Abstract
Arc repressor mutants containing from three to 15 multiple-alanine substitutions have spectral properties expected for native Arc proteins, form heterodimers with wild-type Arc, denature cooperatively with Tms equal to or greater than wild type, and, in some cases, fold as much as 30-fold faster and unfold as much as 50-fold slower than wild type. Two of the mutants, containing a total of 14 different substitutions, also footprint operator DNA in vitro. The stability of some of the proteins with multiple-alanine mutations is significantly greater than that predicted from the sum of the single substitutions, suggesting that a subset of the wild-type residues in Arc may interact in an unfavorable fashion. Overall, these results show that almost half of the residues in Arc can be replaced by alanine en masse without compromising the ability of this small, homodimeric protein to fold into a stable, native-like structure.
Keywords: hyper-stable mutants, fast-folding mutants, operator-DNA binding, heterodimer formation, nonadditivity
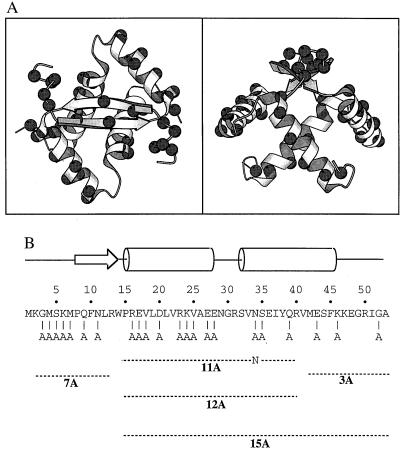
Studies of random and site-directed mutations in proteins as well as design and minimization experiments have shown that many protein folds display significant tolerance to amino acid substitutions (1–17). Our group has been studying the Arc repressor of bacteriophage P22, which folds as a homodimer of 53-residue subunits with a simple architecture consisting of a β-sheet and four α-helices (refs. 18–20; Fig. 1A). Residues 1–7 are disordered in solution (18, 19) but fold and contact the sugar-phosphate backbone in the Arc-operator DNA complex (20). Residues 8–14 of each monomer pair to form the antiparallel β-sheet, which binds in the major groove of operator DNA (20). Residues 16–28 and 32–46 form α-helices A and B, respectively, which pack against each other and the β-sheet to form a roughly globular domain. Dimer dissociation results in denaturation, indicating that monomers of Arc cannot fold in isolation (21).
Figure 1.
(A) Orthogonal views of the Arc repressor dimer prepared by using molscript (41). The Cα atoms at each of the 22 positions (44 in dimer) substituted by alanines are marked by spheres. (B) Arc sequence, secondary structure, and positions of multiple-alanine substitutions in the 3A, 7A, 11A, 12A, and 15A mutants.
In early mutational studies of Arc (1, 22), many residues were shown to tolerate amino acid substitutions without compromising the structural integrity of the protein. Later, a complete set of alanine-substitution mutants was constructed and the stability and folding properties of each of these mutant proteins were determined (23, 24). At almost half of the residues in Arc, individual alanine substitutions caused less than 5°C changes in Tm and less than 1 kcal/mol changes in the free energy of unfolding. The vast majority of these “neutral stability” positions were on the protein surface. One important question that could not be addressed by these studies, however, was whether these residues still might be important for folding but in a way in which one or a few could be changed without consequence. Here, we describe the properties of a set of Arc mutants that contain from three to 15 of the alanine substitutions, which previously were shown to have neutral or minor stability effects. Remarkably, some of these mutant proteins are significantly more stable than wild-type Arc and show both increased folding rates and decreased unfolding rates.
METHODS
Mutagenesis and Construction.
The multiple-alanine mutants were constructed by standard methods of site-directed mutagenesis in the expression plasmid pET800 (25). The structure of the arc gene in each mutant plasmid was verified by DNA sequencing. For each mutant, the st11 tag sequence (HHHHHHKNQHE) also was added to the C terminus to allow affinity purification and to prevent intracellular proteolysis (26). This sequence does not substantially alter the stability or folding properties of Arc. Plasmids were propagated in Escherichia coli strain X90(λDE3), and mutant proteins were purified by chromatography on Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) and SP-Sephadex (Pharmacia) as described (26). An additional Pep-RPC chromatography step was used for purification of the 15A mutant. Arc concentrations in monomer equivalents were determined by using an extinction coefficient at 280 nm of 6756 M−1⋅cm−1. Mutants were stored in buffer S (50 mM Tris⋅HCl, pH 7.5/0.2 mM EDTA/250 mM KCl).
