Abstract
NF-κB is a key transcriptional inducer of the inflammatory response in mature myeloid cells, and also stimulates cell survival, but its role in immature myeloid cell development has not been well characterized. C/EBPα is required for the development of monocytic and granulocytic myeloid cells from early progenitors, and NF-κB and C/EBPβ cooperatively induce several inflammatory mediators. Having found that C/EBPα binds NF-κB p50 preferentially compared with NF-κB p65, we have now investigated myelopoiesis in nfkb1−/− mice lacking NF-κB p50. Absence of p50 leads to a significant reduction in the number of granulocytic progenitors, CFU-G, obtained with G-CSF or GM-CSF in vitro and reduces neutrophil production in vivo in response to G-CSF, with preservation of monopoiesis in vitro in response to cytokines or LPS. To gain insight into the mechanism underlying reduced granulopoiesis in the absence of NF-κB p50, we assessed the expression of several myeloid regulatory proteins in lineage-negative, immature myeloid cells. Although PU.1, C/EBPβ, and STAT3 levels were unchanged, C/EBPα protein and RNA levels were reduced approximately 3-fold in the absence of NF-κB p50. In addition, NF-κB p50 and C/EBPα bound the endogenous C/EBPα promoter in a chromatin immunoprecipitation assay, and NF-κB p50 trans-activated the C/EBPα promoter, alone or in cooperation with C/EBPα. Despite reduction of C/EBPα, GCSFR and MCSFR levels were maintained or total marrow and in lineage-negative cells. Together, these data indicate that acute inflammation not only activates mature myeloid cells but also stimulates neutrophil production via NF-κB p50 induction of C/EBPα transcription.
Keywords: neutrophils, inflammation, cell differentiation, hematopoiesis, gene regulation
NF-κB was first described as an activity in B cells that binds the κ light chain gene enhancer (1, 2). Five NF-κB family members are found in mammalian cells: c-Rel, RelA or p65, RelB, NF-κB1 or p50 and NF-κB2 or p52. Each possesses a 300 amino acid Rel homology domain (RHD)4 that mediates dimerization, DNA binding, and nuclear localization upon degradation of IκB. The most abundant and ubiquitous NF-κB dimers are p50:p65, p50:p50, and p65:p65, with RelB:p52 largely restricted to lymphoid cells. c-Rel, p65, and RelB mediate trans-activation via their C-terminal domains. p50:p50 homodimers repress transcription via interaction with bcl-3 and HDACs, but in some contexts are capable of gene activation (3, 4). Inflammatory mediators and pathogen-associated molecular patterns, such as tumor necrosis factor or lipopolysaccaride (LPS), are potent mediators of NF-κB stimulation via the classical pathway of IKKβ activation and IκB degradation, allowing nuclear translocation of p65 and thereby formation of nuclear p50:p65 or p65:p65 dimers.
In mature myeloid cells, NF-κB and C/EBPβ mediate the inflammatory response in part via cooperative activation of the IL-6, IL-8, G-CSF, serum amyloid, ICAM-1, superoxide dismutase, and Mediterranean fever promoters (5–9). C/EBP proteins dimerize via their C-terminal leucine zipper (LZ) motifs, bind DNA via the adjacent basic regions (BR), and activate transcription via N-terminal domains (10, 11). Within hematopoiesis, C/EBP expression is largely restricted to myeloid cells, with C/EBPα most prominent in immature cells and C/EBPβ and C/EBPδ induced upon maturation to neutrophils or monocytes (12). Consistent with this expression pattern, C/EBPα(−/−) mice demonstrate markedly reduced formation of the granulocyte-monocyte progenitor (GMP) from the common myeloid progenitor (CMP), whereas C/EBPβ(−/−) retain all the hematopoietic lineages (13, 14).
The BR-LZ (or bZIP) domains of C/EBPα and C/EBPβ are highly similar and directly interact with the RHD of p50 or p65 (15, 16). Moreover, we found that mutation of the C/EBPα BR obviates its interaction with p50 but not p65 and that endogenous C/EBPα preferentially binds endogenous p50 compared with p65 in myeloid cells (17). This biochemical link between C/EBPα and p50 prompted us to compare marrow cells from nfkb1(−/−) mice lacking p50 with strain-matched controls for their ability to generate myeloid cells in vitro or in vivo and for their expression of C/EBPα. We confirm that absence of p50 does not alter basal numbers of neutrophils (18) but find that nfkb1(−/−) mice have reduced in vitro production of the CFU-G neutrophil progenitor in response to GM-CSF or G-CSF, reduced in vivo formation of neutrophils in response to G-CSF, and reduced C/EBPα protein and RNA, reflecting the ability of NF-κB p50 to bind and activate the C/EBPα promoter.
