Abstract
CYP1B1-null mice, created by targeted gene disruption in embryonic stem cells, were born at the expected frequency from heterozygous matings with no observable phenotype, thus establishing that CYP1B1 is not required for mouse development. CYP1B1 was not detectable in cultured embryonic fibroblast (EF) or in different tissues, such as lung, of the CYP1B1-null mouse treated with the aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin whereas the equivalent wild-type EF cells express basal and substantial inducible CYP1B1 and lung expresses inducible CYP1B1. CYP1A1 is induced to far higher levels than CYP1B1 in liver, kidney, and lung in wild-type mice and is induced to a similar extent in CYP1B1-null mice. 7,12-dimethylbenz[a]anthracene (DMBA) was toxic in wild-type EFs that express CYP1B1 but not CYP1A1. These cells effectively metabolized DMBA, consistent with CYP1B1 involvement in producing the procarcinogenic 3,4-dihydrodiol as a major metabolite, whereas CYP1B1-null EF showed no significant metabolism and were resistant to DMBA-mediated toxicity. When wild-type mice were administered high levels of DMBA intragastrically, 70% developed highly malignant lymphomas whereas only 7.5% of CYP1B1-null mice had lymphomas. Skin hyperplasia and tumors were also more frequent in wild-type mice. These results establish that CYP1B1, located exclusively at extrahepatic sites, mediates the carcinogenicity of DMBA. Surprisingly, CYP1A1, which has a high rate of DMBA metabolism in vitro, is not sufficient for this carcinogenesis, which demonstrates the importance of extrahepatic P450s in determining susceptibility to chemical carcinogens and validates the search for associations between P450 expression and cancer risk in humans.
Cytochromes P450 (P450) are a superfamily of heme-containing monooxygenases. A limited number of P450s participate in pathways of steroid hormone synthesis whereas the majority of these enzymes are involved in oxidative metabolism of drugs, other foreign compounds, and endogenous substrates, including steroids (1). These xenobiotic-metabolizing P450s mostly fall within the CYP1, CYP2, and CYP3 families and exhibit broad and sometimes overlapping substrate specificity. A limited number of P450s within these families are responsible for the metabolic activation of chemical carcinogens. In the CYP1 family, CYP1A1 and CYP1B1 metabolically activate polycyclic aromatic hydrocarbons and CYP1A2 participates in the metabolic activation of arylamine, heterocylic amines, and aflatoxin B1. CYP2E1 activates a large number of low molecular weight carcinogens including benzene and N-nitrosodimethylamine. These carcinogen-metabolizing P450s are also among the most well conserved of the P450 superfamily and can be found in several mammalian species, including mouse and human (2). CYP1B1 is a conserved member of the P450 superfamily that was first identified and purified from mouse embryonic fibroblasts (EFs) (3) and rat adrenals (4). This form was characterized by its ability to metabolically activate polycyclic aromatic hydrocarbons, including benzo[a]pyrene and 7,12-dimethylbenz[a]anthracene (DMBA), but with a product distribution that is distinct from CYP1A1 (3, 5, 6). CYP1B1 is also inducible by the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), similar to the other members of the CYP1 family, CYP1A1 and CYP1A2 (7, 8).
The complete sequence of CYP1B1 identified from human keratinocytes and from mouse EFs showed 40% identity to CYP1A1 and CYP1A2 (8–10). The gene structure is distinct from other mammalian P450s comprising only three exons and having a very long 3′ untranslated region in exon 3 that results in a 5.2-kilobase (kb) mRNA (11, 12). CYP1B1 has a pattern of expression that differs from the other two CYP1 family P450s; it is expressed constitutively in steroidogenic tissues like the adrenal, ovary, and testes and is inducible by adrenocorticotropin, cAMP, peptide hormones, and aryl hydrocarbon receptor ligands (4, 8, 10, 13, 14). CYP1B1 also is expressed in steroid-responsive tissue of mesodermal origin, such as the uterus, breast, and prostrate. In contrast, CYP1A2 is expressed primarily in liver whereas CYP1A1 is not expressed constitutively in any tissues but is inducible by dioxins and polycyclic aromatic hydrocarbons in almost all tissues and cell lines examined (15). The conservation of CYP1B1 and its expression in reproductive tissue would suggest that it has an important developmental or physiological role in mammals. In addition, the expression of CYP1B1 in many important cancer-producing tissues and the ability of CYP1B1 to metabolically activate polycyclic aromatic hydrocarbon carcinogens indicate that it might be pivotal in chemical carcinogenesis. To investigate these possibilities, a CYP1B1-null mouse was produced.
MATERIALS AND METHODS
Construction of the Targeting Vector.