Biophysical Measurements.
CD spectra were taken by using 10 μM protein at 25°C in buffer S and an Aviv 60 HDS CD spectrometer (Aviv Associates, Lakewood, NJ). Fluorescence emission spectra from 300 to 400 nm (excitation 280 nm) were taken under the same conditions by using 5 μM protein. Equilibrium denaturation was performed in buffer S and monitored by CD ellipticity at 222 nm (thermal melts) or 234 nm (GuHCl melts). Denaturation curves were fit by using nonlinear-least-squares methods and equations for a two-state, unfolded monomer to native dimer transition (21, 26). Kinetic unfolding and refolding experiments were performed at 25°C by using an Applied Photophysics DX17.MV stopped-flow instrument to monitor changes in fluorescence; refolding data were fit to a hyperbolic function for a homodimeric association and refolding reaction, and unfolding data were fit to a single exponential (27, 28). NMR spectra were collected at 20°C by using a Bruker AMX 500 MHz Spectrometer. Free induction decays were averaged over 16,384 scans with 8 K complex data points. Samples contained 95 μM protein in 10 mM potassium phosphate (pH 7.4) and 10% D2O. Chemical shifts were referenced to an internal trimethylsilyl-propionate standard.
DNA-Binding and Heterodimerization Assays.
Footprinting of the wild-type arc operator with 1,10-phenanthroline copper was performed as described (29). To assay heterodimer formation, a 1-ml solution containing 15 μM untagged wild-type Arc and 15 μM mutant protein in buffer A (10 mM Tris⋅HCl/100 mM potassium phosphate/250 mM KCl, pH 8.0) was heated to 70°C for 3 min and then cooled to room temperature. In a separate microcentrifuge tube, 0.5 ml of a 50% slurry of Ni-NTA agarose (Qiagen) was added and centrifuged for 30 s, and the supernatant was withdrawn. The protein mixture then was added to the Ni-NTA resin, mixed by several inversion steps, and left for 5 min at room temperature. The resin was centrifuged for 30 s and the supernatant (flow-through fraction) was withdrawn. One milliliter of buffer A was added to the resin, mixed by inversion, and centrifuged for 30 s, and the supernatant (wash fraction) was withdrawn. One milliliter of buffer A plus 250 mM imidazole (final pH 7.0) was added, allowed to sit for 5 min, and then withdrawn (elution fraction) after centrifugation. Forty microliters of the load, flow-through, wash, and elution fractions were then electrophoresed on 15% Laemmli SDS gels and stained with Coomassie blue, and protein bands were quantified by densitometry.
RESULTS
Multiple-Alanine Mutants.
Fig. 1B shows the residues mutated to alanine in five Arc mutants, which are named 3A, 7A, 11A, 12A, and 15A based on the total number of alanine substitutions in each monomer. Note that because Arc is a homodimer, the mutant proteins actually contain a total of 6, 14, 22, 24, and 30 aa substitutions, respectively. The 3A, 7A, and 12A mutants initially were constructed because they include all of the neutral alanine mutations in Arc except MA1 and KA2 (which alter translational initiation or processing sites) and RA13 (which removes a restriction site used for construction). The 11A mutant is identical to 12A without the NA34 mutation. The 15A mutant contains all of the changes in both the 3A and 12A mutants.
Spectral Properties of Mutants.
