Abstract
Several lines of evidence indicate that microorganisms in the meso- and bathypelagic ocean are metabolically active and respiring carbon. In addition, growing evidence suggests that archaea are fixing inorganic carbon in this environment. However, direct quantification of the contribution from deep ocean carbon sources to community production in the dark ocean remains a challenge. In this study, carbon flow through the microbial community at 2 depths in the mesopelagic zone of the North Pacific Subtropical Gyre was examined by exploiting the unique radiocarbon signatures (Δ14C) of the 3 major carbon sources in this environment. The radiocarbon content of nucleic acids, a biomarker for viable cells, isolated from size-fractionated particles (0.2–0.5 μm and >0.5 μm) showed the direct incorporation of carbon delivered by rapidly sinking particles. Most significantly, at the 2 mesopelagic depths examined (670 m and 915 m), carbon derived from in situ autotrophic fixation supported a significant fraction of the free-living microbial community (0.2–0.5 μm size fraction), but the contribution of chemoautotrophy varied markedly between the 2 depths. Results further showed that utilization of the ocean's largest reduced carbon reservoir, 14C-depleted, dissolved organic carbon, was negligible in this environment. This isotopic portrait of carbon assimilation by the in situ, free-living microbial community, integrated over >50,000 L of seawater, implies that recent, photosynthetic carbon is not always the major carbon source supporting microbial community production in the mesopelagic realm.
Keywords: microbial metabolism, particle flux, particulate organic carbon (POC), chemoautotrophy
Marine microbes play a critical role in transforming inorganic and organic carbon in the ocean (1), and their interactions with the carbon reservoir, in its variety of phases, have important implications for the sequestration of CO2 in either the deep ocean or sediments. However, the role that microorganisms play in the carbon cycle of the deep ocean, the mechanisms they employ, and the extent to which they employ them, are poorly understood. Most of the ocean's reduced carbon accumulates in the meso- and bathypelagic realm as dissolved organic carbon [(DOC); ≈700 Pg] (2), and here is where the fate of a significant fraction of organic carbon leaving the euphotic zone is decided (3). Therefore, microbial transformations of carbon in the “twilight zone” (defined here as depths between the base of the euphotic zone and 1,000 m) (3) play an essential role in determining the ocean's ability to sequester atmospheric CO2.
Below the euphotic zone, first light, and then labile organic matter, become increasingly scarce. Although DOC is present in this environment, it has long been assumed that this reservoir is unavailable to support heterotrophic production. For example, depth profiles of DOC (4), dissolved combined amino acids (5, 6), and dissolved carbohydrates (6, 7) appear to be static below 600 m, consistent with findings that DOC from the deep ocean resists microbial degradation (8). The most convincing evidence of the refractory nature of deep ocean DOC is its average radiocarbon age, which lies between 4,000 and 6,500 radiocarbon years (4, 9, 10).
Despite the apparently static nature of the largest carbon reservoir available to support heterotrophic production in the meso- and bathypelagic zones, the abundance and activity of microorganisms are still measurable (11–14). One study suggests that 20–33 Pg C yr−1 are respired to CO2 in the dark ocean (15), and the majority of this carbon is most likely derived from particulate organic carbon (POC) that sinks out of the euphotic zone. Measurements using particle traps have shown that this flux of POC is strongly attenuated in the mesopelagic ocean (3, 16); and a parameterization of this attenuation—the Martin Curve (16)—demonstrated that 90% of the organic carbon leaving the euphotic zone is lost from the particle phase within the upper 1,500 m.
Bacteria attached to sinking particles and marine snow not only respire carbon but also produce hydrolytic enzymes that transform POC into the dissolved phase at a rate faster than it is assimilated (17, 18). It is hypothesized that this decoupling of hydrolysis and uptake rates produces a “plume” of dissolved organic matter and other nutrients that trail these particles as they sink through the water column (19, 20). Therefore, in addition to respiration, carbon loss from POC may occur through DOC release as well. Strong correlations have been observed between deep ocean free-bacterial abundance and both episodic and long-term particle flux out of the overlying water column (21–23), further indicating that carbon is transferred from sinking POC to DOC to free-living microorganisms.
High-resolution depth profiles show that suspended POC contains bomb 14C at abyssal depth (24) and support the hypothesis that rapidly sinking particles disaggregate to smaller size fractions. Additionally, several geochemical observations suggest that there is a “fresh” or new component within the mesopelagic DOC pool that is masked by the much larger refractory fraction identified above (25, 26), and sinking POC is the likely conduit for the rapid delivery of this new DOC to the deep ocean. Together these independent lines of evidence imply that some fraction of free-living heterotrophic microbes in the deep ocean is supported by inputs of recently-fixed carbon ultimately delivered by sinking particles.
The carbon cycle now appears even more dynamic as a result of the discovery that archaea contribute up to 50% of microbial abundance in the deep ocean (27). As a domain, these organisms demonstrate the ability to take up amino acids as well as fix inorganic carbon (28–32). For example, the recently cultured crenarchaeote Nitrosopumilus maritimus is an ammonia-oxidizing chemoautotroph (33), and the gene coding for subunit A of an archaeal ammonia monooxygenase (amoA) has been found to be ubiquitous throughout the marine environment (34). Crenarchaeaota are now believed to be important players in the marine nitrogen cycle and the deep ocean carbon cycle through chemoautotrophic nitrification (35). Studies have shown that distributions and apparent metabolisms of planktonic archaea vary with depth and across water masses (29, 36). However, the contribution of chemosynthesis (by these planktonic archaea and other organisms) to the total production of the microbial community and how that might change throughout the water column have not been fully determined (29, 32).
