Abstract
YidC/Oxa/Alb3 family proteins catalyze the insertion of integral membrane proteins in bacteria, mitochondria, and chloroplasts, respectively. Unlike gram-negative organisms, gram-positive bacteria express 2 paralogs of this family, YidC1/SpoIIIJ and YidC2/YgjG. In Streptococcus mutans, deletion of yidC2 results in a stress-sensitive phenotype similar to that of mutants lacking the signal recognition particle (SRP) protein translocation pathway, while deletion of yidC1 has a less severe phenotype. In contrast to eukaryotes and gram-negative bacteria, SRP-deficient mutants are viable in S. mutans; however, double SRP-yidC2 mutants are severely compromised. Thus, YidC2 may enable loss of the SRP by playing an independent but overlapping role in cotranslational protein insertion into the membrane. This is reminiscent of the situation in mitochondria that lack an SRP pathway and where Oxa1 facilitates cotranslational membrane protein insertion by binding directly to translation-active ribosomes. Here, we show that OXA1 complements a lack of yidC2 in S. mutans. YidC2 also functions reciprocally in oxa1-deficient Saccharomyces cerevisiae mutants and mediates the cotranslational insertion of mitochondrial translation products into the inner membrane. YidC2, like Oxa1, contains a positively charged C-terminal extension and associates with translating ribosomes. Our results are consistent with a gene-duplication event in gram-positive bacteria that enabled the specialization of a YidC isoform that mediates cotranslational activity independent of an SRP pathway.
Keywords: membrane protein insertion, mitochondria, Streptococcus mutans, gram-positive bacteria, ribosomes
Members of the YidC/Oxa/Alb3 family of proteins are involved in the insertion of proteins into membranes of bacteria, mitochondria, and chloroplasts, respectively, but are absent from the endoplasmic reticulum and the plasma membrane of eukaryotes (1–3). Whereas their primary sequences are not well conserved, all members of this family are comprised of a core region of 5 transmembrane domains flanked by N- and C- terminal regions of variable length (4). This core region represents the catalytically active domain of the protein and functions as a protein insertase: that is, an enzyme that promotes the integration of hydrophobic stretches into the membrane (5, 6). In addition to its integrase activity, the core region contributes to the folding and assembly of membrane proteins, perhaps by helping the proteins to reach their correct topologies in the membrane (7, 8). The molecular mechanism by which members of the YidC/Oxa/Alb3 family facilitate the insertion of their client proteins still remains elusive.
The mitochondrial protein Oxa1 (Oxidase assembly 1) is the founding member of this family. Oxa1 plays an essential role during diverse steps of inner membrane biogenesis, in particular during the membrane insertion of mitochondrial translation products, the assembly of cytochrome oxidase, and of the membrane sector of the F1Fo-ATPase (8–11). In addition to Oxa1, mitochondria contain a second member of the YidC/Oxa/Alb3 family named Cox18 or Oxa2 (12–14). Cox18 functions in a posttranslational manner downstream of Oxa1 in cytochrome oxidase assembly (12). In contrast to Cox18, Oxa1 contains a C-terminal positively charged extension that binds to mitochondrial ribosomes so that Oxa1 can insert nascent chains in a cotranslational process (15, 16). This physical association of the ribosome with the membrane-embedded insertion machinery presumably allowed the loss of a signal recognition particle (SRP) from mitochondria during evolution. Cox18 lacks such a ribosome-binding domain and interacts with translation products only after their Oxa1-mediated membrane insertion (12). Nevertheless, Cox18 also exhibits insertase activity because it functionally complements Escherichia coli mutants lacking YidC (17).
Gram-negative bacteria contain only one YidC/Oxa/Alb3 protein family member. The E. coli homolog is the most well-characterized representative (reviewed in ref. 3). YidC is indispensable for viability in E. coli. YidC of E. coli lacks a ribosome-binding domain but facilitates cotranslational protein insertion in collaboration with the bacterial SRP system. It functions both in concert with the Sec translocon as part of the cotranslational machinery, as well as independently to insert proteins via a posttranslational mechanism (18, 19).
In contrast to gram-negative bacteria, the genomes of most gram-positive bacteria encode 2 YidC proteins. For example, 2 paralogs known as YidC1 and YidC2 were identified in Streptococcus mutans (20), the major causative agent of human dental caries. Both proteins are functional orthologs of E. coli YidC and each complements multiple defects of YidC-deficiency in E. coli and can mediate insertion of both Sec-dependent and Sec-independent YidC-only substrates (21). However, the effects of introducing YidC1 or YidC2 into YidC-depleted E.coli are not identical, suggesting that the proteins are functionally distinct. This is supported by observations in S. mutans, where deletion of yidC1 alone has little obvious effect and the deletion mutant appears robust, whereas the phenotype of a yidC2-deletion strain is strikingly similar to that of SRP pathway mutants, including stress-sensitivity and diminished genetic competence (20). Interestingly, double mutants lacking both YidC2 and SRP components are not viable and cannot be propagated, even in the absence of environmental stressors. This suggests that YidC2 overlaps functionally with the SRP pathway to mediate cotranslational protein insertion.
