Abstract
All-trans and 9-cis retinoic acids (RA) signals are transduced by retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers that act as functional units controlling the transcription of RA-responsive genes. With the aim of elucidating the underlying molecular mechanisms, we have developed an in vitro transcription system using a chromatin template made up of a minimal promoter and a direct repeat with 5-spacing-based RA response element. RARα and RXRα were expressed in and purified from baculovirus-infected Sf9 cells, and transcription was carried out by using naked DNA or chromatin templates. Transcription from naked templates was not affected by the presence of RA and/or RAR/RXR heterodimers. In contrast, very little transcription occurred from chromatin templates in the absence of RA or RAR/RXR heterodimers whereas their addition resulted in a dosage-dependent stimulation of transcription that never exceeded that occurring on naked DNA templates. Most importantly, the addition of synthetic agonistic or antagonistic retinoids to the chromatin transcription system mimicked their stimulatory or inhibitory action in vivo, and activation by a RXR-specific retinoid was subordinated to the binding of an agonist ligand to the RAR partner. Moreover, the addition of the p300 coactivator generated a synergistic enhancement of transcription. Thus, the dissection of this transcription system ultimately should lead to the elucidation of the molecular mechanisms by which RAR/RXR heterodimers control transcription in a ligand-dependent manner.
Keywords: chromatin template, RXR subordination, synergistic transactivation, p300 coactivator, histone acetylation
Retinoic acids (RAs) exert their pleiotropic effects on vertebrate development and homeostasis by binding to nuclear receptors (NRs) (ref. 1 and references therein). These receptors belong to a gene superfamily that includes the receptors for steroid hormones, thyroid hormones, vitamin D3, and a growing number of so-called orphan receptors (for reviews, see refs. 2 and 3). Two families of receptors, the retinoic acid receptor isotypes (RARα, RARβ, and RARγ) and the retinoid X receptors isotypes (RXRα, RXRβ, and RXRγ), are implicated in the transduction of the RA signal (ref. 4 and references therein). RARs bind all-trans RA (tRA) and 9-cis RA (9cRA) whereas RXRs respond exclusively to 9cRA (ref. 5 and references therein). The C-terminal region of RARs and RXRs contains both the ligand binding domain, which functions as a ligand-dependent transactivation domain [activation function 2 (AF-2)], and surfaces for both homo- and heterodimerization as well as for interaction with other factors (see below). An additional ligand-independent activation function, AF-1, is present within the N-terminal region (reviewed in ref. 4).
RARs and RXRs can bind as dimers to RA response elements (RAREs) consisting of two hexameric motifs [PuG(G/A)(T/A)CA] usually arranged as direct repeats. However, RXRs readily heterodimerize with RARs, and RAR/RXR heterodimers bind to and transactivate from RAREs made up of direct repeat motifs separated by 5 (DR5) and 2 (DR2) bp much more efficiently than RAR homodimers on their own. This indicates that RAR/RXR heterodimers might be the functional units transducing the retinoic acid signals in vivo (refs. 4 and 6 and references therein). Several lines of evidence support this possibility: (i) Genetic studies have established the functionality of RXR/RAR heterodimers in the RA-responsive F9 embryonal carcinoma cell line (7–9) as well as in the mouse (refs. 1 and 10–12 and references therein), and (ii) synergistic effects of RXR- and RAR-selective synthetic retinoid on target gene expression, proliferation, apoptosis, and/or differentiation have been observed in a variety of cultured cell lines, including the embryonal carcinoma cell lines F9 and P19 (7–9, 13–23). However, in all cases, the liganded RXR was transcriptionally inactive, unless its RAR partner was itself liganded. This intraheterodimeric subordination of the RXR AF-2 activity to the binding of a RAR agonistic ligand could be caused by an allosteric effect of the unliganded RAR on its liganded RXR partner (24).
Transfection studies have suggested that the AF-2 activation function of NRs is mediated through coactivators (intermediary factors) (25). Numerous proteins that interact directly with NRs in a agonistic ligand-dependent manner have been cloned and characterized, and several of them have been shown to enhance the activity of NR AF-2s when coexpressed in transiently transfected mammalian cells (refs. 4 and 26 and references therein). Some of these putative coactivators, SRC-1 (27), CBP/p300 (28, 29), and ACTR (30), can interact with the histone acetyltransferase p/CAF (31) and also possess an intrinsic histone acetyltransferase activity. Moreover, CBP and p300 also interact with RNA helicase A, which in turns binds RNA polymerase II (32).