Materials and Methods
Mice and marrow cell culture
Marrow cells obtained from the hind limbs of 8–12 week old nfkb1(−/−) and B6;129P strain-matched control mice (Jackson Laboratories) were subjected to red cell lysis with NH4Cl. For colony-forming unit (CFU) assays, cells were plated directly in methylcellulose (Stem Cell Technologies) at 20,000 cells/ml in Iscove’s modified Dulbecco’s medium (IMDM) with 15% heat-inactivated fetal bovine serum (HI-FBS) and either murine GM-CSF (10 ng/ml), murine M-CSF (10 ng/ml) with murine SCF (50 ng/ml) (Peprotech), or human G-CSF (100 ng/ml, Amgen) with SCF (50 ng/ml). CFUs were enumerated 8 days later. For in vivo cytokine exposure, mice were injected intraperitoneally (i.p.) with 1 μg G-CSF or GM-CSF daily for 4 days, and blood samples (50 μl) obtained on days 0 and 5 from the facial vein were analyzed for blood counts and white cell differentials using a Hemavet automated counter (MWI, Inc.). For LPS exposure, cells were cultured in IMDM with 10% HI-FBS with murine SCF (20 ng/ml), murine Flt3 ligand (FL, 100 ng/ml, Peprotech) with or without E. coli 0111:B4 LPS (10 μg/ml, List Biologicals). For protein or RNA preparation from lin− marrow cells, cells obtained from mice treated five days earlier with 5-fluorouracil (5-FU, 150 mg/kg i.p.) were cultured in IMDM with 10% HI-FBS and IL-3, IL-6, and SCF for 5 days, followed by lineage-depletion using immunomagnetic beads and a cocktail of biotinylated lineage antibodies, B220, CD5, Mac-1, Ter119, Gr-1, and 7-4 (Stem Cell Technologies). Studies with mice have been reviewed and approved by our institutional animal care and use committee.
FACS analysis
To quantify hematopoietic stem cells (HSC), megakaryocyte-erythroid progenitors (MEP), CMP, and GMP, marrow was lineage-depleted and also depleted for IL7R by addition of biotin-anti-mouse IL7Rα (Pharmingen) to the lineage cocktail before immunomagnetic depletion. Depleted cells were then stained with FITC-anti-CD34, PerCP-anti-Sca-1, APC-anti-c-kit, and PE-anti-FcγRII/III (Pharmingen), or IgG control antibodies, followed by FACS analysis (19). To quantify common lymphoid progenitors (CLP), lineage-depleted cells were stained with PE-anti-IL7Rα, APC-anti-c-kit, and PerCP-anti-Sca-1 and then subjected to FACS (19). To assess G-CSF receptor (GCSFR) or M-CSF receptor (MCSFR) expression in total marrow mononuclear cells, cells were stained with biotinylated G-CSF or biotinylated M-CSF followed by streptavidin-PE, in the absence or presence of 30-fold excess unlabeled G-CSF or M-CSF (20). To assess GCSFR or MCSFR expression in lineage-negative marrow cells, cells were stained similarly and also with Alexa Fluor 488-linked Mouse Lineage antibody Mixture (Caltag Laboratories), followed by enumerating PE-staining cells within the Alexa Fluor 488-negative gate.
Western blotting and RNA analysis
Total cellular proteins prepared from lineage-depleted marrow cells were prepared in Laemmli sample buffer and samples corresponding to 5 × 105 cells were subjected to Western blotting as described (21), using C/EBPα (14AA), C/EBPβ (C19), PU.1 (T-21) (Santa Cruz Biotechnology) STAT3 (79D7) (Cell Signaling), and β-actin (AC-15) (Sigma) antibodies. Total cellular RNA was prepared from lineage-depleted cells using Trizol reagent (Invitrogen). First-strand cDNA was prepared using AMV reverse transcriptase (Promega) and random primers at 42°C for 1 hr. Quantitative PCR was carried out using 100 ng of each cDNA using iQ SYBR Green supermix (Bio-Rad). Oligonucleotides used were:
CEBPA-F: 5′-TGGATAAGAACAGCAACGAG,
CEBPA-R: 5′-TCACTGGTCAACTCCAACAC,
PU.1-F: 5′-CAGAAGGGCAACCGCAAGAA,
PU.1-R: 5′-GCCGCTGAACTGGTAGGTGA,
mS16-F: 5′- CTTGGAGGCTTCATCCACAT, and
mS16-R: 5′- ATATTCGGGTCCGTGTGAAG.
Chromatin immunoprecipitation and promoter assays
Ten million mouse whole bone marrow cells were used in each chromatin immunoprecipitation (ChIP) reaction. Cells were incubated with 1% formaldehyde at 37°C for 10 minutes, and the reaction was quenched with glycine at a final concentration of 0.125 M. Cells were washed twice with ice-cold phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and Protease Inhibitor Cocktail (Sigma) and resuspended in 1 ml ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0, 1x protease inhibitors). Lysates were sonicated on ice using a Branson Sonifier 250, and the chromatin was sheared to 500–1000 bp fragments. Lysates were centrifuged for 10 min at 16000 × g to remove insoluble cell debris, and ChIP dilution buffer (167 mM NaCl, 16.7 mM Tris pH 8.0, 1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA, 1x protease inhibitors) was added to a final volume of 4 ml. Lysates were precleared with 65 μl blocked protein-A/G sepharose beads (Upstate Biotechnologies) for 1 hour at 4°C. They were briefly centrifuged, and input was obtained before incubating the resulting supernatant with the antisera against C/EBPα, NF-κB p50 or rabbit IgG (Santa Cruz) overnight at 4°C with rocking. Sixty μl of blocked protein-A/G sepharose beads were added, followed by incubation at 4°C for 2 hours with gentle agitation. The beads were precipitated by centrifugation, and the bead-bound complexes were washed for 5 min in each of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8.0, 150 mM NaCl), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8.0, 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% IGEPAL CA-630, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris pH 8.0) and two washes in TE (10 mM Tris pH 8.0, 1 mM EDTA). DNA-protein complexes were eluted from beads using 200 μl of 1% SDS, 0.1 M NaHCO3 at room temperature. Cross-links were reversed by adding 10 μl of 4 M NaCl to the eluates and incubation at 65°C overnight. Each sample was treated with RNAse and Proteinase K, and DNA was isolated with UltraClean DNA Purification Kit (Mo Bio Laboratories). Precipitation of promoters of interest was detected by PCR, and the PCR products were resolved on agarose gels and visualized with ethidium bromide. The following primers were used:
CEBPApr-F: 5′-GAACACTTGACTAGAGTGCTC,
CEBPApr-R: 5′-CTCGTCCATCGCCTAGG,
CEBPAup-F: 5′-CTGTAACCGCAGTGGAAGC, and
CEBPAup-R: 5′-CTCTAGGGTCGCAGGTCAAG.