S. mutans YidC2 is predicted to possess a charged cytoplasmic tail similar to that of yeast Saccharomyces cerevisiae Oxa1. This region of Oxa1 interacts with mitochondrial ribosomes and supports cotranslational membrane protein insertion in the absence of an SRP (15, 16). Here we show by genetic analyses in S. mutans and in yeast that YidC2 and mitochondrial Oxa1 can partially complement each other. Employing mitochondria as a specialized system that lacks an SRP pathway, we show that upon expression in yeast, YidC2 binds ribosomes and promotes cotranslational integration of proteins into the mitochondrial inner membrane in the absence of Oxa1. Our results indicate that independent gene duplications in gram-positive bacteria and in organelles have led to specialized co- and posttranslational functions of YidC/Oxa/Alb3 proteins.
Results
Independent Gene Duplications in the YidC/Oxa/Alb3 Family.
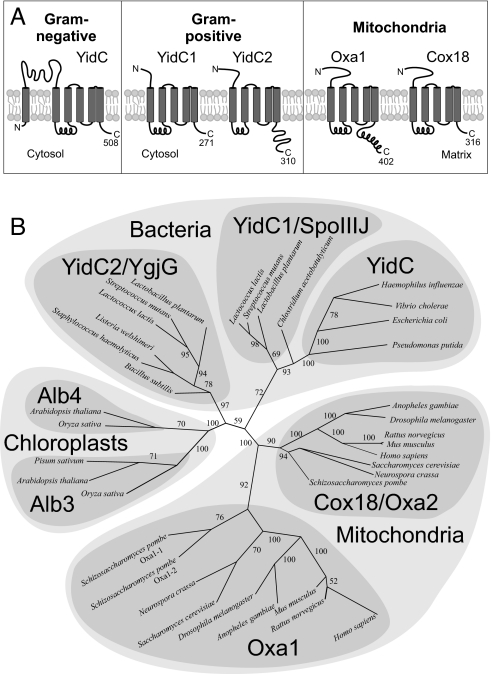
Gram-positive bacteria contain 2 YidC paralogs. Both lack the periplasmic-loop domains that are characteristic for YidC proteins of gram-negative bacteria (Fig. 1A). In gram-positive bacteria, the N-terminal regions of YidC are significantly shorter. They contain consensus signatures for signal peptidase II, which in the case of YidC1 and YidC2 proteins of S. mutans are located at amino acid positions 18 to 21 and 21 to 24, respectively. Thus, YidC homologs of gram-positive bacteria resemble in topology and membrane orientation the Oxa1 and Cox18 proteins of mitochondria. Although YidC1 and YidC2 are of similar topology and overall structure, they differ considerably in the length of their C-termini. YidC2, but not YidC1, contains a hydrophilic tail domain. This hydrophilic stretch would protrude into the bacterial cytosol and has a predicted isoelectric point of 11.29. This highly positively charged sequence resembles the tails found in yeast and human Oxa1 (pI = 11.25 and 11.23, respectively).
Fig. 1.
Gram-positive bacteria express 2 members of the YidC/Oxa/Alb3 family of proteins. (A) Schematic representation of the topology of bacterial and mitochondrial Oxa/YidC proteins. Transmembrane domains are indicated as gray boxes. The numbers of amino acid residues are depicted for the homologs of E. coli, S. mutans, and S. cerevisiae. (B) An unrooted neighbor-joining tree was calculated on the basis of pairwise alignments of all sequences. See Materials and Methods for details. Bootstrap numbers are indicated as indication for the confidence of the individual branches. Sequences used and the corresponding accession numbers are listed in Table S1.
The fact that gram-negative bacteria contain one YidC/Oxa/Alb3 protein, whereas gram-positive bacteria, chloroplasts, and mitochondria contain 2 paralogs, could have two explanations. Either there was an initial gene duplication event early in evolution followed by a selective loss in recent gram-negative bacteria for which sequence information exists, or alternatively, 3 independent gene duplications in gram-positive bacteria, mitochondria, and chloroplasts resulted in the presence of the different subbranches of the family. To distinguish between both scenarios, we analyzed carefully the sequence of various family members [see supporting information (SI) Table S1]. Comprehensive alignments of multiple sequences by using classical algorithms were unsuited to construct reliable trees because of the very low identity of the sequences. To avoid artificial clustering and problems that arise from long-branch attractions, we decided to follow a different strategy. We made individual alignments of all sequences and constructed from these a similarity matrix that was used to calculate a phylogenetic tree and bootstrap support numbers as a measure of reliability. In Fig. 1B, the relationship of YidC/Oxa/Alb3 homologs is depicted as an unrooted tree in which mitochondrial, plastidic, and bacterial branches are clearly distinct. This tree shows early duplications in mitochondria and chloroplasts, and also in bacteria. The gram-positive sequences clearly fall into 2 separate groups that we named YidC1 and YidC2 according to the proteins in S. mutans. Consistent with known phylogenetic relationships for most proteins analyzed, it is unlikely that the YidC/Oxa paralogs in bacteria and mitochondria resulted from horizontal gene transfer, but arose rather from independent gene duplications in bacteria and in mitochondria followed by convergent evolution. Based on sequence, YidC1 proteins appear more closely related to the YidC proteins of gram-negative bacteria. It should be noted that this tree is a result both of the evolutionary relationship among YidC/Oxa/Alb3 proteins and the similarities of the sequences that result from common functional constraints.