Thus, among other possible mechanisms, remodeling of the chromatin template by histone acetylation and recruitment of the general transcription machinery to hormone-responsive promoters appear to be involved in transcriptional activation by the ligand-dependent AF-2 of NRs. However, the lack of transcription systems faithfully reproducing in vitro the effects of retinoids on activation of transcription of responsive genes in vivo has precluded a systematic biochemical dissection aimed at identifying the molecular mechanisms by which retinoid receptors mediate retinoid signals to control the expression of target genes. Using a chromatin-assembled template, we describe here an in vitro transcription system that, in the presence of RAR/RXR heterodimers, mimics the effects of retinoids on gene transactivation as observed in vivo.
MATERIALS AND METHODS
DNA and Chromatin Templates.
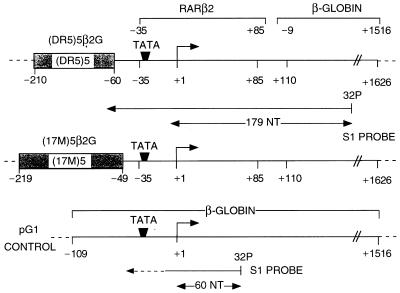
The plasmids (DR5)5β2G and (17M)5β2G (≈5.2 kilobases) were constructed by inserting five copies of the DR5 RA response element from the mouse RARβ2 promoter or the 17-mer GAL4 binding site, respectively, upstream of the mouse RARβ2 core promoter [−35 to +85], which previously had been linked to the −9 to +1516 chicken β-globin gene sequence (Fig. 1). The most 3′ DR5 element is positioned at approximately the same distance from the TATA box as the DR5 RARE found in the natural RARβ2 promoter (33).
Figure 1.
DNA templates and S1 nuclease probe. The structures of the (DR5)5β2G, (17 m)5β2G, and internal control pG1 reporter templates are schematically represented with the positioning of the response elements relative to the transcription start site.
Chromatin assembly extracts were prepared from Drosophila embryos (0–6 hr) as described (34). Chromatin was assembled on supercoiled circular DNA (see Fig. 2B), as described by Becker et al. (35) for 4 hr at 27°C. Determination of supercoiling within (DR5)5β2G chromatin template by using topoisomerase I and/or chloroquine (35) indicated the presence of at least 25 nucleosomes whereas micrococcal nuclease digestion (36) showed that they had a periodicity of ≈160 bp (see Fig. 2B). DNase I footprinting was performed essentially as described (34). DNA fragments generated by DNase I digestion were amplified with VentR (exo-) (New England Biolabs) by using a primer complementary to a sequence located between −280 and −250 upstream of the RARβ2 promoter start site.
Figure 2.
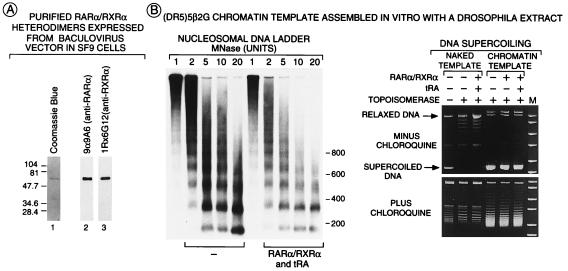
Analysis of RARα/RXRα heterodimers and chromatin structure. (A) Purification of RARα/RXRα heterodimers. FhRARα and HmRXRα were coexpressed in Sf9 cells and were affinity-purified by using a Ni2+ column followed by anti-Flag agarose column that bind the HmRXR moiety and the FhRAR moiety of the heterodimer, respectively. Purified heterodimers (100 ng of protein) were separated on a 10% SDSPAGE gel before staining with Coomassie blue (lane 1) or Western blot analysis using mAbs recognizing either human RARα (lane 2) or mouse RXRα (lane 3). (B) Overall chromatin structure was not affected by RARα/RXRα heterodimers. Chromatin or naked (DR5)5β2G templates (200 pM) incubated in the presence or absence of FhRARα/HmRXRα (1 nM) and tRA (10−6 M) were digested with varying concentrations of micrococcal nuclease in a final volume of 80 μl, were separated on a 1.5% agarose gel, and were Southern blotted by using a [32P] probe corresponding to the −40 to +5 region of the (DR5)5β2G promoter. DNA supercoiling was estimated as described (35) on DNA (200 ng) treated (or not treated) by topoisomerase I (10 units; final volume of 45 μl). DNA was separated on a 1% agarose gel in the presence or absence of 1.2 μM chloroquine. Migration of relaxed and supercoiled template DNA is indicated.
Protein Expression and Purification.
The Spodoptera frugipenda cell line Sf9 was coinfected with baculoviruses expressing His-tagged mouse RXRα (HmRXRα) and Flag-tagged human RARα (FhRARα) for 48 hr. FhRARα/HmRXRα heterodimers were purified from Sf9 cell extracts by affinity chromatography using a Ni2+ column (Amersham Pharmacia), followed by an anti-Flag M2 agarose column (Sigma) as specified in the manufacturer’s instructions. Western blot analysis was performed by using the mAbs anti-RARα 9α-9A6 (37) and anti-RXRα 1RX-6G12 (38) (see Fig. 2A). The DNA and tRA binding properties of the heterodimers were examined by electrophoretic mobility-shift analysis and ligand binding assays as described (39, 40).
