lung primary metastatic disease. Seventy-four patients were included in the analysis (see Supplementary Table 1 online).
To determine the frequency of the SNP alleles, 2433 individuals were genotyped from a global sample of 46 populations. According to population ancestry and geographic locations, these 46 populations are categorized into 4 groups: European (including West Asia), African, Asian (including the Pacific) and Native American. Sample descriptions and samples sizes can be found in the ALlele FREquency Database (ALFRED)19 by searching for the population names (http://alfred.med.yale.edu/). DNA samples were extracted from lymphoblastoid cell lines established and/or grown in the Yale University laboratory of K.K.K. The methods of transformation, cell culture, and DNA purification have been described.20 All volunteers were apparently normal and otherwise healthy adult males or females and samples were collected after receipt of appropriate informed consent.
Lung cancer cases (n=325) for the New Mexico case-control study were recruited beginning in 2004 from Albuquerque through two local hospitals, the Veterans hospital and the University of New Mexico (UNM) hospital. All stages and histological types of lung cancer were included. Controls (n=325) with no history of any prior cancer were recruited from two ongoing local smoker cohorts, the Veterans Smokers Cohort (mainly veterans from Albuquerque) and the Lovelace Smokers Cohort (general residents in Albuquerque). Those two cohorts started to recruit participants in 2001 to conduct longitudinal studies on molecular markers of respiratory carcinogenesis in biological fluids such as sputum from people at risk for lung cancer. A standardized questionnaire was used to collect information on medical, family, and smoking exposure history, and quality of life for both lung cancer cases and control cohort members. Controls were randomly matched to lung cancer cases after categorization into different age groups (5-year differences) by sex and cohort (see Supplementary Table 2A online). Cases with small cell lung cancer were excluded to more precisely assess the effect of the LCS6 SNP on risk for NSCLC. Cases over 82 years old (the maximum age in the control group), cases with any prior cancer history, never smokers or cases with missing data on smoking-related covariates were also excluded in the data analysis, resulting in 218 cases in the analysis.
A second lung cancer case-control study was conducted to validate findings from the New Mexico study. The study population was derived from a large ongoing molecular epidemiological study in Boston, MA, that began in 1992 and now has more than 2205 NSCLC patients. Details of this case-control population have been described previously.21–23 For this study over 3700 samples were analyzed, which included smokers and non-smokers. This study was approved by the Human Subjects Committees of MGH and Harvard School of Public Health, Boston, MA. Briefly, all histologically confirmed, newly diagnosed patients with NSCLC at MGH were recruited between December 1992 and February 2006. Before 1997, only early stage (stage I and II) patients were recruited. After 1997, all stages of NSCLC cases were recruited in this study. Controls were recruited at MGH from healthy friends and non-blood-related family members (usually spouses) of several groups of hospital patients: (a) patients with cancer, whether related or not related to a case; or (b) patients with a cardiothoracic condition undergoing surgery. No matching was performed. Importantly, none of the controls were patients. Potential controls with a previous diagnosis of any cancer (other than non-melanoma skin cancer) were excluded from participation. Over 85% of eligible cases and over 90% of controls participated in this study and provided blood samples. A research nurse administered questionnaires on demographic information and detailed smoking history (see Supplementary Table 2B online). To reduce potential variation in allele frequency by ethnicity, only Caucasians were considered in the analysis.
Evaluation of 3′UTR Sequences and the LCS6 SNP
DNA was isolated from frozen and FFPE lung tissue using the DNeasy Blood and Tissue Kit (Qiagen). Segments of the KRAS 3′UTR were amplified using PfuTurbo DNA polymerase (Stratagene) and DNA primers specific to this sequence (see Supplementary Table 3 online). PCR products were purified using the QIAquick PCR Purification Kit or 96 PCR Purification Kit (Qiagen) and sequenced using the same primers. The NRAS 3′UTR was sequenced in the same manner.
For high-throughput genotyping, the DNA isolated from lymphocytes, blood, or tumor samples was amplified using TaqMan PCR assays designed specifically to identify the T or G allele of the LSC6 SNP (Applied Biosciences).
