Abstract
The carboxyterminal region of activation-induced deaminase (AID) is required for its function in Ig class switch recombination (CSR) and also contains a nuclear-export sequence (NES). Here, based on an extensive fine-structure mutation analysis of the AID NES, as well as from AID chimeras bearing heterologous NESs, we show that while a functional NES is indeed essential for CSR, it is not sufficient. The precise nature of the NES is critical both for AID stabilization and CSR function: minor changes in the NES can perturb stabilization and CSR without jeopardizing nuclear export. The results indicate that the AID NES fulfills a function beyond simply providing a signal for nuclear export and suggest the possibility that the quality of exportin-binding may be critical to the stabilization of AID and its activity in CSR.
Keywords: activation-induced deaminase, antibody diversification, immunoglobulin class switching, exportin, nuclear transport
Activation-induced deaminase (AID) plays a central role in antibody gene-diversification, triggering both somatic hypermutation (SHM) and class-switch recombination (CSR) by deaminating deoxycytidine residues within the Ig locus to yield deoxyuridine (1, 2).
AID is largely found in the cytoplasm of activated B cells but shuttles between cytoplasm and nucleus (3–6). The C-terminal amino acids of AID comprise a nuclear-export sequence (NES): removal of this NES causes AID to be restricted to the nucleus, whereas its addition to a heterologous nuclear protein drives export of the fusion protein into the cytoplasm (3–5). The sequence of the AID C-terminus conforms well to the consensus established for substrates of the Crm1-dependent export machinery: in keeping with this, export mediated by the AID NES is sensitive to leptomycin B.
Analysis of both clinical and contrived mutations in AID have revealed that the AID C-terminal region is also essential for its function in CSR, although not in SHM (7–9). The C-terminal mutations that interfere with AID's ability to potentiate CSR do not compromise its ability to deaminate deoxycytidine in biochemical or bacterial genetic assays. Thus, as previously suggested by others, either the nuclear/cytoplasmic shuttling of AID must be specifically required for CSR or, alternatively, factors specific for CSR must interact with the C-terminal region of AID in an area overlapping the NES (4, 5, 7–9).
It was with a view to discriminating these possibilities that we embarked on a refined mutagenic dissection of the AID NES and investigated the ability of heterologous NESs to substitute for the AID NES. Our results suggest that although nuclear export is likely necessary for CSR, this is not the sole function of the AID C-terminal region, with the data raising the possibility that successful CSR depends on the precise quality of AID binding to exportin or a related protein.
Results
NES Function and CSR Correlate in Alanine-Scanning Mutagenesis.
To ascertain whether there was a correlation between export activity and CSR, we performed an alanine-scanning mutagenesis screen of the C-terminal 10 amino acids of human AID, which comprise its NES (Fig. 1A). Activity in promoting CSR was tested by monitoring CSR to IgG1 in LPS+IL4 cultures of splenic B cells from AID−/− mice that had been transduced with a retrovirus encoding AID (or AID variants), which also carried a GFP reporter translated from an internal ribosome entry site (IRES). Intracellular localization was assessed in NIH 3T3 cells that had been transfected with vectors directing expression of the relevant GFP-AID-mutant fusion protein, as well as in transfected COS and 293 cells.
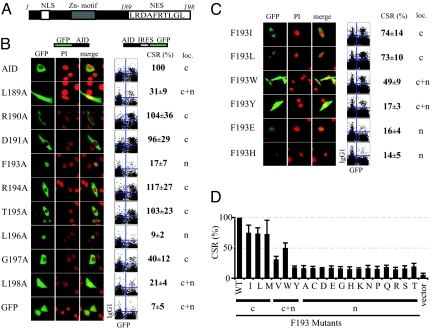
Fig. 1.