Genomic clones corresponding to Cyp1b1 were obtained by screening a 129/Sv genomic library (CLONTECH) by using the rat CYP1B1 cDNA as a probe. A clone spanning ≈10.5 kb and containing two of the three exons of the murine Cyp1b1 gene (exon 1 is noncoding) was subcloned as a SalI fragment into pGEM-3Z (Promega). Both coding exons were targeted to disrupt the gene. The targeting vector was created by inserting a 1.7-kb cassette containing the bacterial phosphoribosyltransferase II gene under control of the phosphoglycerate kinase-1 promoter (PGK-NEO), which confers resistance to the aminoglycoside G418 (Life Sciences, St. Petersburg, FL), into the HincII site of exon 3. This cassette was excised from pPNT (16) and was inserted in the opposite transcriptional orientation as the Cyp1b1 gene. As a negative selection against random integration of the construct, the herpes simplex virus thymidine kinase gene was inserted at the 5′ end of the targeting vector (17). The construct containing ≈4.4 kb of 5′- and 1.5 kb of 3′-genomic DNA flanking the PGK-NEO cassette (see Fig. 1A) was made in four cloning steps: (i) The genomic clone, designated 24.7, in the SalI site of the polylinker region of pGEM-3Z was digested with BamHI, which cleaves once in the pGEM-3Z polylinker region and once in the genomic clone, and was gel-purified and religated. (ii) The shortened genomic clone then was digested with HindIII, which also cleaves once in the pGEM-3Z polylinker region and once in the genomic clone, and was gel-purified and religated. Doing so also removed the HincII site in the polylinker region of pGEM-3Z. (iii) The 5′- and 3′-shortened genomic clone (24.7B−H−) was digested with HincII and was ligated with a PGK-NEO cassette that was isolated from pPNT with XhoI-XbaI and made blunt-ended with Klenow polymerase. This ligation recreated a XbaI site 5′ of the PGK-NEO cassette that was used in a diagnostic Southern blot. (iv) The targeted gene (24.7B−H−/NEO) was excised with BamHI and HindIII from pGEM-3Z and was gel-purified and ligated to pMC1TK (18), which was digested with BamHI and HindIII. The second exon was targeted by a vector (not shown) in which a 600-bp XhoI fragment spanning part of exon 2 and intron 2 was deleted and replaced by pMC1NeopolyA (Stratagene) in the opposite transcriptional orientation as the Cyp1b1 gene. The herpes simplex virus thymidine kinase gene also was added 5′ of this construct as described above.
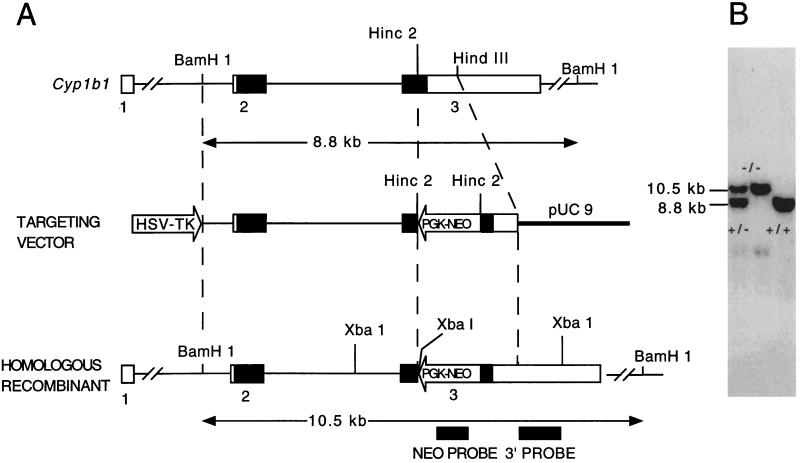
Figure 1.
(A) Strategy for the inactivation of the murine Cyp1b1 gene by homologous recombination. The genomic probes are indicated as solid boxes. The coding regions of the exons are indicated as solid boxes. The 5′- and 3′-untranslated regions of the gene are represented by open boxes. BamHI digestion of genomic DNA generated 8.8- and 10.5-kb bands for the wild-type (+) and Cyp1b1-null (−) alleles, respectively, indicated by horizontal arrows. (B) Southern blot analysis of mouse tail genomic DNA. Genomic DNA was isolated from tails and was digested with BamHI.
Production of Chimeric Mice.
The plasmid DNA used for targeting was purified by banding twice on cesium chloride. After linearization by ClaI, 5, 10, 20 or 40 μg of DNA were electroporated into Genome System (St. Louis) embryonic stem (ES) cells by using conditions previously described (19). The cells were grown on γ-irradiated G418-resistant mouse EFs in the presence of 1,000 units of lymphocyte inhibitory factor/ml (ESgro, Life Sciences). ES cell clones resistant to G418 and ganciclovir (a gift from Syntex, Palo Alto, CA) were selected and screened for homologous recombination. Clones having the expected Southern blot pattern for homologous recombination (see below) were regrown and injected into C57BL/6N blastocysts. The blastocysts were transferred into the uterus of a pseudopregnant recipient NIH Swiss mouse to produce animals showing chimerism. Male chimeras presenting >75% 129/Sv contribution, as determined by coat color, were bred with C57BL/6N females to determine whether the disrupted allele was transmitted to the germ line. Homozygotes were produced by crossing the F1 generation.
