Abstract
Syrbactins, a family of natural products belonging either to the syringolin or glidobactin class, are highly potent proteasome inhibitors. Although sharing similar structural features, they differ in their macrocyclic lactam core structure and exocyclic side chain. These structural variations critically influence inhibitory potency and proteasome subsite selectivity. Here, we describe the total synthesis of syringolin A and B, which together with enzyme kinetic and structural studies, allowed us to elucidate the structural determinants underlying the proteasomal subsite selectivity and binding affinity of syrbactins. These findings were used successfully in the rational design and synthesis of a syringolin A-based lipophilic derivative, which proved to be the most potent syrbactin-based proteasome inhibitor described so far. With a Ki′ of 8.65 ± 1.13 nM for the chymotryptic activity, this syringolin A derivative displays a 100-fold higher potency than the parent compound syringolin A. In light of the medicinal relevance of proteasome inhibitors as anticancer compounds, the present findings may assist in the rational design and development of syrbactin-based chemotherapeutics.
Keywords: enzyme inhibitor, natural product, X-ray analysis, syrbactins
The eukaryotic proteasome represents the central degradation machinery of the ubiquitin proteasome pathway (UPP). The UPP controls many biological processes by adapting cellular protein levels through regulated proteolysis. Accordingly, malfunctions of this essential system are correlated with various severe pathological states such as cancer, Alzheimer's disease, inflammation, or autoimmune disorders (1).
The proteolytic sites of the proteasome reside in an internal chamber of a hollow barrel-shaped multimer commonly referred to as 20S proteasome. It consists of four stacked ring systems that are arranged in a α7β7β7α7 manner (2). Although the α-subunits in combination with flanking protein complexes regulate access to the proteolytic sites, proteolysis is performed by catalytically active N-terminal threonine residues, which are located at three of the seven β-subunits (β1, β2, β5). Fluorogenic peptides were used to assign a distinct substrate specificity to each of these sites, revealing a chymotrypsin-like activity at the β5-subunit, a trypsin-like activity at the β2-subunit and a caspase-like activity at the β1-subunit (3).
Low-molecular-weight inhibitors of the human 20S proteasome exhibit promising anticancer activity. To date, the peptide boronic acid PS341 (bortezomib) is the only U.S. Food and Drug Administration (FDA)-approved proteasome inhibitor (Velcade) and is used for the treatment of relapsed and/or refractory multiple myeloma (4). Meanwhile, three more chemical entities, i.e., NPI-0052, CEP-18770, and carfilzomib, are evaluated in clinical trials (5, 6). Treatment with bortezomib, however, often causes severe side effects, probably as a consequence of its low selectivity or is hampered by bortezomib resistance (7). Consequently, new selective and bioavailable proteasome inhibitors are needed, and various screening programs have already been performed and led to the discovery of several diverse and potent natural product-based proteasome inhibitors (8–14).
We have recently reported the elucidation of syrbactins as a class of highly potent proteasome inhibitors (15). Syrbactins are natural products that share biosynthetic pathways by related gene clusters (16, 17). They exhibit an α,β-unsaturated carbonyl system located within a macrocycle that reacts irreversibly with the proteasomal active site Thr1Oγ by a Michael-type 1,4-addition. The class of syrbactins represents a collective term for two structurally closely related, nevertheless distinct natural product families: the syringolins (18, 19) and the glidobactins (20).
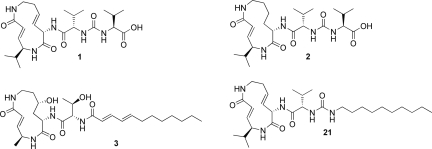
Syringolins are produced by strains of the plant pathogen Pseudomonas syringae pv. syringae (Pss). They are characterized by a 12-membered macrocyclic lactam attached to an exocyclic dipeptide urea moiety. In the major metabolite syringolin A (SylA, 1, Fig. 1), the 12-membered ring system contains two (E)-configured double bonds, resulting in a highly strained macrocycle. SylA acts as a virulence factor in the Pss–plant interaction (15) and has demonstrated anticancer activity and induction of apoptosis in human neuroblastoma and ovarian cancer cells (21).
Fig. 1.
Structures of SylA (1), SylB (2), GlbA (3), and the lipophilic SylA derivative 21.
Pss produces additional minor metabolites such as syringolin B (SylB, 2, Fig. 1) with strong structural similarity to SylA (19). SylB differs from SylA only by the substitution of the SylA 3,4-dehydrolysine residue with a lysine moiety. The resulting macrocycle, however, presents an alternative scaffold structure with less ring strain. To date, no further characterization of the biological activity of SylB has been performed.