Effect of point mutations in the AID NES on protein localization and CSR. (A) Schematic depiction of AID, highlighting the NES, the nuclear localizing signal, and the Zn-coordination motif. (B) Analysis of AID mutants carrying alanine substitutions in the NES. On the left, representative confocal micrographs show the localization of GFP-AID-fusion proteins (green) transiently transfected into NIH 3T3 cells with the nucleus highlighted by costaining with propidium iodide (red). The predominant localization [n: AID staining is predominantly (> 90% of cells) nuclear; c: staining is predominantly cytoplasmic; c+n: staining is clearly seen in both compartments] with the conclusion agreeing well with that obtained from COS (see Table S1) as well as 293 cell (not shown) transfectants. Similar localizations were observed with critical mutants when analyzed as AID-GFP (rather than GFP-AID) fusions (not shown). With respect to CSR, the mutant AIDs were introduced by transduction into splenic B cells from AID−/− mice using a GFP-expressing retrovirus; CSR to IgG1 was analyzed following culture in the presence of LPS+IL4. The extent of CSR is shown in representative flow cytometry plots [IgG1 staining on the y-axis; GFP fluorescence (a marker of cells infected with the AID-GFP retrovirus) on the x-axis] with the column “CSR (%)” giving the percentage (mean ± SD of 5 experiments) of transfected (GFP+) cells that have switched to IgG1 following infection with the mutant AID relative to that obtained with wild-type human AID. (C) Analysis of AID mutants in which F193 has been mutated to each of the other 19 possible amino acids. Mutants were tested for localization and CSR in 5 experiments, as described in the legend to (B). Representative data obtained with 6 of the amino-acid substitutions are displayed. The sIgG1+ GFP+ cells observed in the AID-transduced population may reflect switched B cells that fail to express the IRES-GFP translation unit. (D) The CSR results from the entire set of the 19 different AID F193 substitutions are shown as a histogram, with the pattern of localization of the AID-GFP chimeras in NIH 3T3 transfectants indicated below the line. The results regarding localization in transfectants of COS7 cells are summarized in Table S1.
As shown in Fig. 1B and further supported in Table S1, the most severe effects on intracellular localization were observed following mutation of any of the 4 hydrophobic amino acids (L189, F193, L196, or L198). This is in keeping with the fact that these 4 hydrophobic residues constitute the conserved consensus that is characteristic of Crm1-dependent export sequences (10). The F193A and L196A mutations had the most striking effect, yielding fusion proteins whose expression was largely restricted to the nucleus. The L189A and L198A mutants of human AID were located in both nucleus and cytoplasm, consistent with the similar distribution observed by McBride et al. (5) with the mouse F198A AID mutant. Mutation of any of these 4 hydrophobic amino acids led to a dramatic decrease in CSR activity. In contrast, individual mutation to alanine of most of the intervening amino acids that separate the hydrophobic residues (R190, D191, R194, and T195) did not have a major effect on either intracellular localization or on CSR activity. The only exception to a simple correlation between export and CSR activity observed in these assays was noted with the G197A mutant, where we detected a cytoplasmic localization similar to that of the wild type, whereas its activity in CSR was reduced to 40% that of the wild-type parent.
NES Function and CSR Correlate in a Panel of F193 Mutants.
These results (revealing a broad correlation between CSR and export) were consistent with the idea that export is required for CSR, rather than that the AID C terminus contains distinct but overlapping sites for exportin-binding and the recruitment of a CSR-specific factor. However, to extend this study, we asked whether CSR and export activity also correlated in a family of AID mutants in which F193 had been replaced with each of the 19 other possible amino acids. Again, there was a correlation between AID export and CSR (Fig. 1C). Export was best retained in those AID mutants in which F193 was replaced by the hydrophobic amino acids I, L, M, and W, and these gave the best retention of CSR. Other mutant AIDs were less efficiently exported (exhibiting greater degrees of nuclear retention) and also gave less effective CSR. Thus, CSR was always reduced in mutants that exhibited compromised nuclear export.
Heterologous NESs Restore Export but not CSR.
A straightforward interpretation of the data would appear to be that CSR requires AID to shuttle between nucleus and cytoplasm (for example, to acquire some partner or cytoplasmically generated posttranslational modification) and that the function of C-terminal amino acids in AID is to provide an NES that allows such shuttling. One might therefore conjecture that the ability to potentiate CSR might be readily restored to an AIDΔ[NES] mutant by fusion with a heterologous NES. This is not the case.