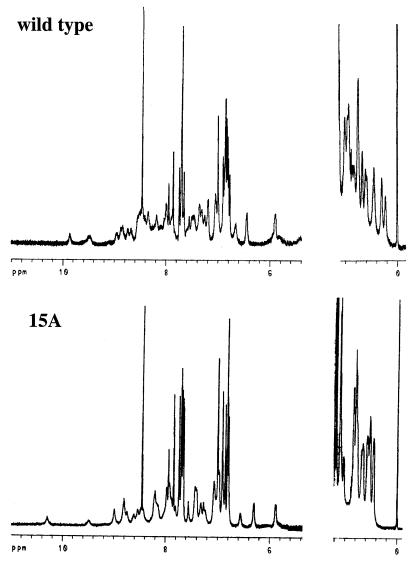
The purified 3A, 7A, 11A, 12A, and 15A mutants had CD and fluorescence spectra that were similar to those of wild-type Arc (data not shown). For example, each of the mutants had CD ellipticities at 222 nm within 5% of the wild-type value (Table 1) and had centers of fluorescence spectral mass that were blue-shifted by 11–15 nm relative to denatured Arc. The former data indicate that the multiple-alanine mutants have α-helical contents similar to wild type; the latter data suggest that the sole tryptophan in Arc (Trp-14) is largely buried in the hydrophobic core of each mutant. One-dimensional NMR spectra were taken for the 15A mutant and wild-type Arc (Fig. 2). The 15A spectrum shows good chemical dispersion in the amide-aromatic and methyl regions as expected for a well-folded protein. Phe-10, Trp-14, Tyr-38, and Phe-45 are the only aromatic residues in Arc and each is an important component of the hydrophobic core. Most aromatic resonances in the wild-type and 15A spectra have similar chemical shifts, suggesting that the aromatic side chains in the cores of these two proteins have similar environments. Taken together, the different spectral probes suggest that the 3A, 7A, 11A, 12A, and 15A mutants are able to adopt native structures with many similarities to wild-type Arc.
Table 1.
Spectroscopic, stability, and kinetic properties of mutants
| Mutants | Ellipticity 222 nm, mdeg | Fluorescence center of mass, nm |
Tm, °C
|
ΔGu, kcal/mol
|
Reduction in unfolding rate2 | Increase in refolding rate3 | ||
|---|---|---|---|---|---|---|---|---|
| Observed | Predicted1 | Observed | Predicted1 | |||||
| 3A | −73.1 | 336.5 | 57.7 ± 0.6 | 56.6 ± 1.1 | – | 10.9 ± 0.9 | 0.92 | – |
| 7A | −69.4 | 339.5 | 59.8 ± 0.6 | 54.9 ± 1.6 | – | 12.1 ± 1.3 | 2.9 | – |
| 11A | −70.5 | 338.2 | 69.2 ± 0.6 | 54.7 ± 2.0 | – | 7.3 ± 1.7 | 15.4 | – |
| 12A | −70.5 | 339.9 | 71.1 ± 0.6 | 59.8 ± 2.1 | 16.0 ± 0.5 | 7.3 ± 1.7 | 31.0 | 34.2 |
| 15A | −70.8 | 340.1 | 72.3 ± 0.6 | 59.1 ± 2.3 | 15.2 ± 0.5 | 7.2 ± 1.9 | 53.5 | 26.6 |
| Arc-native | −69.4 | 336.7 | 57.9 ± 0.6 | 57.9 ± 0.6 | 11.0 ± 0.5 | 11.0 ± 0.5 | 1.0 | 1.0 |
| Arc-denatured4 | 351.4 | |||||||
Figure 2.
NMR spectra in the amide-aromatic and methyl regions of wild-type Arc-st11 and the 15A mutant.
Operator-DNA Binding.
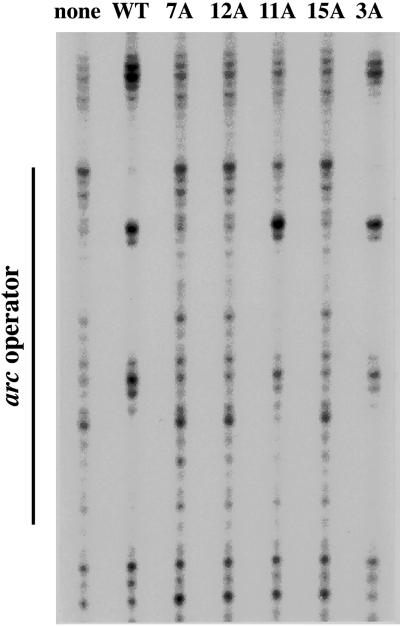
The ability of the mutant proteins to bind operator DNA was probed by footprinting (Fig. 3). The 3A and 11A mutants gave footprints similar to wild-type Arc, whereas the remaining mutants failed to protect the operator. The operator-binding activity of the 3A and 11A mutants indicates that these proteins have structures similar to wild-type Arc. As discussed below, the inactivity of the 7A, 12A, and 15A mutants is likely to result from alterations in critical DNA-binding residues.