NIH 3T3 cells in 60 mm dishes were co-transfected with CMV, CMV-C/EBPα, or CMV-p50 and bcl2 P2(−1278)-Luc using Lipofectamine 2000 (Invitrogen) as described (17). pCMV-βGal was included as an internal control, and samples were collected and assayed 48 hrs post-transfection. The Student t test was used for statistical comparisons.
Results
NF-κB p50 is required for granulopoiesis in vitro
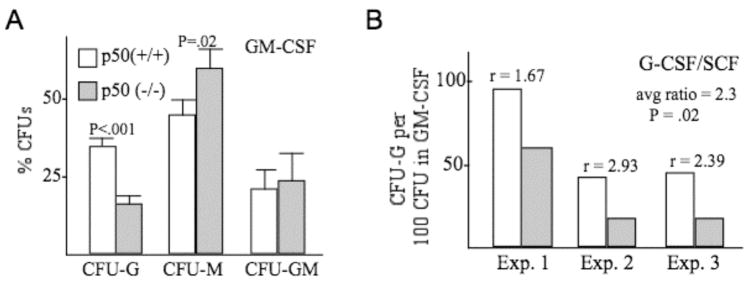
Marrow cells isolated from NF-κB p50(−/−) mice or strain-matched controls were plated in methylcellulose cultures either with GM-CSF or G-CSF/SCF, and CFU-G, CFU-M, and CFU-GM myeloid progenitors were enumerated eight days later (Fig. 1). In GM-CSF culture, CFU-G numbers were significantly reduced in the absence of p50, with a significant increase in CFU-M and no change in CFU-GM. Total CFU obtained in GM-CSF averaged 128/ml plated from p50(+/+) marrow and 110 CFU/ml from p50(−/−) marrow, whereas total CFU-G averaged 44/ml from p50(+/+) marrow and 18/ml from p50(−/−) marrow, and colony sizes were similar between the two genotypes. In G-CSF/SCF, which predominantly yields CFU-G, CFU-G numbers were also consistently reduced in three independent experiments, even when normalized for the mildly reduced total number of CFU obtained with the more permissive CFU-GM cytokine. Reduced CFU-G was not likely due to increased apoptosis, as culture of lineage-negative marrow cells from p50(+/+) or p50(−/−) mice in GM-CSF for 48 hrs led to 28% vs. 27% Annexin-V-positive cells in one experiment and 17% vs. 15% Annexin-V-positive cells in a second, independent experiment (not shown).
Figure 1.
Absence of NF-κB p50 reduces formation of granulocytic progenitors in vitro. A, Equivalent numbers of mononuclear marrow cells from NF-κB p50(+/+) or (−/−) mice were plated in methylcellulose with GM-CSF. CFU-G, CFU-M, and CFU-GM were enumerated eight days later. Shown is the percentage of total CFUs represented by each colony type (mean and SE from four determinations). B, Mononuclear marrow cells from p50(+/+) or (−/−) mice were plated in methylcellulose with G-CSF and SCF, and CFU-G numbers were assessed eight days later. The number of CFU-G, normalized against total CFU obtained in GM-CSF culture, is shown for three separate experiments. The ratios of normalized CFU-G obtained from p50(+/+) versus (−/−) marrow are shown, as is the average ratio.
NF-κB p50 is not required for induction of monopoiesis by LPS in vitro
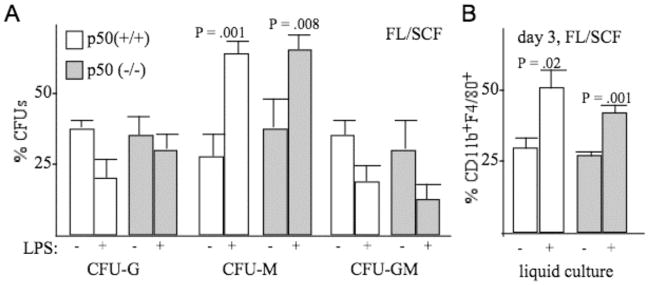
A recent study demonstrated that myeloid progenitors express TLR4 and CD14 and that culture in the presence of FL/SCF and LPS, the ligand for TLR4/CD14, stimulated their development into monocytic cells (22). As TLR signaling activates NF-κB, we sought to determine whether absence of NF-κB p50 impacted on monocytic development in response to LPS. Lineage-negative marrow cells from p50(+/+) or p50(−/−) mice were cultured in IMDM with FBS and FL/SCF, with or without LPS, and cultured either in methylcellulose to assess CFUs or in liquid culture to assess monocyte formation (Fig. 2). Addition of LPS stimulated formation of CFU-M and of CD11b+F4/80+ monocytes similarly, irrespective of the presence or absence of NF-κB p50. This finding extends the conclusion that absence of NF-κB p50 reduces granulopoiesis but not monopoiesis in vitro. Of note, FACS analysis of wild-type vs. p50(−/−) marrow indicated an identical 3.0% Mac1+Gr-1− monocytic cells and an average of 61% versus 57% Mac1+Gr-1+ granulocytic cells in two experiments (not shown).
Figure 2.