S. mutans YidC2 can be Functionally Replaced by Oxa1.
Independent gene duplications in the gram-positive bacterial and organellar systems might have been driven by similar specializations of the proteins. To test the functional similarity of the streptococcal and yeast homologs we used a complementation approach. S. mutans is a remarkably resilient organism and normally survives acidic and high-salt environments; however, cells lacking YidC2 show an inability to contend with environmental stressors and slower growth under nonstress conditions (20). We constructed expression plasmids that allowed the synthesis of Oxa1 or Cox18 in the S. mutans ΔyidC2 mutant background. Expression of Oxa1 restored the growth rate of the mutant under nonstress conditions, whereas introduction of Cox18 had a lesser effect on this phenotypic property (Table 1). When cultured under stress conditions, growth of the ΔyidC2 mutant was virtually nondetectable. The ability to grow under stress was partially complemented by Oxa1 and less well by Cox18. The greater ability of Oxa1 compared to Cox18 to complement the lack of YidC2 led us to examine the relevance of the C-terminal tail of YidC2. Deletion of the C-terminal 52-aa residues of YidC2 reduced the growth rate of S. mutans under stress and nonstress conditions, but not to the extent as elimination of the entire yidC2 gene. This implies that there are distinct functions of YidC2 that do and do not depend on an intact C terminus.
Table 1.
Summary of mean generation times of S. mutans wild-type and mutant strains and the ΔyidC2 mutant complemented with S. cerevisiae Oxa1 or Cox18
| Strain description | Growth condition |
||
|---|---|---|---|
| Nonstress | pH 5.0 | 4% NaCl | |
| Wild-type (NG8) | 57.00 ± 0.000 | 133.0 ± 0.001 | 211.0 ± 0.006 |
| ΔyidC2 | 156.0 ± 0.003* | No growth | No growth |
| ΔyidC2 + Oxa1 | 54.00 ± 0.001 | 181.0 ± 0.008* | 222.0 ± 0.017 |
| ΔyidC2 + Cox18 | 100.0 ± 0.001*† | 194.0 ± 0.001*† | 325.0 ± 0.004*‡ |
| yidC2ΔC | 78.00 ± 0.001* | 201.0 ± 0.004* | 264.0 ± 0.001 |
Values were derived from triplicate growth curves for each strain and expressed in minutes ± SD.
A 2-tailed t-test (alpha = 0.05) was used to determine significance.
*P < 0.001 relative to the wild-type strain and †P < 0.05 and ‡P < 0.005 relative to the ΔyidC2 + Oxa1 strain.
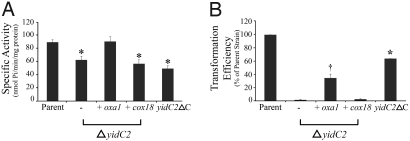
Acid tolerance in S. mutans is mediated in part by the F1Fο-ATPase that hydrolyses ATP to pump protons across the plasma membrane into the extracellular space. YidC and Oxa1 are known to play crucial roles in assembly of the F1Fο-ATPase complex (6, 8, 10, 11). In S. mutans, elimination of YidC2 or signal recognition particle components have comparable negative effects on membrane associated ATPase activity (20). Although both YidC1 and YidC2 can functionally replace E. coli YidC to facilitate membrane insertion of the Fo subunits a and c in the gram-negative background (21), deletion of yidC2, but not yidC1, results in acid sensitivity and diminished F1Fο-ATPase activity in S. mutans (20). This suggests that in this organism, YidC2 functions to support the biogenesis of the F1Fο-ATPase complex, particularly in the absence of the SRP cotranslational membrane protein-insertion pathway. Western blot analysis of membrane preparations of S. mutans mutant strains demonstrates that disruption of the SRP pathway by removal of ffh does not preclude insertion of YidC1 or YidC2, and elimination of yidC1 or yidC2 does not impede membrane localization of the other paralog (Fig. S1). Therefore, inadvertent effects on membrane insertion of the other proteins under study do not cause the phenotypes of these deletion mutants. The expression of Oxa1, but not of Cox18, completely restored the ATPase activity of the ΔyidC2 mutant (Fig. 2A). Hence, Oxa1 can take over this function of YidC2, whereas Cox18 cannot. Cells expressing YidC2ΔC showed a similar diminution in ATPase activity as mutants lacking YidC2 entirely (see Fig. 2A). From this we conclude that YidC2 requires its C terminus to function in the biogenesis of the F1Fο-ATPase complex in S. mutans.