Determining the Impact of the LCS6 Variant Allele on KRAS Expression
We generated a pGL3 derivative containing almost the entire KRAS 3′UTR (KRAS wild-type) as follows. KRAS wild-type includes 3,910 base pairs of the KRAS 3′UTR, which was amplified from human genomic DNA using the forward primer SMJ104 and reverse primer LCJ5 (see Supplementary Table 3 online). NheI restriction sites were included on the 5′ ends of the primers for convenient cloning. The product was first cloned into the TOPO cloning vector (Invitrogen) and then sub-cloned into pGL3 (Ambion) for use in subsequent luciferase assays. The luciferase reporter with the variant LCS6 KRAS 3′UTR (KRAS mLCS6) was constructed through site-directed mutagenesis of KRAS wild-type using GeneTailor (Invitrogen). A549 cells were cultured in DMEM with 10% FBS and penicillin/streptomycin (Invitrogen). A549 cells were transfected with 500 ng KRAS wild-type or KRAS mLCS6 and 50 ng pRL-TK (Promega) using Lipofectamine 2000 (Invitrogen) for 24 hours. Reporter expression was analyzed using the Dual-Luciferase Reporter Assay (Promega) and Wallac Victor2 1420 (PerkinElmer). Two-tailed t-tests were done to verify statistical significance of differences in luciferase expression using GraphPad Prism.
Measuring let-7 in vivo
RNA was isolated from normal or tumor tissue specimens using Ambion isolation kits. RTPCR was done for let-7a, b, d and g using the Ambion RTPCR specific primers for eight tumor samples with the variant allele and eight samples without the variant allele, using the ABI 7900.
Statistical Analysis
To calculate significance a Chi-Square test was used for categorical variables, a t-test was used for continuous variables and in some cases a two-sided Fisher’s exact test was used. Two-sided two-sample t-tests, Chi-Square analyses and two-sided Wilcoxon rank-sum tests were performed, as appropriate to compare the demographic variables between cases and controls. An unconditional logistic regression model was used to calculate odds ratio and confidence intervals (CI) for the KRAS LCS6 SNP in moderate and heavy smokers (defined by below and above the median pack years for the study population, respectively) with adjustment for selected covariates. A likelihood ratio test was used to assess the association with the allele and the pack-year interaction for NSCLC. The dominant model was used for all genetic association analysis due to the low frequency of the rare allele.
Results
A SNP in a let-7 Complementary Site in the KRAS 3′UTR
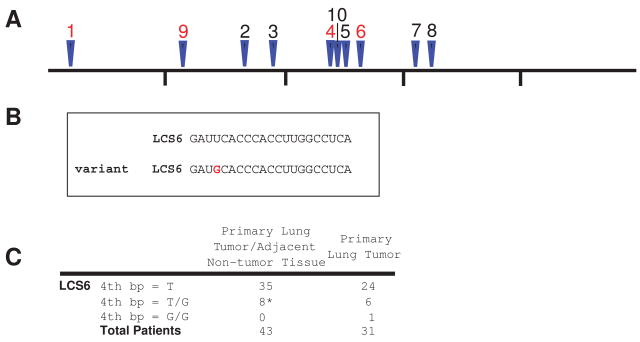
Human KRAS expression is regulated in a 3′UTR and let-7-dependent manner through ten putative let-7 complementary sites (LCSs) in its 3′UTR12 (Fig. 1A). Based on data from the HapMap24 and dbSNP25 databases, only one SNP, rs712(-), is reported in an LCS. Tumors (and adjacent normal tissue when available) from 74 patients with NSCLC were evaluated for sequence variations in these ten LCSs. SNPs in LCS1, LCS9, and LCS4 (see Supplementary Table 4 online) were randomly observed at low frequency in tumor or adjacent tissue, suggesting no specific involvement in NSCLC. However, a SNP (T to G, with G the less frequent variant) identified at the fourth nucleotide in LCS6 was found in 20.3% of tumor and corresponding adjacent normal tissues from NSCLC patients (Fig. 1B, 1C)(Supplementary Fig. 1 online). Because this was a previously unreported SNP that was found frequently in our NSCLC patients, we hypothesized that this SNP might be a marker of an increased risk for NSCLC.
