Abstract
The coupled binuclear “type 3” Cu sites are found in hemocyanin (Hc), tyrosinase (Tyr), and the multicopper oxidases (MCOs), such as laccase (Lc), and play vital roles in O2 respiration. Although all type 3 Cu sites share the same ground state features, those of Hc/Tyr have very different ligand-binding properties relative to those of the MCOs. In particular, the type 3 Cu site in the MCOs (LcT3) is a part of the trinuclear Cu cluster, and if the third (i.e., type 2) Cu is removed, the LcT3 site does not react with O2. Density functional theory calculations indicate that O2 binding in Hc is ≈9 kcal mol−1 more favorable than for LcT3. The difference is mostly found in the total energy difference of the deoxy states (≈7 kcal mol−1), where the stabilization of deoxy LcT3 derives from its long equilibrium Cu–Cu distance of ≈5.5–6.5 Å, relative to ≈4.2 Å in deoxy Hc/Tyr. The O2 binding in Hc is driven by the electrostatic destabilization of the deoxy Hc site, in which the two Cu(I) centers are kept close together by the protein for facile 2-electron reduction of O2. Alternatively, the lack of O2 reactivity in LcT3 reflects the flexibility of the active site, capable of minimizing the electrostatic repulsion of the 2 Cu(I)s. Thus, the O2 reactivity of the MCOs is intrinsic to the trinuclear Cu cluster, leading to different O2 intermediates as required by its function of irreversible reduction of O2 to H2O.
Keywords: density functional theory, laccase, binuclear copper proteins, oxygen binding, oxygen reduction
A number of important biological systems contain coupled binuclear copper active sites that play important roles in O2 binding, activation, and reduction to H2O. The term “coupled” refers to the exchange interaction between two Cu(II) (S = 1/2) centers caused by bridging ligation, leading to antiferromagnetic coupled, diamagnetic, and thus, electron paramagnetic resonance (EPR) silent ground states. They have been historically categorized as “type 3” Cu sites in biology (1) and consistently referred to as one class of related sites. The simplest of the metalloproteins containing type 3 sites are the binuclear Cu proteins, hemocyanins (Hc), catechol oxidase (CatO), and tyrosinase (Tyr) that reversibly bind O2; CatO and Tyr further activate O2 for substrate hydroxylation or oxidation (2, 3). Hc/CatO/Tyr have been shown to have structurally equivalent active sites. The type 3 sites are also found in the multicopper oxidases (MCOs), which include tree and fungal laccase (Lc), ceruloplasmin, Fet3p (the MCO found in yeast with ferroxidase activity), and ascorbate oxidase. MCOs use a minimum of four Cu centers: a “blue,” type 1 Cu site and a trinuclear Cu cluster composed of a “normal,” type 2 Cu and a binuclear type 3 Cu site that together catalyze the 4-electron reduction of O2 to water with concomitant oxidation of substrates (2, 3). Crystal structures indicate that both the type 3 Cu active sites in Hc/CatO/Tyr (4–10) and the MCOs (11–25) are similarly held in the protein by three His ligands on each Cu center. No additional ligands are present in the deoxy forms, whereas oxygen-derived ligands bridge and exchange couple the two Cu(II)s in the oxidized forms.
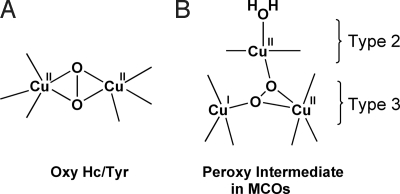
O2 binding to the type 3 Cu sites of deoxy Hc/Tyr generates a side-on μ-η2:η2 peroxo-bridged Cu2O2 oxy structure with Cu–Cu distance of ≈3.6 Å (Fig. 1A) (7, 8, 10). The side-on geometry promotes a large overlap between the O22− π* and the two Cu dx2−y2 orbitals and provides a direct pathway for 2-electron reduction of O2 to peroxide (26). Detailed spectroscopic and theoretical studies have shown that the side-on bridged geometry in oxy Hc/Tyr leads to a large energy splitting between the lowest-unoccupied molecular orbital (LUMO) and the highest-occupied molecular orbital (HOMO), which results in the strong antiferromagnetic coupling between the two Cu(II) centers (2, 3). Evaluation of the O2-binding reaction coordinate by using density functional theory (DFT) calculations has shown that the spin-forbidden transition in the formation of the antiferromagnetically coupled (i.e., singlet) oxy Hc from triplet O2 involves a simultaneous 2-electron transfer, where the initial ferromagnetically coupled coordination modes change along the O2-binding coordinate with increasing metal–ligand overlap to turn on antiferromagnetic coupling to generate the final planar singlet Cu2O2 structure (Fig. 1A).
Fig. 1.
O2 binding in the type 3 Cu sites of Hc/Tyr and MCOs.
The type 3 Cu sites in the MCOs are part of the trinuclear Cu cluster where the 4-electron reduction of O2 to water occurs. Reaction of the fully reduced form of MCO with O2 proceeds via two sequential 2-electron steps generating, first, the peroxy intermediate and then, the native intermediate. The spectral features of the peroxy intermediate are very different from those of oxy Hc/Tyr, indicating that the peroxy intermediate acquires a very different geometry (27). In our recent DFT study, we have generated a spectroscopically relevant structure, where the peroxide is bridged internally in a μ3-1,1,2 geometry with the type 2 and one of the type 3 coppers oxidized (Fig. 1B) (28). Importantly, this structure allows for the irreversible binding of O2 at the trinuclear Cu site that leads to efficient O O bond cleavage in the subsequent 2-electron reduction step to generate the native intermediate.
O bond cleavage in the subsequent 2-electron reduction step to generate the native intermediate.