Fig. 2.
S. cerevisiae Oxa1 complements a lack of YidC2 in S. mutans. (A) ATPase-specific activities of membrane fractions prepared from parent, mutant, and complemented mutant strains were calculated and expressed as nanomoles of phosphate released per minute per milligram of total protein. Statistical significance relative to the parent is indicated by an asterisk, with P < 0.001 using a 2-tailed t test (alpha = 0.05). (B) Genetic competence was assessed by natural transformation using plasmid pDC123 (Cmr). The transformation efficiency is indicated as the percentage of transformants of the mutant or complemented mutant strains compared to the S. mutans parent strain. Statistical significance relative to the ΔyidC2 and ΔyidC2 + Cox18 strains was P < 0.001(*) and P < 0.05(†) using a 2-tailed t test (alpha = 0.05).
Like mutants lacking the SRP pathway, YidC2-deficient strains show impaired genetic competence (20). Competence development in S. mutans is associated with a complex machinery, including membrane-localized and secreted components and a quorum-sensing signaling system (22). This machinery is specific for prokaryotes and is not present in mitochondria. We therefore asked whether Oxa1 is able to substitute for YidC2 in the membrane biogenesis necessary for competence (Fig. 2B). Introduction of Oxa1, but not of Cox18, was able to partially restore competence in the absence of YidC2. This again points to a specific property of Oxa1 that enables its broader degree of functional complementation of YidC2 in S. mutans. Transformation efficiency was diminished by elimination of the C-terminal tail of YidC2, but not to the extent as elimination of the entire yidC2 gene. Similar to the effects on bacterial growth, this result is consistent with a complex multifactorial phenotype and C terminus-dependent and -independent substrates of YidC2, and suggests why Oxa1 did not completely restore competence in the yidC2-deletion strain as it did for ATPase activity.
S. mutans YidC2 Partially Complements Δoxa1 Mutants.
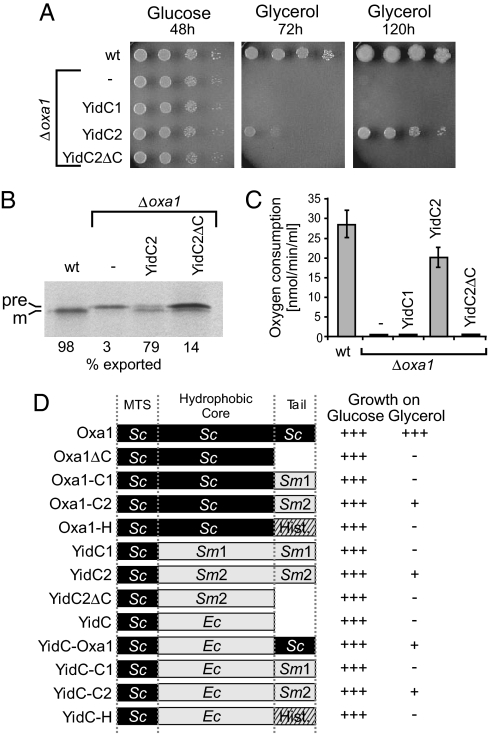
Next, we asked whether YidC1 or YidC2 could take over the function of Oxa1 in mitochondria. To this end we constructed fusion proteins consisting of the mitochondrial-targeting sequence of Oxa1 fused to full-length YidC1 and YidC2, or to the C-terminally truncated variant of YidC2 (Fig. S2A). These fusion proteins were expressed from the OXA1 promoter in an Oxa1-deficient yeast strain. YidC2 and YidC2ΔC were efficiently expressed in yeast cells and targeted to mitochondria (Fig. S2B). Because of the absence of a functional respiratory chain, Oxa1-deficient strains are unable to grow on nonfermentable carbon sources. This defect was partially compensated for by expression of YidC2 but not by YidC2ΔC (Fig. 3A) and is consistent with a common function of the C-terminal tails of YidC2 and Oxa1. YidC1 was found in whole-cell extracts but not in isolated mitochondria, suggesting a lack of import (not shown); therefore, its ability to complement Oxa1 in this system could not be assessed.
Fig. 3.