Full length p300 was prepared from Sf9 cells infected with a p300-expressing baculovirus (41), its purification was monitored by using a rabbit polyclonal anti-p300 (C-20) antibody (Santa Cruz Biotechnology), and the histone acetyltransferase activity of the purified protein was confirmed as described (29). His-tagged GAL(1–147) and GAL-VP16 (42) were expressed from pET3 expression vectors in the BL-21 pLysS bacterial strain and were purified by Ni2+ column chromatography.
In Vitro Transcription.
Transcription was performed by using a HeLa cell nuclear extract (43) as described (34). Chromatin or naked templates were incubated with 1 nM FhRARα/HmRXRα heterodimers (in the presence of ligand or vehicle) for 30 min at 27°C before transcription initiation. The plasmid pG1 (ref. 44; see Fig. 1) was added to each reaction as an internal control. Transcription was quantitated by S1 nuclease analysis (42) using a 32P-labeled probe (S1 probe) that hybridizes with transcripts from the (DR5)5β2G, (17 m)5β2G, and pG1 plasmids through their transcription start sites to yield fragments of 179, 179, and 60 nt, respectively (see Fig. 1). RAR- and RXR-specific agonists or antagonists (BMS 614, BMS 649, BMS 753, and BMS 961) were all gifts from Bristol-Myers Squibb.
RESULTS
The supercoiled plasmid (DR5)5β2G that contains the RARβ2 core promoter (−35 to +85) and five copies of the RARE of the RARβ2 gene were used to study activation of transcription by RAR/RXR heterodimers (see Materials and Methods and Figs. 1 and 2B). To determine whether a chromatin-assembled template was important, we analyzed the transcriptional activity of purified RARα/RXRα heterodimers (see Materials and Methods and Fig. 2A), using both naked and chromatin DNA templates. Periodic nucleosomal arrays (see Fig. 2B) were generated by using supercoiled (DR5)5βG plasmid and a chromatin-assembly extract (see Materials and Methods). Note that the nucleosomal organization of the chromatin template was not grossly affected by the addition of RARα/RXRα heterodimers and tRA (Fig. 2B).
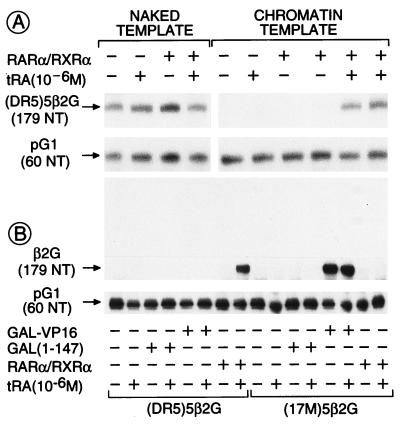
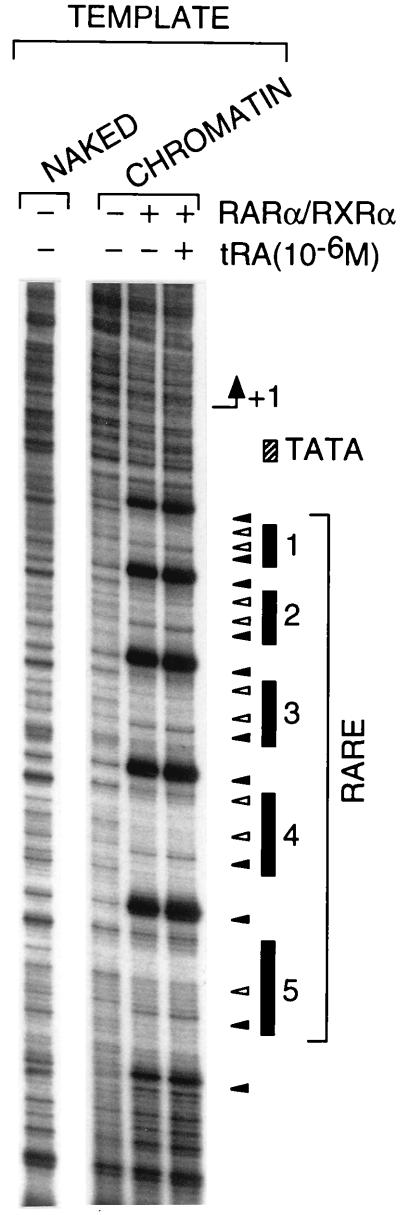
When expressed relative to basal transcription from the internal control naked pG1 template (see Fig. 1), “constitutive” transcription on the naked (DR5)5β2G template was not affected by the presence of RA ligand and/or receptor heterodimers (Fig. 3A). In marked contrast, very little transcription was observed from the corresponding chromatin template in the absence of RARα/RXRα heterodimers, irrespective of the presence of RA. However, the addition of both heterodimers and tRA resulted in a potent activation of transcription (between 30- and 100-fold; Fig. 3A). Note that little ligand-dependent activation of transcription by RARα/RXRα heterodimers was observed when exogenous histones were not added to the Drosophila extract during chromatin assembly on the (DR5)5β2G plasmid (data not shown). Optimal activation of transcription from the chromatin template was achieved by