The different roles of type 3 Cu sites in Hc/Tyr and MCOs are closely related to their distinctive ligand-binding properties. In particular, by using a derivative of tree Lc where the type 2 Cu is reversibly removed [i.e., type 2-depleted (T2D) Lc], we have demonstrated that the reduced type 3 Cu center alone cannot bind O2 in the MCOs, (29, 30). However, no detailed account of the origin of this O2 reactivity difference between the two type 3 Cu sites has been available. In this work, DFT calculations are systematically performed to compare the two classes of type 3 Cu sites in biology. Focus is on evaluating the different O2-binding properties of Hc/Tyr and the type 3 site in the MCOs and elucidating their origins. This work provides molecular insight into the critical role of the protein environment in directing the O2 reactivity of these catalytic active sites.
Results and Analysis
Reduced Structures.
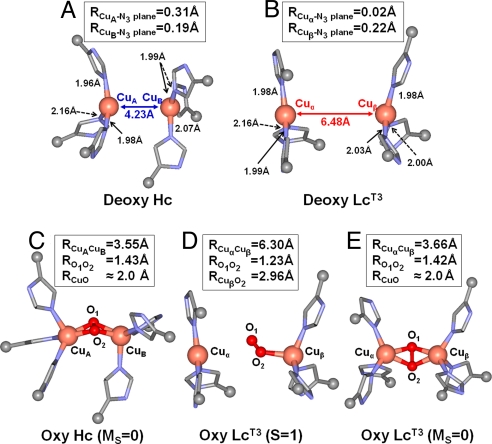
As a starting point for the evaluation of O2 binding to the type 3 sites in Hc/Tyr and MCOs, optimized structures of Hc and Lc [the type 3 Cu center in the absence of the type 2 Cu (LcT3)] with reduced (i.e., deoxy) binuclear Cu centers were obtained (Fig. 2 A and B). Copper atoms are labeled as CuA and CuB for Hc and Cuα and Cuβ for LcT3 for clarity in further descriptions of these sites. Both type 3 sites were modeled with three His ligands on each Cu center; Cu centers were coordinated to εN atoms except for one His ligand of the Cuα center in LcT3, which was coordinated to the δN atom as found in all available crystal structures of MCOs. Each model was geometry optimized with frozen αC positions to reflect the different protein structures at the type 3 sites in Hc and LcT3.
Fig. 2.
Optimized structures of deoxy and oxy forms of Hc and LcT3. (A) Deoxy Hc. (B) Deoxy LcT3. (C) Oxy Hc (MS = 0). (D) Oxy LcT3 (S = 1). (E) Oxy LcT3 (MS = 0). Relevant bond lengths are shown in Å.
The geometry-optimized structures of deoxy Hc and deoxy LcT3 are shown in Fig. 2 A and B. In general, the optimized Cu centers have trigonal planar geometry, typical for d10-ML3 units (31). However, the Cu atoms in the CuA and CuB centers in Hc and Cuβ center in Lc are ≈0.2–0.3 Å out of the plane formed by the N3 ligands, whereas the Cu atom in the Cuα site in Lc remains in the plane. The planar geometry of the Cuα site is largely caused by the δN coordination of one of its His ligands because the αC constraint of the δN-His ligand is not along the Cu N bond and therefore, does not contribute in orienting the Cuα atom out of the N3 plane. The pyramidal distortion in Hc and the in-plane geometry in the Cuα site of Lc is consistent with Cu K-edge X-ray absorption spectroscopy (XAS) data (29, 30, 32, 33) (see Discussion).
N bond and therefore, does not contribute in orienting the Cuα atom out of the N3 plane. The pyramidal distortion in Hc and the in-plane geometry in the Cuα site of Lc is consistent with Cu K-edge X-ray absorption spectroscopy (XAS) data (29, 30, 32, 33) (see Discussion).
The most conspicuous difference between the two deoxy structures is found in the equilibrium Cu–Cu distances, in which deoxy Hc is 4.23 Å and deoxy LcT3 is 6.48 Å (Fig. 2 A and B). For deoxy Hc, the calculated R(Cu–Cu) is in good agreement with those of known crystallographic data of Hc and Tyr, which range from 3.5 to approximately 4.6 Å (5–7, 10). For LcT3, the calculated R(Cu–Cu) is ≈1.2 to ≈1.5 Å larger than those of the known crystal structures of reduced ascorbate oxidase (13), CotA (the MCO found in bacteria Bacillus subtilis) (11) and Fet3p (24), which range from 5.0 to ≈5.3 Å. The deviation in the calculated Cu–Cu distance is reduced when two hydrogen bonds involving His ligands and backbone carbonyl groups† are included in the calculation [supporting information (SI) Fig. S1]. The resulting Cu–Cu distance is 5.80 Å, which is in reasonable agreement with those of the crystal structures. However, the additional hydrogen bonds have virtually no effect on the overall Cu geometries in deoxy LcT3 aside from the Cu–Cu distance. Moreover, the energy difference between deoxy LcT3 at R(Cu–Cu) of 6.48 Å and 5.4 Å is only ≈2 kcal mol−1 (≈1.4 kcal mol−1 if a dielectric with ε = 4.0 is included using the polarizable continuum model) caused by the shallow potential energy surface (PES) along R(Cu–Cu), implicating the flexible nature of the LcT3 site. Thus, despite the long-calculated Cu–Cu distance, the deoxy LcT3 structure shown in Fig. 2B is a reasonable representation of the type 3 site in the MCOs, in particular, in providing energetic descriptions of O2 binding.
Reaction Coordinate for O2 Binding.
As a second step in evaluating O2 binding in the type 3 sites of Hc and LcT3, an O2 molecule was introduced to the deoxy Hc and deoxy LcT3 structures. Geometry optimizations of deoxy Hc+O2 and deoxy LcT3+O2 complexes were performed with frozen αC positions, in both the spin-unrestricted triplet (S = 1) and broken-symmetry (MS = 0) states; the energies of the pure-singlet (S = 0) states were then obtained from the spin-projection method (34–36) by using the broken-symmetry state energies (see Computational Methods). Calculations were performed both with unconstrained Cu–Cu distances and with these distances systemically varied.