In this study, natural variations in the abundance of the radioactive isotope of carbon (14C) were exploited to identify carbon sources fueling production of the total microbial community. The goal was to explicitly test the hypothesis that the microbial community of the mesopelagic ocean is maintained primarily by particle flux from overlying waters. The 3 pools of carbon available to free living microorganisms, dissolved inorganic carbon (DIC), refractory DOC, and fresh DOC injected from particles, have unique 14C signatures (Δ14C) in the deep ocean that can be traced into the intracellular biochemicals of organisms that use these carbon sources. Fresh DOC delivered by POC leaving the surface ocean has a modern Δ14C value > +50‰, whereas DIC in the mesopelagic zone has prebomb Δ14C values and is depleted in 14C because of radioactive decay (≈−200 to −100‰). The bulk DOC pool is the “oldest,” with an average Δ14C value of −525‰ in the deep North Pacific Ocean (9). Total nucleic acids (hereafter referred to as DNA) are used as a biomarker for the total microbial community as they are synthesized de novo in cells by using carbon acquired from the surrounding environment. DNA is expected to be primarily present in viable cells, so the 14C content of DNA provides a snapshot of the living community at the time of sampling. This technique represents a direct method by which to evaluate carbon metabolism in situ, and has been previously used to trace assimilated carbon through aquatic microbial communities (e.g., 37–39).
Results of the study directly support the hypothesis that respiration of organic carbon delivered from rapidly sinking particles represents an important energy source for free-living microorganisms in the subsurface ocean. However, the study also revealed the significance of chemoautotrophy in supporting total community production in this environment. Together, these data provide a geochemical glimpse of the diverse roles microorganisms play in the carbon cycle of the ocean's “twilight zone.”
Results
Table 1 reports the Δ14C of DNA in each size fraction and at each depth. At the surface (21 m), DNA isolated from the 0.2–0.5 μm microbial community is modern, with Δ14C = +60 ± 3‰. The Δ14C value of DNA (Δ14C-DNA) from the 0.2–0.5 μm size fraction at 670 m was 140 ± 17‰, significantly lower than the value for the >0.5 μm fraction at this same depth (−73 ± 4‰; Table 1). At 915 m, Δ14C values for both size fractions were higher than those at 670 m, −15 ± 15‰ for the 0.2–0.5 μm size fraction and +69 ± 6‰ for the >0.5-μm fraction. In 2 cases replicate samples were run (670 m 0.2–0.5 μm and 915 m >0.5-μm samples), and differences in measured Δ14C values ranged from 5 to 20‰ between replicates. δ13C values of all DNA samples at all depths are between −21 and −19‰, within the expected range for samples of marine origin (Table 1).
Table 1.
Collection dates, isotopic composition, size of 14C samples, and C/N of DNA samples
| Sample | Collection date, month/yr | δ13C, ‰ | measured Δ14C, ‰ | Size, μgC | UCIG# | C/N | corrected Δ14C, ‰ |
|---|---|---|---|---|---|---|---|
| 21 m 0.2–0.5 μm | 12/03 & 5/04 | −19.0 | +36 ± 2 | 450 | 5397 | 4.05* | +60 ± 3 |
| 670 m 0.2–0.5 μm | 2/06 | −20.1 | −187 ± 14 | 22, 24 | 11272, 10539 | n.a. | −140 ± 17† |
| 670 m > 0.5 μm | 2/06 | −20.0 | −106 ± 4 | 150 | 38657 | 4.31 | −73 ± 4 |
| 915 m 0.2–0.5 μm | 2/06 | −19.7 | −87 ± 10 | 28 | 10540 | 5.06 | −15 ± 15 |
| 915 m > 0.5 μm | 2/06 | −19.4 | −27 ± 4 | 80, 110 | 38658, 38659 | 5.32 | +69 ± 6 |
All samples were collected at the Natural Energy Laboratory of Hawaii Authority in Kona, HI located at 19.74°N, 156.05°W. Measured Δ14C errors are propagated AMS errors and blank corrections, and in the case of corrected Δ14C values, C/N measurement errors are further incorporated into the error propagation. n.a., not available (amount of N in submitted sample too small for accurate measurement).
*Average surface Pacific 0.2–0.5 μm DNA C/N (n = 3).
†Δ14C-DNA corrections for this sample were calculated using the highest C/N ratio (5.32 for 915 m > 0.5-μm-size fraction) as an upper bound.
Δ14C values for DNA reported above are corrected for possible contributions from non-DNA carbon on the basis of the measured C/N ratio of each sample and reagent blanks. Both measured and corrected Δ14C values are presented in Table 1. Sample C/N values ranged from 4.05 to 5.32 (Table 1) higher than pure DNA, which has an average C/N ratio of 3 (confirmed by measured ratios in a pure E. coli standard). Variations in the sample C/N ratio from pure DNA could represent coisolation of carbon-rich intracellular molecules such as carbohydrates and lipids that should provide the same radiocarbon information as pure DNA and would not represent a contamination of the radiocarbon sample. However, it is possible that elevated C/N ratios also result from carbon contamination associated with either reagents or adsorption of bulk marine DOC to filters followed by coprecipitation with DNA in the extraction method.