Genotyping of ES Cells and Mice.
DNA was isolated from ES cells and mouse tail clips as described (20) and was digested with either BamHI or XbaI. The digested DNAs were subjected to electrophoresis in 0.6% agarose gels and were transferred to Porablot NY plus nylon membranes (Macherey & Nagel) by using 0.4 M NaOH. The conditions for hybridization and washing have been described (19). A 3′-flanking probe derived from a HindIII-XbaI fragment (700 bp) of the wild-type Cyp1b1 allele was used. Fragments of 10.5 or 2.5 kb corresponding to the digestions with BamHI or XbaI, respectively, were diagnostic for the homologous recombinant allele (see Fig. 1B). Mice homozygous for the disrupted Cyp1b1 allele were designated CYP1B1−/− or CYP1B1-null.
Analysis of CYP1B1 Expression.
Microsomes were prepared from passage 2 mouse EFs according to a previously optimized protocol (3), and tissues were from 6-month-old male CYP1B1-null and wild-type mice administered 40 μg/kg TCDD (dissolved in corn oil) via i.p. injection and killed 24 hours later. Microsomes also were prepared from tissues of mice 1 day after the final gavage treatment with DMBA (see below). Protein concentrations were determined following the Pierce BCA kit manufacturer’s protocol (21). Microsomal proteins were separated by SDS/PAGE, and individual P450 proteins were visualized by Western immunoblotting using polyclonal antibodies against mouse CYP1B1 (22) and NADPH-P450 oxidoreductase (23) and mAbs against CYP1A1 (24) with enhanced chemiluminescence detection (Amersham Pharmacia). Northern blotting was carried out by using Poly(A)-containing mRNA prepared according to established protocols (25). RNA was subjected to electrophoresis on 1.3% agarose gels containing 1.1 M formaldehyde and blotting to Nytran maximum strength nylon membranes (Schleicher & Schuell). After UV crosslinking, the RNA was hybridized with the mouse CYP1B1 cDNA (9) and was exposed to a Molecular Dynamics PhosphorImager.
Clinical Chemical Parameters.
EDTA decoagulated blood, taken periorbitally, was centrifuged at 800 × g for 10 min, and plasma was recovered and analyzed on a Hitachi 717 automated plasma analyzer (Neuharlaching Hospital, Munich, Germany). Statistical differences were analyzed with the Student’s t test.
Preparation of EFs and DMBA Metabolism and Cytotoxicity.
To determine cytotoxicity of DMBA, primary mouse EFs at passage 2 were used. Pregnant mice at 14 days of gestation (determined by visual identification of vaginal plugs as day 1) were killed by carbon dioxide asphyxiation (26). The embryos were placed in PBS, and the internal organs and heads were removed. The remaining torsos were minced and placed into 2 ml of 0.25% trypsin for 30 min at 37°C. The torsos were suspended by pipetting, and the reaction was stopped with incubation medium (DMEM with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine). The cells were grown at 37°C in 5% carbon dioxide until confluent and were trypsinized and stored at −80°C. For each incubation, the cells were thawed and regrown for 1 day and were trypsinized and seeded into 96-well plates at a density of 3,000 cells/well in 100-μl of medium. After attaching and growing for 1 day, dilutions of DMBA (Aldrich Chem, Metuchen, NJ) in 100 μl of medium were added to the cells. The cells were incubated for 3 days (maximum linear growth), were fixed with 25% trichloroacetic acid, and were washed and stained with sulforhodamine as described (27). Control cells on the same 96-well plate were incubated with vehicle. Metabolism of DMBA was performed by using intact passage 2 cells incubated at 104 cells/cm2 in medium containing 5 μM DMBA. Controls were incubated in 0.1% dimethyl sulfoxide used as the vehicle for DMBA. Medium was collected after 1 hour of incubation, and DMBA metabolites were resolved by HPLC analysis by using a C18 reverse phase column (5).
Carcinogenesis Study.
Six-week-old female wild-type and CYP1B1-null mice were dosed intragastrically with 200 μg/kg DMBA in 50 μl of corn oil once daily for 5 days. After 3 days, the regimen was repeated until three rounds were completed. Mice were killed by carbon dioxide asphyxiation after either a sudden weight loss of >20% of body weight or a tumor >1 cm. Some animals died without these observable changes. All mice were analyzed by histopathology for the presence and identity of tumors.
RESULTS
Production of the CYP1B1-Null Mouse.