Figure 3.
Copper-phenanthroline footprinting of the arc operator at 22°C by wild-type Arc-st11 and the multiple-alanine mutants at protein concentrations of 1 μM in Tris⋅HCl (pH 7.5), 3 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 100 μg/ml BSA, and 0.02% Nonidet P-40.
Heterodimer Formation with Wild-Type Arc.
Each of the multiple-alanine mutants was able to form heterodimers with wild-type Arc. In these experiments (Table 2), the 3A, 7A, 11A, 12A, and 15A mutants, which contain 6His sequences within the st11 C-terminal tags, were individually mixed with untagged wild-type Arc, and the retention of this latter protein on Ni-NTA resin was used to assay heterodimer formation. Control experiments showed that untagged wild-type Arc was not retained by the Ni-NTA resin by itself or when mixed with a λ-repressor N-terminal variant containing a 6His sequence. Because folding and dimerization are the same process for Arc (21), these data provide further evidence that the 3A, 7A, 11A, 12A, and 15A mutants are capable of adopting an essentially wild-type fold.
Table 2.
Heterodimerization assay; ability of 6His-tagged multiple-alanine mutants to bind untagged, wild-type Arc to Ni-NTA agarose
| 6His-tagged protein | Fraction untagged Arc in
|
|
|---|---|---|
| Ni-NTA flow-through + wash | Ni-NTA imidazole elution | |
| 3A | 0.89 | 0.11 |
| 7A | 0.78 | 0.22 |
| 11A | 0.73 | 0.27 |
| 12A | 0.66 | 0.34 |
| 15A | 0.56 | 0.44 |
| λN-domain-H6 | 1.00 | 0.00 |
| None | 0.99 | 0.01 |
Equilibrium Stability.
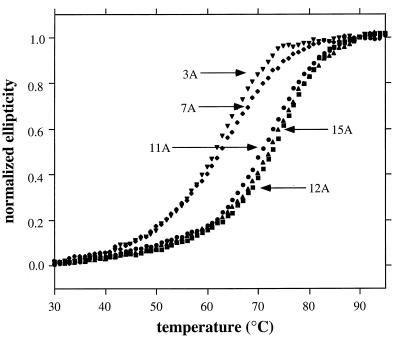
All of the multiple-alanine mutants displayed cooperative thermal denaturation transitions (Fig. 4). The 3A and 7A mutants had Tms similar to wild-type Arc, whereas the 11A, 12A, and 15A mutants were significantly more stable, with Tms increased by 11–14°C (Table 1). In GuHCl denaturation studies, the 12A and 15A mutants were also more stable than wild type, in this case, by 4–5 kcal/mol (Table 1); GuHCl denaturation of the 3A, 7A, and 11A mutants was not performed.
Figure 4.
Thermal denaturation of the multiple-alanine mutants (10 μM) in buffer S. Normalized ellipticity is calculated as (ɛ − ɛ25)/(ɛ95 − ɛ25) where ɛ, ɛ25, and ɛ95 are the CD ellipticities at 222 nm at any temperature, 25°C, and 95°C, respectively.
Unfolding and Refolding Kinetics.
Part of the enhanced stability of the 7A, 11A, 12A, and 15A mutants results from a reduction in the rate of dimer unfolding and dissociation (Fig. 5A; Table 1). These rates were determined after jumps to high concentrations of urea for 3A and 7A or GuHCl for the hyper-stable 11A, 12A, and 15A mutants. The 3A mutant unfolds slightly faster than wild-type Arc, whereas the 7A, 11A, 12A, and 15A mutants unfold from 3- to 53-fold more slowly than wild type at reference concentrations of 4.6 M urea or 4.5 GuHCl (Table 1). We did not attempt to estimate unfolding rates for the mutants in the absence of denaturant, as previous studies have shown that curvature in the log(k) versus denaturant plots leads to significant extrapolation errors at low denaturant concentrations (30).