Figure 1. Identification of a SNP in the KRAS 3′UTR.
(A) Location of the putative LCSs in the KRAS 3′UTR. LCS1-LCS8 had been previously identified.12 LCSs where mutations are found are shown in red. The KRAS 3′UTR is 5016 bp, and the markers are positioned every 1000 bp. (B) The sequence of LCS6 with either the reference allele or the variant allele (red). (C) The variant allele in LCS6 was seen in 20.3% of the primary lung tumors and was present in the adjacent tissue when available (marked with an *).
As a control, we sequenced (in the same NSCLC patients) the 3′UTR of NRAS, which is not associated with lung cancer but contains 9 putative LCSs,12 to look for similar SNPs. No SNPs were identified within the LCSs of the NRAS 3′UTR (data not shown), further supporting the idea that the identified SNP in the KRAS 3′UTR is likely an important change with respect to NSCLC.
Frequency of the Variant Allele Across World Populations
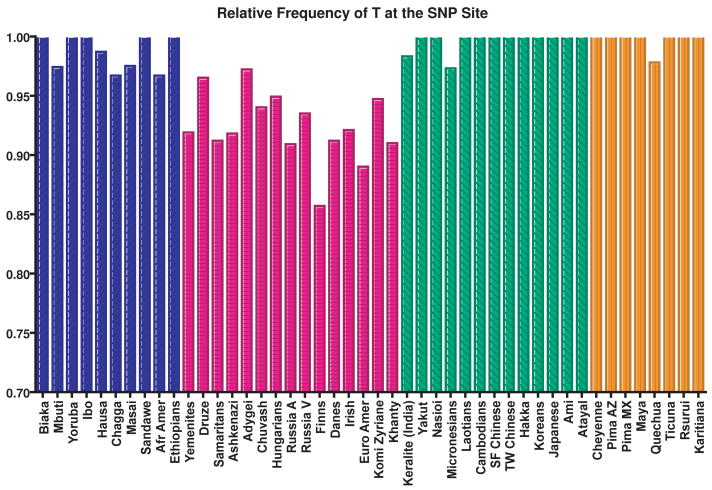
To determine if the prevalence of the variant allele in our NSCLC population was higher than expected, we determined the allele frequencies of the LCS6 SNP in the general population with a unique human genetic resource found at Yale University, a collection of genomic DNA from 2433 healthy individuals from a global set of 46 populations.26 An extensive data base of genetic variations in these samples exists and can be found, along with the population descriptions in ALFRED, the ALlele FREquency Database (http://alfred.med.yale.edu).19 Using a TaqMan assay, we found that <3% of the 4866 chromosomes, or 5.8% of the people tested, had the G allele (variant) at the LCS6 SNP site (Fig. 2). The frequency of this allele varied across geographic populations, with “European” populations exhibiting the variant allele most frequently (7.6% of the chromosomes tested); African populations less frequently (<2.0% of chromosomes tested); and “Asian” and Native American populations infrequently (<0.4% of chromosomes tested). Of note, over 85% of the patients in our retrospective patient cohort were of European descent. Importantly, these findings indicate that the prevalence of the variant allele (20.3%) in our NSCLC patient cohort is significantly higher than expected in any existing geographic population, further supporting the hypothesis that that this variant allele is a marker of an increased risk to NSCLC.
Figure 2. Frequency of the reference allele across the world.
Frequency of the reference allele (T) at the SNP locus was examined in 2433 people. They represent 46 populations from around the world, which were categorized based on geography: Africa (blue), Europe, Southwest Asia and Western Siberia (pink), South central Asia, East Asia and the Pacific (green), and the Americas (orange). The frequency of the reference allele (T) across all chromosomes tested is 97.1% and the frequency of the alternative allele (G) is 2.9%. The frequency of the G allele in African populations is 1.3%, in European populations is 7.6%, in Asian populations is 0.3%, and in Native American populations is 0.3%. The allele frequencies in the individual populations are in ALFRED.