Chimeras of human AID were generated in which its C-terminal 10 amino acids have been removed and replaced by an NES from a different source [Protein kinase inhibitor alpha (PKI), HIV-1 Rev, HTLV-1 Rex, MAP kinase kinase (MAP), or Ran binding protein 1 (Ran-BP1)], all of which have been shown by others to mediate export through the Crm1-dependent pathway as judged by sensitivity to leptomycin B (LMB) (11, 12). These heterologous NESs were all able to mediate export of chimeric AID, as assayed as GFP fusion proteins. Thus, the various fusion proteins were localized in the cytoplasm of transfected fibroblasts (Fig. 2A; see Table S1) but retained in the nucleus following incubation with LMB (not shown). A similar cytoplasmic localization (with nuclear retention following LMB treatment) was observed when an anti-AID mAb was used to detect untagged AID-NES chimeras in transfected DT40 B cells. (Fig. 2B). However, none of the heterologous NESs was able to restore efficient CSR as judged by in vitro CSR assays (see Fig. 2 A and B, Table S1). This inability to restore CSR also extended to several other Crm1-dependent NESs analyzed (from p53, transcription factor TFIIIA, IκB, adenovirus type 5 EIB-55K protein, and fragile X mental retardation protein) (Table S2).
Fig. 2.
NESs from heterologous proteins restore nuclear export but not CSR. (A) AID chimeras in which C-terminal 10 amino acids of AID were replaced by NESs from other Crm1-dependent genes (PKI, protein kinase inhibitor; Rev, HIV1-Rev protein; Rex, HTLV1-Rex; Ran, RanBP1; MAP, MAP Kinase Kinase) were tested for their intracellular localization and ability to potentiate CSR as described in Fig. 1. The CSR obtained with the various chimeric AIDs is given as a percentage of that obtained with wild type, as determined in 3 independent experiments. (B) Untagged AID chimeras carrying the PKI or MAP NES localize to the cytoplasm in DT40 B cell transfectants, but LMB treatment of the MAP chimera causes nuclear accumulation. (Similar results were obtained with other chimeras). (C) An ability to potentiate CSR is retained by chimeras in which the NES of human AID is replaced by the NESs of AIDs from other species. Ch, chicken; Da, Dario; Fr, frog; Fu, fugu. (D) Analysis of chimeras in which the MAP NES is fused to the C terminus of full-length AID (thereby generating a polypeptide with 2 sequential NESs) or to the C terminus of full length AID[F193A].
The fusion of a heterologous NES to full-length AID does not actually inhibit CSR. Switching is retained using a full-length human AID to which the MAP NES has been appended at the C terminus, although there is no rescue of CSR if the same MAP NES fragment is fused to the C terminus of AID[F193A] (Fig. 2D).
Thus, the AID NES appears to possess features that uniquely allow it to sustain CSR, as the heterologous NESs do not provide this function. It is therefore notable that the sequence of the C-terminal region of AID has not been fully conserved during evolution. Nevertheless, despite such sequence variation, the AID C-terminal NES-consensus sequences from chicken, frog, and fish all exhibit the ability to activate CSR effectively when tested as chimeras with human AID (Fig. 2C).
Sensitivity of CSR to the Interdigitating Hydrophilic Residues in the NES.
Although the initial alanine-scanning mutagenesis revealed that disruption of the critical hydrophobic amino acid residues in the AID NES led to both a disruption of export function and a substantial reduction in CSR activity, the fusions with the heterologous NESs indicate that export itself is not sufficient for CSR. In particular, they suggest that some of the interdigitating amino acid residues in the NES are critical for CSR activity, but not for export function. Informed primarily by a comparison of the AID and PKI export sequences, we generated a new set of AID NES mutants carrying a variety of individual or clustered amino acid substitutions. Although analysis of these mutant AIDs in immunolocalization and CSR assays does not yield a simple conclusion as to a unique characteristic required for CSR, the results (Fig. 3) do reveal that CSR is indeed more fastidious than export in its dependence on the identity of the interdigitating amino acids, indicating for example that CSR is especially sensitive to an A->K substitution at position 192.
Fig. 3.
Importance of the interdigitating hydrophilic residues in the AID NES. The sequences of human AID mutants carrying substitutions of the interdigitating hydrophilic amino acids in the NES are depicted on the right (dashes indicate identity with the corresponding position in wild-type human AID). Results are presented as in Fig. 1, with the percentages being given as the average of 3 independent experiments (except with M5 and M6, where n = 5). Data on some further mutants of the interdigitating hydrophilic residues are included in Table S2.
Reduced CSR Correlates with Reduced AID Abundance.