S. mutans YidC2 can partially complement an Oxa1-deficient yeast mutant. (A) Yeast wild-type cells (wt) and Δoxa1 mutants carrying plasmids encoding the indicated chimeric S. mutans proteins were grown to exponential phase. Tenfold serial dilutions of the cultures were spotted on YP plates containing glucose or glycerol. (B) Translation products were radiolabeled for 20 min at 30 °C in isolated mitochondria from the indicated strains. Cox2 was isolated by immunoprecipitation and visualized by autoradiography. The precursor and mature forms of Cox2 are indicated as “pre” and “m,” respectively. (C) Isolated mitochondria from the indicated strains were incubated in 400 μl of 0.6 M sorbitol, 1 mM MgCl2, 5 mM EDTA, and 20 mM Hepes pH 7.4 in an oxygen electrode (Oxygraph, Hansatech Instruments). Respiration was started by addition of 15 μl of 0.2 M NADH and the consumption of oxygen was recorded over time. (D) The chimeric fusion proteins represented on the left were expressed in Δoxa1 cells. The strains were grown in liquid YPD (glucose) and YPG (glycerol) medium and doubling times were calculated. “+++” indicates doubling times of less than 8 h. “+” indicates doubling times between 20 and 35 h. Strains indicated with “-” did not grow or have doubling times of more than 150 h.
Subunit 2 of the cytochrome oxidase (Cox2) is inserted into the inner membrane of mitochondria in an Oxa1-dependent, cotranslational process (9, 23). Cox2 is synthesized as a precursor protein that is matured by the Imp1 protease of the intermembrane space. In the absence of Oxa1, the N terminus of Cox2 is not translocated across the inner membrane and Cox2 accumulates in the matrix in its precursor form. We radiolabeled translation products in mitochondria isolated from wild-type and mutant strains, purified Cox2 by immunoprecipitation, and analyzed the samples by autoradiography (Fig. 3B). In wild-type mitochondria, newly synthesized Cox2 was completely processed to its mature form. In contrast, in Δoxa1 mitochondria, Cox2 remained in the precursor form. Expression of YidC2 again allowed the efficient maturation of Cox2, confirming that YidC2 can replace Oxa1 in cotranslational protein insertion in mitochondria. As now expected, this activity required the presence of the C terminus of YidC2 and Cox2 was not efficiently processed in the YidC2ΔC-expressing mitochondria.
To assess the ability of YidC2 to mediate assembly of mitochondrial respiratory chain complexes more generally, we also measured the respiration-dependent oxygen consumption of isolated mitochondria. Because of the severely reduced levels of complex III and complex IV in Δoxa1 mitochondria, no respiratory activity was detected in this mutant. The expression of YidC2, but not YidC1 or YidC2ΔC, almost completely restored the respiratory activity of the Δoxa1 mutant (Fig. 3C), again indicating that YidC2 can serve as a functional equivalent and substitute for Oxa1 in the insertion and assembly of proteins of the mitochondrial inner membrane.
To identify the critical YidC sequences more systematically, we constructed an array of fusion proteins in which the different domains of Oxa1, E. coli YidC, and YidC1 and YidC2 of S. mutans were combined (Fig. 3D) and stably expressed in yeast cells (see Fig. S3). In addition, we replaced the C-terminal tail of Oxa1 with the N-terminal 86 residues of histone H3 (Hht1) that resembles the Oxa1 ribosome-binding region in that it has an α-helical structure and positive net charge (pIHht1 = 12.33; pIOxa1 = 11.25). All constructs that contained either the C terminus of Oxa1 or YidC2 allowed the growth on the nonfermentable carbon-source glycerol. The C terminus of YidC1 or the histone-derived sequence did not (data not shown). This confirmed that the C terminus of YidC2 can functionally replace the C-terminal ribosome-binding domain of Oxa1 in yeast mitochondria, and that this result does not stem simply from a nonspecific electrostatic interaction with the ribosome.
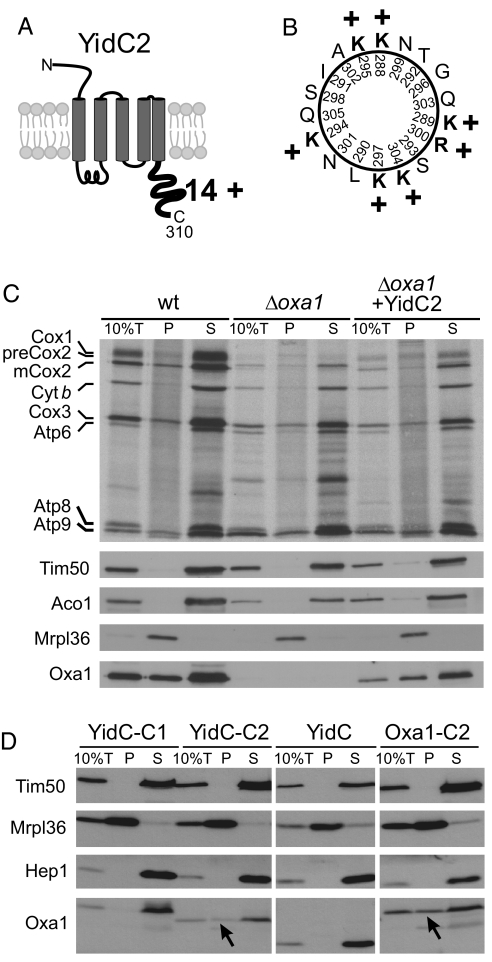
YidC2 Binds to Mitochondrial Ribosomes.