Cleaned, blank filters (n = 2) subjected to the entire method yielded a negligible 0.7 ± 0.4 μgC with an average Δ14C value of −379 ± 300‰. The small yield of carbon indicates that blanks, because of either reagents or filter material, are not solely responsible for the differences identified above in C/N ratios. All values shown in Table 1 have been corrected for this experimentally determined blank, incorporating errors of both the blank size and isotopic composition. If DOC was coprecipitated with our DNA, the extremely 14C-depleted DOC at these depths in the ocean would contaminate our measured Δ14C-DNA values. Accounting for this possible source of contamination (see SI Text and Table S2) shifted our measured Δ14C-DNA values toward modern (Fig. 1 and provided as corrected values in Table 1), and brought them closer to the Δ14C value of DIC in the surface ocean and at 670-m depth.
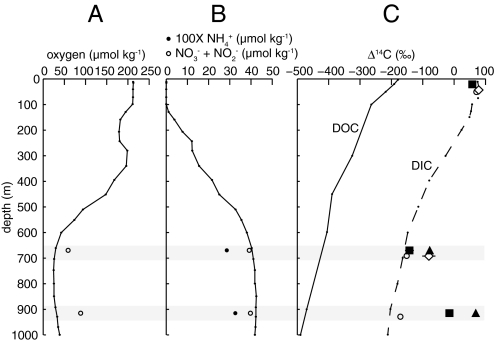
Fig. 1.
Vertical profiles of oxygen (A) and nitrate + nitrite (B) data are for nearby station ALOHA (http://hahana.soest.hawaii.edu). Data (○) are specific to the NELHA sampling site (at 19.74°N, 156.05°W) (32, 41). (C) Corrected Δ14C values for 0.2–0.5 μm (■) and >0.5-μm (▴) size fractions of DNA extracted from 21 m, 670 m, and 915 m (see SI Text for correction calculations), along with vertical profiles of Δ14C of DOC (at 31°N, 159°W) (9) and DIC (at 35°N, 155°E) (40). Δ14C-DNA error bars are propagated AMS errors and blank corrections, and if not visible, are less than the width of the marker. Also included are abundance-weighted averages and error of archaeal lipid Δ14C from 21 m and 670 m (◇) (32).
Proton NMR (1H-NMR) spectroscopy was also used to assess the quantity and quality of extracted sample DNA. The NMR spectra verified that there was no contamination of DNA samples from process chemicals, which would appear as sharp, well-defined resonances. In addition, the presence of significant 1H-NMR resonances in the region between 7–8.5 ppm was indicative of aromatic groups not typically abundant in marine detritus, but found in DNA. However, NMR data could not conclusively rule out the presence of either intracellular biochemicals other than DNA or small amounts of ambient DOC in our precipitated samples. For these reasons we relied primarily on the measured C/N ratio of each DNA sample to estimate possible carbon contamination and apply the Δ14C correction reported in Table 1.
A set of 14C process blanks was also developed to demonstrate that Δ14C-DNA was indeed reflective of available carbon sources. GF/F-filtered seawater collected off the Scripps Institution of Oceanography (SIO) pier was amended with 883 μM nitrate, 36.3 μM phosphate, and a carbon source with a known Δ14C value (glucose, Δ14C = +87 ± 2‰; or acetate, Δ14C = −987 ± 4‰). These carbon sources were chosen to provide a range of Δ14C values from modern (Δ14C >0‰) to essentially radiocarbon dead (Δ14C = −1,000‰). The natural, marine microbial community was then incubated in the dark for 3 days until biomass was ≈109 cells mL−1 and then subjected to the same filtration, lysis, and extraction method as all environmental samples. Isolated DNA was processed for Δ14C and δ13C analyses with the results shown in Table 2. 14C sample sizes were comparable to the amount of carbon analyzed for environmental Δ14C-DNA (Tables 1 and 2).
Table 2.
Isotopic composition and sample sizes of carbon sources and microbial DNA from 14C process blanks
| Sample | Date, month/yr | Carbon source | Culture volume, L | Δ14C, ‰ | Size, μg C | UCIG# | δ13C, ‰ |
|---|---|---|---|---|---|---|---|
| glucose | +87 ± 2 | 819 | 3411 | −10.7 | |||
| microbial DNA | 10/03 | glucose | 20 | +63 ± 5 | 1015 | 1565 | −11.7 |
| microbial DNA | 6/07 | glucose | 8 | +64 ± 4, +62 ± 4 | 120, 200 | 38655, 38656 | −11.7 |
| acetate | −988 ± 4, −986 ± 4 | 800, 250 | 38653, 38654 | −35.3 | |||
| microbial DNA | 6/07 | acetate | 8 | −821 ± 4 | 110 | 39810 | −34.5 |
Δ14C-DNA values, +63 ± 2‰ (average ± SD; n = 3) for the glucose culture and −821‰ for the acetate culture, showed rapid incorporation of the Δ14C signature of carbon sources by the microbial community. Measured Δ14C-DNA values do deviate to different extents from the Δ14C signature of each respective carbon source. This observed deviation could be caused by a combination of several factors: utilization of ambient DOC present in the collected seawater by cometabolism with the added glucose or acetate (42), selective utilization if the added carbon source was isotopically inhomogeneous, and contamination from either process chemicals or passive adsorption of ambient bulk DOC to filters during cell isolation as discussed previously. It is clear that no consistent carbon contamination from either a modern or 14C-dead source is introduced through our extraction procedure. This finding was expected based on previous results showing negligible carbon contamination from reagents and filter materials. Nevertheless, contamination from an exogenous component, such as ambient DOC, with an intermediate Δ14C value (between modern and 14C-dead) could cause the observed deviation. However, source carbon and bacterial DNA had very similar δ13C values after the incubation (Δδ13C < ±1‰; Table 2) and suggest that contamination from an exogenous source, if present, was relatively small (see SI Text).