The O2-binding mechanism in Hc has been studied in detail, and the structure of Hc+O2 (i.e., oxy Hc) is well established (26). Therefore, the optimized oxy Hc structure, in the broken-symmetry (MS = 0) state, is first obtained to validate our method and model systems (Fig. 2C). Our calculations indicate that O2 binding in Hc is exothermic by −2.9 kcal mol−1, which is in reasonable agreement with experiment (ΔH = −11.5 to −6.0 kcal mol−1 and ΔG = −5.0 kcal mol−1 at 25 °C) (37–41). Key structural features are consistent with experimental data, yielding the side-on μ-η2:η2 O22− core structure with R(Cu–Cu) of 3.55 Å (experimental ≈3.6 Å). The Cu2O2 structure is slightly butterflied, and the μ-η2:η2 O22− bridge provides a strong superexchange pathway for a large antiferromagnetic coupling of the Cu(II) centers with a calculated −2J (Ĥ = −2J Ŝ1·Ŝ2) of ≈1,280 cm−1, also consistent with experiment (>600 cm−1). A reaction coordinate was further generated by varying the Cu–Cu distance from the starting R(Cu–Cu) of 4.2 Å (i.e., that of reduced Hc) (Fig. 3Lower Left) and optimizing the rest of the structure with frozen αC positions (note that the S = 0 energies shown in Fig. 3 are obtained from spin-projection method by using the broken-symmetry state energies and geometries). As a result, a spin-state transition from S = 1 to S = 0 occurs with decreasing Cu–Cu distance (at ≈3.9 Å). Associated with this spin-state transition is a structural change from a μ-η1:η2 peroxy bridge at large distances (Fig. S2) to the final μ-η2:η2 Cu2O2 structure. These results are consistent with our previous study on O2 binding to deoxy Hc, where a reaction coordinate was generated by systematically varying the distance of the peroxide above the molecular plane (26). It was shown that the charges on both copper centers change at the same rate even in the asymmetric bridged structure, indicating that O2 binding involves the simultaneous transfer of 2 electrons. Likewise, it is found here that 2 electrons from both Cu centers have already been transferred to O2 to form peroxide at an R(Cu–Cu) of ≈4.0 Å, as indicated by spin-density distribution among Cu and O atoms in the broken-symmetry wave functions: At R(Cu–Cu) = 4.0 Å, spin densities on CuA, CuB, O1, O2 atoms are +0.46, −0.46, +0.09, and −0.11, whereas at the equilibrium R(Cu–Cu) = 3.55 Å, these are +0.48, −0.48, −0.03, and +0.04, respectively. Thus, the Cu–Cu distance in deoxy Hc is prepositioned for facile 2-electron transfer upon O2 binding.
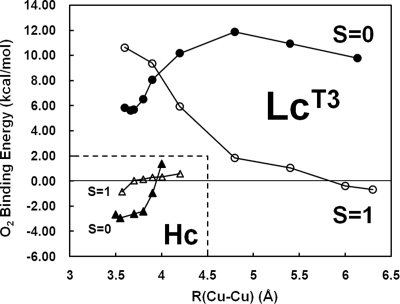
Fig. 3.
Reaction coordinates for O2 binding in Hc and LcT3 along Cu–Cu. The O2-binding energies were obtained by subtracting the sum of the energies of triplet O2 molecule and the most stable deoxy forms (i.e., deoxy forms in Fig. 2) from that of the oxy forms at each R(Cu–Cu). The S = 0 energies are obtained from the spin-projection method by using the broken-symmetry state energies and geometries.
In contrast to oxy Hc, the most stable structure for oxy LcT3 is found in the triplet (S = 1) state. As shown in Fig. 2D, the triplet oxy LcT3 structure has R(Cu–Cu) of 6.30 Å. Consequently, O2 is unable to bridge the two Cu centers but is only very weakly bound to the Cuβ center with an end-on geometry and a low O2-binding energy of −0.7 kcal mol−1 (note that we were not able to generate a structure with O2 bound to the Cuα atom). Bond lengths, R(Cuβ O) = 2.96 Å and R(O
O) = 2.96 Å and R(O O) = 1.23 Å, are also indicative of a very weak Cuβ
O) = 1.23 Å, are also indicative of a very weak Cuβ O2 bond that lacks any electron transfer between Cu and O2 (i.e., no superoxide or peroxide formation). At shorter Cu–Cu distances, the side-on μ-η2:η2 Cu2O2 structure in the broken-symmetry spin state is also obtained (Fig. 2E). The singlet oxy LcT3 structure has R(Cu–Cu) of 3.66 Å and R(O–O) of 1.42 Å, indicative of a 2-electron reduced peroxo-species. However, the singlet oxy LcT3 is found to have O2-binding energy of +5.6 kcal mol−1 (6.3 kcal mol−1 higher than the triplet oxy LcT3), making its formation thermodynamically unfavorable.
O2 bond that lacks any electron transfer between Cu and O2 (i.e., no superoxide or peroxide formation). At shorter Cu–Cu distances, the side-on μ-η2:η2 Cu2O2 structure in the broken-symmetry spin state is also obtained (Fig. 2E). The singlet oxy LcT3 structure has R(Cu–Cu) of 3.66 Å and R(O–O) of 1.42 Å, indicative of a 2-electron reduced peroxo-species. However, the singlet oxy LcT3 is found to have O2-binding energy of +5.6 kcal mol−1 (6.3 kcal mol−1 higher than the triplet oxy LcT3), making its formation thermodynamically unfavorable.