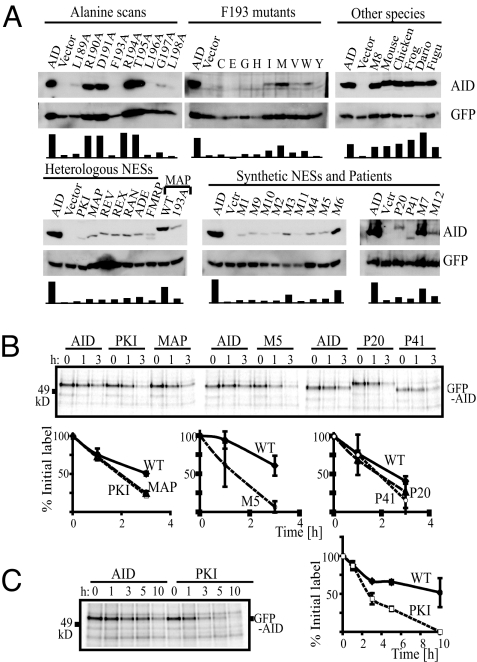
The half-life of AID differs considerably, dependent on whether it is located in nucleus and cytoplasm (13). Indeed, fibroblasts transfected with the nuclear-restricted AID-GFP chimeras were usually less bright than those expressing the export-proficient proteins (see Fig. 1). However, we were interested in obtaining more quantitative estimates of the abundance of the different AID NES mutants, especially assayed in untagged form and in the context of the retrovirally transduced B cells in which CSR is monitored. The AID protein levels were monitored in cell extracts by Western blotting: a remarkable correlation was observable between the levels of AID polypeptide and the ability of the same AID mutant to potentiate CSR (Fig. 4A).
Fig. 4.
Abundance and stability of mutant AIDs. (A) AID abundance was monitored by Western blot analysis of extracts of AID−/− B cells made 3 days after they had been retrovirally transfected with the relevant AID-expressing pMx-GFP construct (directing the independent expression of GFP and untagged AID) and cultured in the presence of LPS+IL4. Blots were reprobed with anti-GFP antibodies to control for loading and infection. Histograms show the ratio of AID:GFP band intensities for individual lanes, normalized for each blot. (B) The turnover of GFP-AID fusion proteins in 293 cell transfectants was monitored by labeling cells for 45 min with L-[35S]methionine before chasing with unlabeled methionine for the indicated length of time and then performing autoradiography following SDS/PAGE of anti-GFP immunoprecipitates. Densitometry analysis of autoradiographs was used to deduce the amount of residual labeled GFP-AID at each time-point, expressed as a percentage of that present at the beginning of the chase. The data shown are the mean ± SD of the results from 2 experiments. In the AID P41 mutant, the R190 codon has been mutated to a stop codon [as found in hyperIgM patient P41 of Imai et al. (9)], resulting in a deletion of the NES. (C) The turnover of GFP-AID fusion proteins in 293 cell transfectants was monitored as in (B), except that the chase was performed over 3 h.
Thus, with regard to the alanine-scanning mutagenesis and consistent with the recent work of Aoufouchi et al. (13), mutation of the hydrophobic residues (L189, F193, L196, L198) that resulted in reduced nuclear export and CSR also led to reduced abundance; in contrast, mutation of R190, D191, R194, or T195 to alanine had little effect on localization, abundance, or CSR (see Fig. 4A). Similarly, of the various substitutions at F193, those that retained CSR and export exhibited the greatest abundance.
However, what is striking and unexpected is that AID abundance was greatly diminished with those AID mutants that retained functional NESs but which exhibited reduced CSR. All of the NESs from heterologous genes that restored export but not CSR failed to restore high abundance: of the synthetic NESs that restored export, only those that restored CSR (M3 and M6, as well as M7) gave good abundance (see Fig. 4). Thus, the major correlation is between abundance and CSR rather than between abundance and cytoplasmic localisation per se.
These observations led us to investigate a clinical mutant of AID (P20) described by Ta et al. (8), in which an in-frame insertion of 34 heterologous amino acids at AID position 182 destroyed AID's ability to potentiate CSR but did not prejudice nuclear export. We find that, similarly to what is observed with many of the NES swaps, the P20 and P41AID mutants shows reduced abundance (see Fig. 4A). In view of the fact that P41-type truncation mutations can yield a dominant-negative hyperIgM phenotype in patients (8, 9), it is possible that the truncations could lead to destabilization of complexes containing multiple AID polypeptides.