Further evidence that the Natural Energy Laboratory of Hawaii Authority (NELHA; Kona, HI) samples primarily contain native DNA is their ability to be amplified by PCR. Because of the nature of the seawater supply system, we were motivated to analyze the microbial community to confirm that we were not isolating DNA from organisms that were unique to the seawater intake pipe. 16S rRNA bacterial and archaeal clone libraries establish that the microbes sampled are representative of those found in other mesopelagic marine environments (including nearby Station ALOHA) (Table S1 and Fig. S1), further confirming the presence of native DNA in our samples. No absolute trends in community composition based on depth or size fraction are apparent. The majority of archaeal clones from all depths and size fractions are Group I Crenarchaeota (>85%), with the remainder belonging to the Group II Euryarchaeota. Of note are a number of clones with 98–99% identity to the ammonia-oxidizing N. maritimus (33).
Significant differences were not observed for inorganic nitrogen between the 2 depths as NO3− + NO2− concentrations were 39.6 μmol kg−1 at 670 m and 40.1 μmol kg−1 at 915 m (41). NH4+ concentrations were also comparable with values of 0.29 μmol kg−1 and 0.33 μmol kg−1 at 670 m and 915 m, respectively. There were measurable O2 variations as the oxygen concentration at 670 m of 60.9 μmol kg−1 was depleted relative to the 90.1 μmol kg−1 at 915 m (Fig. 1) (41).
Discussion
Measurements of the 14C content of microbial DNA directly demonstrate significant metabolic stratification in the mesopelagic water column. As expected, microorganisms in the surface ocean derived their carbon primarily from DIC and fresh DOC. In the mesopelagic realm, the free-living (0.2–0.5 μm) microbial population at 915 m incorporated carbon delivered primarily by sinking particles, but the same microbial size-fraction at 670 m relied more extensively on in situ chemoautotrophy to satisfy community carbon demand. Existing measurements do not predict this dynamic view of the deep “twilight zone” (between 670 m and 915 m). For example, the DOC reservoir is static between these depths (4–7), the geochemical environment at both depths is similar (41), the POC flux at each depth predicted by the parameterization of Buesseler et al., (3), is not consistent with the observed variability, and similar bacterial numbers are expected for both depths (43). Also significant is the demonstration that a fraction of the free-living microbial community had adapted to a metabolic lifestyle that was distinct from microorganisms found in larger size fractions, such as those associated with particles.
A primarily autotrophic microbial community in the surface ocean would have a Δ14C signature that is identical to surface DIC (+71 ± 3‰ at the time of sampling) (32). Archaeal lipids conformed to this expected trend—surface glycerol dialkyl glycerol tetraether (GDGT) lipids of the Crenarchaeota had an average Δ14C value of +82 ± 12‰ (32). Sterols from autotrophs and/or heterotrophs isolated in the >0.5-μm fraction at this site had values of +56 ± 9‰ and +69 ± 9‰ (32). Additionally, sugars isolated from the DOC reservoir, a possible substrate for heterotrophic microorganisms, had modern Δ14C values between 40 and 67‰ at this site in 2001 (25). Therefore, heterotrophs consuming fresh DOC, such as dissolved carbohydrates, and autotrophs should have similar Δ14C-DNA values in the surface ocean. The surface DNA sample (0.2–0.5 μm) isolated in the present study included both archaeal and bacterial DNA (Table S1) and had a Δ14C signature of +60 ± 3‰. This value fell within the reported Δ14C range for modern carbon sources confirming that some combination of DIC and recently produced organic carbon was fueling the surface free-living microbial community at the time of sampling. The only previous 14C study of marine microbial DNA (0.2–0.7 μm) reported depleted Δ14C values (−34 ± 24‰) in surface waters of the eastern North Pacific Ocean (34°50′ N, 123°00′ W; Station M) (38) and suggested microbial incorporation of an old DOC component (Δ14C = −549‰). In contrast, DNA isolated from the surface microbial community at NELHA did not indicate utilization of any refractory DOC.
Utilization of refractory DOC in the mesopelagic zone also appeared to be negligible, and the relative importance of ambient DIC-fixation varied markedly with depth. The depleted Δ14C-DNA value (−140 ± 17‰) for the 0.2–0.5 μm size-fraction at 670 m showed that microbes are incorporating carbon with a Δ14C value that is similar to in situ DIC (−151 ± 3‰) (Fig. 1) (32), either directly through chemoautotrophy or through secondary production. The depleted Δ14C-DNA value (Table 1) could have also resulted from consumption of a fraction of the ambient DOC pool (Fig. S2). However, archaeal lipids produced in situ at 670 m had an average Δ14C value of −112‰ and showed that the archaeal community derived 83% of their carbon from autotrophy at this depth (32). This latter finding favors the hypothesis that the observed 14C-depletion of total DNA results from chemoautotrophic production supplying the majority of carbon (as much as 95%) (Fig. S2) incorporated by the microbial community at this site and depth. The available phylogenetic data (Fig. S1 and Table S1) identified microorganisms other than chemoautotrophs in the 0.2–0.5 μm size-fraction from 670 m, so Δ14C-DNA signatures must further reflect some heterotrophy coupled to in situ chemoautotrophy (i.e., both primary and secondary production). For example, dissolved neutral sugars isolated from high-molecular-weight (>1,000 Da) DOC at this site had an average Δ14C value of −123 ± 10‰ at 670 m (25), indicating that a major fraction of these carbohydrates could have been freshly produced in situ through chemoautotrophy. These sugars and other compounds produced in situ would provide a labile, 14C-depleted carbon source for heterotrophs at 670 m. If heterotrophic free-living bacteria at this depth rely primarily on 14C-depleted, fresh DOC produced from in situ autotrophy, then both autotrophic and heterotrophic microorganisms would have a similar Δ14C signature.