We have also generated a reaction coordinate for the formation of singlet oxy LcT3 by varying R(Cu–Cu), in both the triplet (S = 1) and singlet (S = 0) states (Fig. 3). The PES for the triplet state is uphill from R(Cu–Cu) of 6.30 Å with decreasing Cu–Cu distance, whereas that for the singlet state is downhill until the equilibrium R(Cu–Cu) of 3.66 Å is reached. At R(Cu–Cu) > 4.5 Å, the broken-symmetry optimized structures (representing singlet states) are characterized by the side-on η2 superoxo structures formed between O2 and the oxidized Cuβ center via internal 1-electron transfer from Cuβ to O2 (see Fig. S3), in contrast to the end-on structure of the triplet state (Fig. 2D). When R(Cu–Cu) decreases below ≈4.5 Å in the broken-symmetry state, a bridged μ-η1-η2 geometry is initially formed, then the final μ-η2-η2 peroxo structure, with transfer of the second electron from the Cuα center to the bridging O2 moiety (spin density at the Cuα atom changes from 0.0 to −0.33 to −0.45 with change in R(Cu–Cu) from 6.13 Å to 4.20 Å to 3.66 Å in the broken-symmetry state). Notably, the spin state transition from S = 1 to S = 0 occurs at R(Cu–Cu) ≈4.0 Å, where an energy barrier is found to be ≈9 kcal mol−1 relative to that of the triplet oxy LcT3. The presence of this energy barrier makes the O2 binding to deoxy LcT3 kinetically difficult, in contrast to Hc where the O2 binding to the deoxy Hc is uniformly downhill (Fig. 3 Lower Left). Thus, our calculations indicate the O2 binding in LcT3 is inhibited thermodynamically, as well as kinetically, and therefore the long Cu-Cu distance in deoxy LcT3 creates a large energy barrier for the singlet oxy LcT3 formation.
At this point, we evaluate the origin of different O2 reactivities of Hc and LcT3. As mentioned above, the O2-binding energies of Hc and LcT3 are different by ≈8.5 kcal mol−1 (−2.9 kcal mol−1 for Hc and +5.6 kcal mol−1 for Lc). However, the total energy of the oxy LcT3 structure itself is only ≈1.6 kcal mol−1 higher than that of the oxy Hc structure, which does not account for the difference in O2-binding energies. Rather, the remaining 6.9 kcal mol−1 difference is found in the energy difference of the deoxy structures of Hc and LcT3. As shown below, the relative destabilization of the deoxy Hc structure leads to the thermodynamically favorable O2 binding in Hc, whereas the relative stabilization in the deoxy LcT3 structure results in unfavorable O2 binding in LcT3.
Potential Energy Surfaces of Deoxy Sites.
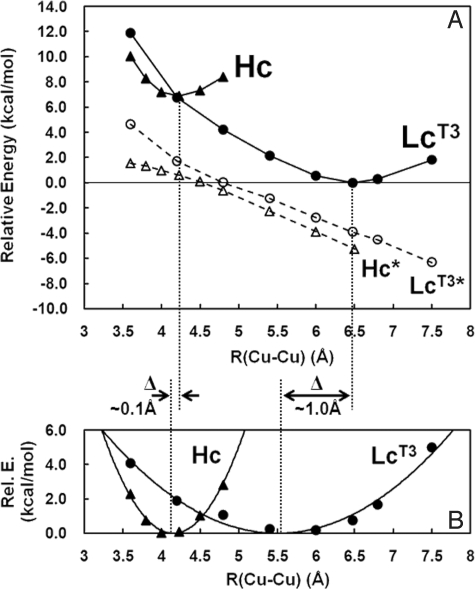
To gain insight into the origin of the difference in deoxy Hc and deoxy LcT3, PESs along Cu–Cu were generated, where geometry optimizations of the deoxy Hc and deoxy LcT3 were performed with frozen αC positions (Fig. 4A, filled symbols), as well as without the αC constraints (Fig. 4A, open symbols, Hc* and LcT3*). As described above, the deoxy LcT3 (with αC constraints) energy minimum is 6.9 kcal mol−1 lower in energy than that for deoxy Hc and has an equilibrium Cu–Cu distance of 6.48 Å, in contrast to the 4.23 Å optimized distance of deoxy Hc. Alternatively, unconstrained PESs for deoxy Hc and deoxy LcT3 (Hc* and LcT3* in Fig. 4A) are both effectively linear, and the slopes are very similar (approximately −2.5 kcal mol−1Å−1), indicating that the Cu(I)–Cu(I) electrostatic repulsion of the two deoxy forms are the same (note that the difference between the two unconstrained PESs are from the small additional contribution caused by a decrease in steric interactions of the three His ligands between Cu centers, which are eclipsed in Lc and staggered in Hc). Notably, the energy difference of the unconstrained deoxy LcT3 at R(Cu–Cu) = 6.5 Å and 4.2 Å is ≈5.8 kcal mol−1 (i.e., the electrostatic energy difference), which accounts for most of the 6.9 kcal mol−1 energy difference between deoxy Hc and deoxy LcT3. Thus, the decrease in energy of the deoxy LcT3 site reflects the decrease in electrostatic repulsion of the two Cu(I) centers in the low dielectric of the protein. The lack of O2 reactivity of the type 3 site in LcT3 reflects its electrostatic and structural stabilization at its long-calculated equilibrium Cu–Cu distance of 6.48 Å.
Fig. 4.
PESs of deoxy Hc and deoxy LcT3 along Cu–Cu. (A) PESs of deoxy Hc and deoxy LcT3 with αC constraints are shown as Hc and LcT3 in filled symbols, whereas PESs of deoxy Hc and deoxy LcT3 without αC constraints are shown as Hc* and LcT3* in open symbols. (B) PESs of deoxy Hc and deoxy LcT3, where the energies of Hc* and LcT3* in A are subtracted from those of Hc and LcT3 in A. Symbols are the results of the subtraction, whereas the solid lines are the fit curves using a second-order parabolic function. For comparison of the two PES components, the minima are set as zero in relative energies.