The Cyp1b1 gene was isolated from a 129/Sv genomic library and was used to prepare a targeting construct. The gene consists of three exons with a noncoding first exon (Fig. 1A). The PGK-NEO cassette was inserted into the coding region of exon 3 by using a HincII restriction site. Probes were prepared from outside the targeting construct region to identify specific recombinants. About 12% of the clones that survived the double selection with G418 and ganciclovir were homologous recombinants. Targeting exon 2 did not yield any homologous recombinants in 700 clones screened. An ES cell line was isolated that contained one allele in which specific recombination had occurred, as determined by Southern blot analysis, and was used to generate a mouse line. An 8.8-kb fragment was diagnostic for the wild-type Cyp1b1 allele, and the recombinant disrupted allele was identified by a 10.5-kb fragment (Fig. 1B). The larger size of the latter fragment is attributable to the addition of the 1.7-kb PGK-NEO cassette. The absence of nonspecific integration of the construct into the original ES cell clone was confirmed by the presence of a single 10.5-kb fragment detected by use of PGK-NEO as a probe (data not shown).
Characterization of the CYP1B1-Null Mouse.
On breeding of heterozygotes, mice homozygous for the disrupted allele were born at the expected frequency of ≈25%. They were indistinguishable from their wild-type and heterozygous litter mates, and gross pathological examination revealed no apparent abnormalities. The mice were fertile and produced normal-sized litters. Histopathological analysis of the breasts, cerebrum, cerebellum, thyroid, salivary glands, thymus, heart, lung, adrenal glands, liver, kidney, pancreas, intestine, prostate, urine bladder, testicles, uterus, or ovaries revealed no abnormalities. Clinical chemical parameters, including salts, glucose, urea, creatinine, alkaline phosphatase, triglycerides, and cholesterol, were in the normal ranges found in mice and were not different between wild-type and CYP1B1-null mice (P < 0.1, n = 10, data not shown).
Characterization of the EFs from Wild-Type and CYP1B1-Null Mice.
CYP1B1 is by far the predominant P450 in mouse EF (28). To evaluate the expression and possible function of CYP1B1 in these cells, we generated primary EF from CYP1B1-null and wild-type mice. CYP1B1 protein was detected in TCDD-treated EF from wild-type C57BL/6N and 129/Sv mice and in untreated and TCDD-treated 10T1/2 cells but was not found in untreated and TCDD-treated EF derived from the CYP1B1-null mice (data not shown). As shown in an earlier report, CYP1A1 was not detected in untreated or TCDD-treated EF from either wild-type or CYP1B1-null mice (28). Intact CYP1B1 mRNA also was not seen in untreated and TCDD- treated CYP1B1-null cells but was detected readily in untreated and TCDD-treated EF from wild-type mice.
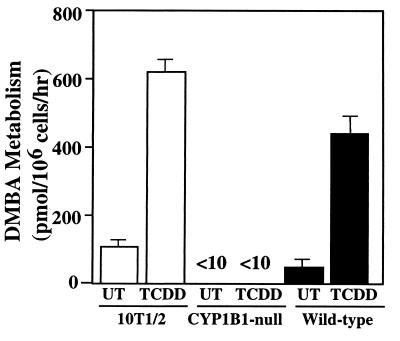
As a more sensitive assay for CYP1B1 expression, DMBA metabolism by EF was examined. These cells do not express CYP1A1 (28). EF derived from CYP1B1-null mice had lower DMBA metabolism than wild-type EF, and TCDD induction did not result in any additional DMBA metabolism (Fig. 2). In contrast, wild-type EF had both constitutive and TCDD-inducible metabolism, consistent with a CYP1B1 metabolism profile in which 3,4-, 9,10- and 10,11-dihydrodiols were major products (3, 5). Analysis of DMBA metabolites formed by CYP1B1-null EF revealed no 3,4-diol metabolite that was readily detected in the wild-type EF incubations. These data establish that CYP1B1 is not expressed in cells derived from the CYP1B1-null mouse and that this P450 is solely responsible for DMBA metabolism in wild-type EF, confirming previous studies (3, 28).
Figure 2.
Total metabolism of DMBA. 10T1/2 cells and CYP1B1-null and wild-type EFs were incubated with 5 μM DMBA, and the extent of total metabolism was measured by HPLC analysis. The distribution of products was similar for 10T1/2 and wild-type cells. The results are mean ± SD of three experiments.
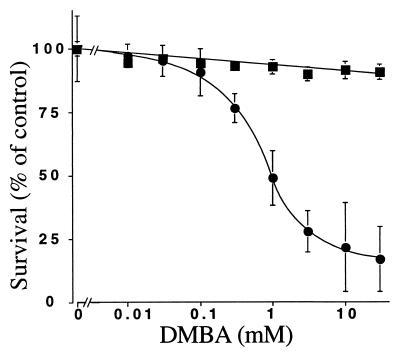
To determine whether the lack of CYP1B1 was of functional significance to DMBA toxicity, EF from wild-type and CYP1B1-null mice were treated with the carcinogen, and cell growth inhibition was measured. DMBA was highly toxic to wild-type cells with an IC50 of ≈1.0 μM DMBA (Fig. 3). In contrast, cells from CYP1B1-null mice were highly resistant to DMBA toxicity. This observation establishes that the toxicity in these cells is caused by the metabolism of DMBA by CYP1B1 to toxic metabolites, notably the mutagenic precursor 3,4-dihydrodiol.