Surprisingly, DNA from the 670 m 0.2–0.5-μm sample was more depleted in 14C than the same size fraction at 915 m (Fig. 1). At 915 m, DNA in the 0.2–0.5-μm size fraction contained more 14C (−15 ± 15‰) than DIC (Δ14C-DIC = −171 ± 3‰) indicating that sinking POC was more important for sustaining the free-living microbial population at this depth. Although some chemoautotrophy is occurring at 915 m, only up to 36% of DNA carbon in the 0.2–0.5-μm size fraction at 915 m ultimately could be derived from ambient DIC fixation as compared to up to 95% at 670-m depth (Fig. S2).
The >0.5-μm size fractions at 670 m and 915 m followed a depth-trend similar to the smaller size fraction but had higher Δ14C values (Fig. 1) than their free-living counterparts. Particles would be isolated into this larger size fraction and POC metabolism by particle-attached cells is the likely cause of these enriched Δ14C-DNA values. As mentioned previously, particle sinking rates are high enough that little 14C-decay occurs between surface waters and the mesopelagic zone, so POC and microbes that consume this POC will retain an enriched surface ocean Δ14C signature relative to ambient C pools (i.e., DIC and total DOC) at mesopelagic depths. Therefore, DNA isolated from this larger size fraction should have Δ14C values similar to DIC and DNA in surface waters. However, DNA in the >0.5-μm fraction at 670 m was significantly depleted in 14C relative to surface C pools, and this depletion reflects the coisolation of free-living microorganisms onto the >0.5-μm filter. Larger filters such as GF/F filters (0.7-μm nominal pore size) have been shown to collect over 50% of marine bacteria (44, 45), and studies have observed that archaea are larger than bacteria in some marine environments (46). Therefore, Δ14C-DNA values for the >0.5-μm size fraction must integrate the 14C signature of DNA from both free-living, mesopelagic microorganisms and those attached to particles sinking out of surface waters.
The greater 14C-depletion of the >0.5-μm sample at 670 m compared to 915 m indicates the coisolation of a free-living community that incorporates more in situ DIC at 670 m and is consistent with results obtained from smaller size fractions (Table 1 and Fig. 1). Together, data from the 2 different size-fractions demonstrate that some free-living microorganisms use carbon sources that are significantly different from those used by their particle-attached counterparts.
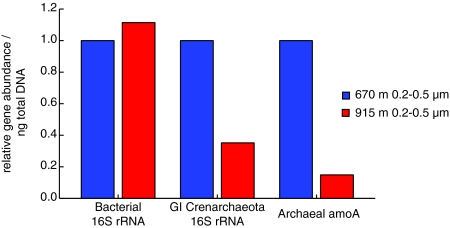
Spatial variations in the extent of autotrophy implied by the Δ14C-DNA signature of both size fractions are consistent with the variability in archaeal amoA gene copies observed in other marine studies (35, 47–49). The latter suggest spatial gradients in the extent of archaeal ammonia oxidation and by extension, autotrophy. In an effort to explore the possible metabolisms that may have contributed to the assimilation of carbon present within the extracted DNA, quantitative PCR (qPCR) was used to determine the number of marine group I Crenarchaeota 16S rRNA and archaeal amoA gene copies present per ng total DNA in the same samples isolated for radiocarbon analyses. Although this analysis is only an indication of community composition and metabolic potential and not gene expression, a correlation has been shown between both marine Crenarchaeota 16S rRNA and archaeal amoA copy number and ammonia oxidation rates in the Gulf of California (48). qPCR assays with the 0.2–0.5 μm DNA samples showed comparable numbers of bacterial 16S rRNA genes at 670 m and 915 m, but more of both marine group I Crenarchaeota 16S rRNA and archaeal amoA gene copies per ng total DNA from 670 m than 915 m (Fig. 2). These trends are consistent with the Δ14C data and indicate that DIC fixation fuels a greater proportion of free-living microbial community production at 670-m depth. In addition, a comparison of the 2 size fractions from 915 m showed greater abundance of both genes in the smaller size fraction (0.2–0.5 μm), again consistent with radiocarbon results. Although the focus here has been on archaeal autotrophy, carbon fixation by other members of the microbial community cannot be ruled out. For example, so far planktonic archaea appear to be primarily nitrosifiers, able to perform only the first step in the process of nitrification; therefore, the contribution to autotrophy by bacterial nitrifiers must also be considered (33, 47).
Fig. 2.
qPCR data for 0.2–0.5-μm-size fraction DNA samples showing relative abundances of bacterial 16S rRNA, group I Crenarchaeota 16S rRNA, and archaeal amoA gene copies per ng total DNA, as quantified by PicoGreen. For ease of sample comparison the qPCR data are normalized to the 670-m 0.2–0.5-μm sample.