The glidobactins feature more structural differences to SylA (20). Their lactam ring system is similar to SylB although incorporating a 3-hydroxy lysine residue at the position of the SylB lysine moiety. The exocyclic side chains, however, are more diverse, lacking the characteristic urea function of syringolins and exhibiting a supplemental N-terminal lipophilic alkyl chain. Among the known glidobactins, only glidobactin A (GlbA, 3, Fig. 1) has yet been elucidated as a proteasome inhibitor, although several glidobactins were recognized for their anticancer activity (20).
With a Ki′ of 49 ± 5.4 nM for the chymotryptic activity, GlbA is the most potent syrbactin proteasome inhibitor reported so far and is 15-fold more active than SylA for the chymotryptic and the tryptic activity. In contrast, GlbA does not inhibit the caspase-like activity whereas SylA moderately affects this activity. Strikingly, the cocrystallization of SylA or GlbA with the yeast 20S proteasome confirmed our observation and revealed similar binding affinities: whereas SylA binds to all three catalytic subunits, GlbA occupies only the active site clefts of the chymotryptic and tryptic activities.
To gain further insights into the binding determinants of SylA and GlbA, we developed a strategy for the chemical synthesis of syrbactins and their derivatives. In addition, enzyme kinetics and the crystal structure elucidation of the proteasome:syringolin B complex provided further knowledge about the enhanced and different binding affinity of GlbA with regard to the distinct proteasomal active sites compared with SylA.
Results
Synthesis of SylB.
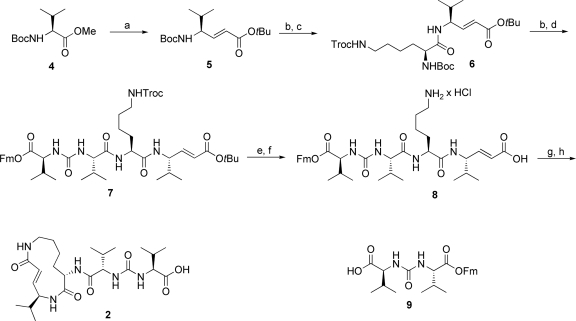
We first addressed the synthesis of SylB because this compound represents a less strained ring system compared with SylA, therefore acting as a potential model system for syrbactins. Synthetic studies of GlbA have elucidated a macrolactamization approach as the most efficient synthetic strategy (22–24) to establish the macrocycle, and, therefore, we decided to follow a similar approach (Fig. 2).
Fig. 2.
Synthesis of SylB. (a) (i) DIBAL-H (2 eq), toluene, −78 °C, 4 h. (ii) Ph3PCHCO2tBu (1.5 eq), DCM, room temperature, 12 h, 84% (both steps). (b) HCl (2 M) in dioxane/tBuOAc (1:1), −5 °C to 10 °C, overnight, 84% or 83%. (c) Boc-Lys(Troc)-OH (1.7 eq), PyBOP (3 eq), HOAt (3 eq), DIEA (8 eq), DCM, room temperature, overnight, 85%. (d) 9 (1.2 eq), PyBOP (1.5 eq), HOAt (1.5 eq), DIEA (3 eq), DCM, 0 °C to room temperature, overnight, 75%. (e) Zn (150 eq), THF/AcOH (1:1), 3 h, 97%. (f) (i) Wet HCO2H. (ii) HCl (0.1 M) in water >98% (both steps). (g) PyBOP (3 eq), HOAt (3 eq), DIEA (6 eq), DMF, 24 h, 30%. (h) Piperidine/DMF (1:4), 1 h, 73%.
To accomplish this, we converted Boc-valine methyl ester (4) into the (E)-configured α,β-unsaturated derivative 5 by a DIBAL-H reduction followed by a Wittig reaction. Selective cleavage of the Boc-protecting group and subsequent peptide coupling of a suitable protected lysine building block yielded dipeptide 6. An adjacent attachment of the exocyclic urea dipeptide 9 generated a linear precursor peptide 7 that was selectively cleaved to yield the macrolactamization precursor 8. The following key ring closure was achieved under high-dilution conditions by PyBOP/HOAt in DMF and produced a satisfying yield of 30%, followed by the removal of the remaining fluorenylmethyl ester protecting group with piperidine in DMF. Final HPLC purification afforded the desired product SylB (2) in 9 steps with an overall yield of 7.8%. The NMR spectra of synthetic SylB and of a mixture of natural SylB isolated as described in ref. 19 and synthetic SylB were almost completely identical. In addition, a coinjection experiment on a chiral HPLC system (SI Appendix) of synthetic SylB with natural SylB revealed no significant differences, thus verifying our initial stereochemical assignment of SylB (2).