Ribosome binding is critical for functionality of Oxa1 (15, 16). Its positively charged helical C-terminal tail is both necessary and sufficient for the interaction with the ribosome. The hydrophilic C-terminal tail domain of S. mutans YidC2 has a net charge of + 14 (Fig. 4A). Prediction programs suggest that the C-terminal half of this region forms a helical structure that would expose many positive charges to one face (Fig. 4B). Similar positive regions are often found in proteins that interact with the negatively charged RNA on the surface of ribosomes. To test whether YidC2 is able to bind to ribosomes in the complete absence of any SRP components, we radiolabeled mitochondrial translation products in mitochondria before ribosomes were isolated by centrifugation through a high-density step gradient (Fig. 4C). In wild-type mitochondria, a large fraction of Oxa1 is released from the ribosomes because of the high salt concentration in the lysis buffer (350 mM KCl); however, a significant amount of Oxa1 is still recovered with the ribosomes. Likewise, when YidC2 was expressed instead of Oxa1 in mitochondria, a fraction of this bacterial homolog was also recovered with mitochondrial ribosomes. To confirm the suspected ribosome-binding activity enabled by the C terminus of YidC2, we further tested the ribosome association of chimeric proteins of E. coli YidC fused to the C termini of S. mutans YidC1 and YidC2. E. coli YidC cannot bind to yeast mitochondrial ribosomes unless the C-terminal region of Oxa1 is appended to it (24). The YidC-YidC2 fusion, but not the YidC-YidC1 fusion was recovered with the mitochondrial ribosomal pellet (Fig. 4D). Likewise, when the C terminus of Oxa1 was replaced with that of YidC2, this fusion protein was also recovered from the mitochondrial ribosomal pellet. Taken together, we conclude that YidC2, like Oxa1, has the ability to bind to ribosomes and to independently promote cotranslational protein insertion without assistance from the SRP machinery.
Fig. 4.
YidC2 binds to mitochondrial ribosomes. (A) Schematic representation of the predicted topology and the cytoplasmic extension of S. mutans YidC2. The C-terminal tail domain is very basic and has a net charge of + 14 as indicated. (B) Helical-wheel representation of YidC2 C-terminal residues 288–305. Positively charged residues are shown in bold. (C) Translation products were radiolabeled for 15 min at 25 °C in isolated mitochondria of the indicated strains. Ten percent of the sample total (10%T) was directly applied to the gel. The residual extract was lysed in the high salt buffer and loaded on a layer of 1.2-M sucrose and centrifuged for 60 min at 190,000 × g. Proteins of the ribosome-containing pellet (P) and the supernatant (S) were analyzed by autoradiography and Western blotting. Oxa1 and YidC2 were detected with Oxa1-specific antibodies that recognize the N-terminal region that is shared by both proteins. Signals of the ribosomal protein Mrpl36, and of the non-ribosomal proteins Tim50, Hep1, and Aco1 are shown for control. Please note that the “smeary” background in the pellet lanes of the autoradiography is because of the presence of nascent chains that are associated with the translation-active ribosomes. Cyt b, cytochrome b. (D) Mitochondria were isolated from Δoxa1 strains expressing the fusion proteins indicated. The samples were fractionated as described in (C). Arrowheads indicate fusion proteins that comigrate with the ribosomal pellet.
Discussion
Protein secretion and membrane insertion systems have been more extensively classified and characterized in gram-negative (25) compared to gram-positive organisms, and it is coming to light that important differences exist (26). In gram-negative bacteria such as E. coli, the SRP components Ffh and 4.5S RNA are essential for viability, as they exhibit indispensable functions in cotranslational protein insertion (27, 28). It therefore came as a surprise that the gram-positive bacteria S. mutans (20) and Streptococcus pyogenes (29) tolerate loss of SRP pathway components. That the phenotypes of S. mutans SRP pathway and ΔyidC2 mutants are highly similar suggests that these systems might serve similar overlapping functions and operate in concert to ensure cotranslational membrane-protein insertion in this organism. Here we showed that YidC2 and Oxa1 can be exchanged between S. mutans and yeast cells and partially complement one another. Like Oxa1, YidC2 has a positively charged C-terminal tail and demonstrated an ability to bind to ribosomes and mediate cotranslational translocation upon expression in yeast mitochondria. This property is not shared by E. coli YidC but was conferred by appending YidC2's tail to it. Interestingly, the tail domains of Oxa1 and YidC2 do not show primary sequence homology, although they share the physical property of strong enrichment in positive charges. Our phylogenetic analysis suggests that these domains developed independently during evolution. Thus, the hydrophobic core domain of the core insertase appears to have been combined at least twice with a ribosome-binding site, presumably to increase the efficiency of cotranslational protein insertion (Fig. 5).Apparently, this specialization was accompanied by a gene duplication that allowed the other paralog to operate independently of the translation machinery. In mitochondria the SRP complex was lost; however, gram-positive bacteria, such as the streptococci, still contain this cotranslational pathway, now known experimentally to be dispensable in several species. Thus, the situation in gram-positive bacteria appears to resemble an intermediate state between the systems found in the cytosol of eukaryotes and gram-negative bacteria in which the SRP is still required, and that found in mitochondria that relies exclusively on direct ribosome binding of the insertase.