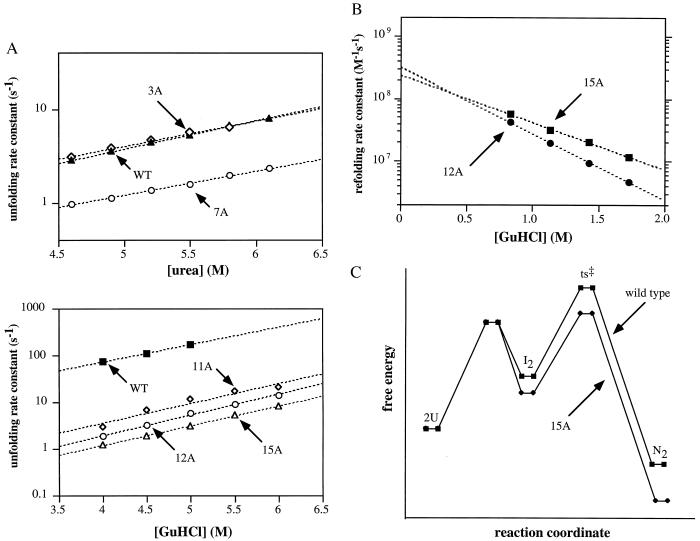
Figure 5.
(A) Unfolding and dissociation rate constants determined after jumps into different concentrations of urea or GuHCl (25°C, buffer S). (B) Refolding and association rate constants determined after dilution from 6 M GuHCl. (C) Reaction-coordinate diagrams for wild-type Arc and the 15A mutant.
Rates of refolding and association were determined for the 12A and 15A mutants at different GuHCl concentrations after dilution from 6 M GuHCl (Fig. 5B). Extrapolation of these data to 0 M denaturant indicates that each of these proteins refolds and dimerizes with a second-order rate constant of 2.5–3.5⋅108 M−1⋅s−1, more than 20-fold faster than wild-type Arc (kf ≈ 107 M−1⋅s−1; ref. 27). Thus, the enhanced stabilities of the 12A and 15A mutants arise both from faster rates of refolding and from slower rates of unfolding (Table 1).
DISCUSSION
As a set, the five multiple-alanine mutants studied here contain substitutions for 22 of the 53 residues of wild-type Arc. Previous work had shown that these residue positions could be individually mutated to alanine with only small changes in protein stability. Here, we have demonstrated that these residues can be substituted by alanine en masse without compromising Arc stability or structure. The 3A, 7A, 11A, 12A, and 15A mutants have CD and fluorescence spectra expected for native Arc proteins, display cooperative thermal denaturation with Tms equal to or greater than wild-type Arc, and are capable of forming heterodimers with wild-type Arc. These properties suggest strongly that each of these mutants can stably adopt a structure similar to that of wild-type Arc. Moreover, the 3A and 11A mutants footprint arc operator DNA in vitro and the 15A mutant has an NMR spectrum with many similarities to wild type, providing additional evidence of a native-like Arc fold for these mutants.
Why do the 7A, 12A, and 15A mutants fail to bind operator DNA? The inactivity of the 7A mutant is easily rationalized as each of the individual mutations in this protein affect DNA-binding residues and cause substantial decreases in operator-binding affinity (20, 29). The inactivity of the 12A and 15A mutants must, in part, be caused by the NA34 substitution, as this sequence change is the only difference between the inactive 12A and the active 11A mutant. The NA34 single mutant causes a large decrease in operator affinity, by uncoupling phosphate backbone contacts from base contacts mediated by β-sheet residues (20, 29). Mutations other than NA34 also must contribute to the inability of 12A and 15A to bind operator DNA, as the single NA34 mutant does footprint the operator (29). The RA23 and SA35 mutations are good candidates as these sequence changes also reduce operator affinity in single mutants (29).