The Variant Allele is Associated with Increased Risk for NSCLC in Moderate Smokers
Lung cancer cases and controls from smokers enrolled in a lung cancer case-control study in New Mexico were genotyped for the LCS6 variant allele. The frequency of the variant allele in the NSCLC cases was 18.8%, which was not significantly different from the frequency in the lung cancer patients studied at Yale (p=0.20). While the presence of the LCS6 variant allele did not predict NSCLC risk for the entire patient cohort, the variant allele was associated with increased NSCLC risk in smokers with a < 41 pack-year smoking history (Table 1A, OR=2.3, 95% confidence interval (CI) = 1.1–4.6, p = 0.02), meaning a person has smoked the equivalent of one pack of cigarettes per day for 41 years, the median level of smoking in this population. The OR was adjusted for age, gender, smoking status, pack-years of smoking, and years since smoking cessation. This finding was then validated in a larger independent study of lung cancer cases and controls enrolled in Boston, MA. In that study, the frequency of the variant allele was 18.1%, and again no association was seen between the allele and lung cancer risk in the entire population. However, after stratifying by the median smoking level (40 pack-years), a 1.4-fold increase in risk for lung cancer was seen in persons who smoked <40 pack-years (Table 1B, CI = 1.1–1.7, p = 0.01). These studies did not find an association between the LCS6 variant allele and tumor histology.
Table 1.
| Table 1A. Association between KRAS variant allele and non-small cell lung cancer in the New Mexico case-control study | |||||
|---|---|---|---|---|---|
| Group | Genotype | Crude OR | p | Adjusted ORa | p |
| Overall (218 cases vs. 325 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 1.4 (0.9–2.3) | 0.13 | 1.5 (0.9–2.4) | 0.15 | |
| Pack-yearsb <41 (75 cases vs. 195 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 2.3 (1.1–4.4) | 0.02 | 2.3 (1.1–4.6) | 0.02 | |
| Pack-years ! 41 (143 cases vs. 130 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 1.0 (0.5–1.9) | 0.99 | 0.9 (0.5–1.8) | 0.86 | |
| Table 1B. Association between KRAS variant allele and non-small cell lung cancer in the Boston case-control study | |||||
|---|---|---|---|---|---|
| Group | Genotype | Crude OR | p | Adjusted ORc | p |
| Overall (2205 cases vs. 1497 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 1.11 (0.93–1.32) | 0.23 | 1.17 (0.97–1.44) | 0.15 | |
| Pack-yearsd <40 (956 cases vs. 1214 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 1.28 (1.03–1.60) | 0.03 | 1.36 (1.07–1.73) | 0.01 | |
| Pack-years ! 40 (1249 cases vs. 283 controls) | |||||
| TT | 1.0 | ||||
| TG+GG | 0.85 (0.61–1.29) | 0.34 | 0.89 (0.63–1.25) | 0.49 | |
Age, race, gender and smoking status were adjusted in an unconditional logistic regression model.
41 pack-years is the median of 543 smokers.
Age, gender, smoking status, pack-years of smoking and years since smoking cessation (if ex-smoker) were adjusted in an unconditional logistic regression model.
40 pack-years is the median of 3702 smokers.
These findings confirm that the variant allele is a marker for an increased risk of NSCLC in patients with less cigarette exposure, which in these studies was less than the median smoking exposure of ~ 40 pack-years. Our finding that the LCS6 SNP only impacts cancer risk for those with less cigarette exposure agrees with other studies showing a dose-dependent gene-environment interaction for smoking-induced lung cancer risk;23, 27, 28 with higher smoking exposure any genetic predisposition is hypothesized to be overwhelmed by the extent of smoking-related damage.