Synthesis of SylA.
The chemical structure of SylA was originally disclosed without stereochemical information. An analysis of the SylA synthetase gene cluster, however, suggests an l-configuration of the amino acid residues because no isomerase modules are found (16). Because the structurally and functionally related natural product GlbA is unambiguously based on l-configured amino acids, we focused our synthetic studies on a SylA derivative with l-configured amino acids.
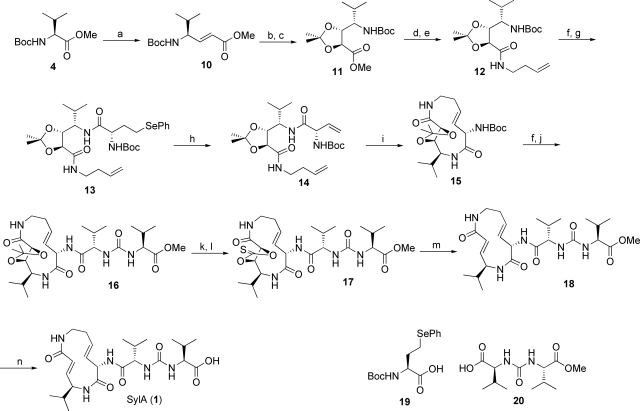
Surprisingly, SylA synthesis by the macrolactamization strategy as described for SylB did not reveal the desired product. We therefore changed our synthetic approach to a ring-closing metathesis (RCM)-based strategy, generating the 3,4-dehydrolysine residue during ring closure (25) (Fig. 3).
Fig. 3.
Synthesis of SylA. (a) (i) DIBAL-H (2 eq), toluene, −78 °C, 4 h. (ii) Ph3PCHCO2Me (1.15 eq), DCM, room temperature, 12 h, 60% (both steps). (b) OsO4 (0.05 eq), NMO (1.5 eq), acetone/H2O (2:1), room temperature, 48 h, 85%. (c) 2,2-Dimethoxypropane (30 eq), PPTS (0.05 eq), DCM, reflux, >98%. (d) 1 M aq LiOH (3 eq), MeOH/H2O (1:1), 0 °C to room temperature, 30 min, >98%. (e) 3-Butenylamine hydrochloride (1.2 eq), PyBOP (1.5 eq), HOAt (1.5 eq), DIEA (2 eq), DCM, 0 °C to room temperature, overnight, 82%. (f) 2,6-Lutidine (2 eq), TMSOTf (1.5 eq), DCM, room temperature, 15 min, >98%. (g) 19 (1.3 eq), PyBOP (1.5 eq), HOAt (1.5 eq), DIEA (2 eq), DCM, 0 °C to room temperature, overnight, 89%. (h) 30% aq H2O2/DIEA (1:1), DCM, 50 °C, 3 h, 93%; (i) Grubbs II catalyst (0.15 eq), toluene, 90 °C, 18 h, 49%. (j) 20 (1.4 eq), PyBOP (1.5 eq), HOAt (1.5 eq), DIEA (2 eq), DCM, room temperature, overnight, 95%. (k) MW, 45W, 110 °C, 90 min, formic acid/methanol (3:2), >98%. (l) Thiocarbonyl diimidazole (10 eq), DMAP (10 eq), THF, 80 °C, overnight, 89%. (m) P(OMe)3, 130 °C, 3 h, 76%. (n) AlCl3 (8 eq), methylethylsulfide, room temperature, 1 h, 92%.