The reduced abundance of the various AID mutants in retrovirally transduced B cells correlates with faster degradation of the polypeptide as judged by biosynthetic labeling experiments: GFP-AID has a considerably longer half-life than a GFP-AID mutant with a deleted NES (AID-P41) (see Fig. 4 B and C). However, diminished stability was also evident using AID mutants in which the NES functioned in export but not in CSR (MAP, PKI, M5, and P20). Thus, instability is not simply a consequence of nuclear retention: it will be interesting to ascertain whether, in light of the findings of Aoufouchi et al. (13), it is also associated with ubiquitination
Somatic Mutation.
Mutant AIDs that have nonfunctional NESs do not support CSR but have nevertheless been shown in previous work to be able to potentiate SHM both in transfected cell-lines and in vivo (7, 8). We were therefore interested in monitoring SHM in cells transfected with AID mutants carrying an NES that drove nuclear export but did not potentiate CSR. For the assay, we transfected the mutant AIDs into AID−/− DT40 cells carrying a deletion of the IgVλ pseudogenes. As a result of this ϕV deletion, AID-triggered dC deamination in the rearranged IgVλ gene leads to SHM (rather than gene conversion): this can be read out by monitoring the consequent generation of sIgM-loss variants (14).
As shown in Fig. 5, the F193A mutant (which is present in much lower abundance than the wild-type control) nevertheless gives a significantly increased frequency of SHM. This presumably reflects the wholly nuclear localization of this mutant with its nuclear abundance being higher than that of wild-type AID (although its overall abundance is less). This agrees with the conclusion reached by McBride et al. (5), using a somewhat different assay regarding SHM by mouse AID[F198A]. In contrast to the enhanced SHM observed with AID[F193A], the AID mutants carrying heterologous or altered NESs that function in export but not in CSR give reduced SHM. [This is a reduction and not a complete ablation of SHM, consistent with the fact that patient P20 harbors B cells expressing mutated IgV genes (8).] These same AID mutants showed diminished abundance in the stably transfected DT40 cells (see Fig. 5B). Thus, as analyzed with mutant AIDs and in contrast to what was observed with CSR, there seems to be a broad correlation between extent of IgV SHM in these assays and the abundance of AID in the nucleus. Similarly, whereas AID[F193A] also targets the pre-Sμ region for mutation but not CSR, the AID[MAP] chimera gives only a low level of pre-S mutation, consistent with its low nuclear abundance (see Fig. 5C).
Fig. 5.
Somatic mutation by the mutant AIDs. (A) Somatic mutation of the IgV was assayed by monitoring surface IgM-loss in AID−/− ϕV−/− sIgM+ DT40 cells that had been stably transfected with expression constructs expressing the indicated AID mutants. The percentage of sIgM-loss variants is expressed as the mean of that in multiple (n) independent clonal transfectants as determined 3 weeks after sorting and subculturing (PKI, n = 18; MAP, 16; M5, 24; F193A, 16; P20, 2; AID WT, 4). (B) The abundance of AID in the various DT40 transfectants was determined by Western blot analysis of cell extracts. A band that cross-reacts with the anti-AID mAb and which is detected in untransfected AID−/− DT40 cells is indicated by an asterisk and serves as a loading control. (C) Somatic mutation of the pre-Sμ region was analyzed by sequencing a 700 nucleotide region extending from nucleotide 136679 in GenBank sequence accession number AC073553 in DNA that had been PCR amplified and cloned from splenic B cells from AID−/− mice 3 days following infection with pMx-GFP retroviruses directing the expression of AID[F193A] or AID[MAP]. The mutations identified are provided in Fig. S1.
Discussion
The detailed dissection and substitution analysis of the AID NES described here was performed to discover whether its functions in CSR and nuclear export can be dissociated. They can, in that export is not sufficient for CSR. Comparison of a panel of mutant AIDs carrying altered or heterologous NESs revealed that all of a large number of individual amino acid substitutions that compromise nuclear export also compromise CSR. Thus, a functional NES appears essential for CSR. However, it is not sufficient. Many of the mutant/heterologous NESs studied that allowed export did not potentiate CSR. Thus, NESs can be divided into 2 classes: one class supports CSR and export whereas the other merely supports export. We find that these 2 classes of NES also differ in their ability to confer stability on AID.