Fig. 5.
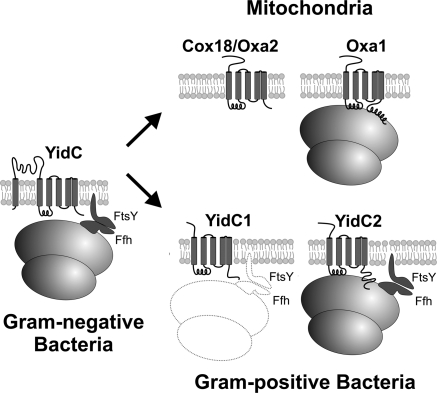
Independent gene duplications allowed a specialization of Oxa/YidC proteins. As illustrated in the schematic diagram, gram-negative bacteria contain one YidC homolog. Cotranslational protein insertion into the inner membrane is facilitated by interaction of the SRP complex and its receptor with the ribosome. In E. coli, Ffh and FtsY are essential for viability, but in streptococci they are not. In mitochondria, a gene duplication event led to 2 insertases: Oxa1 that interacts with ribosomes and is specialized in the cotranslational insertion of translation products and Cox18 that serves a posttranslational role and functions downstream of Oxa1. Our phylogenic analysis indicates that a similar gene duplication event occurred in the evolution of gram-positive bacteria, giving rise to 2 YidC variants. By virtue of its charged C-terminal tail, S. mutans YidC2 acquired the additional ability to mediate cotranslational insertion of membrane proteins independent of the SRP pathway, explaining the unexpected dispensability of the SRP pathway in this gram-positive organism. The primary function of YidC1 is not yet known, but the lack of obvious detrimental consequences of its elimination from S. mutans indicates that it plays a limited physiologic role. The inability to simultaneously delete yidC1 and yidC2 from S. mutans (20) suggests that YidC1 retains the functional potential (hence the illustration in outline form) to serve as a back up to support SRP-mediated cotranslational membrane protein insertion when YidC2 is absent.
If such gene duplications were successful, why didn't they occur in gram-negative bacteria? Given the fundamental difference in cell-envelope structure, a double membrane and periplasmic space in gram-negative organisms (compared to a single membrane in gram-positive organisms), it is likely that gram-positive bacteria have different functional requirements than gram-negative organisms with regard to membrane protein insertion and secretion, and the presence of 2 YidCs allows greater flexibility and easier simultaneous use of co- and posttranslational pathways. In gram-positive bacteria the substrates of co- and posttranslational pathways are as yet largely unexplored, and an understanding of membrane biogenesis in these organisms is just at the beginning. Therefore, continued dissection of the similarities and unique features of the gram-positive membrane protein insertion and secretion machineries compared to other prokaryotic and organellar systems promises to be enlightening in the future.
Materials and Methods
Sequence Analysis.
A neighbor-joining tree was calculated on the basis of a distance matrix for all possible pairwise sequence combinations. See SI Materials and Methods for details.
Strains and Growth Media.
See Table S2. All of the yeast strains used in this study are isogenic to the wild-type W303–1A (MAT a, ade2 ura3 leu2 his3 trp1). The plasmids expressing the various chimeric proteins used in this study were introduced into a Δoxa1 strain (30). For details see SI Materials and Methods. Yeast cultures were grown at 30 °C in YP medium (1% yeast extract, 2% peptone) supplemented with 2% galactose, or in minimal medium supplemented with 20 μg/ml adenine, histidine, and tryptophan, 30 μg/ml of leucine and lysine, and 2% galactose.
Streptococcal mutant strains are isogenic to S. mutans wild-type NG8 and were grown in Todd-Hewitt broth (BBL, Becton Dickson Microbiology System) supplemented with 0.3% (wt/vol) yeast extract (THYE) at 37 °C with 5% CO2. Kanamycin (500 μg/ml plates or 100 μg/ml broth), erythromycin (10 μg/ml), and chloramphenical (10 μg/ml) were used as antibiotics. E. coli strain Top10F‘ (Invitrogen) was used for cloning and maintenance of plasmid constructs. E. coli was grown in LB broth supplemented with erythromycin (300 μg/ml) or kanamycin (50 μg/ml), when appropriate. For construction of strains and plasmids see SI Materials and Methods.