One issue that arises in considering the effects of the multiple mutations on protein stability is additivity. Table 1 shows the Tms and ΔGus predicted for the multiple-alanine mutants based on summing the effects of the single-alanine mutations (23). Clearly, the 11A, 12A, and 15A mutants are more stable than expected based on the effects of the single substitutions. Nonadditivity of this type suggests either that the wild-type residues that were replaced interact unfavorably or that the mutant residues that were introduced interact favorably. It is unlikely, based on the wild-type structure, that the mutant alanines would be close enough to interact directly. As a result, we favor the model that the nonadditivity arises from unfavorable interactions among a subset of the wild-type residues that are replaced in the 11A, 12A, and 15A mutants. Alternatively, differences in interactions between the wild-type and mutant residues might destabilize the mutant unfolded state, as has been suggested to explain nonadditivity of multiple-alanine mutations that stabilize myoglobin (31). We note, however, that this latter model by itself could not explain the decreased unfolding rates of the 11A, 12A, and 15A mutants relative to wild type.
Together, the 7A and 15A mutants contain substitutions at all 22 of the residue positions chosen for mutation (Fig. 1B). The 7A mutations alter residues that do not serve important structural roles in Arc’s N-terminal arm and β-sheet and thus it is not surprising that this mutant folds stably. The 15A mutations, by contrast, change numerous residues that appear to be make stabilizing interactions in helix A and helix B. For example, the RA16, EA17, DA20, RA23, EA28, EA43, and KA46 mutations remove hydrogen-bonding and salt-bridge interactions, and the VA18 and VA25 mutations alter residues that are more than 85% buried in the hydrophobic core. Because 15A not only folds but is substantially more stable than wild-type Arc, it is clear that the wild-type residues at these positions are needed neither singly nor in aggregate for Arc folding. Alanine is one of the best helix-forming residues (32, 33) and because 14 of the residues replaced in 15A are α-helical, it is possible that this compensates for many of the wild-type interactions lost in the mutant. Some additional factors appear necessary, however, to account for the observed nonadditivity. Specifically, as noted above, it seems likely that some combination of the side chains replaced in 15A interact unfavorably in the wild-type protein. We note that 18 of the 30 side chains altered in the 15A dimer affect charged residues, and thus it is possible that some of these charged residues have unfavorable electrostatic interactions in wild-type Arc.
The increased stability of the 15A mutant is caused by faster refolding as well as by slower unfolding. The same is true for 11A and 12A, each of which contains a subset of the 15A mutations. In the current model for wild-type Arc refolding (27, 28, 30, 34), two denatured monomers collide at the diffusion-controlled rate (≈109 M−1⋅s−1) to form a partially structured intermediate dimer (I2) that kinetically partitions between dissociation to monomers (99%) and completion of folding (1%). The same model could explain 15A refolding if dissociation of the mutant intermediate dimer was less favored (75%) and completion of folding more favored (25%). This model is illustrated schematically in the reaction-coordinate diagram in Fig. 5C, which postulates that the I2 and native forms of 15A are more stable than the corresponding wild-type states and that the transition-state barrier is lower for 15A than wild type. The enhanced stability of the I2 intermediate in 15A relative to wild type could be explained either by reduced unfavorable electrostatic interactions or increased helix stability. The reduction in the transition-state barrier may occur because fewer hydrogen bonds and salt bridges need to be formed in 15A refolding, as formation of these types of interactions has been shown to raise this barrier for wild-type Arc (34).
Given that the 15A mutant contains an extensive run of nonpolar residues (Trp-Pro-Ala-Ala-Ala-Leu-Ala-Leu-Val-Ala-Ala-Ala-Ala-Ala-Ala) has a dimer mass reduced by 11.5% relative to wild type and contains side-chain truncations of almost 30% of its residues, it is remarkable that this mutant protein folds as well as, if not better, than wild-type Arc. It has been suggested that polar surface residues may contribute to folding specificity by preventing formation of alternative structures in which these residues are buried (2, 35). Indeed, we recently have found that an Arc mutant containing a single surface substitution (NL11) is able, under some conditions, to adopt an alternative fold in which the β-sheet is replaced by helices and Leu-11 is buried (M. Cordes, N. Walsh, R. Burton, J. McKnight, and R.T.S., unpublished work). Nevertheless, en masse substitution of Arg-16, Glu-17, Asp-20, Arg-23, Lys-24, Glu-27, Glu-28, Asn-34, Ser-35, Glu-39, Glu-43, and Lys-46 with alanine in the 15A mutant does not appear to result in misfolding. Some surface positions therefore appear to be far more important than others in helping to specify structure. In a similar vein, some hydrophobic core residues are far more consequential than others. Replacing either of the wild-type core residues, Val-22 or Val-41, with alanine prevents Arc structure formation (23, 36), whereas replacing both of the core residues, Val-18 and Val-25, with alanine in the 11A, 12A, and 15A mutants does not compromise folding. One of the current obstacles to improving understanding of protein folding is our inability to predict a priori which surface and buried side chains are critical determinants of a protein fold and which are ancillary.