As discussed in previous studies (e.g., 32, 35), widespread chemoautotrophy in the deep ocean, if fueled primarily by ammonium oxidation, necessitates a starring role for regenerated, reduced N in the ocean's twilight zone. Therefore, the dominant free-living microbial metabolisms identified at both depths in this study (autotrophy, if supported primarily through nitrification, at 670 m, and heterotrophy of recently produced carbon at 915 m) require fresh organic carbon and nitrogen delivered presumably from sinking particles. The current study estimates that the majority of microbial carbon accumulating at 670 m is ultimately derived from chemoautotrophy. In the deep ocean several factors such as in situ microbial growth rates, and carbon fixation pathways and rates determine the magnitude of the reduced N requirement, and these parameters remain poorly constrained (29). Furthermore, particle flux to the deep ocean is unsatisfactorily defined (3) and may be inadequate to support carbon respiration rates (e.g., 15) or calculated archaeal nitrification rates in the mesopelagic realm. In fact, at nearby Station ALOHA bacterial carbon demand in the mesopelagic zone has been shown to exceed POC flux by 3- to 4-fold (43), and investigators have invoked vertically migrating zooplankton as one additional mechanism of carbon transport to mesopelagic depths at this site (43, 50). In situ carbon-fixation could also alleviate some of the heterotrophic carbon demand at these depths, although it remains enigmatic what process might supply the reductant. One possibility is a decoupling of dissolved nitrogen from dissolved carbon delivery to the mesopelagic zone. Nitrogen is a major waste product of vertically migrating zooplankton (e.g., 51), which could impact the nitrogen cycle at depth (52) and could mediate the depth-dependent requirement for excess reduced N, as would be necessary for intensive nitrification. The results of this study argue for a concerted focus on delineating the in situ microbial physiology of the deep ocean and constraining carbon and nitrogen fluxes into this environment.
Our approach provides a true in situ measurement with no tracer amendments or environmental manipulations and integrates production by the total microbial community in a significant volume of seawater (≈50,000 L) over approximately 1 week of sampling. This approach differs significantly from the snapshot view captured in other studies via discrete water column sampling, such as those that assess RNA gene expression. The data show conclusively that source(s) of carbon used by the microbial community in the subsurface ocean can vary with depth (670 m vs. 915 m) and between free-living and particle-attached organisms (0.2–0.5 μm fraction vs. >0.5-μm fraction). The direct demonstration of the importance of both rapid DOC delivery to the mesopelagic ocean and chemoautotrophy in maintaining total microbial production in this environment was only possible through the Δ14C-DNA method developed here. This technique could provide a microbial perspective on the shape of the Martin Curve (16)—a relatively high-resolution depth profile of the Δ14C signature of microbial DNA would demonstrate the importance of surface-derived carbon to microbes living in the mesopelagic zone and identify depths where significant respiration of vertical carbon flux occurs.
Methods
Sampling and DNA Extraction.
Cellulose ester filters (0.5-μm) (Millipore Opticap) plumbed in-line with 0.2-μm polyethersulfone filters (Pall Supor) were used to collect particulate organic matter from approximately 50,000 L of surface (21 m), 670-m, and 915-m seawater by using the large-volume pumping capabilities at NELHA (Kona, HI) (19.74°N, 156.05°W). Filters were stored at −80 °C until DNA was extracted. To isolate the nucleic acids, a method modified from Blair et al. (53) was used to minimize organic carbon contamination for 14C analysis. Filters were cut open by using a combusted hacksaw, unfolded, and extracted in 1.5 M NaClO4 at 4 °C for 48 h with shaking to lyse collected cells. The resulting 2-L solution was ultrafiltered (Pellicon MWCO 5,000 Da; Millipore) down to 20 mL and an organic extraction was performed with an equal volume of chloroform and isoamyl alcohol (24:1). Nucleic acids were precipitated from the resulting aqueous fraction with ethanol, pelleted by centrifugation, and re-dissolved in sterile water. All reagents used in the extraction of environmental DNA were the same and used in the same quantities as for the 14C process blanks. Fractions were set aside for elemental and stable isotope analysis (University of California-Davis Stable Isotope Facility; Scripps Institution of Oceanography Analytical Facility), 1H-NMR spectroscopy, and molecular biology procedures, and the majority of the sample was lyophilized in 9-mm quartz tubes, combusted to CO2 at 850 °C with the addition of Ag and CuO, quantified, and converted to graphite according to the methods of Vogel et al. (54) for accelerator mass spectrometry (AMS) analysis at the University of California Irvine Keck Carbon Cycle AMS facility. Δ14C values are reported as per geochemical samples according to Stuiver and Polach (55).
PCR, Cloning, and Sequencing.
Portions of the 16S rRNA gene were amplified from environmental DNA samples by using Takara ExTaq Polymerase under the conditions described by the manufacturer's protocol. Universal bacterial (27F, 1492R) and archaeal (21F, 958R) primers were used at a final concentration of 0.5 μM. Products from 6 reactions were pooled, cleaned by using the Qiagen PCR Purification kit, and eluted in 30-μl water. Four microliters of this cleaned product was used in a TOPO cloning reaction with vector pCR2.1 as described by the manufacturer (Invitrogen). Plasmid DNA was extracted from white colonies by using a Qiagen Miniprep kit and was submitted to SeqXcel for sequencing. The 5′ end of both strands of the bacterial 16S rRNA gene was sequenced with primer 536r and M13f/r, whereas M13f&r were used to sequence entire archaeal inserts. DNA sequences were assembled and edited by using VectorNTI (Invitrogen), checked for chimeras by using the Bellerophon server (56), aligned to the Silva database (57), grouped into operational taxonomic units by using DOTUR (58), and inserted into ARB for phylogenetic analysis (59). Sequences were deposited into GenBank under accession numbers EU817582–EU817654.
Quantitative PCR.