Absence of NF-κB p50 does not reduce induction of monopoiesis by LPS in vitro. A, Mononuclear marrow cells from NF-κB p50(+/+) or (−/−) mice were plated in methylcellulose with FL and SCF, without or with LPS. CFU-G, CFU-M, and CFU-GM were enumerated eight days later. Shown are the percentages of total CFUs represented by each colony type (mean and SE from three determinations). B, Cells cultured similarly in liquid culture were subjected to FACS analysis for CD11b and F4/80 on day 3. Shown are the percentages of CD11b+F4/80+ monocytic cells (mean and SE from three determinations).
NF-κB p50 is required for granulopoiesis in vivo
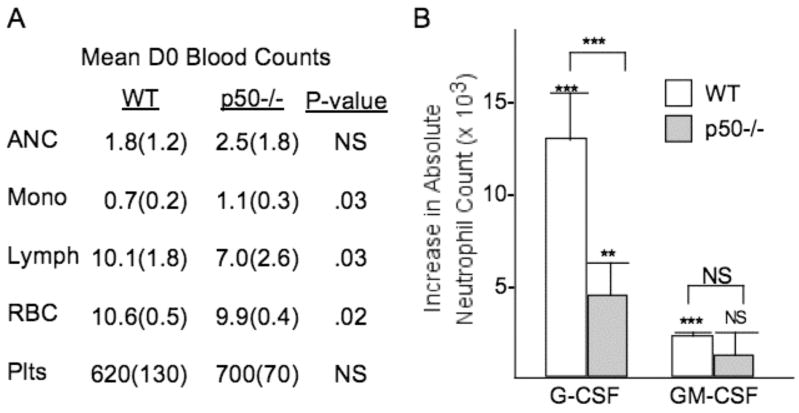
We next sought to determine whether NF-κB p50 is required for G-CSF- or GM-CSF-induced granulopoiesis in vivo. Wild-type p50(+/+) or knockout p50(−/−) mice were injected intraperitoneally with 1 μg of G-CSF or GM-CSF daily for four days. Complete blood counts (CBCs) were obtained on day 0 and day 5. Absolute neutrophil counts (ANC) were similar between p50(+/+) and p50(−/−) mice on day 0 as shown in a summary of blood counts obtained from these mice (Fig. 3A). However, G-CSF induced a significantly greater rise in ANC in wild-type mice by day 5, namely 13.0 vs. 4.6 thousand per μl on average (Fig. 3B). GM-CSF significantly increased the ANC from baseline in wild-type mice but not in p50−/− mice; however, the increase in ANC observed in wild-type mice was not significantly different from that observed in a similar number of p50−/− mice, perhaps reflecting the weaker potency of GM-CSF compared with G-CSF in this assay (Fig. 3B). Of note, monocytes were increased in the day 0 blood counts in the p50(−/−) compared with the p50 (+/+) mice, lymphocytes were decreased, red blood cells were slightly reduced, and platelet numbers were similar (Fig. 3A). Overall, these data indicate an increase in monocytes at baseline without a decrease in granulocytes, the latter potentially due to in vivo compensation, but reduced responsiveness of endogenous myeloid progenitors to G-CSF or GM-CSF, paralleling the in vitro findings.
Figure 3.
Absence of NF-κB p50 reduces formation of granulocytic progenitors in vivo. A, Comparison of mean baseline blood counts in six wild-type (WT) and NF-κB p50(−/−) mice. ANC – absolute neutrophil count, thousand per μl; Mono – monocytes, thousand per μl; Lymph – lympocytes, thousand per μl; RBC – red blood cells, million per μl; Plts – platelets, thousand per μl. Standard errors are shown in parentheses. Each blood cell subset was compared between the two genotypes using the student’s t test. B, NF-κB p50(+/+) or (−/−) mice received 1 μg G-CSF or 1 μg GM-CSF i.p. daily for four days. Shown are the increases in peripheral blood absolute neutrophil counts (ANC) on day 5 compared with day 0 (with mean and SE from 6 mice for G-CSF and 5 mice for GM-CSF). Significance of the increased ANC on day 5 compared to day 0 in each group was analyzed by the paired student’s t test (indicated above each error bar), and comparison between the mean ANC increases observed for WT versus p50−/− mice injected with each cytokine was obtained using the student’s t test (indicated above the brackets); *** P < 0.001; ** P = 0.002.
Absence of NF-κB p50 has minimal effects on progenitors preceding CFU-G
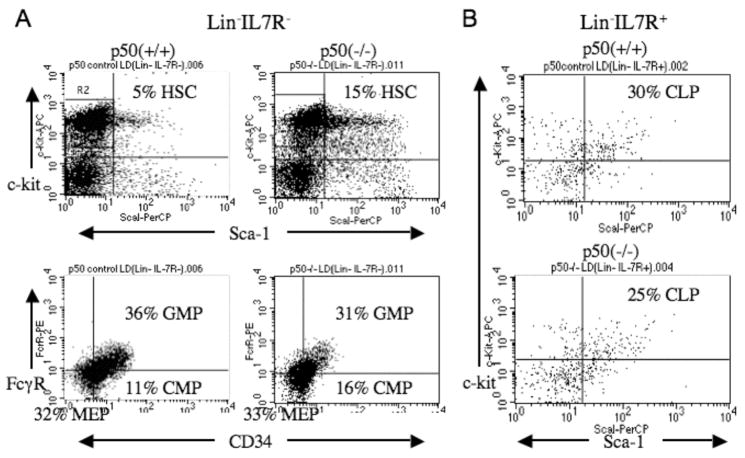
To gain insight into the role of NF-κB p50 in early hematopoiesis, we enumerated HSC, CMP, GMP, MEP, and CLP in p50(+/+) and p50(−/−) marrow (Fig. 4). GMP numbers were mildly reduced whereas CMP and HSC were mildly increased, and CLP were also mildly reduced, in the absence of NF-κB p50. Overall, these changes reflect our finding and the prior demonstration (18) that deletion of NF-κB p50 does not markedly alter basal production of mature lymphoid, myeloid, erythroid, or megakaryocytic blood cells.