using 1 nM RARα/RXRα heterodimer that corresponds to approximately five heterodimers per (DR5)5β2G template molecule (200 pM), i.e., one heterodimer per DR5 response element (Fig. 3A and data not shown). Consistent with this observation, DNase I footprinting analysis showed that all five DR5 RAREs were bound by RARα/RXRα heterodimers at these concentrations, with no RARE being particularly favored (Fig. 4). Note that, in contrast to activation of transcription, the binding of the heterodimers to the chromatin template did not depend on the presence of tRA (Fig. 4). Binding of unliganded RAR/RXR heterodimers to chromatin is therefore clearly not sufficient for transcriptional activation, thus suggesting that the critical step in transactivation is a ligand-dependent transconformation of DNA-bound heterodimers.
Figure 3.
RARα/RXRα heterodimers activate transcription from chromatin templates in a ligand- and template-specific manner. (A) tRA-induced derepression of transcription from chromatin templates by RARα/RXRα. In vitro transcription was performed on chromatin or naked (DR5)5β2G templates (200 pM) by using a HeLa cell nuclear extract (100 μg) for 45 min in the presence or absence of FhRARα/HmRXRα (1 nM) and tRA (1 μM) in a final reaction volume of 50 μl as indicated. S1 nuclease analysis was carried out after deproteinization. (B) Template specificity of activated transcription. Activation of transcription on chromatin (DR5)5β2G or (17M)5β2G templates was determined in the presence of 1 nM of activator [either Gal4(1–147), Gal4-VP16, or FhRARα/HmRXRα] with or without tRA (1 μM) as above. S1 nuclease digestion of RNA transcripts originating from β2G and pG1 templates generated 179- and 60-nt fragments, respectively (see Fig. 1).
Figure 4.
RARα/RXRα heterodimers bind all five RAREs in the promoter region of the (DR5)5β2G chromatin template, irrespective of the presence of tRA. Chromatin or naked (DR5)5β2G templates (250 ng) were incubated in the presence or absence of FhRARα/HmRXRα and tRA (10−6 M) (under the conditions described above for transcription reactions) for 30 min, were subjected to DNase I digestion (5 units; final volume of 50 μl), then were analyzed by primer extension footprinting (see Materials and Methods). Sites of increased (closed triangle) or decreased (open triangle) sensitivity to DNase I are shown.
The response element specificity of transcriptional activation by RARα/RXR heterodimers was examined by comparing transcription from the cognate (DR5)5β2G template and the (17M)5β2G template, in which the five DR5 RAREs have been replaced by five copies of the 17-mer binding site for the DNA binding domain [GAL(1–147)] of the yeast transactivator Gal4 (Fig. 1). RARα/RXRα heterodimers did not activate transcription from the chromatin-assembled (17M)5β2G template whereas, under similar conditions, the chimeric acidic transactivator GAL-VP16 (45) efficiently activated transcription from that template but not from the chromatin-assembled (DR5)5β2G template (Fig. 3B).