Accordingly, Boc-valine methyl ester was converted into the (E)-configured α,β-unsaturated valine methyl ester 10, followed by a diastereoselective dihydroxylation and protection step to obtain a suitable RCM precursor. C-terminal coupling of butenylamine after selective cleavage of the methyl ester resulted in intermediate 12. Selective deprotection at the N terminus and coupling of 19 as a synthetic precursor to the vinylglycine system yielded 13, which upon treatment with H2O2 (26) was transformed into the RCM precursor 14. RCM of 14 by using the Grubbs II catalyst in toluene at 90 °C as the key step in the synthetic sequence resulted in the formation of the desired (E)-configured macrocyclic lactam 15 in 49% yield, whereas the corresponding (Z)-isomer was formed in traces only. Selective cleavage of the Boc group followed by attachment of the urea building block 20 by PyBOP/HOAt led to the formation of 16. The required α,β-unsaturated carbonyl system was restored after cleavage of the acetonide, generation of thiocarbonate 17, and adjacent Corey–Winter elimination. Finally, the methyl ester was removed with aluminum chloride in methylethylsulfide (27), yielding the natural product SylA (1) with an overall yield of 9.1% from 4 in 16 steps. Comparison of the spectral (SI Appendix) and inhibition data (Table 1) and a coinjection experiment of synthetic and natural SylA isolated as described in ref. 18 on a chiral HPLC system indicate that our original stereochemical assignment of 1 is correct.
Table 1.
Apparent Ki′ and kassoc values for the chymotrypsin-, trypsin-, and caspase-like activity of human 20S proteasome
| Inhibitor | Inhibited activity | Ki′, nM | kassoc, M−1 s−1* |
|---|---|---|---|
| Synthetic SylA (1) | C-L† | 1,016 ± 179 | 828 ± 195 (n = 3; 100–200 nM) |
| T-L† | 10,300 ± 1,400 | 83 ± 2.9 (n = 3; 1.25–5 μM) | |
| Cas-L† | ND‡ | 15.8 ± 3.7 (n = 3; 20–40 μM) | |
| SylA§ | C-L | 843 ± 8.4 | 863 ± 106 (n = 6; 100–200 nM) |
| T-L | 6,700 ± 700 | 94 ± 12 (n = 6; 150–600 nM) | |
| Cas-L | ND | 6 ± 0.3 (n = 6; 20–40 μM) | |
| Synthetic SylB (2) | C-L | 7,778 ± 2,259 | 122 ± 31.7 (n = 3; 1.5–3.1 μM) |
| T-L | 107,800 ± 39,200 | 4.6 ± 0.7 (n = 3; 50–100 μM) | |
| Cas-L | NA¶ | NA | |
| GlbA§ | C-L | 49 ± 5.4 | 3,377 ± 341 (n = 6; 40–60 nM) |
| T-L | 2,000 ± 600 | 141 ± 21 (n = 6; 250–500 nM) | |
| Cas-L | NA | NA | |
| SylA methyl ester (18) | C-L | 757 ± 148 | 1,925 ± 433 (n = 3; 100–200 nM) |
| T-L | 18,700 ± 500 | 186 ± 30 (n = 3; 1.25–2.5 μM) | |
| Cas-L | ND | 10.2 ± 1.4 (n = 3; 25–50 μM) | |
| Lipophilic SylA derivative (21) | C-L | 8.65 ± 1.33 | 40,477 ± 2,333 (n = 3; 6.3–12.5 nM) |
| T-L | 79.6 ± 29.3 | 7,947 ± 1,321 (n = 3; 31.3–62.5 nM) | |
| Cas-L | 943 ± 100 | ND |
*kassoc, rates of covalent inhibition over inhibitor concentrations given in parentheses derived from n measuring points.
†C-L, chymotrypsin-like activity; T-L, trypsin-like activity; Cas-L, caspase-like activity.
‡ND, not determined.
§Literature values taken from ref. 15.
¶NA, not active at a concentration up to 150 μM.
Structural and Enzyme Kinetic Studies.
To investigate the inhibitory potential of SylB (2), we used an in vitro assay containing human 20S proteasome (Table 1).
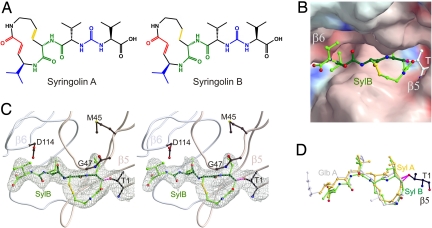
Surprisingly, SylB proved at least 10-fold less potent than SylA. To understand this unexpected result better, the crystal structure of SylB in complex with the yeast 20S proteasome was elucidated, which allowed us to determine its mode of action (Fig. 4). Similar to GlbA, SylB only binds to the subunits β2 and β5, respectively, compared with SylA, which binds to all proteolytically active sites. Interestingly, the spatial arrangement of the lactam ring system of SylB and GlbA in complex with the proteasome was superimposable, whereas SylA displayed a significantly different backbone orientation resulting in an offset of the 3,4-dehydrolysine moiety compared with the lysine or 3-hydroxy lysine residue of SylB and GlbA, respectively. Importantly, the consequential backbone conformation of SylA is more suitable to adopt the characteristic antiparallel β-sheet interaction with the proteasome than SylB and GlbA.