Figure 4.
Absence of NF-κB p50 only minimally alters the number of early hematopoietic stem/progenitor cells. A, Marrow mononuclear cells from NF-κB p50(+/+) or (−/−) mice were depleted using a lineage-antibody cocktail and IL7Rα antibody, followed by staining for Sca-1, c-kit, FcγRII/III, and CD34 to allow enumeration of Sca-1+c-kit+ HSC, Sca-1−c-kit+FcγR+CD34+ GMP, Sca-1−c-kit+FcγRloCD34+ CMP, and Sca-1−c-kit+FcγRloCD34− MEP. Area R2 indicates the Sca-1−c-kit+ cells gated prior to enumeration of GMP, CMP, and MEP in the lower panels. B, Lineage-depleted marrow mononuclear cells from NF-κB p50(+/+) or (−/−) mice were stained for IL7Rα, Sca-1, and c-kit. IL7Rα+ cells represented 4.0% of p50(+/+) and 4.4% of p50(−/−) total mononuclear cells in the experiment shown. Shown is FACS analysis gating on these populations, to enumerate Sca-1+c-kit+IL7Rα+ CLP. Similar findings were obtained in a second, independent experiment.
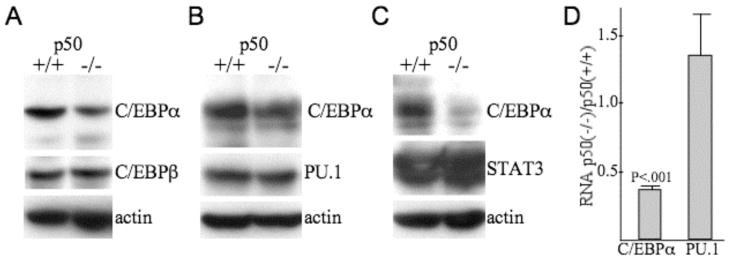
C/EBPα is diminished in immature myeloid cells lacking NF-κB p50
Total cellular proteins prepared from lineage-negative marrow cells from p50(+/+) and (−/−) mice were subjected to Western blotting for C/EBPα, C/EBPβ, PU.1, STAT3, and β-actin (Fig. 5A–C). C/EBPα was diminished 2- to 3-fold in three independent sets of samples, whereas C/EBPβ, PU.1, and STAT3 levels were unchanged. Total cellular RNAs prepared from lineage-negative marrow cells from p50(+/+) or p50(−/−) mice were subjected to quantitative RT-PCR analysis for C/EBPα and PU.1, normalized to mouse ribosomal protein S16 (mS16) RNA levels (Fig. 5D). C/EBPα RNA was reduced 3-fold in the absence of NF-κB p50, whereas PU.1 levels were unchanged. Thus, absence of p50 reduces C/EBPα RNA expression and thereby C/EBPα protein expression in lineage-negative hematopoietic cells obtained from marrow cells that had been cultured with cytokines that favor survival and proliferation of myeloid progenitors (32).
Figure 5.
C/EBPα protein and RNA levels are reduced in immature myeloid cells lacking NF-κB p50. A–C, Marrow cells from 5-FU-treated NF-κB p50(+/+) or (−/−) mice were expanded in culture in IL3, IL6, and SCF and then subjected to lineage-depletion. Total cellular proteins prepared from the resulting lin− population were subjected to Western blotting for C/EBPα, C/EBPβ, PU.1, STAT3, and β-actin. Data in each panel were generated from a separate set of mice. D, RNA prepared from lin− hematopoietic cells obtained using this same experimental protocol from p50(+/+) or p50(−/−) mice were subjected to quantitative RT-PCR analysis for RNAs encoding C/EBPα or PU.1. Shown are the ratios of C/EBPα or PU.1 RNA in p50(−/−) cells: C/EBPα or PU.1 RNA in p50(+/+) cells (means and SE from three determinations).
NF-κB p50 binds and activates the CEBPA promoter
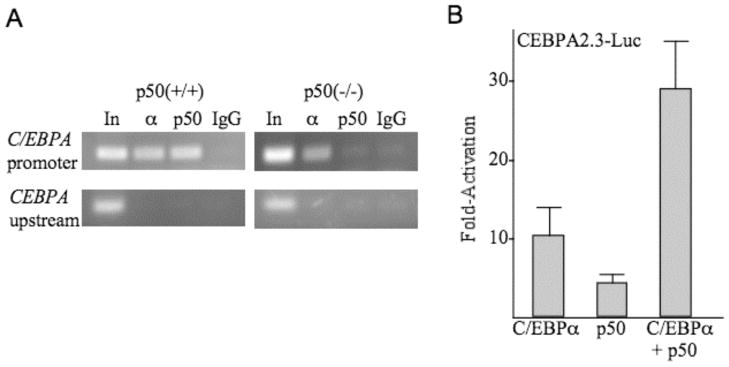
Marrow cells isolated from NF-κB p50(+/+) or p50(−/−) mice were subjected to ChIP analysis using C/EBPα or p50 antiserum or rabbit Ig, followed by PCR for a 133 bp genomic DNA fragment centered at −718 bp of the CEBPA promoter or for a more upstream 132 bp fragment centered at −1462 bp (Fig. 6A). Interaction of NF-κB p50 with the endogenous CEBPA promoter in wild-type cells was evident. Lack of interaction in knockout cells is expected and provides an additional control indicating lack of non-specific interaction of the p50 antiserum with other proteins in the vicinity of the CEBPA promoter. C/EBPα bound the promoter region in the presence or absence of NF-κB p50. Finally, neither protein interacted with the more upstream CEBPA genomic segment, indicating sufficiency of sonication and localizing these interactions to the vicinity of the more downstream primer set.