Figure 3.
Growth inhibition of primary EFs derived from CYP1B1−/− and wild-type mice after exposure to DMBA. The cells were seeded and allowed to attach for 1 day and then were exposed to DMBA for 3 days. Only surviving cells could be fixed with trichloroacetic acid and stained with sulforhodamine. Control cells without DMBA were used as 100%. Mean ± SD (n = 8) for wild-type (●) and CYP1B1−/− (■) cells are given.
CYP1B1 and CYP1A1 Expression in Tissues from Wild-Type and CYP1B1-Null Mice.
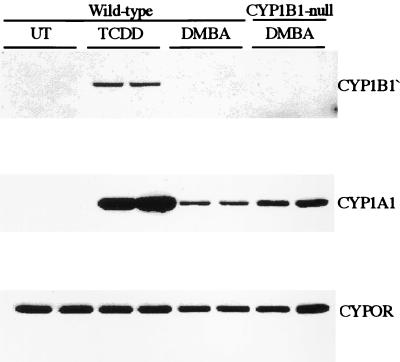
To establish that CYP1B1 is not expressed in the CYP1B1-null mice and to determine whether CYP1A1 was affected, immunoblotting was performed on microsomes isolated from tissues taken from TCDD-treated wild-type and CYP1B1-null mice. The CYP1B1 protein was readily induced in the lung of wild-type mice treated with TCDD (Fig. 4) but was absent in the lungs of CYP1B1-null mice treated under the same conditions (data not shown). Low levels of CYP1B1 were also inducible in uterus and kidney in the wild-type mice but not in CYP1B1-null mice. CYP1B1 is barely detectable, even after induction in liver of wild-type mice treated with TCDD as demonstrated (9). DMBA, when administered by gavage using the same protocol that was used for the carcinogenesis study (see below), also induced CYP1A1 in the lung; however, the extent of induction was less than one-fifth the level detected with TCDD (Fig. 4). Of interest, CYP1A1 induction in lung was 2- to 3-fold higher in CYP1B1-null mice. Again, no measurable CYP1B1 was seen after this treatment, even in wild-type mice, in lung, liver, kidney, or uterus. These data confirm that the Cyp1b1 allele is functionally inactive in the CYP1B1-null mouse line. In contrast, CYP1A1 was found to be induced in wild-type and CYP1B1-null mouse lungs by TCDD and to a lesser extent by DMBA. The NADPH-P450 oxidoreductase (CYPOR) was expressed in the CYP1B1-null at levels similar to that found in wild-type mice.
Figure 4.
Western immunoblot analysis of CYP1B1 expression in lung. Microsomes were isolated from the lungs of wild-type and CYP1B1-null mice treated with TCDD and DMBA and were subjected to Western blot analysis using an antibody directed against mouse CYP1B1, CYP1A1, and NADPH-P450 oxidoreductase (CYPOR).
DMBA Carcinogenesis.
To determine whether the results in vitro were predictive for sensitivities to DMBA in vivo, carcinogen bioassays were performed. DMBA was administered by gavage at a dosage level and frequency that induces tumors in normal mice (29). At 6 months after treatment, ≈60% of the wild-type mice had died from tumor burdens or had been killed because of severe morbidity whereas <10% of the CYP1B1-null mice had died. Autopsies revealed that 70% of wild-type mice developed lymphomas whereas only 7. 5% of the CYP1B1-null mice had these tumors throughout the course of the study (Table 1). A quarter of the mice also exhibited skin abnormalities with hyperplasia and various tumors in the most extreme cases. These abnormalities rarely were seen in the CYP1B1-null mice. Occasionally, tumors in ovaries were seen in wild-type mice. The CYP1B1-null mice had a higher level of lung adenocarcinomas as compared with wild-type mice (12.5 vs. 2.4%). However, it should be noted that these tumors were found at the end of the study, at which time only 40% of the wild-type mice were left for autopsy. Thus, if adenocarcinomas arise, they may be underestimated in the wild-type mice because of early mortality caused by the lymphomas. These data indicate that, for this oral route of administration, CYP1B1 is required for the activation of DMBA to carcinogenic metabolites that produce lymphomas.
Table 1.