If the 25 residues at which alanine mutations have “neutral” stability phenotypes are discounted, which of the remaining residues are most critical for Arc’s ability to fold stably? Eight additional Arc residues also can be substituted without changing structure and often with an increase in stability. Pro-8 can be replaced by Ala or Leu (22, 23, 37), Ala-53 can be replaced by Asp (38), and six residues that form a hydrogen-bond and salt-bridge network (Asn-29, Arg-31, Glu-36, Arg-40, Ser-44, and Glu-48) can be replaced by alanines and hydrophobic residues (39, 40). Of the remaining 20 residues, two are glycines in turns, two are α-helical N-cap positions, two are polar surface residues (although the interaction partners of these residues are replaced in the mutants studied here), and 14 are hydrophobic core residues. This finding indicates that the majority of the sequence information required for Arc structure and stability is encoded by the residues that form the hydrophobic core.
Acknowledgments
We thank Marcos Milla, Tracy Smith, Brigitte Raumann, Brenda Schulman, Alan Davidson, Peter Kim, and Paul Schimmel for advice, materials, and use of equipment. This work was supported by National Institutes of Health Grant AI-15706.
ABBREVIATION
- NTA
nitrilotriacetic acid
References
- 1.Bowie J U, Sauer R T. Proc Natl Acad Sci USA. 1989;86:2152–2156. doi: 10.1073/pnas.86.7.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie J U, Reidhaar-Olson J F, Lim W A, Sauer R T. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 3.Rennell D, Bouvier S, Hardy L, Poteete A R. J Mol Biol. 1991;222:67–87. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 4.Basse W A, Eriksson A E, Zhang X J, Heinz D W, Sauer U, Blaber M, Baldwin E P, Wozniak J A, Matthews B W. Faraday Discuss Chem Soc. 1992;93:173–181. doi: 10.1039/fd9929300173. [DOI] [PubMed] [Google Scholar]
- 5.Michael S F, Kilfoil V J, Schmidt M H, Amann B T, Breg J M. Proc Natl Acad Sci USA. 1992;89:4796–4800. doi: 10.1073/pnas.89.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer R T, Lim W A. Curr Opin Struct Biol. 1992;2:46–51. [Google Scholar]
- 7.Hu J C, Newell N E, Tidor B, Sauer R T. Protein Sci. 1993;2:1072–1084. doi: 10.1002/pro.5560020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregoret L, Sauer R T. Proc Natl Acad Sci USA. 1993;90:4246–4250. doi: 10.1073/pnas.90.9.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markiewicz P, Kleina L, Cruz C, Ehret S, Miller J H. J Mol Biol. 1993;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 10.Jin L, Wells J A. Protein Sci. 1994;3:2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang Z, Issac V E, Li H, Patel L, Catron K M, Curran T, Montelione G T, Abate C. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaber M, Baase W A, Gassner N, Matthews B W. J Mol Biol. 1995;246:317–330. doi: 10.1006/jmbi.1994.0087. [DOI] [PubMed] [Google Scholar]
- 13.Yu M H, Weissman J S, Kim P S. J Mol Biol. 1995;249:388–397. doi: 10.1006/jmbi.1995.0304. [DOI] [PubMed] [Google Scholar]
- 14.Desjarlais J R, Handel T M. Protein Sci. 1995;4:2006–2018. doi: 10.1002/pro.5560041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes M H J, Davidson A R, Sauer R T. Curr Opin Struct Biol. 1996;6:3–10. doi: 10.1016/s0959-440x(96)80088-1. [DOI] [PubMed] [Google Scholar]