Fig. 4.
X-ray analysis of the complex of SylB (2) and the 20S proteasome and comparison with other syrbactins. (A) Structure of syringolin A (1) and B (2). (B) Electrostatic potential surface [contoured from +15 kT/e (intense blue) to −15 kT/e (intense red)] of SylB covalently bound to β5. (C) Stereo representation of SylB (2) bound to the chymotryptic like active site in complex with 20S proteasome (rose, subunit β5; gray, subunit β6). (D) Structural superimposition of SylA (1, yellow), SylB (2, green), and GlbA (3, light gray).
Synthesis and Enzyme Kinetics of a SylA Lipid Derivative.
To probe the influence of the N-terminal alkyl chain on proteasome inhibition, we envisioned synthesizing a suitable SylA derivative. Therefore, we first tested the impact of the SylA free carboxylic acid moiety on proteasome inhibition because we rationalized that this group is predestined for further modification. As expected from the X-ray analysis of SylA in complex with the yeast 20S proteasome (15), the free carboxylic acid moiety is not required for potent inhibition because both SylA and SylA methyl ester (18, Table 1) inhibit all proteolytic activities of the proteasome in a similar range. After this positive result, we started the synthesis of a suitable modified SylA derivative 21 (Fig. 1), which bears a lipophilic alkyl-chain analogously to GlbA.
This derivative 21 proved to be the most potent inhibitor of the syrbactin derivatives synthesized so far, inhibiting the chymotryptic activity of the human 20S proteasome with a Ki′ of 8.65 ± 1.33 nM, which is >100-fold higher than SylA and >6-fold higher than GlbA (Table 1). Similar inhibition improvements were observed for the trypsin- (Ki′ of 79.6 ± 29.3 nM) and for the caspase-like activity (Ki′ of 943 ± 100 nM, Table 1), ranking this derivative among the most potent proteasome inhibitors described so far.
Discussion
Nature has evolved the biosynthesis of a whole family of structurally related proteasome inhibitors, commonly referred to as syrbactins. These compounds differ in the structure of their macrocyclic lactam systems and their exocyclic chains. All syrbactins investigated so far inhibit the eukaryotic proteasome in a substrate-like binding mode, however, with different potencies and subsite selectivities.
To gain insight into their binding determinants, we developed the total syntheses of the proteasome inhibitors SylA and SylB. The total synthesis of SylA and SylB allowed a verification of its stereochemical assignment, indicating an l-amino acid configuration of all residues. The robustness of the developed route was further demonstrated by the synthesis of a lipophilic SylA derivative 21, using an essentially similar synthesis route. The synthesis of SylB delivered the required material for the enzyme kinetic and structural studies.
To our surprise, SylB displayed a much weaker proteasome inhibition in our biochemical activity assays. The X-ray analysis of SylB in complex with the yeast 20S proteasome suggests several reasons that could explain the higher binding affinity of SylA compared with SylB. First, the 3,4-dehydrolysine double bond induces rigidity into the ring system, which might lower entropic losses during binding. Furthermore, the higher ring strain of the SylA macrocyclic system might contribute to a more reactive α,β-unsaturated carbonyl system or could be preorganized into a more suitable conformation for nucleophilic attack by the proteasome.
The X-ray analysis revealed that the macrocyclic lactam moieties of SylB and GlbA adopt an almost identical binding mode (Fig. 4). Because no significant binding contribution is seen from the supplemental hydroxyl group at the lysine residue of GlbA, the exocyclic lipophilic alkyl chain seems to account mainly for the much higher potency of GlbA. The lipophilic chain shows well-defined electron density in the cocrystal structure of GlbA with the yeast 20S proteasome, which is in agreement with the tight binding to a distinct hydrophobic pocket. This hydrophobic binding patch is built up from the residues Phe-92, Pro-94, Phe-96, Leu-115, and Ile-116 of the β3-subunit for GlbA binding to β2 (tryptic activity) and the residues Tyr-96, Val-97, His-98, Pro-115, and Val-116 of the β6-subunit for GlbA binding to β5 (chymotryptic activity).