Tumors in wild-type and CYP1B1-null mice after DMBA administration
| Pathology | Wild-type
|
CYP1B1-null
|
||||||
|---|---|---|---|---|---|---|---|---|
| Males, n = 20 | Female, n = 21 | Total, n = 41 | Percent | Males, n = 21 | Female, n = 19 | Total, n = 40 | Percent | |
| Lymphoblastic lymphoma | 13 | 16 | 29 | 70.0 | 1 | 2 | 3 | 7.5 |
| Hyperplasia of the skin, verruciform, follicular | 2 | 3 | 5 | 12.2 | 1 | 0 | 1 | 2.5 |
| Keratoakanthoma, skin | 2 | 2 | 4 | 9.8 | 0 | 1 | 1 | 2.5 |
| Squamous cell carcinoma of the skin | 0 | 1 | 1 | 2.4 | 0 | 0 | 0 | |
| Adenoma of the lung | 0 | 0 | 0 | 3 | 1 | 4 | 10.0 | |
| Adenocarcinoma of the lung | 0 | 1 | 1 | 2.4 | 3 | 2 | 5 | 12.5 |
| Hemangioendothelioma of the liver | 1 | 1 | 2 | 4.9 | 0 | 0 | 0 | |
| Hemangiosarcoma of the liver | 0 | 0 | 0 | 1 | 0 | 1 | 2.0 | |
| Cystic endometrial hyperplasia, uterus | 0 | 1 | 1 | 2.4 | 0 | 0 | 0 | |
| Histiocytic sarcoma, uterus | 0 | 0 | 0 | 0 | 1 | 1 | 2.5 | |
| Hemangioma, uterus | 0 | 1 | 1 | 2.4 | 0 | 0 | 0 | |
| Kystoma of the ovary | 0 | 2 | 2 | 4.9 | 0 | 0 | 0 | |
| Granulosa cell tumor, ovary | 0 | 1 | 2 | 4.9 | 0 | 0 | 0 | |
The mice were administered 200 μg of DMBA/mouse (30 g) by gavage in corn oil 5 days a week for 3 weeks. The observation period was 6 months.
DISCUSSION
The absence of any obvious change in phenotype for the CYP1B1-null mice shows that CYP1B1 is not required for mammalian development and physiological homeostasis. This does not exclude physiological functions because it remains a possibility that other processes can replace such functions through gene compensation. This was unexpected because CYP1B1 is well conserved across mammalian species as compared with other xenobiotic-metabolizing P450s. Although CYP1B1 is expressed in a number of extrahepatic reproductive tissues such as the ovary, testis, uterus, and mammary gland (4, 8, 10, 14), it did not appear to have a major impact on reproductive vitality. Thus, the normal reproductive capacity and lack of any phenotype in these mice suggest that it is not required for physiological homeostasis. The absence of detectable CYP1A1 in EFs from CYP1B1-null mice, which is also typical of wild-type cells (28), indicates that this cell type does not have a compensatory increased expression of CYP1A1 that results from loss of CYP1B1. Western immunoblotting of microsomes from cultured whole mouse EFs and also from mouse tissues in vivo showed that constitutive and TCDD-induced CYP1B1 expression was lost in CYP1B1-null mice.
CYP1B1 has been shown to be inducible by TCDD and polycylic aromatic hydrocarbons and is involved in the metabolic activation of DMBA and benzo[a]pyrene (3, 5, 6). Recent work revealed that CYP1B1 is the predominant P450 in fibroblasts in which CYP1A1 is not significantly expressed (28). In the mouse, both basal expression and inducible expression of CYP1B1 are regulated by the aryl hydrocarbon receptor. CYP1A1 has similar properties to CYP1B1, but their relative contributions to chemical carcinogenesis are likely to depend on the tissue distributions of each form, which are very different, and on their regioselective metabolism profiles for polycyclic aromatic hydrocarbons. Embryonic fibroblasts from CYP1B1-null mice were far more resistant to DMBA toxicity than cells from wild-type mice, indicating that CYP1B1 is mainly responsible for polycyclic aromatic hydrocarbon activation in these cells, presumably via conversion to dihydrodiol epoxides. This is not surprising because previous work has shown that CYP1B1 is both constitutively expressed and induced by aryl hydrocarbon receptor agonists like TCDD in these cells whereas CYP1A1 is barely detectable, even after induction (28). CYP1B1 is also very effective in converting DMBA to its toxic and procarcinogenic metabolite, 3,4-dihydrodiol. Because CYP1B1 is by far the dominant P450 form in C3H10T1/2 cells, the activation of numerous polycyclic aromatic hydrocarbons that has been studied in these cells is presumably through CYP1B1. Recent work has shown that human CYP1B1 readily converts polycyclic aromatic hydrocarbon dihydrodiols to mutagenic metabolites, presumably the dihydrodiol epoxides (30, 31).