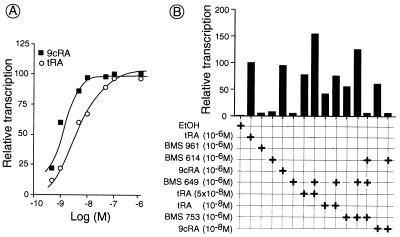
The above results demonstrate that activation of transcription by RAR/RXR heterodimers depends on packaging of the template into a nucleosomal structure and that it is specific, in that it requires the heterodimer, a cognate ligand, and a cognate response element. Because tRA binds RARs, but not RXRs, the effect of ligands that bind to RXRs then was investigated. Of interest, 9cRA that binds both RARs and RXRs was more efficient than tRA at limiting concentrations, with ED50 of ≈9 × 10−10 M and 4 × 10−9 M for 9cRA and tRA, respectively (Fig. 5A). Because these differential effects of 9cRA and tRA suggested that synergistic activation of transcription might occur when both RARα and RXRα are liganded, we further investigated transcriptional activation by RARα/RXRα heterodimers on addition of receptor-specific synthetic retinoids (Fig. 5B). As expected, a stimulation was observed in the presence of the RARα-specific agonist BMS753 (16) but not on addition of either the RARγ-specific agonist BMS961 (16) or the RARα antagonist BMS614 (16) (Fig. 5B). Of interest, the RXR-specific panagonist BMS 649 (identical to SR11237; see ref. 16) did not activate transcription on its own (Fig. 5B). However, a synergistic stimulation was observed on concomitant addition of BMS649 and limiting concentrations of RAR agonists (Fig. 5B; compare 10−8 and 5 × 10−8 M tRA in the presence and absence of BMS649 and also BMS753 in the presence and absence of BMS649). In contrast, no stimulation resulted from the simultaneous addition of the RARα antagonist BMS614 and the RXR agonist BMS649 (data not shown). It appears, therefore, that the AF-2 activation function of RXRα can act synergistically with that of RARα but that the binding of an agonist to RARα is a prerequisite for effective activation of transcription by agonist-bound RXRα. This conclusion was further supported by the observation that the RARα-specific antagonist BMS614 abrogated the synergistic effect of the RARα-specific agonist BMS753 and RXR agonist BMS649 (Fig. 5B). Similarly, BMS 614 abrogated the 9cRA-induced transcriptional activation by RARα/RXRα heterodimers (Fig. 5B), even though 9cRA binds to both RARs and RXRs.
Figure 5.
Dose-dependent synergistic effects of specific retinoids on activation of transcription by RARα/RXRα heterodimers. (A) Dose-dependent activation by tRA and 9cRA. Transcription reactions were performed as described in Fig. 3 on a (DR5)5β2G template by using FhRARα/HmRXRα in the presence of varying concentrations (5 × 10−10 to 10−6 M) of tRA (open circles) or 9cRA (closed squares). (B) Receptor-selective and synergistic activation of transcription. Transcription reactions were performed as described above by using synthetic retinoid agonists and antagonists at the concentrations indicated. The receptor specificity of retinoids used are as follows: tRA (panRAR-specific ligand), 9cRA (panRAR- and panRXR-ligand), BMS 753 (RARα-specific agonist), BMS961 (RARγ-specific agonist), BMS649 (panRXR-specific agonist), and BMS614 (RARα-specific antagonist). Transactivation by FhRARα/HmRXRα is expressed relative to that observed from the internal control template (pG1). Induction by tRA (10−6 M) was arbitrarily set to 100%. All points are the average of at least two independent experiments run in duplicate.
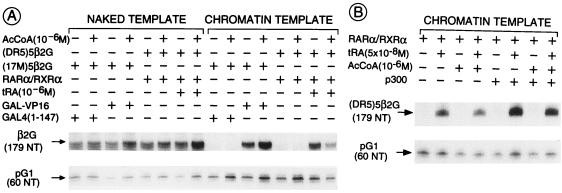
Acetylation and deacetylation of nucleosomal histones in transcriptionally active (euchromatin) and inactive (heterochromatin) chromatin, respectively, is well documented (reviewed in ref. 46). The facilitating role of histone acetylation in transcriptional activation also has been demonstrated recently in vitro (47, 48). We therefore examined the effect of the addition of acetyl CoA to our in vitro transcription system. No effect could be evidenced by using either RARα/RXRα heterodimers or the Gal-VP16 activator, in the presence of either naked or chromatin-assembled cognate templates (Fig. 6A). Because certain coactivators are thought to mediate transactivation by nuclear receptors at least in part through their intrinsic histone acetyltransferase activities (e.g., SRC-1, ACTR, CBP, and p300; see Introduction for references), we also investigated whether addition of purified baculovirus-expressed p300 could stimulate transcriptional activation by RARα/RXRα in our in vitro system. p300 enhanced the activation of transcription by the heterodimers ≈4-fold in the presence of tRA whereas transcription of the chromatin template remained repressed in the absence of the agonistic ligand, irrespective of the presence of the heterodimers (Fig. 6B). No p300 effect was seen on naked DNA templates (data not shown). The further addition of acetyl CoA had no effect on the extent of transcriptional enhancement, even though the purified p300 coactivator exhibited histone acetyltransferase activity (data not shown). Note, however, that our in vitro system contains some endogenous histone acetyltransferase activity that was not enhanced further by the addition of p300 to the transcription reaction (data not shown).