The LCS6 Variant Allele Impacts KRAS Regulation and is Associated with Lower let-7
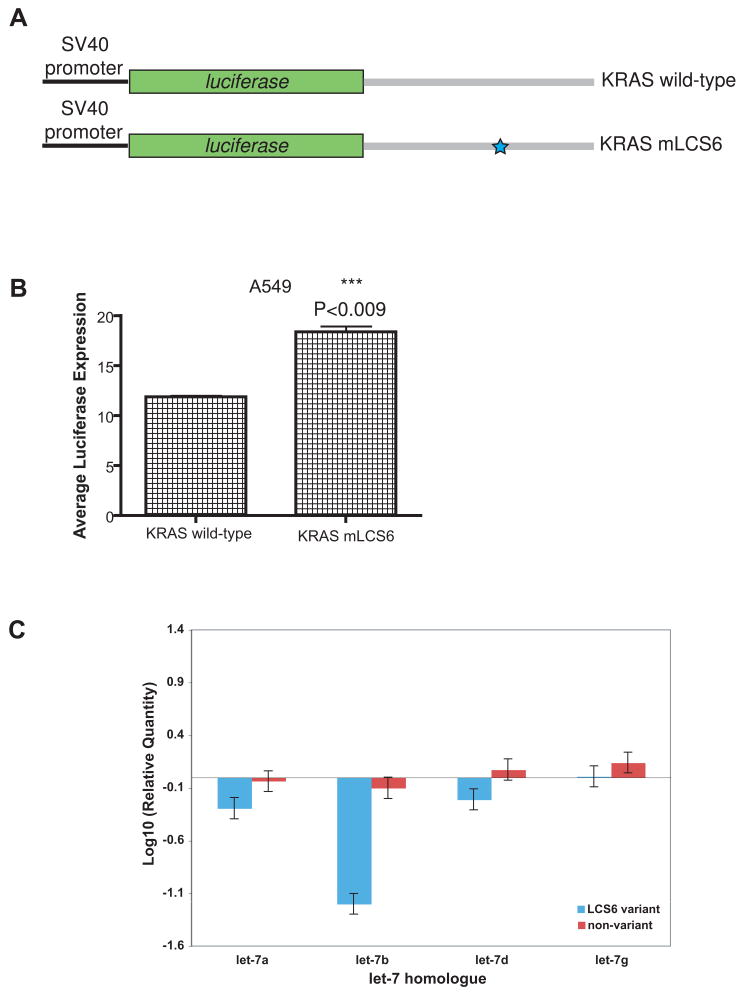
To determine if LCS6 impacts KRAS regulation, we used a previously described luciferase reporter for KRAS expression to determine the effect of the LCS6 variant on expression.12 We transfected a luciferase reporter with a full length KRAS 3′UTR containing the variant allele at LCS6 (KRAS mLCS6) into A549 cells, a lung cancer cell line, with known low let-7 levels 14, and compared luciferase expression to A549 cells transfected with the luciferase reporter with a wild-type KRAS 3′UTR (KRAS wild-type). We found that there was a significant increase in luciferase activity in cells transfected with KRAS mLCS6 versus the KRAS wild-type (Fig. 3A, 3B). These findings support the hypothesis that the variant allele has altered the dependence of the KRAS 3′UTR on LCS6, and allows increased KRAS expression in the presence of this variant allele.
Figure 3. The effect of the variant allele on KRAS and let-7 levels.
(A) Luciferase reporter constructs containing the KRAS 3′UTR. Grey boxes signify the 3′UTR used in the reporter constructs. A blue star represents the variant allele in LCS6. (B) Representative graph of luciferase activity of KRAS wild-type and KRAS mLCS6 (P<0.009) in A549 cells. Triplicate repeats were conducted showing similar results. (C) let-7a, b, d and g levels in eight tumors with and eight without the variant allele. Samples were normalized to two non-cancerous patients, whose let-7 levels were similar. Error bars represent variation between PCR reactions for each sample.
We were unable to measure KRAS levels in patient samples due to limited tissue and only DNA access for case control samples. However, we did evaluate the KRAS gene in patient samples harboring the SNP for common activating mutations (in codons 12, 13, and 61), and did not find any activating mutations (data not shown). As KRAS is rarely activated in squamous tumors and in only 30% of lung adenocarcinomas, these findings are not surprising, and additional studies will be required to fully understand KRAS expression and/or mutations in tumors harboring the variant allele.
We next measured the levels of let-7 in available patient tumors with or without the variant allele, using RT PCR. On average, we found that the levels of let-7a, b, d and g were lower in patients with the variant allele compared to patients without the variant allele (Fig. 3C). These findings suggest that the variant allele is associated with lower let-7, at least in the NSCLC tumors in this study.