The role of CYP1B1 in DMBA carcinogenesis was established by using a bioassay with a high dose exposure. CYP1B1-null mice were protected against DMBA-induced tumors whereas their wild-type counterparts developed malignant lymphomas. These data suggest that CYP1B1 is an essential enzyme for metabolic activation and thus the carcinogenic potential of DMBA. This is particularly remarkable because CYP1B1 is only expressed at very low levels in the liver and kidney, even after polycyclic aromatic hydrocarbon induction (4, 9), whereas CYP1A1 is present at substantial levels after exposure to polycyclic aromatic hydrocarbons, including 3-methylcholanthrene, benzo[a]pyrene, or β-napthoflavone (32). Indeed, under the route of administration used in the present study to induce tumors in wild-type mice, CYP1A1 is induced in major organs (liver, lung, and kidney) after DMBA administration, although the extent of induction is less than that achieved with TCDD. Of interest, the extent of CYP1A1 induction by DMBA was increased in the lungs of CYP1B1-null mice compared with wild-type mice. Because CYP1B1 expression is essentially absent in each of the major metabolic organs, removal of CYP1B1 is unlikely to alter DMBA pharmacokinetics in the CYP1B1-null mice. Perhaps, deletion of CYP1B1 may enhance the potency of CYP1A1 induction in the lung through increasing DMBA concentrations within the tissue. Although this modest difference in CYP1A1 in the lung may cause the apparent increase in lung tumorigenesis in the CYP1B1-null mice, it seems very unlikely that this can impact the dramatic differences in generation of the lymphoblastomas. CYP1A1 is severalfold more active than CYP1B1 in metabolizing DMBA (33) and therefore must contribute to the vast majority of the metabolism of DMBA in both wild-type and CYP1B1-null mice. However, CYP1A1 produces only a small amount, on a percentage of total metabolite basis, of the putative proximate carcinogenic metabolite 3,4-dihydrodiol (1 vs. 25% for CYP1B1) and therefore may be relatively ineffective in activating this hydrocarbon. Nevertheless, most of the circulating 3,4-dihydrodiol should arise from the high levels of CYP1A1 induced in the major organs after DMBA administration. This difference in activation is less marked for benzo[a]pyrene in which 7,8-dyhydrodiol is effectively produced by CYP1A1 (3). Thus, if regioselective differences are a key factor, benzo[a]pyrene should be similarly carcinogenic in wild-type and CYP1B1-null mice. Whether this is so remains to be investigated. The resistance of CYP1B1-null mice to DMBA-induced lymphomas is consistent with the recent finding of both constitutive and DMBA-inducible CYP1B1 in bone marrow cells (34). At present, we cannot exclude the possibility that CYP1B1 plays an additional role in the progression of tumorigenesis. In this regard, CYP1B1 deficiency was found to accelerate fibroblast differentiation in vitro (data not shown).
It recently was established that an autosomal recessive mutation in CYP1B1 was found to be associated with development of primary congenital glaucoma in humans (35, 36). Gross examination of eyes from the CYP1B1-null revealed no evidence for glaucoma, and the animals were not blind, as assessed by standard behavioral comparisons with wild-type mice in their response to light and dark. It remains a possibility that differences in substrate specificity and tissue distribution between the human and mouse CYP1B1 may account for this difference. In the absence of this, these data indicate that lack of CYP1B1 may be only one factor required for the development of glaucoma and that other genes may contribute to progression of the disease. Nevertheless, the human studies confirm that CYP1B1 indeed has a role in developmental processes, as originally suggested from the unusual tissue distribution of this form. Further studies should be carried out to establish firmly the role of CYP1B1 in the eye.
Further studies using different routes of administration need to be carried out to determine whether CYP1B1 is required for carcinogenesis of polycyclic aromatic hydrocarbons in skin, lung, and other epithelial tissues. Because this study establishes that CYP1B1 is critical for carcinogenesis, it may be important to determine whether a genetic differences in expression of this enzyme plays a role in cancer susceptibility in humans.
Acknowledgments
This work was supported in part by Deutsche Forschungsgemeinschaft Grant DO 242/9-1 (to J.T.M.B. and J.D.) and National Institutes of Health Grants T32 ES07015 and CA 16265 (to C.R.J.).
ABBREVIATIONS
- DMBA
7,12-dimethylbenz[a]anthracene
- PGK-NEO
phosphoglycerate kinase1-phosphoribosyltransferase II gene cassette