Figure 6.
p300 enhances transactivation by RARα/RXRα heterodimers in vitro. (A) Addition of exogenous acetyl CoA (AcCoA) does not effect ligand-dependent transactivation by RARα/RXRα. Transcription reactions were performed in the presence or absence acetyl CoA (1 μM) on naked or chromatin (17M)5β2G or (DR5)5β2G templates plus or minus 1 nM activator [either Gal4(1–147), Gal4-VP16, or FhRARα/HmRXRα] and/or tRA (1 μM), as described in Fig. 3. (B) Addition of acetyl CoA does not further enhance p300-activated transcription. Transcription was performed on (DR5)5β2G templates in the presence or absence of FhRARα/HmRXRα and/or tRA (5 × 10−8 M). Where indicated, the coactivator p300 (0.5 nM) and acetyl CoA (1 μM) were added.
DISCUSSION
Numerous studies aimed at reproducing transactivation by nuclear receptors in vitro have been reported over the last years. Most of these studies were performed with naked DNA templates, and the reported transcriptional activations were either ligand-independent (49–51) or only modestly dependent on addition of agonistic ligands (52–54). Moreover, these studies did not or only poorly reproduced the effects of agonistic and antagonistic ligands, as observed on responsive genes in vivo. In one case (55), a greater stimulation was observed with salt dialysis-reconstituted chromatin templates than with naked DNA templates, but the ligand-dependency of transcriptional activation was not established. With the recent finding that several putative transcriptional coactivators that interact in an agonist-ligand-dependent manner with NRs possess histone acetyltransferase activity, it became evident that more physiologically relevant templates might be required to faithfully reproduce in vitro the essential features of ligand-dependent transcriptional activations as observed in vivo. Estrogen- and anti-estrogen-regulated transcriptional activation by the estrogen receptor α, resembling the natural mechanism of action of estrogen receptor α in vivo, was, in fact, recently achieved in vitro with chromatin but not with naked DNA templates (41). Using a similar approach, we have shown here that constitutive transcription from a naked DNA template containing a RA-responsive promoter [(DR5)5β2G] is not affected by RARα/RXRα heterodimers, irrespective of the presence of the tRA agonist. In contrast, there is very little transcription when the same promoter is present in chromatin-assembled templates, unless tRA is bound to the RAR/RXR heterodimers, which results in activation of transcription to a level similar to that achieved with naked DNA templates. These observations clearly establish that the tRA-induced transcriptional activation mediated by RAR/RXR heterodimers corresponds to the relief of a repression generated by the chromatin organization of the template. Moreover, our results show that this relief does not correspond to the binding of RAR/RXR heterodimers to the chromatin template because it is clear from DNase I footprinting data (Fig. 4) that unliganded heterodimers specifically bind to the RA response elements. Thus, the critical events underlying ligand-induced transcriptional activation by RAR/RXR heterodimers must occur subsequent to their binding to the chromatin template.
Previous studies of the effects of retinoids on transcription of RA-responsive genes, differentiation, and apoptosis of mouse embryonal carcinoma cells (7–9, 15, 17, 19, 20, 22) and human acute promyelocytic leukemia cells (16), as well as genetic studies in the mouse (refs. 10 and 12 and references therein), have established that RAR/RXR heterodimers are the main functional units mediating the effect of retinoids in vivo (see Introduction for additional references). Furthermore, these studies have shown that the ligand-dependent activation function AF-2 of both RAR and RXR partners are instrumental in this mediation. However, in all cases there is a subordination of the activity of RXR AF-2 to the binding of an agonistic ligand to the RAR partner (see Introduction). Most remarkably, our present in vitro system reproduces these in vivo features of retinoid action. Although ligands binding to RARα but not to RXRα (tRA, BMS753) can activate transcription on their own, a ligand binding to RXRα but not to RARα (BMS649) is inactive on its own. Consistent with the subordination of RXR AF-2 activity to that of RAR AF-2, the transcriptional activation brought about by 9-cis RA that induces both RAR and RXR AF-2s is abrogated by the addition of the RARα antagonist BMS614. Furthermore, in agreement with previous in vivo observations (7, 15, 19–22, 56), we observe here synergistic effects between limiting amounts of RAR ligands and a RXR-specific ligand.
Structural studies (refs. 57–60 and references therein) have demonstrated that binding of an agonistic ligand triggers a transconformation of the ligand binding domain. This generates an interaction surface for coactivators of the activation function AF-2 that are thought to recruit factors of the general transcription machinery and/or act on chromatin remodeling through histone acetyltransferase activities (see Introduction). Of interest, Kraus and Kadonaga (41) have recently reported that the p300 coactivator (61, 62) acts synergistically with ligand-activated estrogen receptor α to stimulate transcription in vitro from a cognate chromatin template. Similarly, on addition of exogenous p300, we report here a further 4-fold enhancement in ligand-dependent transcription from chromatin templates in the presence of RARα/RXRα heterodimers. Note that the actual level of enhancement by p300 is likely to be higher because endogenous p300/CBP is already present in the HeLa cell extract used in the transcription system. However, though our purified p300 exhibits intrinsic histone acetyltransferase activity, the addition of acetyl CoA to the present transcription system has no further effect on p300-activated transcription. A similar observation was made by Naar et al. (63) in a study of Sp1/SREBP-1-activated transcription on a chromatin template in the presence of CBP. Thus, p300 may further enhance ligand-induced activation of transcription on chromatin templates by bridging RARα/RXRα heterodimers to RNA polymerase II through its interaction with RNA helicase A (32), rather than by locally remodeling the chromatin structure through histone acetylation.