Figure 6.
NF-κB p50 binds and activates the CEBPA promoter. A, Marrow cells from NF-κB p50(+/+) and (−/−) were subjected to ChIP analysis using C/EBPα or p50 rabbit antiserum and control, normal rabbit Ig, followed by PCR for a 133 bp CEBPA promoter segment located at −718 bp from the transcription start site and for a 132 bp more upstream CEBPA genomic segment located at −1462. Amplified bands were visualized by agarose gel electrophoresis followed by ethidium bromide staining. B, NIH 3T3 cells in 60 mm dishes were transiently transfected with 1.5 μg CEBPA2.3-Luc together with 100 ng of pCMV, pCMV-C/EBPα, pCMV-p50, or both pCMV-C/EBPα and pCMV-p50. 10 ng pCMV-βGal was included in each transfection as an internal control. Luciferase and β-galactosidase activities were assessed 48 hrs post-transfection. Shown is fold activation relative to activity obtained with pCMV (mean and SE from three determinations).
CEBPA2.3-Luc, containing 2.3 kb of the murine CEBPA promoter linked to a luciferase reporter, was co-transfected with empty CMV vector or with CMV expression plasmids encoding C/EBPα, NF-κB p50 or both C/EBPα and NF-κB p50, along with CMV-βGal as an internal control (Fig. 6B). Relative to promoter activity in the presence of the empty vector, C/EBPα activated its own promoter 10-fold, as previously described (23), p50 activated the promoter 4-fold, and the combination activated the promoter 28-fold. Together, these data indicate that NF-κB p50 binds and activates the CEBPA promoter cooperatively with C/EBPα. Co-expression of C/EBPα and p50 activated a 341 bp CEBPA promoter fragment 12-fold, on average, and activated a 125 bp promoter fragment 7-fold (not shown), indicating that there may be several relevant κB cis elements, which will require elucidation by future detailed biochemical and functional studies.
GCSFR and MCSFR levels are not reduced in NF-κB p50 (−/−) marrow cells
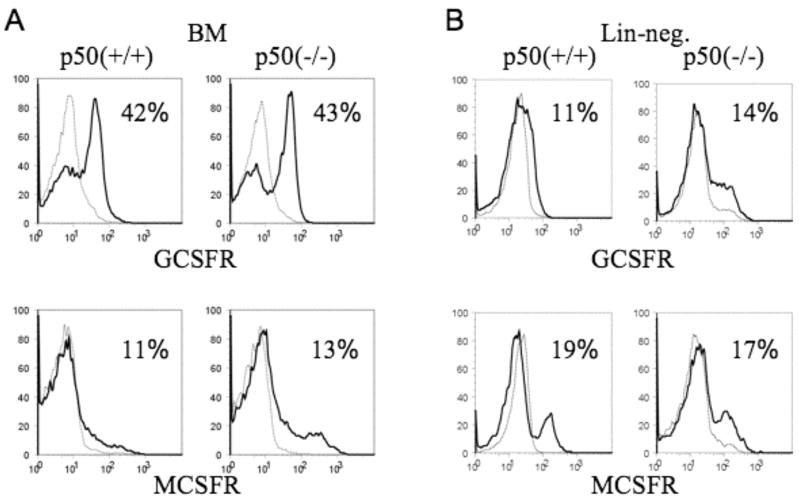
C/EBPα binds and activates the GCSFR promoter (24). Having found that absence of NF-κB p50 reduces C/EBPα protein expression, we therefore considered the possibility that reduced GCSFR levels in NF-κB p50(−/−) hematopoietic cells might account for their reduced responsiveness to G-CSF in vitro and in vivo. Total marrow mononuclear cells from NF-κB p50(+/+) or (−/−) mice were subjected to FACS analysis for GCSFR and also for MCSFR, another C/EBPα target (25). Both receptors were expressed similarly in the presence or absence of NF-κB p50 (Fig. 7A). Comparable findings were obtained using cells from a second set of mice (not shown). GCSFR and MCSFR levels were also evaluated lineage-negative p50(+/+) versus (−/−) marrow cells (Fig. 7B), and again similar levels were detected.
Figure 7.
Absence of NF-κB p50 does not reduce GCSFR or MCSFR expression. A, Total marrow mononuclear cells from NF-κB p50(+/+) or (−/−) mice were subjected to FACS analysis for GCSFR (top) or MCSFR (bottom) expression, using biotinylated G-CSF or M-CSF and streptavidin-PE. The darker curves represent staining in the absence of unlabelled G-CSF or M-CSF, and the lighter curves represent staining in the presence of excess cold G-CSF or M-CSF. B, Lineage-negative marrow mononuclear cells from NF-κB p50(+/+) or (−/−) mice were analyzed similarly. Representative histograms from triplicate analysis and the percentage of positive cells are shown.