In Fig. 4B, the components of the PESs in Fig. 4A where the electrostatic contributions (i.e., Hc* and LcT3* in Fig. 4A) are removed are given. The resultant PESs provide important insight into the elasticity of the two type 3 sites imposed by the protein constraints (through the αC atoms in our models) in the absence of electrostatic contributions. The PES components (Fig. 4B, filled symbols) were fitted to second-order parabolic functions (Fig. 4B, solid lines) to obtain spring force constants, k. It is found that k for deoxy LcT3 is 2.3 kcal mol−1Å−2, whereas that for deoxy Hc is 14.1 kcal mol−1Å−2, which is ≈7 times larger than that of deoxy LcT3. Notably, in Fig. 4B, the minimum in the deoxy LcT3 PES component is ≈1 Å shorter than that of the original PES in Fig. 4A (from 6.48 Å to 5.51 Å), reflecting the low value of k and thus, the flexible nature of the Cu geometry in deoxy LcT3. In contrast, the minimum in the deoxy Hc PES is shifted by only 0.08 Å (from 4.23 Å to 4.15 Å). Thus, the large k in deoxy Hc demonstrates that the Cu–Cu distance is kept short by the protein environment, whereas it is not in deoxy LcT3.
Discussion
Generally, the binuclear Cu sites in Hc/Tyr and MCOs (i.e., LcT3) have been classified into the same type 3 group because both have antiferromagnetically coupled diamagnetic ground states and thus, no EPR feature. Despite the same active-site components of [Cu(His)3]2, however, the two type 3 sites have very different ligand-binding properties. In particular, the reduced type 3 Cu site in T2D Lc is stable to O2 binding, in strong contrast to Hc/Tyr, which reversibly bind O2 to form oxy Hc/Tyr (29, 30). Moreover, exogenous ligands (carbon monoxide and azide) were shown to bind to the type 3 site of T2D Lc, demonstrating that small molecules are not blocked from having access to this active site. However, these ligands bind only to one Cu center in T2D Lc, further demonstrating the different ligand-binding properties compared with Hc/Tyr, in which exogenous ligands bridge the two-Cu center (42).
We have calculated that O2 binding to Hc is exothermic by 2.9 kcal mol−1. This is in contrast to O2 binding to the deoxy LcT3 in the formation of singlet oxy LcT3, which is found to be endothermic by 5.6 kcal mol−1 (Fig. 3), consistent with the fact that T2D Lc does not bind O2.‡ The origin of this 8.5 kcal mol−1 destabilization of O2 binding in LcT3 relative to Hc reflects the relative stabilization of the deoxy LcT3 structure in the protein environment of the MCOs. The 6.9 kcal mol−1 stabilization of deoxy LcT3 relative to deoxy Hc derives from the decrease in electrostatic repulsion of the two Cu(I)s at the long Cu–Cu distance in deoxy LcT3 in the low dielectric of the protein. This increase in Cu–Cu distance is allowed by the combined effects of the protein constraints and the flexibility in the active-site environment.
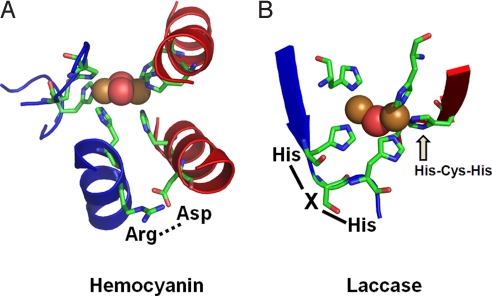
From Fig. 5, the large structural differences between the coupled binuclear site in Hc and the type 3 site in MCOs relate to the very different structural constraints in the two protein environments. The binuclear Cu(I) site of deoxy Hc is kept at its 4.2 Å Cu–Cu distance because of the constraints of the protein. The most conspicuous constraint is associated with two His ligands, one on each Cu, for which the αC–αC distance is only ≈5 Å compared with >9 Å for other αC–αC distances between His ligands on the two Cu centers. The origin of the close αC–αC distance is attributed to an Arg(Lys)–Asp salt bridge between two helices that is conserved in Hc/CatO/Tyr (see sequence alignment in Fig. S4). Alternatively, in the LcT3 Cu site, the two Cu(I)s are kept at an electrostatically stable distance of ≈6.5 Å (calculated) by two sets of two His ligands (αC–αC distances of the two pairs of His ligands are ≈7 Å), where each set is from an H–X–H motif that is on a loop extending from a β-sheet, leaving the Cu(I)s relatively unconstrained. Importantly, this shows that the different ligand architecture in Hc and LcT3 account for the large difference in equilibrium Cu–Cu distances in deoxy Hc and deoxy LcT3. This is a critical factor in the O2-binding properties of Hc and LcT3, where the close Cu–Cu distance in deoxy Hc leads to simultaneous 2-electron transfer from both Cus to O2, whereas the long Cu–Cu distance in deoxy LcT3 leads to stabilization of this state and energetically unfavorable O2 binding.
Fig. 5.
Features of Cu active sites Hc (A) and LcT3 (B) that affect the equilibrium Cu–Cu distances. Structures are adapted from crystal structure database 1JS8 (oxy Hc from O. dofleini Hc) (8) and 1GYC (resting oxidized fungal Lc from T. versicolor) (19).
In addition, the protein constraints further contribute to the very different PESs for deoxy Hc and LcT3 along Cu–Cu (Fig. 4B), where the binuclear force constant k in deoxy LcT3 is ≈7 times smaller than that of deoxy Hc. This large force constant difference results in the shift in the equilibrium Cu–Cu distance by ≈0.1 Å in deoxy Hc and ≈1 Å in deoxy LcT3 because of the linear distorting force from the electrostatic repulsion (Fig. 4). Note that the ≈1 Å shift in deoxy LcT3 results in an energy stabilization of ≈1.4 kcal mol−1, whereas the energy of deoxy Hc is lowered by only ≈0.2 kcal mol−1 by the ≈0.1 Å shift.§ Thus, the difference in force constants between deoxy LcT3 and deoxy Hc contributes ≈20% of the total energy difference (i.e., ≈7 kcal mol−1) between these sites. Importantly, the large force constant in deoxy Hc indicates that the active site is tightly controlled to keep the Cu–Cu distance short for facile reversible O2 binding. Alternatively in deoxy LcT3, the low force constant allows a large change in Cu–Cu distance with little change in energy, which is required in the reaction coordinate for O O bond cleavage at the trinuclear Cu cluster in the MCOs (43).