In conclusion, we have established an in vitro chromatin template-based transcription system to study the molecular events underlying activation of transcription by RAR/RXR heterodimers that essentially mimics the in vivo synergistic effect of RAR- and RXR-selective retinoids and the subordination of RXR AF-2 activity to binding of an agonist to RAR. This presently crude system can now be dissected biochemically to ultimately provide us with a thorough molecular view of the events that occur during ligand-dependent transcriptional activation by retinoid receptors.
Acknowledgments
We thank P. Becker for advice on chromatin assembly and J. T. Kadonaga and W. L. Kraus for providing p300-expressing baculovirus and for advice on DNase I footprinting using chromatin templates. We are grateful to R. Gopalkrishnan for providing GAL(1–147) and GAL-VP16 expression vectors and to M. Sporn and Bristol-Myers Squibb chemists for gifts of 9-cis RA and synthetic retinoids, respectively. We also thank C. Pernot for technical help, I. Kolb-Cheynel for performing baculovirus infections, M. Brand for critically reading the manuscript, and L. Tora, I. Davidson, and R. Losson for useful discussions. This work was supported by grants from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Collège de France, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur la Cancer, and Bristol-Myers Squibb. F.J.D. is a fellow of the Natural Science and Engineering Research Council and the Fondation pour la Recherche Médicale. A.B. is the recipient of a Marie-Curie fellowship from the European Commission.
ABBREVIATIONS
- NR
nuclear receptor
- RA
retinoic acid
- RARα
RA receptor α
- RXRα
retinoid X receptor α
- FhRARα
Flag-tagged human RARα
- HmRXRα
His-tagged mouse RXRα
- tRA
all-trans RA
- 9cRA
9-cis retinoic acid
- RARE
RA response element
- DR5
DNA response element with directly repeated motifs spaced by 5 bp
- AF-2
activation function 2
References
- 1.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 2.Gronemeyer H, Laudet V. Protein Profile. 1995;2:1173–1236. [PubMed] [Google Scholar]
- 3.Perlmann T, Evans R M. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 4.Chambon P. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 5.Allenby G, Bocqel M-T, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo J F, Chambon P, et al. Proc Natl Acad Sci USA. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leid M, Kastner P, Chambon P. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 7.Clifford J, Chiba H, Sobieszczuk D, Metzger D, Chambon P. EMBO J. 1996;15:4142–4155. [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba H, Clifford J, Metzger D, Chambon P. J Cell Sci. 1997;139:735–747. doi: 10.1083/jcb.139.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba H, Clifford J, Metzger D, Chambon P. Mol Cell Biol. 1997;17:3013–3020. doi: 10.1128/mcb.17.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastner P, Mark M, Ghyselinck N, Krezel W, Dupé V, Grondona J M, Chambon P. Development (Cambridge, UK) 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- 11.Kretzel W, Ghyselinck N, Samad T A, Dupe V, Kastner P, Borrelli E, Chambon P. Science. 1998;279:863–867. doi: 10.1126/science.279.5352.863. [DOI] [PubMed] [Google Scholar]
- 12.Mascrez B, Mark M, Dierich A, Ghyselinch N B, Kastner P, Chambon P. Development (Cambridge, UK) 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- 13.Apfel C M, Kamber M, Klaus M, Mohr P, Keidel S, Le Motte P K. J Biol Chem. 1995;270:30765–30772. doi: 10.1074/jbc.270.51.30765. [DOI] [PubMed] [Google Scholar]
- 14.Lotan R, Dawson M I, Zou C C, Jong L, Lotan D, Zou C P. Cancer Res. 1995;55:232–236. [PubMed] [Google Scholar]
- 15.Roy B, Taneja R, Chambon P. Mol Cell Biol. 1995;15:6481–6487. doi: 10.1128/mcb.15.12.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J-Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Nature (London) 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 17.Horn V, Minucci S, Ogryzko V V, Adamson E D, Howard B H, Levin A A, Ozato K. FASEB J. 1996;10:1071–1077. doi: 10.1096/fasebj.10.9.8801169. [DOI] [PubMed] [Google Scholar]
- 18.La Vista-Picard N, Hobbs P D, Pfahl M, Dawson M I. Mol Cell Biol. 1996;16:4137–4146. doi: 10.1128/mcb.16.8.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taneja R, Roy B, Plassat J-L, Zusi C F, Ostrowski J, Reczek P R, Chambon P. Proc Natl Acad Sci USA. 1996;93:6197–6202. doi: 10.1073/pnas.93.12.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taneja R, Rochette-Egly C, Plassat J-L, Penna L, Gaub M-P, Chambon P. EMBO J. 1997;16:6452–6465. doi: 10.1093/emboj/16.21.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botling J, Castro D S, Oberg F, Nilsson K, Perlmann T. J Biol Chem. 1997;272:9443–9449. doi: 10.1074/jbc.272.14.9443. [DOI] [PubMed] [Google Scholar]