The results raise a number of questions. What are the differences in the molecular interactions mediated by the 2 categories of NES, which allow members of one category but not the other to potentiate both CSR and protein stability? What is the connection between protein stabilization and the ability to trigger CSR: is CSR simply a consequence of increased AID stability or are stabilization and CSR independently reliant on a common molecular interaction? And why is a functional NES required at all for CSR, given that, as shown here and previously demonstrated by others (7, 8), removal of the C terminus does not restrict AID activity in various assays of SHM?
The fact that protein stabilization and CSR exhibit a similar pattern of dependence on the detailed nature of the hydrophobic-interdigitating amino acids in the AID NES suggests that both features might depend on a common molecular interaction. Indeed, it is possible that the diminished CSR obtained with some of the export-proficient mutant NESs might at least in part be the direct result of their diminished stability: the abundance of AID in normal B cells is known to be tightly controlled, with the efficiency of CSR being very sensitive to AID expression levels (15–19). Such a requirement for both export and abundance might nicely explain the CSR-deficiency exhibited by the AID P20 mutant (8), which carries a 34-aa insertion upstream of the NES, as we find that this mutant exhibits diminished stability although retaining a functional export sequence.
The nature of the molecular interactions mediated by the NES that allow CSR and confer protein stability remains to be determined. However, the fact that both CSR and stability require maintenance of key hydrophobic positions in the NES and show a similar dependence on the identity of the interdigitating hydrophilic residues suggests that they might both depend on the quality of the AID-exportin interaction. Changes in the hydrophilic interdigitating amino acids in an NES have been shown in other systems to affect the quality of exportin-binding and kinetics of intracellular transport (20, 21). The dependence of CSR and protein stability on the nature of the NES could therefore simply reflect a requirement for particular kinetics of nucleocytoplasmic shuttling. A more intriguing possibility is that CSR (and stabilization) occur while AID is directly complexed with components of the export machinery. We can imagine a scenario in which CSR is mediated by AID when it is complexed with exportin and other factors. Participation in such a complex might confer stability on AID. The abundance of AID in the complex could also be critical to achieve co-ordinated deamination in both donor and acceptor switch regions; in the absence of this, S region-restricted deamination could simply lead to localized hypermutation. This might explain why CSR is much more dependent than SHM on AID stability and the NES.
Although such a model envisages a role for exportin-binding that goes beyond its function in nuclear export, it is interesting that somewhat analogous roles as chaperones for basic proteins have already been proposed for other karyopherins, such as importin β (22–24). Furthermore, a very recent study in yeast indicates that DNA that has been damaged by strand breaks is targeted to nuclear pores for an E3-ligase-controlled repair pathway (25). Thus, although the model presented is speculative and goes considerably beyond the data, our demonstration here that both CSR and AID stabilization depends not just on a functional export sequence but also on particular characteristics of the NES suggest that further investigation of AID-Crm1 interaction may well give significant insight into molecular processes in antibody-gene diversification.
Materials and Methods
Class Switching.
B cells were purified from AID−/− mice and cultured in the presence of LPS+IL4, as previously described (26). The mice were infected (24-h incubation) with hAID-encoding retroviruses that had been harvested from Plat-E cells 36 h after they had been transfected using Genejuice (Novagen) with pMx-GFP-derived plasmids in which hAID expression (along with that of GFP from an IRES, to provide an internal control) is directed from the retroviral LTR (27). CSR to IgG1 was analyzed by flow cytometry 3 days after retroviral infection, gating on live B cells that amounted to 5 to 7% of events, regardless of the identity of the retroviral construct (26). Procedures involving the use of animals were carried out under UK Home Office Project License 80/2226.
Somatic Mutation.
AID-induced IgV gene somatic mutation was monitored by measuring the frequency of sIgM-loss variants in AID−/− ϕV−/− sIgM+ DT40 cells (14) transfected with hAID-encoding vectors based on pExpressPuro2 (gift from J.-M. Buerstedde, Munich, Germany). From transfectants that had been expanded under selection (0.25 μg/ml puromycin) and checked for AID expression by Western blot analysis, flow-cytometrically sorted GFP+/sIgM+ cells were seeded (about 100 cells per well) into the wells of a 96-well plate, expanded for 3 weeks, and analyzed by flow cytometry for the percentage of cells retaining surface IgM expression. For analysis of mutations in the pre-Sμ region, genomic DNA was prepared from GFP+-sorted splenic B cells 3 days after retroviral infection. The pre-Sμ region was PCR-amplified from about 10,000 sorted-cell equivalents and cloned and sequenced as described in Xue et al. (28).
AID Localization.