The cocrystal structures of SylA, SylB, and GlbA with the yeast 20S proteasome provide a possible explanation for their distinct subsite selectivities. Although GlbA inhibits the chymotrypsin- and trypsin-like activity much more potently than SylA, it does not affect the caspase-like activity that is inhibited by SylA. In principle, this different subsite selectivity could be caused by the amino acid residues of SylA and GlbA that occupy the S1–S3 pockets at the β1-subunit. However, neither in the case of SylA nor GlbA, the P1–P3 residues show conspicuous preferences or clashes for β1. Furthermore, SylB, which binds like GlbA with its macrocyclic lactam moiety but shares the P1–P3 residues with SylA, was not found to occupy the β1-subunit, despite the high SylB concentrations used during crystallization. These findings suggest that the macrocyclic lactam structure in SylB and GlbA indeed is the major determinant for the observed subsite selectivity. A structural superimposition of SylA bound to the β5-subunit with SylA bound to β1 reveals an identical binding conformation for SylA in both subsites (Fig. S1 in the SI Appendix). A docking of the structural conformation of SylB and GlbA bound to β5 into the β1-subunit, however, indicates a disturbed antiparallel β-sheet interaction, displacing the peptide backbone from the ideal alignment with the active-site cleft and thereby significantly lowering binding affinity and thus subsite occupancy. A similar effect of subsite selectivity as a consequence of strained ring conformations has been elucidated with TMC-95A derivatives (28, 29).
These results prompted us to develop the lipophilic SylA derivative 21, the inhibitory properties of which are in accord with these suggestions. This derivative is currently one of the most potent proteasome inhibitors known to date, displaying a Ki′ of 8.65 ± 1.33 nM for the chymotryptic-like active site. Interestingly, 21 also blocks the caspase-like activity with a Ki′ of 943 ± 100 nM as opposed to GlbA and SylB. Thus, syrbactins, decorated with a lipophilic alkyl side chain, can be significantly improved in their proteasomal binding affinity, which is in agreement with the recently described lipophilic natural product proteasome inhibitor fellutamide B (11). Furthermore, the observed enhanced inhibition of the caspase-like activity of 21 compared with SylA demonstrates that the lipid chain has no negative influence on binding to the caspase-like activity. Our data indicate that although the SylA scaffold structure is able to address the caspase-like activity, the macrocyclic lactam core structure of SylB and GlbA hampers binding to this subsite. The syrbactin family therefore is a nature-derived example for the influence of the spatial arrangement of the reactive group on subsite proteasome selectivity. Interestingly, studies with mechanism-based derived inhibitors employing different reactive group warheads have shown similar effects (30, 31).
Because of the significance of proteasome inhibitors as potential anticancer chemotherapeutics, this work describes the chemical synthesis of syrbactin-based proteasome inhibitors and provides an important platform for the design of a plethora of new syrbactin-based proteasome inhibitors. Moreover, the elucidation of the chemical synthesis of syrbactins will allow the production of large compound quantities, which are needed for studies in animal models and, ultimately, for the further development of leads into viable anticancer drugs.
Materials and Methods
Chemical Synthesis.
Unless otherwise noted, all reagents and solvents were purchased from Acros, Fluka, Sigma–Aldrich, or Merck and used without further purification. Dry solvents were purchased as anhydrous reagents from commercial suppliers. LC-MS analyses were performed on an HPLC system from Agilent (1200 series) with an Eclipse XDB-C18, 5-μm (column dimensions: 150 × 4.60 mm) column from Agilent and a Thermo Finnigan LCQ Advantage Max ESI-Spectrometer. The corresponding gradients are described in the SI Appendix. The chiral purity of syringolin A was checked with the chiral column Chiralcel OD-R (column dimensions: 250 × 4.60 mm) from Daicel/Chiral Technologies. Preparative HPLC was conducted on a Varian HPLC system (Pro Star 215) with a VP 250/21 Nucleosil C18 RP column from Macherey–Nagel. The corresponding gradient is described in the SI Appendix. NMR spectra were recorded on a Varian Mercury 400 system (400 MHz for 1H and 100 MHz for 13C NMR), a Bruker Avance DRX 500 system (500 MHz for 1H and 125 MHz for 13C NMR), or a Varian Unity Inova 600 system (600 MHz for 1H and 150 MHz for 13C NMR). TLC analyses were performed with TLC aluminum sheets 20- × 20-cm silica gel 60 F254 from Merck. HRMS measurements were performed on a LC-HR/ESI-FTMS machine from Thermo Electron Corporation. The microwave-assisted reactions were conducted by using a focused microwave unit (discover reactor from CEM Corporation).
Full experimental details and characterization data for all synthesized compounds are included in the SI Appendix.
Human 20S Proteasome Assays.