Discussion
NF-κB is central mediator of the inflammatory response. A key finding of this study is that NF-κB also enables granulopoiesis in response to inflammatory cytokines. NF-κB p50:p65 and p50:p50 are the most ubiquitous and abundant NF-κB dimers. NF-κB p65(−/−) mice display embryonic lethality due hepatocyte apoptosis (26), an effect rescued by deletion of TNFRI (27). TNFRI(−/−);p65(−/−) murine marrow cells contain normal numbers of neutrophils and monocytes, but their myeloid progenitor numbers and response to inflammatory cytokine stimulation has not been assessed. NF-κB p50(−/−) mice also generate normal numbers of blood neutrophils, as well as normal numbers of peritoneal macrophages and B and T cell subsets (18); however, our further characterization of hematopoiesis in these mice has uncovered reduced in vitro formation of CFU-G in response to either GM-CSF or G-CSF and reduced in vivo granulopoiesis in response to G-CSF. Moreover, we provide mechanistic insight into the link between NF-κB p50 expression and granulopoiesis by demonstrating that NF-κB p50 binds and activates the CEBPA promoter, accounting for the striking several-fold reduction in C/EBPα protein and mRNA we detect in immature myeloid cells in the absence of NF-κB p50. Although the hybrid genetic background of available NF-κB p50(−/−) mice precludes transplantation into B6;129 strain-matched controls, the similar GMP numbers and yet increased response of wild-type mice to exogenous G-CSF suggests that the granulopoietic defect observed in the absence of NF-κB p50 is cell intrinsic.
C/EBPα is a key mediator of granulopoiesis. Adult C/EBPα(−/−) mice obtained by deletion of floxed CEBPA alleles have a block in the CMP to GMP transition, leading to reduced granulocytes and monocytes and their progenitors (14). Interestingly, monopoiesis at baseline or in response to GM-CSF or LPS was preserved or even mildly increased in NF-κB p50(−/−) marrow cells in our study, suggesting that the reduction of C/EBPα observed in immature myeloid cells from p50(−/−) mice, to about 33% of normal levels, mainly impacted granulopoiesis. We previously demonstrated that exogenous C/EBPα-ER increases monopoiesis in transduced lineage-negative marrow myeloid progenitors (28), and this effect was preserved in the absence of NF-κB p50 (not shown), again indicating that p50 is not critical for monopoiesis. We assessed marrow progenitors from C/EBPα(+/−) murine marrow, which have approximately 50% of normal C/EBPα, and found no defect in granulopoiesis or monopoiesis (not shown), indicating that absence of NF-κB p50 either further reduces C/EBPα below a threshold critical for optimal granulopoiesis or that NF-κB has a separate activity beyond regulation of the CEBPA gene that contributes to neutrophil production. Perhaps analogous to our findings, it is interesting to note that PU.1(+/−) mice display normal hematopoiesis, whereas PU.1(kd/kd) mice, with 20% of normal PU.1 levels due to deletion of a distal enhancer, have greatly reduced monopoiesis (29). Maintenance of GCSFR and MCSFR levels in p50(−/−) marrow cells does not preclude a role for C/EBPα in additional events required for granulopoiesis, as dominant-inhibition of C/EBPα to an extent sufficient to diminish GCSFR expression prevents granulopoiesis even in the presence of exogenous GCSFR (30). In addition, other cytokine receptor levels might be reduced; we attempted to assess GM-CSF receptor levels using biotinylated GM-CSF we generated and validated using Ba/F3 cells expressing exogenous receptor, but could not detect GM-CSF receptor above background levels in total or lineage-depleted marrow cells, suggesting low receptor numbers (not shown).
NF-κB p50 directly interacts with C/EBPα to facilitate activation of the genes encoding blc-2 or FLIP (17, 31). Our finding that NF-κB p50 stimulates C/EBPα expression uncovers an additional layer of cooperation between these two transcription factors. These two levels of synergy underscore the evolutionary importance of NF-κB p50:C/EBPα functional interaction. Our finding that C/EBPα binds the endogenous CEBPA promoter is consistent with previous in vitro DNA-binding studies (23) and lends further support to the conclusion that C/EBPα auto-regulates its own expression. C/EBPα interaction with the bcl-2 or FLIP promoters is markedly reduced in the absence of NF-κB p50, whereas C/EBPα binding to its own promoter or to the neutrophil elastase (NE) promoter does not require the presence of p50 (31). Thus, C/EBPα regulates genes that control differentiation, such as CEBPA and NE, independent of p50 but cooperates with p50 to activate genes that inhibit apoptosis. Nevertheless, we herein demonstrate functional cooperation between C/EBPα and NF-κB p50 during granulopoiesis, as NF-κB p50 directly binds and activates the CEBPA promoter. Exogenous C/EBPα enhances monocytic development in lineage-negative wild type murine progenitors dependent upon zippering with AP-1 proteins (28, 32); the same phenomenon occurs upon transduction of p50 (−/−) cell with C/EBPα (not shown), consistent with a greater role for p50 in granulocyte lineage commitment. The ability of directed C/EBPα homodimers to rescue granulopoiesis in p50(−/−) progenitors will be pursued in future experiments.
The finding that GMPs express TLR2 and TLR4 provides one avenue for stimulation of myeloid cell production during an inflammatory response (22), and in fact those investigators demonstrate that LPS stimulates monopoiesis. Absence of NF-κB p50 did not blunt this response in our study, suggesting a role for additional signaling molecules activated by TLR4. Future investigations may find that ligation of additional TLRs stimulates granulopoiesis, with a specific role for NF-κB p50.
Overall, this study indicates that NF-κB p50 activation by inflammatory cytokines induces C/EBPα to stimulate “emergency” granulopoiesis. Future investigations will seek to identify additional regulatory proteins that cooperate in this process to gain additional insight into mechanisms of granulopoiesis under basal and stress conditions.
Acknowledgments
We thank Li Li for guidance regarding enumeration of murine peripheral blood counts.