For the qPCR assays, each reaction consisted of 12.5 μL Power SYBR® Green PCR Master Mix (Applied Biosystems), 0.4 μM each primer, 2 ng BSA, and 1–8 ng, 670-m and 915-m 0.2–0.5-μm size fraction DNA as determined by PicoGreen (Molecular Probes) in a final volume of 25 μL. Archaeal amoA assays were run in triplicate by using primers and cycling parameters from Beman et al. (48), although run for 40 cycles with the marine group I Crenarchaeota assays similarly adapted from Mincer et al. (47). Bacterial 16S rRNA genes were quantified by using primers 932F and 1062R (60). PCR products generated by using the qPCR primers were used as standards in tenfold dilution series from 107 to 102 copies based on PicoGreen quantification. PCR efficiencies and correlation coefficients for standard curves were as follows: 96% and r2 = 0.997 for archaeal amoA; 82% and r2 = 0.994 for GI Crenarchaeota 16S rRNA; and 94% and r2 = 0.999 for bacterial 16S rRNA. Results are presented as the relative abundance of gene copies per ng total DNA normalized to the 670-m 0.2–0.5-μm DNA sample.
Supplementary Material
Acknowledgments.
We thank Dan Repeta for sample collection; Jan War and the staff at Natural Energy Laboratory of Hawaii Authority for site access and nutrient data; Greg Dick for cloning and sequencing advice; Michael Beman for qPCR protocols and assistance; and Bruce Deck and the Scripps Institution of Oceanography Analytical Facility as well as the University of California-Davis Stable Isotope Facility for elemental and stable isotope analyses. Funding was provided by National Science Foundation OCE02-42160, Geisel Award, and University of California San Diego Faculty Career Development Award Program (to L.I.A.); National Science Foundation OCE02-41363 (to A.P.); and R.L.H. was supported by a National Defense Science and Engineering Graduate Fellowship and Lawrence Livermore National Laboratory Student Employee Graduate Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU817582–EU817654).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810871106/DCSupplemental.
References
- 1.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2008;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Hedges JI. Global biogeochemical cycles: Progress and problems. Mar Chem. 1992;39:67–93. [Google Scholar]
- 3.Buesseler KO, et al. Revisiting carbon flux through the ocean's twilight zone. Science. 2007;316:567–570. doi: 10.1126/science.1137959. [DOI] [PubMed] [Google Scholar]
- 4.Williams PM, Druffel ERM. Radiocarbon in dissolved organic matter in the central North Pacific Ocean. Nature. 1987;330:246–248. [Google Scholar]
- 5.Lee C, Bada JL. Dissolved amino acids in the equatorial Pacific, the Sargasso Sea, and Biscayne Bay. Limnol Oceanogr. 1977;22:502–510. [Google Scholar]
- 6.Williams PM. In: Plankton Dynamics of the Southern California Bight. Eppley RW, editor. Berlin: Springer; 1986. pp. 53–83. [Google Scholar]
- 7.Mopper K. Sugars and uronic acids in sediment and water from the Black Sea and North Sea with emphasis on analytical techniques. Mar Chem. 1977;5:585–603. [Google Scholar]
- 8.Barber RT. Dissolved organic carbon from deep waters resists microbial oxidation. Nature. 1968;220:274–275. doi: 10.1038/220274a0. [DOI] [PubMed] [Google Scholar]
- 9.Druffel ERM, Williams PM, Bauer JE, Ertel JR. Cycling of dissolved and particulate organic matter in the open ocean. J Geophys Res. 1992;97:15639–15659. [Google Scholar]
- 10.Druffel ERM, Bauer JE. Radiocarbon distributions in Southern Ocean dissolved and particulate organic matter. Geophys Res Lett. 2000;27:1495–1498. [Google Scholar]
- 11.Azam F, Smith DC, Carlucci AF. Bacterial transformation and transport of organic matter in the Southern California Bight. Prog Oceanogr. 1992;30:151–166. [Google Scholar]
- 12.Massana R, et al. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol Oceanogr. 1998;43:607–617. [Google Scholar]
- 13.Perez MT, Pausz C, Herndl GJ. Major shift in bacterioplankton utilization of enantiomeric amino acids between surface waters and the ocean's interior. Limnol Oceanogr. 2003;48:755–763. [Google Scholar]
- 14.Teira E, Lebaron P, van Aken H, Herndl GJ. Distribution and activity of bacteria and archaea in the deep water masses of the North Atlantic. Limnol Oceanogr. 2006;51:2131–2144. [Google Scholar]
- 15.Arístegui J, Agustí S, Duarte CM. Respiration in the dark ocean. Geophys Res Lett. 2003;30:1041–1044. [Google Scholar]
- 16.Martin JH, Knauer GA, Karl DM, Broenkow WW. VERTEX: Carbon cycling in the Northeast Pacific. Deep-Sea Res. 1987;34:267–285. [Google Scholar]
- 17.Karner M, Herndl GJ. Extracellular enzymatic activity and secondary production in free-living and marine snow associated bacteria. Mar Biol. 1992;113:341–347. [Google Scholar]
- 18.Smith DC, Simon M, Alldredge AL, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 19.Azam F, Long RA. Sea snow microcosms. Nature. 2001;414:495–498. doi: 10.1038/35107174. [DOI] [PubMed] [Google Scholar]
- 20.Kiørboe T, Jackson GA. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr. 2001;46:1309–1318. [Google Scholar]
- 21.Cho BC, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature. 1988;332:441–443. [Google Scholar]
- 22.Nagata T, Fukuda H, Fukuda R, Koike I. Bacterioplankton distribution and production in deep Pacific waters: Large scale geographic variations and possible coupling with sinking particle fluxes. Limnol Oceanogr. 2000;45:426–435. [Google Scholar]