The biochemical proteasome assays were performed as described in ref. 15, with commercially available human erythrocyte 20S proteasomes from Biomol. DMSO stock solutions were prepared from SylA (1, 10 mM), SylA methylester (18, 20 mM), SylB (2, 10 mM), and SylA lipophilic derivative (21, 9 mM), and a dilution series in DMSO was prepared for determining the corresponding Ki′ values. Each data point has been determined in three independent experiments (n = 3).
X-Ray Analysis.
Crystals of 20S proteasome from Saccharomyces cerevisiae were grown in hanging drops at 24 °C (32) and incubated for 60 min with syringolin B (2, 10 mM in DMSO). The protein concentration used for crystallization was 40 mg/mL in Tris·HCl (10 mM, pH 7.5) and EDTA (1 mM). Drops contained 3 μL of protein and 2 μL of reservoir solution [30 mM MgOAc, 100 mM Mes (pH 7.2), and 10% 2-methyl-2,4-pentanediol (MPD)]. The space group of proteasomal complex crystals belongs to P21 with cell dimensions of a = 133.5 Å, b = 301.6 Å, c = 143.4 Å and β = 112.6°. Data to 2.7 Å were collected by using synchrotron radiation with λ = 1.00 Å at the X06SA beamline at SLS/Villingen/Switzerland. Crystals were soaked in a cryoprotecting buffer [30% MPD, 20 mM MgOAc, 100 mM Mes (pH 6.9)] and frozen in a stream of liquid nitrogen gas at 90 K (Oxford Cryo Systems). X-ray intensities and data reduction were performed by using the XDS program package (33). Anisotropy of diffraction was corrected by an overall anisotropic temperature factor, comparing observed and calculated structure amplitudes with the program CNS (34). A total of 944,365 reflections yielded 282,923 unique reflections (99.2% completeness). The corresponding Rmerge was 15.4% at 2.7-Å resolution (42.4% for the last resolution shell). Electron density was improved by averaging and back-transforming 10 times over the 2-fold noncrystallographic symmetry axis with the program package MAIN (35). Conventional crystallographic rigid body, positional, and temperature factor refinements were carried out with CNS using the yeast 20S proteasome structure as starting model (2). For model building, the program MAIN was used. The structure was refined to a crystallographic R-factor of 24.9% (free R-factor 26.4%) with rms deviations from target values of 0.008 Å for bonds and 1.35° for angles. Modeling experiments were performed by using the coordinates of yeast 20S proteasome with the program MAIN.
Supplementary Material
Acknowledgments.
Financial support from the Swiss National Science Foundation (Grant 3100A0-115970, to R.D.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID Code 3GPJ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901982106/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 3.Nussbaum AK, et al. Cleavage motifs of the yeast 20S proteasome β-subunits deduced from digests of enolase 1. Proc Natl Acad Sci USA. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voorhees PM, Orlowski RZ. The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- 5.Moore BS, Eustáquio AS, McGlinchey RP. Advances in and applications of proteasome inhibitors. Curr Opin Chem Biol. 2008;12:434–440. doi: 10.1016/j.cbpa.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sterz J, et al. The potential of proteasome inhibitors in cancer therapy. Exp Opin Invest Drugs. 2008;17:879–895. doi: 10.1517/13543784.17.6.879. [DOI] [PubMed] [Google Scholar]
- 7.Naujokat C, Fuchs D, Berges C. Adaptive modification and flexibility of the proteasome system in response to proteasome inhibition. Biochim Biophys Acta. 2007;1773:1389–1397. doi: 10.1016/j.bbamcr.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Feling RH, et al. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chem Int Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 9.Fenteany G, et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 10.Meng, et al. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hines J, Groll M, Fahnestock M, Crews CM. Proteasome inhibition by fellutamide B induces nerve growth factor synthesis. Chem Biol. 2008;15:501–512. doi: 10.1016/j.chembiol.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asai A, et al. A new structural class of proteasome inhibitors identified by microbial screening using yeast-based assay. Biochem Pharmacol. 2004;67:227–234. doi: 10.1016/j.bcp.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 13.Koguchi Y, et al. Structures of TMC-95A-D: Novel proteasome inhibitors from Apiospora montagnei sacc. TC 1093. J Antibiot. 2000;53:105–109. doi: 10.1021/jo991375+. [DOI] [PubMed] [Google Scholar]