Constructs encoding hAID fused to the C terminus of GFP were generated by PCR-based mutagenesis, cloned into pEGFPc3 (Clontech), transfected into fibroblasts in a 6-well format using Genejuice and replated onto glass slides 24 h after transfection. Cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and, after incubation (2 h, 37 °C) with 10 μg/ml DNase-free RNase A, mounted in PI-containing mounting medium (Vectashield). For localization in B cells, pExpressPuro2 constructs expressing the relevant untagged AID chimera were introduced into AID−/− ϕV−/− sIgM+ DT40 cells and immunofluoresence analysis performed using biotinylated mouse anti-hAID mAb hAnp52–1 (raised against a peptide corresponding to residues 5–19 of human AID; Mason Lu and M.S.N., unpublished data), developing with Streptavidin-AlexaFluor 680 (Invitrogen) (red), counterstained with DAPI (cyan).
AID Abundance and Turnover.
Extracts prepared by heating 106 cells in 50 μl of reducing SDS-sample buffer were subjected to SDS/PAGE; AID abundance was monitored by Western blot analysis using mAb h52–1 (29). GFP was detected using HRP-conjugated goat anti-GFP antiserum (Abcam). For analysis of AID turnover, 293 cells were transiently transfected with pEGFPc3-hAID and transferred after 36 h into methionine-free RPMI medium supplemented with 25 mM Hepes, 2%-FBS, 4 mM-glutamine. After a 1 h preincubation, L-[35S]methionine (Amersham; 30 μCi and 1.5 × 106 cells in 200 μl per time-point) was added to the culture and, after 45 min, cells were washed with prewarmed medium that had been supplemented to 1 mM in methionine and resuspended in 3 ml methionine-supplemented medium per time-point. Cells were lysed at the indicated timepoints in 0.5%-Nonidet P-40, 20 mM-Tris pH8.0, 125 mM-NaCl, 10%-glycerol. The GFP-AID fusion proteins were immunoprecipitated using rabbit anti-GFP antiserum (Abcam) and protein G Sepharose (Sigma Aldrich) and then subjected to SDS/PAGE and autoradiography.
Supplementary Material
Acknowledgments.
We thank Karuna Ganesh and other members of the group for helpful discussions and suggestions and Richard Grenfell for assistance with cell sorting. We also thank J.M. Buerstedde and T. Honjo for kindly providing ϕV−/− DT40 cells and AID−/− mice, respectively. The FWF Austrian Science Fund provided the Erwin Schrödinger postdoctoral fellowship J2508 (to R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810808106/DCSupplemental.
References
- 1.Alt FW, Honjo T, editors. AID for Immunoglobulin Diversity. Adv Immunol. 2007;94:xiii–xv. [Google Scholar]
- 2.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 3.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 4.Ito S, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci USA. 2002;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 8.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 9.Imai K, et al. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol. 2005;115:277–285. doi: 10.1016/j.clim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 11.Elfgang C, et al. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc Natl Acad Sci USA. 1999;96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 13.Aoufouchi S, et al. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arakawa H, Saribasak H, Buerstedde JM. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang C, Zhao ML, Diaz M. Activation-induced deaminase heterozygous MRL/lpr mice are delayed in the production of high-affinity pathogenic antibodies and in the development of lupus nephritis. Immunology. 2009;126:102–113. doi: 10.1111/j.1365-2567.2008.02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1945–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Yébenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelsma D, Bernad R, Calafat J, Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heger P, Lohmaier J, Schneider G, Schweimer K, Stauber RH. Qualitative highly divergent nuclear export signals can regulate export by the competition for transport cofactors in vivo. Traffic. 2001;2:544–555. doi: 10.1034/j.1600-0854.2001.20804.x. [DOI] [PubMed] [Google Scholar]
- 22.Jäkel S, Mingot JM, Schwarzmaier P, Hartmann E, Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Nakahara S, Hogan V, Inohara H, Raz A. Importin-mediated nuclear translocation of galectin-3. J Biol Chem. 2006;281:39649–39659. doi: 10.1074/jbc.M608069200. [DOI] [PubMed] [Google Scholar]
- 25.Nagai S, et al. Functional targeting of DNA damage to a nuclear-pore-associated SUMO-dependent ubiqutin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Noia JM, et al. Dependence of antibody gene diversification on uracil excision. J Exp Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 28.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conticello SG, et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.