Miscellaneous.
Restriction endonucleases and other DNA-modifying enzymes were obtained from Invitrogen, New England Biolabs, Inc., and Promega Corp., and used according to the specifications of the suppliers. Details regarding the streptococcal competence assay can be found in SI Materials and Methods. The following methods were carried out as described: growth and mean generation time measurements of S. mutans and ATPase assays (20), labeling of mitochondrial translation products and immunoprecipitation (10), and determination of enzyme activities (31).
Supplementary Material
Acknowledgments.
We thank Sandra Esser, Marica Malesic, and Andrea Trinkaus for excellent technical assistance, Martin Prestele for help with some experiments, Monika Oli for assistance with data analysis, Martin Ott for critical comments on the manuscript, and Walter Neupert and Arnold Bleiweis for constant support and stimulating discussions. This work was supported by the Deutsche Forschungsgemeinschaft (SFB530, Teilprojekt C15 and FOR967, Teilprojekt 1), the Stiftung für Innovation in Rheinland-Pfalz, and the National Institute for Dental and Craniofacial Research (DE08007).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809951106/DCSupplemental.
References
- 1.Luirink J, Samuelsson T, de Gier JW. YidC/Oxa1p/Alb3: Evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 2001;501:1–5. doi: 10.1016/s0014-5793(01)02616-3. [DOI] [PubMed] [Google Scholar]
- 2.Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiefer D, Kuhn A. YidC as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol. 2007;259:113–138. doi: 10.1016/S0074-7696(06)59003-5. [DOI] [PubMed] [Google Scholar]
- 4.Yen MR, Harley KT, Tseng YH, Saier MH., Jr Phylogenetic and structural analyses of the Oxa1 family of protein translocases. FEMS Microbiol Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]
- 5.Serek J, et al. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 2004;23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. F1Fo ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagamori S, Smirnova IN, Kaback HR. Role of YidC in folding of polytopic membrane proteins. J Cell Biol. 2004;165:53–62. doi: 10.1083/jcb.200402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia L, Dienhart MK, Stuart RA. Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol Biol Cell. 2007;18:1897–1908. doi: 10.1091/mbc.E06-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: Distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hell K, Neupert W, Stuart RA. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiburek L, et al. Knockdown of human Oxa1l impairs the biogenesis of F(1)F (o) -ATP synthase and NADH:ubiquinone oxidoreductase. J Mol Biol. 2007;374:506–516. doi: 10.1016/j.jmb.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 12.Funes S, Nargang FE, Neupert W, Herrmann JM. The Oxa2 protein of Neurospora crassa plays a critical role in the biogenesis of cytochrome oxidase and defines a ubiquitous subbranch of the Oxa1/YidC/Alb3 protein family. Mol Biol Cell. 2004;15:1853–1861. doi: 10.1091/mbc.E03-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell. 2002;13:1122–1131. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souza RL, Green-Willms NS, Fox TD, Tzagoloff A, Nobrega FG. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J Biol Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- 15.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates cotranslational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia L, et al. Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal hydrophilic region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bloois E, Koningstein G, Bauerschmitt H, Herrmann JM, Luirink J. Saccharomyces cerevisiae Cox18 complements the essential Sec-independent function of Escherichia coli YidC. FEBS J. 2007;274:5704–5713. doi: 10.1111/j.1742-4658.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 18.Beck K, et al. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2001;2:709–714. doi: 10.1093/embo-reports/kve154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuelson JC, et al. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 20.Hasona A, et al. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci USA. 2005;102:17466–17471. doi: 10.1073/pnas.0508778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y, et al. Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J Bacteriol. 2008;190:2458–2469. doi: 10.1128/JB.01366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cvitkovitch DG. Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med. 2001;12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- 23.Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- 24.Preuss M, Ott M, Funes S, Luirink J, Herrmann JM. Evolution of mitochondrial Oxa proteins from bacterial YidC: Inherited and acquired functions of a conserved insertion machinery. J Biol Chem. 2005;280:13004–13011. doi: 10.1074/jbc.M414093200. [DOI] [PubMed] [Google Scholar]
- 25.Saier MH., Jr Protein secretion and membrane insertion systems in gram-negative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 26.Saier MH., Jr Structure and evolution of prokaryotic cell envelopes. Microbe. 2008;3:323–328. [Google Scholar]
- 27.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 28.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 29.Rosch JW, Vega LA, Beyer JM, Lin A, Caparon MG. The signal recognition particle pathway is required for virulence in Streptococcus pyogenes. Infect Immun. 2008;76:2612–2619. doi: 10.1128/IAI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.