Discussion
We have identified a variant allele in a let-7 complementary site in the KRAS 3′UTR that alters let-7-mediated regulation of KRAS expression and is associated with a 1.4–2.3 fold increased risk for NSCLC among moderate smokers. This variant allele is found in 18.1–20.3% of lung cancer cases versus in only 5.8% of the world populations. The association of this allele to lung cancer was identified in the NM case-control study and validated in the MA case-control study, making this finding highly significant.
We find that this variant allele at the LCS6 site indeed affects regulation of KRAS expression in vitro, allowing increased levels of KRAS. Interestingly, tumors containing the variant allele had lower let-7 levels than tumors without the variant allele, and as low let-7 has been associated with a poor prognosis in NSCLC,14, 15, 30 these findings raise the possibility that the subset of NSCLC patients harboring the variant allele may be those with an especially poor prognosis, a hypothesis requiring future studies to validate.
While it is not possible at this time to further define the mechanism of cancer predisposition for patients who harbor this variant allele, the finding that the variant allele appears to allow increased KRAS in vitro suggests that KRAS overexpression might be one plausible mechanism. An additional hypothesis could be that altered let-7 binding in the KRAS 3′UTR could somehow lower cellular levels of let-7, perhaps through a feedback loop, and lower let-7 levels could further result in increased cell growth, as let-7 is known to repress cell growth pathways.16 For example, recent evidence indicates that a negative feedback loop involving lin-28 specifically regulates cellular let-7 levels.31 In the background of the variant allele, a cycle creating lowered let-7 levels and increased KRAS expression could in concert act as the first steps in oncogenesis. These hypotheses need testing in future studies.
Because lung cancer is so deadly when caught at later stages, screening programs have been initiated in current and ex-smokers: The Early Lung Cancer Action Project (ELCAP) found that a chest computed tomography (CT) scan is three times more sensitive in detecting early-stage lung cancer than a chest X-ray in “high-risk” populations (2.4% versus 0.7%).32, 33 Yet there remains considerable controversy over the use of lung CT scans as a global screening approach for lung cancer, because of the expense (estimated cost 2 billion dollars yearly in the US alone) and the very low yield of cancers detected yearly (1.2%).34 One of the primary problems is that although smoking is the number one risk factor for developing lung cancer, only 10% of smokers ever develop lung cancer. With 44.5 million current smokers in the United States (20.9% of the population) and over 1.3 billion smokers worldwide, there is a clear need to identify markers whose genetic variation is associated with lung cancer risk that would help better prioritize who should be screened clinically and be offered chemopreventive agents.
While numerous studies of carcinogen metabolizing and detoxifying genes and DNA repair genes have identified sequence variations associated with risk for lung cancer, a meta-analysis of such polymorphisms in DNA repair pathways concluded that for any increased risk association between a single SNP and lung cancer, the risk fluctuation would likely be minimal, and only panels evaluating a collection of SNPs would ever successfully predict lung cancer risk.35 Supporting this hypothesis, two SNPs associated with lung cancer risk (OR = 1.19–1.8) were recently identified from three large studies examining over 300,000 SNPs. The mechanism of lung cancer predisposition caused by these SNPs is controversial, but is hypothesized to be through affects on a nicotine receptor.36 37
In contrast, in this study, we have identified the first miRNA binding-site SNP that alone can predict a significant increase in NSCLC risk in people with a moderate smoking history. The mechanism may be due to altered regulation of the KRAS oncogene and possibly cellular let-7 levels as well. These findings give insight into a new paradigm, and support the pursuit of 3′UTR sequencing for similar SNPs in all tumor related genes to better understand their role in genetic cancer risk. This strategy can be a complementary, and likely productive, approach to enhance current efforts to define genetic cancer risk.