- 16.Dahiyat B I, Mayo S L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 17.Gregoret L M, Sauer R T. Folding Design. 1998;3:119–126. doi: 10.1016/S1359-0278(98)00017-0. [DOI] [PubMed] [Google Scholar]
- 18.Breg J N, van Ophesden J H J, Burgering M J, Boelens R, Kaptein R. Nature (London) 1990;346:586–589. doi: 10.1038/346586a0. [DOI] [PubMed] [Google Scholar]
- 19.Bonvin A M J J, Vis H, Breg J N, Burgering M J M, Boelens R, Kaptein R. J Mol Biol. 1994;236:328–341. doi: 10.1006/jmbi.1994.1138. [DOI] [PubMed] [Google Scholar]
- 20.Raumann B E, Rould M A, Pabo C O, Sauer R T. Nature (London) 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 21.Bowie J U, Sauer R T. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 22.Vershon A K, Bowie J U, Karplus T M, Sauer R T. Proteins Struct Funct Genet. 1986;1:302–311. doi: 10.1002/prot.340010404. [DOI] [PubMed] [Google Scholar]
- 23.Milla M E, Brown B M, Sauer R T. Nat Struct Biol. 1994;1:518–523. doi: 10.1038/nsb0894-518. [DOI] [PubMed] [Google Scholar]
- 24.Milla M E, Brown B M, Waldburger C D, Sauer R T. Biochemistry. 1995;34:13914–13919. doi: 10.1021/bi00042a024. [DOI] [PubMed] [Google Scholar]
- 25.Brown B M, Bowie J U, Sauer R T. Biochemistry. 1990;29:11189–11195. doi: 10.1021/bi00503a006. [DOI] [PubMed] [Google Scholar]
- 26.Milla M E, Brown B M, Sauer R T. Protein Sci. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milla M E, Sauer R T. Biochemistry. 1994;33:1125–1133. doi: 10.1021/bi00171a011. [DOI] [PubMed] [Google Scholar]
- 28.Milla M E, Brown B M, Waldburger C D, Sauer R T. Biochemistry. 1995;34:13914–13919. doi: 10.1021/bi00042a024. [DOI] [PubMed] [Google Scholar]
- 29.Brown B M, Milla M E, Smith T L, Sauer R T. Nat Struct Biol. 1994;1:164–168. doi: 10.1038/nsb0394-164. [DOI] [PubMed] [Google Scholar]
- 30.Jonsson T, Waldburger C D, Sauer R T. Biochemistry. 1996;35:4795–4802. doi: 10.1021/bi953056s. [DOI] [PubMed] [Google Scholar]
- 31.Lin L, Pinker R J, Phillips G N, Kallenbach N R. Protein Sci. 1994;3:1430–1435. doi: 10.1002/pro.5560030909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaber M, Zhang X J, Matthews B W. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 33.Munoz V, Serrano L. Curr Opin Biotechnol. 1995;6:382–386. doi: 10.1016/0958-1669(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 34.Waldburger C D, Jonsson T, Sauer R T. Proc Natl Acad Sci USA. 1996;93:2629–2634. doi: 10.1073/pnas.93.7.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul C H. J Mol Biol. 1982;155:53–62. doi: 10.1016/0022-2836(82)90491-0. [DOI] [PubMed] [Google Scholar]
- 36.Milla M E, Sauer R T. Biochemistry. 1995;34:3344–3351. doi: 10.1021/bi00010a025. [DOI] [PubMed] [Google Scholar]
- 37.Schildbach J F, Milla M E, Jeffrey P D, Raumann B E, Sauer R T. Biochemistry. 1995;34:1405–1412. doi: 10.1021/bi00004a035. [DOI] [PubMed] [Google Scholar]
- 38.Bowie J U, Sauer R T. J Biol Chem. 1989;264:7596–7602. [PubMed] [Google Scholar]
- 39.Waldburger C D, Schildbach J F, Sauer R T. Nat Struct Biol. 1995;2:122–128. doi: 10.1038/nsb0295-122. [DOI] [PubMed] [Google Scholar]
- 40.Hendsch Z S, Jonsson T, Sauer R T, Tidor B. Biochemistry. 1996;35:7621–7625. doi: 10.1021/bi9605191. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]