- 23.Hansell DA, Ducklow HW. Bacterioplankton distribution and production in the bathypelagic ocean: Directly coupled to particulate organic carbon export? Limnol Oceanogr. 2003;48:150–156. [Google Scholar]
- 24.Druffel ERM, Bauer JE, Williams PM, Griffin S, Wolgast D. Seasonal variability of particulate organic radiocarbon in the northeast Pacific Ocean. J Geophys Res. 1996;101:20543–20552. [Google Scholar]
- 25.Repeta DJ, Aluwihare LI. Radiocarbon analysis of neutral sugars in high-molecular-weight dissolved organic carbon: Implications for organic carbon cycling. Limnol Oceanogr. 2006;51:1045–1053. [Google Scholar]
- 26.Mortazavi B, Chanton JP. Use of Keeling plots to determine sources of dissolved organic carbon in nearshore and open ocean systems. Limnol Oceanogr. 2004;49:102–108. [Google Scholar]
- 27.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 28.Ouverney CC, Fuhrman JA. Marine planktonic Archaea take up amino acids. Appl Environ Micro. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herndl GJ, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Micro. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI. Origins of lipid biomarkers in Santa Monica Basin surface sediment: A case study using compound-specific Δ14C analysis. Geochim Cosmochim Acta. 2001;65:3123–3137. [Google Scholar]
- 31.Wuchter C, Schouten S, Boschker HTS, Sinninghe Damste JS. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 32.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 34.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varela MM, van Aken HM, Sintes E, Herndl GJ. Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ Microbiol. 2008;10:110–124. doi: 10.1111/j.1462-2920.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- 37.Coffin RB, Velinsky DJ, Devereux R, Price WA, Cifuentes LA. Stable carbon isotope analysis of nucleic acids to trace sources of dissolved substrates used by estuarine bacteria. Appl Environ Microbiol. 1990;56:2012–2020. doi: 10.1128/aem.56.7.2012-2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherrier J, Bauer JE, Druffel ERM, Coffin RB, Chanton JP. Radiocarbon in marine bacteria: Evidence for the ages of assimilated carbon. Limnol Oceanogr. 1999;44:730–736. [Google Scholar]
- 39.McCallister SL, Bauer JE, Cherrier JE, Ducklow HW. Assessing sources and ages of organic matter supporting river and estuarine bacterial production: A multiple-isotope (Δ14C, δ13C, and δ15N) approach. Limnol Oceanogr. 2004;49:1687–1702. [Google Scholar]
- 40.Kumamoto Y, Murata A, Saito C, Honda M, Kusakabe M. Bomb radiocarbon invasion into the northwestern North Pacific. Deep-Sea Res II. 2002;49:5339–5351. [Google Scholar]
- 41.War J. Monthly Deep Seawater Analyses. Kona, HI: Nat Energy Lab of Hawaii Authority; 2006. Natural Energy Laboratory of Hawaii Authority Keahole Point Cooperative Environmental Monitoring Program. [Google Scholar]
- 42.Carlson CA, et al. Effect of nutrient amendments on bacterioplankton production, community structure, and DOC utilization in the northwestern Sargasso Sea. Aquat Microb Ecol. 2002;30:19–36. [Google Scholar]
- 43.Steinberg DK, et al. Bacterial vs. zooplankton control of sinking particle flux in the ocean's twilight zone. Limnol Oceanogr. 2008;53:1327–1338. [Google Scholar]
- 44.Lee S, Fuhrman JA. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Micro. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollibaugh JT, Buddemeier RW, Smith SV. Contributions of colloidal and high molecular weight dissolved material to alkalinity and nutrient concentrations in shallow marine and estuarine systems. Mar Chem. 1991;34:1–27. [Google Scholar]
- 46.Kirchman DL, Elifantz H, Dittel AI, Malmstrom RR, Cottrell MT. Standing stocks and activity of archaea and bacteria in the western Arctic Ocean. Limnol Oceanogr. 2007;52:495–507. [Google Scholar]
- 47.Mincer TJ, et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 48.Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME Journal. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- 49.Agogué H, Brink M, Dinasquet J, Herndl GJ. Major nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- 50.Al-Murairi H, Landry MR. Active export of carbon and nitrogen at Station ALOHA by diel migrant zooplankton. Deep-Sea Res II. 2001;48:2083–2103. [Google Scholar]
- 51.Longhurst AR, et al. NFLUX: A test of vertical nitrogen flux by diel migrant biota. Deep-Sea Res. 1989;36:1705–1719. [Google Scholar]
- 52.Steinberg DK, Goldthwait SA, Hansell DA. Zooplankton vertical migration and the active transport of dissolved organic and inorganic nitrogen in the Sargasso Sea. Deep-Sea Res I. 2002;49:1445–1461. [Google Scholar]
- 53.Blair N, et al. Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl Environ Microbiol. 1985;50:996–1001. doi: 10.1128/aem.50.4.996-1001.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel JS, Nelson DE, Southon JR. 14C background levels in an accelerator mass spectrometry system. Radiocarbon. 1987;29:323–333. [Google Scholar]
- 55.Stuiver M, Polach HA. Discussion: Reporting of 14C data. Radiocarbon. 1977;19:355–363. [Google Scholar]
- 56.Huber T, Faulkner G, Hugenholtz P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 57.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schloss PD, Handelsman J. Introduing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allen AE, Booth MG, Verity PG, Frischer ME. Influence of nitrate availability on the distribution and abundance of heterotrophic bacterial nitrate assimilation genes in the Barents Sea during summer. Aquat Microb Ecol. 2005;39:247–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.