Footnotes
This work was supported by National Institutes of Health Grant HL082948 and by a grant from the Children’s Cancer Foundation (to A.D.F.) and by grants from the St. Baldrick’s Foundation and Alex’s Lemonade Stand Foundation (to I.P.P.).
Abbreviations used in this paper: C/EBPα, CCAAT/enhancer binding protein α; CFU, colongy-forming unit; G-CSF, granulocyte colony stimulating factor; GM-CSF, granulocyte-macrophage colony stimulating factor; LPS, lipopolysaccharide; NF-κB, nuclear factor κB; RHD, rel homology domain.
References
- 1.Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 2.Singh H, Sen R, Baltimore D, Sharp PA. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 3.Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, Johnson PF. BCL-3 and NF-κB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem. 2004;279:49995–50003. doi: 10.1074/jbc.M404246200. [DOI] [PubMed] [Google Scholar]
- 4.Kurland JF, Kodym R, Story MD, Spurgers KB, McDonnell TJ, Meyn RE. NF-κB1 (p50) homodimers contribute to transcription of the bcl-2 oncogene. J Biol Chem. 2001;276:45380–45386. doi: 10.1074/jbc.M108294200. [DOI] [PubMed] [Google Scholar]
- 5.Dunn SM, Coles LS, Lang RK, Gerondakis S, Vadas MA, Shannon MF. Requirement for nuclear factor (NF)-κB p65 and NF-interleukin-6 binding elements in the tumor necrosis factor response region of the granulocyte colony-stimulating factor promoter. Blood. 1994;83:2469–2479. [PubMed] [Google Scholar]
- 6.Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-κB p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- 7.Xia C, Cheshire JK, Patel H, Woo P. Cross-talk between transcription factors NF-κB and C/EBP in the transcriptional regulation of genes. Int J Biochem Cell Biol. 1997;29:1525–1539. doi: 10.1016/s1357-2725(97)00083-6. [DOI] [PubMed] [Google Scholar]
- 8.Maehara K, Hasegawa T, Xiao H, Takeuchi A, Abe R, Isobe K. Cooperative interaction of NF-κB and C/EBP binding sites is necessary for manganese superoxide dismutase gene transcription mediated by lipopolysaccharide and interferon-gamma. FEBS Lett. 1999;449:115–119. doi: 10.1016/s0014-5793(99)00408-1. [DOI] [PubMed] [Google Scholar]
- 9.Papin S, Cazeneuve C, Duquesnoy P, Jeru I, Sahali D, Amselem S. The tumor necrosis factor α-dependent activation of the human mediterranean fever (MEFV) promoter is mediated by a synergistic interaction between C/EBPβ and NF-κB p65. J Biol Chem. 2003;278:48839–48847. doi: 10.1074/jbc.M305166200. [DOI] [PubMed] [Google Scholar]
- 10.Landschulz WH, Johnson PF, McKnight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 11.Friedman AD, McKnight SL. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 12.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 13.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, Bistoni F, Frati L, Cortese R, Gulino A, Ciliberto G, Costantini F, Poli V. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Hines JA, Akashi K, Tenen DG. Enhancement of hematopoietic stem cell repopulating ability and self-renewal in the absence of the transcription factor C/EBPα. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein B, Cogswell PC, Baldwin AS., Jr Functional and physical associations between NF-κB and C/EBP family members a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paz-Priel I, Cai DH, Wang D, Kowalski J, Blackford A, Liu H, Heckman CA, Gombart A, Koeffler HP, Boxer LM, Friedman AD. C/EBPα and C/EBPα myeloid oncoproteins induce bcl-2 via interaction of their basic regions with NF-κB p50. Mol Cancer Res. 2005;3:585–596. doi: 10.1158/1541-7786.MCR-05-0111. [DOI] [PubMed] [Google Scholar]
- 18.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–390. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Wang X, Ward A, Touw I, Friedman AD. C/EBPα and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15:779–786. doi: 10.1038/sj.leu.2402094. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Scott E, Sawyers CL, Friedman AD. C/EBPα bypasses G-CSF signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood. 1999;94:560–571. [PubMed] [Google Scholar]
- 22.Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, Takatsu K, Kincade PW. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christy RJ, Kaestner KH, Geiman DE, Lane MD. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1(Spi-1) and C/EBPα regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 25.Zhang DE, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen HM, Hiebert SW, Tenen DG. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 27.Alcamo E, Mizgerd JP, Horwitz BH, Bronson R, Beg AA, Scott M, Doerschuk CM, Hynes RO, Baltimore D. Targeted mutation of TNF receptor I rescues the relA-deficient mouse and reveals a critical role for NF-κB in leukocyte recruitment. J Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, D’Costa J, Civin CI, Friedman AD. C/EBPα directs monocytic commitment of primary myeloid progenitors. Blood. 2006;108:1223–1229. doi: 10.1182/blood-2005-12-008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genetics. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Friedman AD. C/EBPs are required for granulopoiesis independent of their induction of the granulocyte-colony stimulating factor receptor. Blood. 2002;99:2776–2785. doi: 10.1182/blood.v99.8.2776. [DOI] [PubMed] [Google Scholar]
- 31.Paz-Priel I, Ghosal AK, Kowalski J, Friedman AD. C/EBPα or C/EBPα oncoproteins regulate the intrinsic and extrinsic apoptotic pathways by direct interaction with NF-κB p50 bound to the bcl-2 and FLIP gene promoters. Leukemia. 2008 doi: 10.1038/leu.2008.297. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBPα:AP-1 leucine zipper heterodimers bind novel DNA element, activate the PU.1 promoter, and direct monocyte lineage commitment more potently than C/EBPα homodimers or AP-1. Oncogene. 2008;27:2772–2779. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]