Supplementary Material
Acknowledgments
We thank William C. Speed and Dr. Judith R. Kidd for their assistance in setting up and performing the population genetic screen. We thank Welela Tereffe and Lynn Wilson for their critical reading of this manuscript. We acknowledge the assistance of Drs. Kofi Asomaning and Yen-tsung Huang with the statistical analysis for the Boston study. This work was supported by a grant from the Connecticut Department of Public Health to FJS and JBW, Shannon Foundation funds to JBW, grant R01 CA122676 to YZ, grant U01 CA097356 to SB, grant CA074386, CA090578, CA092824 and ES00002 to DC and a grant from the Flight Attendant Medical Research Institute (FAMRI) to RZ.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics. 2006:38. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 3.Saunders MA, Liang H, Wen-Hsiung L. Human polymorphism at microRNAs and microRNA target sites. PNAS. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 6.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 7.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 8.Abelson JF, Kwan KY, O’Roak BJ, et al. Sequence Variants in SLITRK1 Are Associated with Tourette’s Syndrome. Science. 2005;310:317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 9.Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landi D, Gemignani F, Barale R, Landi S. A Catalog of Polymorphisms Falling in MicroRNA-Binding Regions of Cancer Genes. DNA and Cell Biology. 2007;0:1–9. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 11.He H, Jazdzewski K, Wei L, et al. The role of microRNA genes in papillary thyroid carcinoma. PNAS. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 15.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Johnson C, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Research. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008:7. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 18.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. PNAS. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung K, Osier M, Kidd J, Pakstis A, Miller P, Kidd K. ALFRED: an allele frequency database for diverse populations and DNA polymorphisms. Nucleic Acids Res. 2000;28:361–3. doi: 10.1093/nar/28.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson M, JF G. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20:856–8. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 21.Zhai R, Liu G, Zhou W, et al. Vascular endothelial growth factor geneotypes, haplotypes, gender, and the risk of non-small cell lung cancer. Clin Cancer Res. 2008;14:612–7. doi: 10.1158/1078-0432.CCR-07-1655. [DOI] [PubMed] [Google Scholar]
- 22.Su L, Zhou W, Asomaning K, et al. Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis. 2006;27:1024–9. doi: 10.1093/carcin/bgi283. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W, Liu G, Miller D, et al. Gene-environment interaction for the ERCC2 polymorphism and cumulative cigarette smoking exposure in lung cancer. Cancer Research. 2002;62:1377–81. [PubMed] [Google Scholar]
- 24.Consortium TIH. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.Sherry S, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- 26.Speed W, O’Roak BZT, et al. Haplotype Evolution of SLITRK1, a Candidate Gene for Gilles de la Tourette Syndrome. American Journal of Genetics. 2007 doi: 10.1002/ajmg.b.30641. in advance. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, Liu G, Park S, et al. Gene-Smoking Interaction Associations for the ERCC1 Polymorphisms in the Risk of Lung Cancer. Cancer Epidemiology, Biomarkers & Prevention. 2005;14:491–6. doi: 10.1158/1055-9965.EPI-04-0612. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, Wheatley-Price P, Zhou W, et al. Genetic polymorphisms of MDM2, cumulative cigarette smoking and nonsmall cell lung cancer risk. Int J Cancer. 2007 doi: 10.1002/ijc.23178. online. [DOI] [PubMed] [Google Scholar]
- 29.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huncharek M, Muscat J, Geschwind J-F. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: a combined analysis of 881 cases. Carcinogenesis. 1999:20. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 31.Viswanathan SR, Daley GQ, Gregory RI. Selective Blockade of MicroRNA Processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henschke C, McCauley D, Yankelevitz D, et al. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet. 1999:354. doi: 10.1016/S0140-6736(99)06093-6. [DOI] [PubMed] [Google Scholar]
- 33.Henschke CI, Shaham D, Yankelevitz DF, Nasser KA. CT Screening for Lung Cancer: Past and Ongoing Studies. Thoracic and Cardiovascular Surgery. 2005;17:99–106. doi: 10.1053/j.semtcvs.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Investigators TIELCAP. Survival of Patients with Stage I Lung Cancer Detected on CT Screening. NEJM. 2006;355:1763–71. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
- 35.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: A meta-analysis. Int J Med Sci. 2007;4:59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, et al. The variant associated with nicotine dependence, lung cancer and peripheral artierial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amos C, Wu X, Broderick P, GOrlow IP, Gu J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Genetics. 2008 doi: 10.1038/ng.109. online. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.