O bond cleavage at the trinuclear Cu cluster in the MCOs (43).
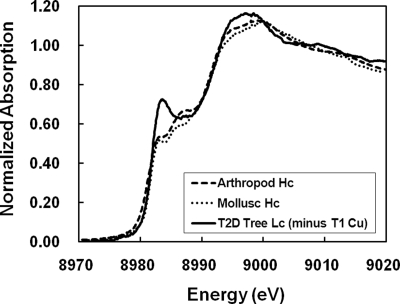
Insight into the origin of the difference in force constants in deoxy Hc and deoxy LcT3 can be gained from Cu K-edge XAS data. As shown in Fig. 6, the preedge features of deoxy Hc and deoxy T2D Lc (i.e., deoxy LcT3) are very different (29, 30, 32, 33). In deoxy T2D Lc, the sharp intense peak at ≈8,983 eV is the Cu 1s → Cu 4pz transition in a trigonal planar geometry, with the Cu 1s → Cu 4px,y transitions shifted up in energy with lower intensities because of the equatorial ligands. In deoxy Hc, the Cu 1s → Cu 4pz transition has lower intensity whereas the Cu 1s → Cu 4px,y transitions shift to lower energy and higher intensity, reflecting the pyramidal distortion of the Cu centers out of the ligand N3 plane. These Cu K-edge XAS data are consistent with the calculated structures of deoxy Hc and deoxy LcT3 (Fig. 2 A and B); in deoxy Hc, both Cu centers show a pyramidal distortion (Cu centers ≈0.2–0.3 Å out of the N3 plane) whereas in deoxy LcT3, only one Cu is distorted, and the other, Cuα in Fig. 2B, retains a trigonal planar geometry, where the planarity derives largely from the δN ligation of one of its His ligands. These different Cu geometries are closely related to the different force constants; in the trigonal planar geometry of the Cuα center in deoxy LcT3, the vibrational mode normal to the N3 plane involves only bends, whereas for the trigonal pyramidal geometries of the Cu centers in deoxy Hc (and Cuβ in deoxy LcT3), both bends and bond stretches are involved. Thus, the larger force constant along Cu–Cu in deoxy Hc largely reflects the pyramidal distortion of the Cu center that is imposed by the protein environment, whereas the low force constant in deoxy LcT3 derives from its relatively unconstrained environment, resulting in the trigonal planar structure typical of Cu(I) d10–ML3 complexes.
Fig. 6.
Normalized Cu K-edge XAS spectra of deoxy Hc and T2D Lc. The two Hc spectra were obtained from Hcs from an arthropod Panulirus interruptus (spiny lobster) (32) and a mollusk Busycon canaliculatum (sea snail) (32), and the T2D Lc spectrum was obtained from Lc from Rhus vernicifera (lacquer tree) (29, 30). The features of the type 1 Cu (T1 Cu) was subtracted from the T2D Lc spectrum by using the XAS spectrum of the blue Cu protein plastocyanin.
In summary, deoxy Hc is electrostatically destabilized to react with O2 to form oxy Hc, and this can be cooperatively regulated by changing the Cu(I)–Cu(I) distance (26) in the protein quaternary structure (i.e., tense and relaxed conformations) (44). Alternatively, the deoxy LcT3 Cu site in the multicopper oxidases is electrostatically stable and does not react with O2 when the T2 Cu is not present. When the type 2 Cu is also present, the trinuclear Cu cluster does react with O2 to form a 2-electron reduced peroxide, bridging between the type 2 and type 3 Cu centers (27, 28). The trinuclear Cu cluster further promotes cleavage of the O O bond by stabilizing the products of peroxide reduction (oxo and hydroxo moieties) through multiple bridging interactions requiring a large change in Cu–Cu distance.
O bond by stabilizing the products of peroxide reduction (oxo and hydroxo moieties) through multiple bridging interactions requiring a large change in Cu–Cu distance.
Computational Methods
Spin-unrestricted DFT calculations were performed on Gaussian 03 (45). All geometry optimizations were performed with B3LYP hybrid functional (46) by using TZVP basis sets for Cu and coordinated N/O atoms and TZV basis sets for the rest (47, 48). The initial geometries of the type 3 sites of Hc and Lc were adapted from the crystal structures of the oxy form of Octopus dofleini Hc [Protein Data Bank (PDB) ID code 1JS8] (8) and the resting oxidized form of Trametes versicolor fungal Lc (PDB ID code 1GYC) (19), where the bridging O atoms were removed. Three His ligands on each Cu center were replaced with methyl-imidazolyl ligands, where Cu centers were coordinated to εN atoms, except for one imidazolyl ligand of the Cuα center in Lc, which was coordinated to δN atom, as shown consistently in all available crystal structures of MCOs. All geometry optimizations were performed with frozen methyl C atoms (i.e., αC atoms; depicted by gray spheres in Fig. 2, except for the deoxy Hc and deoxy LcT3 structures in generating the Hc* and Lc* potential energy surfaces shown in Fig. 4A, in which all of the αC constraints were removed). For oxy Hc and oxy LcT3 structures, geometry optimizations were performed in both the triplet (S = 1) and the broken-symmetry (MS = 0) states. The energies of the pure singlet (S = 0) states were then computed by using the spin-projection method (34–36):
where the triplet energy, ES = 1, is computed for the single-determining high-spin configuration at the broken-symmetry geometry and 〈SBS2〉 is the spin-expectation value of the broken-symmetry calculation. The O2-binding energies were obtained by subtracting the sum of the energies of triplet O2 molecule and the most stable deoxy forms from that of the oxy forms.
Supplementary Material
Acknowledgments.
We thank Dr. Ritimukta Sarangi for assistance with the Cu K-edge XAS data analysis. This work was supported by National Institutes of Health Grant DK-31450.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902127106/DCSupplemental.