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- kb
kilobase
- ES cell
embryonic stem cell
- EF
embryonic fibroblast
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Nelson D R, Koymans L, Kamataki T, Stegeman J J, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R W, et al. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Guengerich F P. Chem Biol Interact. 1997;106:161–182. doi: 10.1016/s0009-2797(97)00068-9. [DOI] [PubMed] [Google Scholar]
- 3.Pottenger L H, Jefcoate C R. Carcinogenesis. 1990;11:321–327. doi: 10.1093/carcin/11.2.321. [DOI] [PubMed] [Google Scholar]
- 4.Otto S, Marcus C, Pidgeon C, Jefcoate C. Endocrinology. 1991;129:970–982. doi: 10.1210/endo-129-2-970. [DOI] [PubMed] [Google Scholar]
- 5.Pottenger L H, Christou M, Jefcoate C R. Arch Biochem Biophys. 1991;286:488–497. doi: 10.1016/0003-9861(91)90070-y. [DOI] [PubMed] [Google Scholar]
- 6.Crespi C L, Penman B W, Steimel D T, Smith T, Yang C S, Sutter T R. Mutagenesis. 1997;12:83–89. doi: 10.1093/mutage/12.2.83. [DOI] [PubMed] [Google Scholar]
- 7.Sutter T R, Guzman K, Dold K M, Greenlee W F. Science. 1991;254:415–418. doi: 10.1126/science.1925598. [DOI] [PubMed] [Google Scholar]
- 8.Sutter T R, Tang Y M, Hayes C L, Wo Y Y, Jabs E W, Li X, Yin H, Cody C W, Greenlee W F. J Biol Chem. 1994;269:13092–13099. [PubMed] [Google Scholar]
- 9.Savas U, Bhattacharyya K K, Christou M, Alexander D L, Jefcoate C R. J Biol Chem. 1994;269:14905–14911. [PubMed] [Google Scholar]
- 10.Bhattacharyya K K, Brake P B, Eltom S E, Otto S A, Jefcoate C R. J Biol Chem. 1995;270:11595–11602. doi: 10.1074/jbc.270.19.11595. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y M, Wo Y Y P, Stewart J, Hawkins A L, Griffin C A, Sutter T R, Greenlee W F. J Biol Chem. 1996;271:28324–28330. doi: 10.1074/jbc.271.45.28324. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Savas U, Alexander D L, Jefcoate C R. J Biol Chem. 1998;273:5174–5183. doi: 10.1074/jbc.273.9.5174. [DOI] [PubMed] [Google Scholar]
- 13.Brake P B, Jefcoate C R. Endocrinology. 1995;136:5034–5041. doi: 10.1210/endo.136.11.7588239. [DOI] [PubMed] [Google Scholar]
- 14.Otto S, Bhattacharyya K K, Jefcoate C R. Endocrinology. 1992;131:3067–3076. doi: 10.1210/endo.131.6.1332854. [DOI] [PubMed] [Google Scholar]
- 15.Kimura S, Gonzalez F J, Nebert D W. Mol Cell Biol. 1986;6:1471–1477. doi: 10.1128/mcb.6.5.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 17.Bradley A. Teratocarcinoma and Embryonic Stem Cells: A Proctical Approach. Oxford: IRL; 1987. [Google Scholar]
- 18.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 19.Lee S S, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 22.Savas U, Carstens C P, Jefcoate C R. Arch Biochem Biophys. 1997;347:181–192. doi: 10.1006/abbi.1997.0339. [DOI] [PubMed] [Google Scholar]
- 23.Yamano S, Aoyama T, McBride O W, Hardwick J P, Gelboin H V, Gonzalez F J. Mol Pharmacol. 1989;36:83–88. [PubMed] [Google Scholar]
- 24.Goldfarb I, Korzekwa K, Krausz K W, Gonzalez F, Gelboin H V. Biochem Pharmacol. 1993;46:787–790. doi: 10.1016/0006-2952(93)90485-f. [DOI] [PubMed] [Google Scholar]
- 25.Badley J E, Bishop G A, St. John T, Frelinger J A. Biotechniques. 1988;6:114–116. [PubMed] [Google Scholar]
- 26.Reznikoff C A, Bertram J S, Brankow D W, Heidelberger C. Cancer Res. 1973;33:3239–3249. [PubMed] [Google Scholar]
- 27.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren J T, Bokesch H, Kenney S, Boyd M R. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 28.Alexander D L, Eltom S E, Jefcoate C R. Cancer Res. 1997;57:4498–4506. [PubMed] [Google Scholar]
- 29.Qing W G, Conti C J, LaBate M, Johnston D, Slaga T J, MacLeod M C. Carcinogenesis. 1997;18:553–559. doi: 10.1093/carcin/18.3.553. [DOI] [PubMed] [Google Scholar]
- 30.Shimada T, Gillam E M, Sutter T R, Strickland P T, Guengerich F P, Yamazaki H. Drug Metab Dispos. 1997;25:617–622. [PubMed] [Google Scholar]
- 31.Shimada T, Wunsch R M, Hanna I H, Sutter T R, Guengerich F P, Gillam E M. Arch Biochem Biophys. 1998;357:111–120. doi: 10.1006/abbi.1998.0808. [DOI] [PubMed] [Google Scholar]
- 32.Whitlock J P, Jr, Chichester C H, Bedgood R M, Okino S T, Ko H P, Ma Q, Dong L, Li H, Clarke-Katzenberg R. Drug Metab Rev. 1997;29:1107–1127. doi: 10.3109/03602539709002245. [DOI] [PubMed] [Google Scholar]
- 33.Christou M, Savas U, Spink D C, Gierthy J F, Jefcoate C R. Carcinogenesis. 1994;15:725–732. doi: 10.1093/carcin/15.4.725. [DOI] [PubMed] [Google Scholar]
- 34.Heidel, S. M., Czuprynski, C. J. & Jefcoate, C. R. (1998) Mol. Pharmacol.54., [DOI] [PubMed]
- 35.Stoilov I, Akarsu A N, Sarfarazi M. Hum Mol Genet. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 36.Sarfarazi M. Hum Mol Genet. 1997;6:1667–1677. doi: 10.1093/hmg/6.10.1667. [DOI] [PubMed] [Google Scholar]