- 22.Minucci S, Leid M, Toyama R, Saint-Jeannet J-P, Peterson V J, Horn V, Ishmael J E, Bhattacharyya N, Dey A, Dawid I B, et al. Mol Cell Biol. 1997;17:644–655. doi: 10.1128/mcb.17.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph B, Lefebvre O, Mereau-Richard C, Danze P M, Belin-Plancot M T, Formstecher P. Blood. 1998;91:2423–2432. [PubMed] [Google Scholar]
- 24.Vivat V, Zechel C, Wurtz J-M, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P. EMBO J. 1997;16:5698–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 26.Glass C K, Rose D W, Rosenfeld M G. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 27.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, et al. Nature (London) 1997;389:191–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 28.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 31.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima T, Uchida C, Anderson S F, Lee C-G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 33.Zelent A, Mendelsohn C R, Kastner P, Krust A, Garnier J-M, Ruffenach F, Leroy P, Chambon P. EMBO J. 1991;9:71–81. doi: 10.1002/j.1460-2075.1991.tb07922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazin M J, Kamakaka R T, Kadonaga J T. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- 35.Becker P B, Tsukiyama T, Wu C. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 36.Bellard M, Dretzen G, Giangrande A, Ramain P. Methods Enzymol. 1989;170:317–346. doi: 10.1016/0076-6879(89)70054-9. [DOI] [PubMed] [Google Scholar]
- 37.Gaub M P, Rochette-Egly C, Lutz Y, Ali S, Matthes H, Scheuer I, Chambon P. Exp Cell Res. 1992;201:335–346. doi: 10.1016/0014-4827(92)90282-d. [DOI] [PubMed] [Google Scholar]
- 38.Rochette-Egly C, Lutz Y, Pfister V, Heyberger S, Scheuer I, Chambon P, Gaub M P. Biochem Biophys Res Commun. 1994;204:525–536. doi: 10.1006/bbrc.1994.2491. [DOI] [PubMed] [Google Scholar]
- 39.Kumar V, Chambon P. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 40.Rochel N, Renaud J-P, Ruff M, Vivat V, Granger F, Bonnier D, Lerouge T, Chambon P, Gronemeyer H, Moras D. Biochem Biophys Res Commun. 1997;230:293–296. doi: 10.1006/bbrc.1996.5787. [DOI] [PubMed] [Google Scholar]
- 41.Kraus W L, Kadonaga J T. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 43.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassone-Corsi P, Duboule D, Chambon P. Cold Spring Harbor Symp Quant Biol. 1985;50:747–752. doi: 10.1101/sqb.1985.050.01.092. [DOI] [PubMed] [Google Scholar]
- 45.Sadowski I, Ma J, Triezenberg S, Ptaschne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 46.Kuo M-H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 49.Shemshedini L, Ji J, Brou C, Chambon P, Gronemeyer H. J Biol Chem. 1992;267:1834–1839. [PubMed] [Google Scholar]
- 50.Schmitt J, Stunnenberg H G. Nucleic Acid Res. 1993;21:2673–2681. doi: 10.1093/nar/21.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Vos P, Schmitt J, Verhoeven G, Stunnenberg H G. Nucleic Acid Res. 1994;22:1161–1166. doi: 10.1093/nar/22.7.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valcarcel R, Holz H, Jimenez C G, Barettino D, Stunnenberg H G. Genes Dev. 1994;8:3068–3079. doi: 10.1101/gad.8.24.3068. [DOI] [PubMed] [Google Scholar]
- 53.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemon B D, Fondell J D, Freedman L P. Mol Cell Biol. 1997;17:1923–1937. doi: 10.1128/mcb.17.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schild C, Claret F-X, Wahli W, Wolffe A P. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 58.Moras D, Gronemeyer H. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 59.Nolte R T, Wisely B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M R, Wilson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–144. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 60.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanstein B, Eckner R, Direnzo J, Halachmi H, Jiu H, Searcy R, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakravarti D, LaMorte V J, Nelson M C, Nakajima I G, Schulman H, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 63.Naar A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tijan R. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]