- 14.Nickeleit I, et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Groll M, et al. A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature. 2008;452:755–758. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- 16.Amrein H, et al. Functional analysis of genes involved in the synthesis of syringolin A by Pseudomonas syringae pv. syringae B301D-R. Mol Plant–Microbe Interact. 2004;17:90–97. doi: 10.1094/MPMI.2004.17.1.90. [DOI] [PubMed] [Google Scholar]
- 17.Schellenberg B, Bigler L, Dudler R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ Microbiol. 2007;9:1640–1650. doi: 10.1111/j.1462-2920.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 18.Wäspi U, Blanc D, Winkler T, Ruedi P, Dudler R. Syringolin, a novel peptide elicitor from Pseudomonas syringae pv. syringae that induces resistance to Pyricularia oryzae in rice. Mol Plant–Microbe Interact. 1998;11:727–733. [Google Scholar]
- 19.Wäspi U, et al. Identification and structure of a family of syringolin variants: Unusual cyclic peptides from Pseudomonas syringae pv. syringae that elicit defense response in rice. Microbiol Res. 1999;154:89–93. [Google Scholar]
- 20.Oka M, et al. Glidobactins A–C, new antitumor antibiotics. I. Production, isolation, chemical properties and biological activity. J Antibiot. 1988;41:1331–1337. doi: 10.7164/antibiotics.41.1331. [DOI] [PubMed] [Google Scholar]
- 21.Coleman CS, et al. Syringolin A, a new plant elicitor from the phytopathogenic bacterium Pseudomonas syringae pv. syringae, inhibits the proliferation of neuroblastoma and ovarian cancer cells and induces apoptosis. Cell Prolif. 2006;39:599–609. doi: 10.1111/j.1365-2184.2006.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka M, et al. Chemical modification of the antitumor antibiotic glidobactin. J Antibiot. 1988;41:1812–1822. doi: 10.7164/antibiotics.41.1812. [DOI] [PubMed] [Google Scholar]
- 23.Meng Q, Hesse M. Synthetic approaches toward glidobamine, the core structure of the glidobactin antibiotics. Tetrahedr Lett. 1991;47:6251–6264. [Google Scholar]
- 24.Schmidt U, Kleefeldt A, Mangold R. The synthesis of glidobactin A. J Chem Soc Chem Commun. 1992:1687–1689. [Google Scholar]
- 25.Gradillas A, Pérez-Castells J. Macrocyclization by ring-closing metathesis in the total synthesis of natural products: Reaction conditions and limitations. Angew Chem Int Ed. 2006;45:6086–6101. doi: 10.1002/anie.200600641. [DOI] [PubMed] [Google Scholar]
- 26.Bartley DM, Coward JK. A stereoselective synthesis of phosphinic acid phosphapeptides corresponding to glutamyl-γ-glutamate and incorporation into potent inhibitors of folypoly-γ-glutamyl synthetase. J Org Chem. 2005;70:6757–6774. doi: 10.1021/jo0507439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Node M, Nishide K, Sai M, Fuji K, Fujita E. Hard acid and soft nucleophile systems. 3. Dealkylation of esters with aluminum halide–thiol and aluminum halide–sulfide systems. J Org Chem. 1981;46:1991–1993. [Google Scholar]
- 28.Kaiser M, Groll M, Renner C, Huber R, Moroder L. The core structure of TMC-95A is a promising lead for reversible proteasome inhibition. Angew Chem Int Ed. 2002;41:780–783. doi: 10.1002/1521-3773(20020301)41:5<780::aid-anie780>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser M, et al. Binding mode of TMC-95A analogues to eukaryotic 20S proteasome. ChemBioChem. 2004;5:1256–1266. doi: 10.1002/cbic.200400096. [DOI] [PubMed] [Google Scholar]
- 30.Verdoes M, et al. Mixing of peptides and electrophilic traps gives rise to potent, broad-spectrum proteasome inhibitors. Org Biomol Chem. 2007;5:1416–1426. doi: 10.1039/b702268a. [DOI] [PubMed] [Google Scholar]
- 31.Groll M, Nazif T, Huber R, Bogyo M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chem Biol. 2002;9:655–662. doi: 10.1016/s1074-5521(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 32.Groll M, Huber R. Purification, crystallization, and X-ray analysis of the yeast 20S proteasome. Methods Enzymol. 2005;398:329–336. doi: 10.1016/S0076-6879(05)98027-0. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 34.Brünger A, et al. Crystallography & NMR system (CNS): A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Turk D. Munich: Technische Universität München; 1992. Improvement of a programme for molecular graphics and manipulation of electron densities and its application for protein structure determination. PhD thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.