†The hydrogen-bonding interactions are present in the two HXH loop motifs in the LcT3 site. In each HXH loop, the δNH of the first His ligand is in position to hydrogen bond with the C O group of the X residue.
O group of the X residue.
‡We have also calculated deoxy LcT3 structures with a fourth His ligand, as found in some of the crystal structures of T2D Lc (14, 15). Our results show that the fourth His ligand does not coordinate with the Cu center, whereas instead, a H2O molecule can coordinate very weakly.
§If the PES is described by E0 = 1/2 k(Q − Q0)2 = 1/2 k(ΔQ)2 and the linear distortion by λ(Q − Q0) = λ(ΔQ), the total energy becomes E = 1/2 k(ΔQ)2 + λ(ΔQ). Therefore, the energy will be lowered −λ2/2k by the distortion, with a shift in Q by −λ/k. With k of 14.1 and 2.3 kcal mol−1Å−2 and the slope of the distorting electrostatic force λ of −2.5 kcal mol−1Å−1, Q will be shifted by +0.2 Å and +1.2 Å (within 10% of those in Fig. 4B) with energies lowered by 0.2 kcal mol−1 and 1.4 kcal mol−1 for deoxy Hc and deoxy LcT3, respectively.
References
- 1.Malkin R, Malmström BG. State and function of copper in biological systems. Adv Enzymol. 1970;33:177–244. doi: 10.1002/9780470122785.ch4. [DOI] [PubMed] [Google Scholar]
- 2.Solomon EI, Chen P, Metz M, Lee SK, Palmer AE. Oxygen binding, activation, and reduction to water by copper proteins. Angew Chem Int Ed. 2001;40:4570–4590. doi: 10.1002/1521-3773(20011217)40:24<4570::aid-anie4570>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 4.Gaykema WPJ, et al. 3.2 Å structure of the copper-containing, oxygen-carrying protein Panulirus interruptus haemocyanin. Nature. 1984;309:23–29. [Google Scholar]
- 5.Volbeda A, Hol WGJ. Crystal structure of hexameric haemocyanin from Panulirus interruptus refined at 3.2 Å resolution. J Mol Biol. 1989;209:249–279. doi: 10.1016/0022-2836(89)90276-3. [DOI] [PubMed] [Google Scholar]
- 6.Hazes B, et al. Crystal structure of deoxygenated Limulus polyphemus subunit II hemocyanin at 2.18 Å resolution: Clues for a mechanism for allosteric regulation. Protein Sci. 1993;2:597–619. doi: 10.1002/pro.5560020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnus KA, et al. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- 8.Cuff ME, Miller KI, van Holde KE, Hendrickson WA. Crystal structure of a functional unit from octopus hemocyanin. J Mol Biol. 1998;278:855–870. doi: 10.1006/jmbi.1998.1647. [DOI] [PubMed] [Google Scholar]
- 9.Klabunde T, Eicken C, Sacchettini JC, Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 10.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 11.Bento I, Martins LO, Lopes GG, Carrondo MA, Lindley PF. Dioxygen reduction by multicopper oxidases: A structural perspective. Dalton Trans. 2005:3507–3513. doi: 10.1039/b504806k. [DOI] [PubMed] [Google Scholar]
- 12.Messerschmidt A, et al. Refined crystal structure of ascorbate oxidase at 1.9 Å resolution. J Mol Biol. 1992;224:179–205. doi: 10.1016/0022-2836(92)90583-6. [DOI] [PubMed] [Google Scholar]
- 13.Messerschmidt A, Luecke H, Huber R. X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini: Reduced, peroxide and azide forms. J Mol Biol. 1993;230:997–1014. doi: 10.1006/jmbi.1993.1215. [DOI] [PubMed] [Google Scholar]
- 14.Ducros V, et al. Crystal structure of the type 2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat Struct Biol. 1998;5:310–316. doi: 10.1038/nsb0498-310. [DOI] [PubMed] [Google Scholar]
- 15.Ducros V, et al. Structure of the laccase from Coprinus cinereus at 1.68 Å resolution: Evidence for different “type 2 Cu-depleted” isoforms. Acta Crystallogr D. 2001;57:333–336. doi: 10.1107/s0907444900013779. [DOI] [PubMed] [Google Scholar]
- 16.Zaitsev VN, Zaitseva I, Papiz M, Lindley PF. An X-ray crystallographic study of the binding sites of the azide inhibitor and organic substrates to ceruloplasmin, a multicopper oxidase in the plasma. J Biol Inorg Chem. 1999;4:579–587. doi: 10.1007/s007750050380. [DOI] [PubMed] [Google Scholar]
- 17.Zaitseva I, et al. The X-ray structure of human serum ceruloplasmin at 3.1 Å: Nature of the copper centers. J Biol Inorg Chem. 1996;1:15–23. [Google Scholar]
- 18.Bertrand T, et al. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41:7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- 19.Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90 Å resolution containing a full complement of coppers. J Biol Chem. 2002;277:37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- 20.Hakulinen N, et al. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat Struct Biol. 2002;9:601–605. doi: 10.1038/nsb823. [DOI] [PubMed] [Google Scholar]
- 21.Roberts SA, et al. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:2766–2771. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enguita FJ, et al. Substrate and dioxygen binding to the endospore coat laccase from Bacillus subtilis. J Biol Chem. 2004;279:23472–23476. doi: 10.1074/jbc.M314000200. [DOI] [PubMed] [Google Scholar]
- 23.Smith AW, Camara-Artigas A, Wang MT, Allen JP, Francisco WA. Structure of phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. Biochemistry. 2006;45:4378–4387. doi: 10.1021/bi0525526. [DOI] [PubMed] [Google Scholar]
- 24.Taylor AB, Stoj CS, Ziegler L, Kosman DJ, Hart PJ. The copper–iron connection in biology: Structure of the metallo-oxidase Fet3p. Proc Natl Acad Sci USA. 2005;102:15459–15464. doi: 10.1073/pnas.0506227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garavaglia S, et al. The structure of Rigidoporus lignosus laccase containing a full complement of copper ions, reveals an asymmetrical arrangement for the type 3 copper pair. J Mol Biol. 2004;342:1519–1531. doi: 10.1016/j.jmb.2004.07.100. [DOI] [PubMed] [Google Scholar]
- 26.Metz M, Solomon EI. Dioxygen binding to deoxyhemocyanin: Electronic structure and mechanism of the spin-forbidden two-electron reduction of O2. J Am Chem Soc. 2001;123:4938–4950. doi: 10.1021/ja004166b. [DOI] [PubMed] [Google Scholar]
- 27.Shin W, et al. Chemical and spectroscopic definition of the peroxide-level intermediate in the multicopper oxidases: Relevance to the catalytic mechanism of dioxygen reduction to water. J Am Chem Soc. 1996;118:3202–3215. [Google Scholar]
-
28.Yoon J, Solomon EI. Electronic structure of the peroxy intermediate and its correlation to the native intermediate in the multicopper oxidases: Insights into the reductive cleavage of the O
 O bond. J Am Chem Soc. 2007;129:13127–13136. doi: 10.1021/ja073947a. [DOI] [PMC free article] [PubMed] [Google Scholar]
O bond. J Am Chem Soc. 2007;129:13127–13136. doi: 10.1021/ja073947a. [DOI] [PMC free article] [PubMed] [Google Scholar] - 29.Lubien CD, et al. Chemical and spectroscopic properties of the binuclear copper active site in Rhus laccase: Direct confirmation of a reduced binuclear type 3 copper site in type 2-depleted laccase and intramolecular coupling of the type 3 to the type 1 and type 2 copper sites. J Am Chem Soc. 1981;103:7014–7016. [Google Scholar]
- 30.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. X-ray absorption-edge determination of the oxidation state and coordination number of copper: Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 31.Albright TA, Burdett JK, Whangbo M-H. Orbital Interactions in Chemistry. New York: Wiley; 1985. [Google Scholar]
- 32.Kau LS. PhD dissertation. Stanford, CA: Stanford Univ; 1988. Chemical and spectroscopic studies on metal active sites in metalloproteins and heterogeneous catalysts. [Google Scholar]
- 33.Brown JM, Powers L, Kincaid B, Larrabee JA, Spiro TG. Structural studies of the hemocyanin active site. 1. Extended X-ray absorption fine structure (EXAFS) analysis. J Am Chem Soc. 1980;102:4210–4216. [Google Scholar]
- 34.Noodleman L. Valence bond description of antiferromagnetic coupling in transition metal dimers. J Chem Phys. 1981;74:5737–5743. [Google Scholar]
- 35.Yamaguchi K, Jensen F, Dorigo A, Houk KN. A spin correction procedure for unrestricted Hartree–Fock and Moller–Plesset wavefunctions for singlet diradicals and polyradicals. Chem Phys Lett. 1988;149:537–542. [Google Scholar]
- 36.Isobe H, et al. Extended Hartree–Fock (EHF) theory of chemical reactions. VI. Hybrid DFT and post-Hartree–Fock approaches for concerted and nonconcerted transition structures of the Diels–Alder reaction. Mol Phys. 2002;100:717–727. [Google Scholar]
- 37.Brunori M, Kuiper HA, Zolla L. In: Life Chemistry Reports. Woods EJ, editor. New York: Harwood; 1983. pp. 239–250. [Google Scholar]
- 38.Zolla L, Kuiper HA, Brunori M, Antonini E. Thermodynamics of Oxygen Binding to Helix pomatia β-Hemocyanin. New York: Dekker; 1979. [Google Scholar]
- 39.Klarman A, Daniel E. Oxygen binding properties of stripped (calcium ion and magnesium ion free) hemocyanin from the scorpion Leirus quinquestriatus. Biochemistry. 1980;19:5176–5180. doi: 10.1021/bi00564a004. [DOI] [PubMed] [Google Scholar]
- 40.Er-El Z, Shaklai N, Daniel E. Oxygen binding properties of hemocyanin from Levantina hierosolima. J Mol Biol. 1972;64:341–352. doi: 10.1016/0022-2836(72)90502-5. [DOI] [PubMed] [Google Scholar]
- 41.Antonini E, Brunori M, Colosimo A, Kuiper HA, Zolla L. Kinetic and thermodynamic parameters for oxygen binding to the allosteric states of Panulirus interruptus hemocyanin. Biophys Chem. 1983;18:117–124. doi: 10.1016/0301-4622(83)85005-4. [DOI] [PubMed] [Google Scholar]
- 42.Spira-Solomon DJ, Solomon EI. Chemical and spectroscopic studies of the coupled binuclear copper site in type 2 depleted Rhus laccase: Comparison to the hemocyanins and tyrosinase. J Am Chem Soc. 1987;109:6421–6432. [Google Scholar]
- 43.Solomon EI, Augustine AJ, Yoon J. O2 reduction to H2O by the multicopper oxidases. Dalton Trans. 2008:3921–3932. doi: 10.1039/b800799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brouwer M, Bonaventura C, Bonaventura J. Analysis of effect of three different allosteric ligands on oxygen binding by hemocyanin of shrimp, Penaeus setiferus. Biochemistry. 1978;17:2148–2154. doi: 10.1021/bi00604a019. [DOI] [PubMed] [Google Scholar]
- 45.Frisch MJ, et al. Gaussian 03, Revision C 01. Pittsburgh: Gaussian; 2004. [Google Scholar]
- 46.Becke AD. Density functional thermochemistry. 3. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 47.Schäfer A, Horn H, Ahlrichs R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J Chem Phys. 1992;97:2571–2577. [Google Scholar]
- 48.Schäfer A, Huber C, Ahlrichs R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J Chem Phys. 1994;100:5829–5835. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.